Abstract
Several studies suggest that cis-regulatory mutations are the favorite target of evolutionary changes, one reason being that cis-regulatory mutations might have fewer deleterious pleiotropic effects than protein-coding mutations. A review of the process also suggests that this bias towards adaptive cis-regulatory variation might be less pronounced at the intraspecific level compared with the interspecific level. In this study, we assessed the contribution of cis-regulatory variation to adaptation at the intraspecific level using populations of wild pearl millet (Cenchrus americanus ssp. monodii) sampled along an environmental gradient in Niger. From RNA sequencing of hybrids to assess allele-specific expression, we identified genes with cis-regulatory divergence between two parental accessions collected in contrasted environmental conditions. This revealed that ∼15% of transcribed genes showed cis-regulatory variation. Intersecting the gene set exhibiting cis-regulatory variation with the gene set identified as targets of selection revealed no excess of cis-acting mutations among the selected genes. We additionally found no excess of cis-regulatory variation among genes associated with adaptive traits. As our approach relied on methods identifying mainly genes submitted to strong selection pressure or with high phenotypic effect, the contribution of cis-regulatory changes to soft selection or polygenic adaptive traits remains to be tested. However our results favor the hypothesis that enrichment of adaptive cis-regulatory divergence builds up over time. For short evolutionary time-scales, cis-acting mutations are not predominantly involved in adaptive evolution associated with strong selective signal.
Keywords: allele-specific expression, cis-regulation, environmental gradient, local adaptation, Cenchrus americanus, selection
Introduction
Dissecting the molecular basis of adaptive evolution is a major goal in evolutionary biology. One debated question in this field is the relative contribution of protein-coding and cis-regulatory mutation to adaptation. It is generally accepted that mutations in cis-regulatory regions are the favorite target of evolutionary changes (King and Wilson 1975; Hoekstra and Coyne 2007; Wray 2007; Stern and Orgogozo 2008; Wittkopp and Kalay 2011). The main argument supporting this assumption (reviewed in Stern and Orgogozo 2008) is that cis-regulatory mutations might have fewer deleterious pleiotropic effects than mutations altering the amino-acid sequence of a protein. Indeed, cis-regulatory regions are composed of noncoding DNA regulating the expression of nearby gene through promoters and enhancers. The modular nature of the architecture of the cis-regulatory element (CRE) implies that changes in CREs may result in specific changes in gene expression limited to a particular tissue, life stage or environmental condition (Prud’homme et al. 2007). Conversely, changes in sequence of coding regions would affect all the subsequent genetic pathways in which the gene product, protein or mature RNA, is involved.
This cis-regulatory hypothesis of phenotypic evolution has been challenged many times (Alonso and Wilkins 2005; Hoekstra and Coyne 2007; Lynch and Wagner 2008; Wagner and Lynch 2008). Using a compilation of studies aiming at identifying mutations underlying phenotypic evolution, Stern and Orgogozo (2008) highlighted a contrasting tendency of protein-coding and cis-regulatory mutations contributions to evolutionary changes depending on the evolutionary time-scale. They found a higher proportion of null coding mutations causing phenotypic variation among studies based on intraspecific comparisons (genes involved in adaptive divergence over short evolutionary time-scales) and higher proportion of cis-regulatory mutations among studies based on interspecific comparisons (long evolutionary time-scales). Stern and Orgogozo (2009) argued that for a short period of evolution, adaptive mutations may be selected even if they have a deleterious pleiotropic effect because nondeleterious mutations are absent. In contrast, after a long period of evolution, adaptive mutations with no pleiotropic deleterious effects have more opportunity to occur and to be selected. However this hypothesis mainly relied on a collection of selected genes across organisms that might be biased in some way (Coolon et al. 2014). Other experimental studies on yeast (Gruber et al. 2012; Metzger et al. 2016) showed that cis-regulatory mutations arose less frequently with more moderate effects on phenotypic trait than other type of mutations potentially explaining their higher contribution to adaptive divergence over a longer period of time. Yet, the impact of selection on transcriptome-wide cis-regulatory variation over short and long time-scales remains to be clarified. Although contribution of allele-specific expression variation (aseQTLs) to purifying selection has been recently addressed in plant (Josephs et al. 2015), much remains to be done to fully understand the relation between adaptation and cis-regulation at the different time-scales of evolution.
The usual way to directly identify genes with cis-regulation variation is to investigate allelic expression in F1 hybrids, as cis-acting polymorphism causes unequal expression of the two parental alleles, or allele-specific expression (ASE). When ASE is observed, it could be due to genetic or methylation changes in CRE or insertion of transposable elements (Wittkopp and Kalay 2011). ASE should reflect only the contribution of cis-regulatory changes and not trans-effects as trans-acting variation should act on the expression of both alleles within an individual. The recent development of next generation sequencing (NGS) makes it possible to distinguish parental alleles from nucleotide variation and hence quantify differential transcript abundance patterns of two parental alleles in hybrids at a genome-wide level. Such an approach using RNA-seq technology has been used in many organisms such as yeast (Emerson et al. 2010), drosophila (McManus et al. 2010; Coolon et al. 2014) and plants (He et al. 2012, 2016; Lemmon et al. 2014; Steige et al. 2015; Arunkumar et al. 2016) to address the question of cis/trans-regulatory contribution to phenotypic divergence focusing mainly on the inter-specific level (Mack et al. 2016). The intraspecific role of cis-regulatory divergence in accessions collected in natural populations has been less explored to date (but see Zhang and Borevitz 2009; Cubillos et al. 2014; Steige et al. 2017), and based on the hypothesis of Stern and Orgogozo (2009), one would expect to observe weaker or no association between cis-regulation and selection.
In this study, we analyzed cis-regulatory divergence between genotypes of the outcrossing pearl millet wild relative (Cenchrus americanus ssp. monodii) sampled in contrasted ecological habitats. Pearl millet produced in the driest environment on earth is a staple crop in Africa and India. Its wild progenitor species can be found in the Sahelian region in Africa, the probable domestication center, distributed along a south–north gradient of humidity (Mariac et al. 2011; Dussert et al. 2013). By sampling populations along this gradient, Berthouly-Salazar et al. (2016) used genome scanning to identify genomic variation correlated with climatic variables and hence to find genomic regions potentially involved in local adaptation. In another study, Ousseini et al. (2016) showed that between-population phenotypic variability correlates with environmental conditions of origin in a common garden experiment and built an association genetics framework to identify the genomic regions involved in variation in adaptive traits.
Here, we used the same data to investigate the contribution of cis-regulatory changes to adaptation at the intraspecific level of wild pearl millet. We first used ASE to infer the presence of cis-regulatory variation affecting gene expression among wild-pearl millet accessions sampled from the most extreme habitats along the environmental gradient. Then, using the results of previous studies on wild pearl millet adaptation, we tested for enrichment of ASE genes among the selected genes and among the genes associated with adaptive traits. Our main results suggest that cis-regulatory variation is not the main driver of short-term intraspecific adaptation.
Materials and Methods
Plant Material
Two accessions of wild pearl millet (Cenchrus americanus (L.) Morrone ssp. monodii) were collected in West Africa from populations growing in the most drastic environmental conditions experienced by the species according to bioclimatic data (see details in Berthouly-Salazar et al. 2016). One accession was collected in the driest environment in Niger (P9A, geographic coordinates: 18°31′48″N 8°38′24″E) and the other was collected in the wettest environment in Niger (8106A, 15°46′12″N 6°30′0″E).
Reciprocal crosses were performed between the two parental accessions under controlled conditions in a greenhouse. Day length was set at 14 h of light followed by 10 h of dark for 1 month and was then switched to 10 h of light followed by 14 h of dark. Humidity was maintained at 70% and temperature at 28 °C during the light period and at 25 °C during the dark period. Each parent being used as pollen donor as well as pollen receiver, we obtained two F1 hybrids (referred to as P9Ax8106A and 8106AxP9A). F1 hybrid seedlings were confirmed using two highly polymorphic and informative microsatellite markers 2201 and 2237 (Oumar et al. 2008).
DNAseq Analysis
Sequencing and Mapping
Genomic DNA was extracted from leaf tissues of the F1 hybrids as previously described in Mariac et al. (2014). Libraries were constructed using 6-bp barcodes to allow for multiplexing (Mariac et al. 2014) and were sequenced at the MGX-Montpellier GenomiX plateform (Montpellier, France) on two Illumina HISeq2500 lanes generating paired-end reads of 2×125 pb.
Demultiplexing based on the 6-bp barcodes was performed using the freely available Perl script Demuladapt (https://github.com/Maillol/demultadapt) with a 1-mismatch threshold. The resulting raw reads were cleaned for remaining adapter sequences using cutadapt (v.1.8, Martin 2011). Reads were filtered based on their length (L > 35 bases) and on their quality mean values (Q > 30) using a freely available Perl script (https://github.com/SouthGreenPlatform/arcad-hts/blob/master/scripts/arcad_hts_2_Filter_Fastq_On_Mean_Quality.pl).
The remaining paired-end reads were mapped to the reference genome of cultivated pearl millet (Varshney RK, personal communication) using BWA software (v.0.7.2, Li and Durbin 2009) with the MEM algorithm. The resulting SAM file was converted to BAM using SAMtools (v.1.2, Li et al. 2009) and duplicated reads were marked using Picard’s MarkDuplicates (v.1.83, http://broadinstitute.github.io/picard). The genome mean coverage was estimated using QualiMap (v.2.0.2, Garcia-Alcalde et al. 2012).
Variant Discovery, SNP Calling, and SNP Filtering
We used the Genome Analysis Toolkit (GATK, v.3.3, McKenna et al. 2010) to perform local realignment around indels using IndelRealigner and to find heterozygous positions in hybrids HaplotypeCaller. Variants were called jointly using the GVCF of two hybrids. The purpose of calling variants was to identify highly confident heterozygous positions. We focused on SNPs found in transcribed regions. Indels and nonbiallelic SNPs were excluded from the final set of variants. Following the hard filtration recommendations of the GATK’s best practices guide, poorly reliable SNPs were excluded based on the following: (1) the quality of the variant divided by the depth of the nonreference allele (QD < 10), (2) the strand bias (FS > 60) and (3) the mapping quality (MQ < 30). Finally, depth of reads criteria at variant positions were considered for SNP filtering to exclude poorly supported positions. SNPs supported by low (DP < 10) or high (DP > 240) depth of reads over the two hybrids were excluded from the final set of variants, as were variants with an alternate allele supported by less than four reads. At the end, a VCF file including only the heterozygous SNPs remaining after filtration was obtained for each hybrid separately and used for the following steps of the analyses.
RNAseq Analysis
Sequencing and Mapping
RNAs were extracted from fresh tissue of immature ears at the stigma emergence state collected from the two F1 hybrid plants in three technical replicates. RNA was extracted using the RNAeasy Plant Mini kit (ref: 74904, Qiagen, Basel, Switzerland). The average RIN (RNA integrity number) was 7.3 for the P9Ax8106A hybrid and 7.4 for the 8106AxP9A hybrid. Libraries were constructed using the Illumina TruSeq RNA Library Preparation Kit (Ref: RS-122-2001 et RS-122-2002) and were sequenced at the GeT-PlaGe platform (Toulouse, France) on three Illumina HISeq2000 lanes generating paired-end reads of 2×100 pb. In the following, we only used the R1 read to avoid nonindependence in read counts.
Like for the DNA-seq reads, remaining adapter sequences were removed from the resulting RNA-seq raw reads, filtered based on their length (L > 35 bases) and their quality mean values (Q > 30). The remaining reads were mapped to the reference genome of cultivated pearl millet using the spliced alignment program STAR (v 2.4.1.c, Dobin et al. 2013) according to the 2-pass protocol (Engström et al. 2013). In the first alignment pass, previously known spliced junctions of cultivated pearl millet were provided to the software through a GFF annotation file (Varshney RK, personal communication) to map the reads to the reference genome and to identify new spliced junctions. Using the options –outSJfilterCountUniqueMin and –outSJfilterCountTotalMin, only junctions supported by at least 5 reads for canonical junctions or 10 reads for noncanonical junctions were retained. In the second alignment pass, a list of known filtered and novel spliced junctions was provided to map the reads. The resulting SAM file was then converted into BAM using SAMtools (v.1.2, Li et al. 2009). We used the GATK software (v.3.4, McKenna et al. 2010) to perform local realignment around indels using IndelRealigner tool.
Analysis of Allele-Specific Expression
Counting Allele-Specific Reads
Analyses of ASE were based on allele-specific read counts at heterozygous SNPs specifically identified for each hybrid from DNA sequencing. RNA-seq read counts at each position were obtained from BAM files using the ASEReadCounter tool in GATK v3.4. Only positions supported by at least 10 RNA-seq reads in each repetition were kept. The fraction of reads carrying the reference allele over the total number of reads mapped at that position, i.e. the reference ratio, was calculated at each heterozygous SNP. As the correlations of reference ratios between RNA-seq repetitions were high (coefficient correlation of Pearson R > 0.70, P values < 0.0001, supplementary fig. S1, Supplementary Material online), allelic read counts were summed between replicates of the same hybrids to calculate the reference ratio.
ASE Analysis
Following the method implemented in the ASEQ tool (Romanel et al. 2015), we first performed a binomial test to identify the positions at which the ratio of the two alleles in the transcriptomic data is significantly different from 0.5. Then we assessed the reference ratio derived from DNA sequencing and tested the difference between the allelic ratios obtained from the DNA and the RNA data using a Fisher exact test. A position was considered as ASE if both the two tests were significant. The rationale of this methodology is as follows: (1) to identify positions showing disequilibrium in read count for one allele compared with the other in the transcriptomic data and (2) among those positions, to exclude sites showing the same pattern of read count disequilibrium at the genomic level. Indeed, deviation from the expected 0.5 value of allelic ratio can be observed in genomic data as well as in transcriptomic data due to biases occurring during the mapping step either associated with copy number variation (CNV) or higher mappability of reads carrying the reference allele compared with the one carrying the alternate allele. To avoid no-independence between counts, we only considered SNPs separated by >100 bp. The whole analysis was performed separately for each hybrid.
Our analysis enabled the identification of ASE at the level of SNPs. Using the intersect function of bedtools (v2.25) (http://bedtools.readthedocs.io/; last accessed January 27, 2017), SNPs were assigned to genes identified in the GFF file of cultivated pearl millet (Varshney RK, personal communication). Genes were considered ASE if at least one of their SNPs was identified as ASE.
Gene Ontology Enrichment Analysis
Considering genes annotated in the pearl millet genome only, we tested for enrichment of gene ontology (GO) biological process terms within the ASE genes using Fisher exact tests implemented in the R package TopGO v.2.22 (Alexa et al. 2006). We performed both classical and weight01 algorithms, the latter taking the hierarchical relationships between terms into account. To reduce statistical artifacts of GO terms with few annotated genes, terms supported by less than five annotated genes were excluded from the enrichment analyses.
Joint Analysis of ASE, Selection, and Association
We tested whether there was an excess of genes with evidence for ASE among genes identified as selected in a previous related study (Berthouly-Salazar et al. 2016). That study was based on a RNA seq experiment in which reads were mapped to the wild pearl millet reference transcriptome to identify SNPs and to detect the selected contigs. Intersecting the ASE gene set from our experiment with the gene set detected as selected in the previous study required localizing the selected contigs with the SNP used for the ASE experiment on the genome. To that end, contigs from the pearl millet reference transcriptome were mapped to the reference genome using BWA to extract the contig coordinates on the reference genome. On the 50,313 contigs of the reference transcriptome, 49,107 were mapped in 134,167 fragments with a minimal size of 30 bp. We discarded the contigs mapped on different chromosomes and the contigs with fragments on the same chromosome separated by an interval exceeding 1 mega bp. Finally we intersected the contig coordinates on the reference genome with the previously identified SNPs coordinates. We discarded ambiguous SNPs located on two or more different contigs and those separated by <100 bp. We identified ASE contig containing at least one ASE SNP as described in the previous section. We tested for enrichment of ASE contigs among the selected contigs using a one-sided Fisher exact test in R v. 3.2 for each hybrid separately. To estimate the ability of our approach to find enrichment of ASE genes among the selected genes, we first tested the power of our tests to detect an excess of a given percentage of ASE genes among the selected one using simulations. We also inferred the posterior distribution of ASE genes proportion among the selected genes fitting our data, i.e. with a higher P value than observed for the tests carried on our data. Finally, as the methods used in Berthouly-Salazar et al. (2016) to detect loci under selection could result in the identification of false positive, we redid the enrichment analysis considering as selected the contig that were detected in at least two methods of detection for selection in Berthouly-Salazar et al. (2016).
In the same way, we tested whether there was an excess of genes with evidence for ASE among genes identified as associated with phenotypic traits in a previous related study (Ousseini et al. 2016). In that work, a total of 11 phenotypic traits related to flowering time, flowering abundance and vegetative architecture were measured. Association with those traits was tested for 216 SNPs localized in 148 contigs of the wild pearl millet reference transcriptome. As previously described, we tested for enrichment of ASE contigs among the associated contigs using a one-sided Fisher exact test for each hybrid separately.
Results
Identification of Heterozygous SNPs for ASE Analysis
In order to identify heterozygous SNPs in the hybrids, we sequenced the whole genome generating an average of 320 million paired-end reads per hybrid mapped after filtration for quality (see “Material and Methods” section). The resulting mean coverage genome was 20× for P9Ax8106A and 22× for 8106AxP9A, with respectively 60% and 63% of the reference genome covered by at least 10 reads. A total of 552,544 genomic heterozygous SNPs were identified in the expressed regions for the two hybrids. After filtration, 379,746 SNPs remained, 314,237 SNPs are found in the P9Ax8106A cross and 320,725 in the 8106AxP9A cross. A total of 83% of the SNPs were common between crosses. As the initial parents were heterozygote, we expect some SNPs to be specific of a given hybrid.
Allele-Specific Expression Analysis
A total of 213 million reads were generated from the RNA sequencing experiment with an average of 44 million single-end reads per technical replicate for P9Ax8106A and 27 million for 8106AxP9A. More than 83% of those reads were mapped to a single position on the reference genome (supplementary table S1, Supplementary Material online). Considering only the SNPs covered by at least 10 RNA-seq reads per replicate and spaced by at least 100 bp, allele-specific reads counts were retrieved at 30959 and 24862 heterozygous positions for P9Ax8106A and 8106AxP9A hybrids, respectively (supplementary table S2, Supplementary Material online).
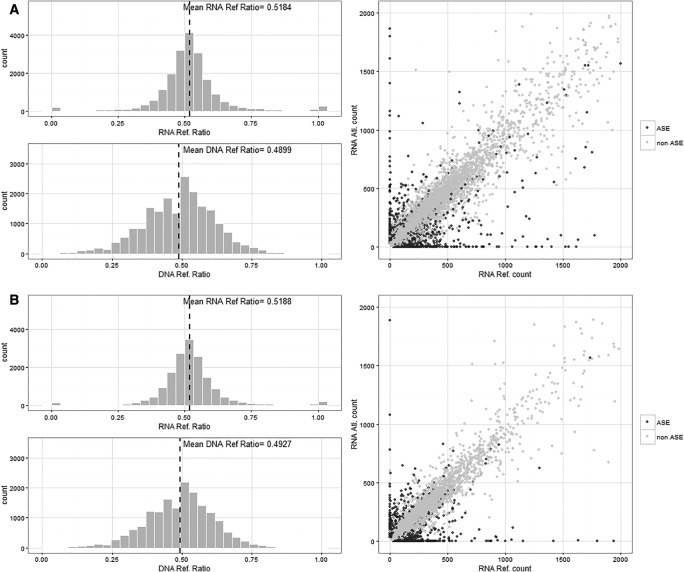
To obtain an estimate of the proportion of ASE genes, here we only considered the positions located within the annotated pearl millet genes, therefore corresponding to 19,924 and 16,652 positions for P9Ax8106A and 8106AxP9A hybrids, respectively (table 1). The total number of genes considered in the ASE analysis (i.e. with at least one informative heterozygous position) was 8,789 for the P9Ax8106A hybrid and 7,491 for the 8106AxP9A hybrid. The mean reference ratio of both hybrids was 0.518 when estimated from RNA-seq data and 0.490 from the DNA-seq data (fig. 1).
Table 1.
ASE Statistics for SNPs and Genes Analyzed in the Two Wild Pearl Millet Hybrids
| No. of SNPs a analyzed | Mean RNA Ref. Ratio | Mean DNA Ref. Ratio | No. of ASE SNP b | ASE SNP Proportion | No. of Genes c Analyzed | No. of ASE Gene d | ASE Gene Proportion | |
|---|---|---|---|---|---|---|---|---|
| P9Ax8106A | 19,924 | 0.518 | 0.490 | 1,838 | 0.092 | 8,789 | 1,391 | 0.158 |
| 8106AxP9A | 16,652 | 0.519 | 0.493 | 1,418 | 0.085 | 7,491 | 1,066 | 0.142 |
Total number of heterozygous SNP analyzed for ASE, located in annotated genes of the pearl millet genome, covered by at least 10 RNAseq reads and separated from other identified SNP by at least 100 pb.
Number of SNP found as ASE according to the combined binomial–Fisher test.
Number of annotated genes with at least one heterozygous SNP analyzed for ASE.
Number of annotated genes with at least one SNP found as ASE according to the combined binomial–Fisher test.
Fig. 1.—
Distribution of the reference ratio measured from the RNA-seq read counts (upper left panel) and DNA-seq read counts (lower left panel) and plot of the reference allele read count versus alternative allele read count (right panel) from the two wild pearl millet hybrids: P9Ax8106A (A) and 8106AxP9A (B).
Using our Binomial–Fisher combined test (referred to as BFC test in the following), we identified 1,838 (9.2%) and 1,418 (8.5%) heterozygous positions showing differential allelic expression for P9Ax8106A and 8106AxP9A hybrids, respectively. This led to the estimation of 15.8% and 14.2% ASE genes, respectively (table 1). Taking into account the 6,615 common genes tested for ASE in the two hybrids, 495 were found ASE in both hybrids.
GO Terms Analysis
Among the 8,789 P9Ax8106A hybrid genes testable for ASE, 3,640 were annotated with sufficiently supported GO terms belonging to the biological process (BP) category. A total of 586 of those genes were identified as ASE. The same analysis was conducted with the second hybrid with 3,099 usable genes among which 432 were found as ASE (supplementary table S3, Supplementary Material online). We found significant enrichment for the GO term related to defense response (GO:0006952) among ASE genes in both hybrids. More specific to the P9Ax8106A hybrid, we found enrichment of genes associated with the oxidation–reduction process (GO:0055114) involving ion transmembrane transport (GO:0034220) and with the electron transport chain (GO:0022900). ASE genes from the 8106AxP9A hybrid were found more specifically enriched in GO terms associated with DNA changes (GO:0006265 and GO:0006284) and amino acid processes (GO:1901606).
Joint Analysis of ASE, Selection, and Association
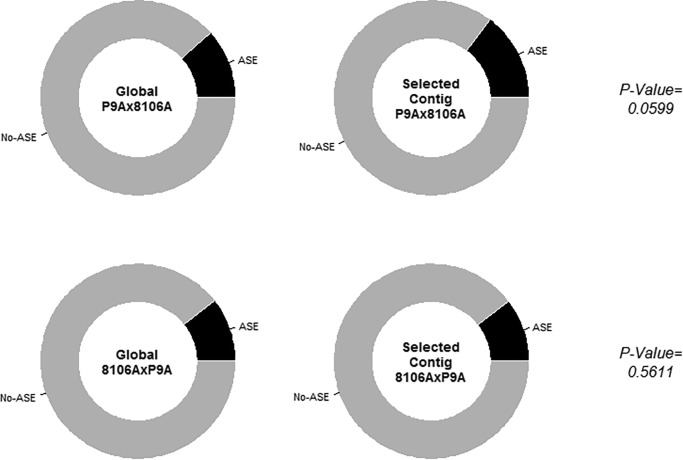
In a previous study, Berthouly-Salazar et al. (2016) used an RNA-seq approach based on reference transcriptome mapping to detect outlier loci in four wild pearl millet populations evolving in contrasted environmental conditions. They identified 540 selected contigs among the 11,155 contigs of the reference transcriptome tested. After positioning the heterozygous SNPs within contigs of the transcriptome reference to identify ASE contigs (supplementary table S4, Supplementary Material online), we intersected the set of contig tested for ASE with the set of contig tested for selection. We were able to retain 5,981 contigs tested both for selection and ASE for the P9Ax8106A hybrid. Among them, 292 were found selected, 697 were found as ASE, and 43 were found both selected and ASE. After testing for enrichment, we found a nonsignificant greater proportion of ASE contigs among the selected ones compared with all the tested contigs (one-sided Fisher exact test, P value = 0.0599, fig. 2a ). The same proportion of ASE contigs among the selected ones compared with all the tested contigs (P value = 0.5611, fig. 2b ) was found for the 8106AxP9A hybrid with 5,785 tested contigs among which 30 were both found as selected and ASE. Same results were found when considering as selected only the contigs detected by at least two methods in Berthouly-Salazar et al. (2016) (one-sided Fisher exact test, P value = 0.13 for P9Ax8106A hybrid, P value= 0.11 for 8106AxP9A hybrid). Using simulations, we assessed the ability of our approach to find enrichment of ASE genes among the selected genes. We found a probability of 79% to detect an excess of 5% of ASE genes among the selected one compared with the other genes. This probability reached almost 100% to detect an excess of 10%. So our approach has good power to detect even low increase of ASE among the selected genes. Additionally, our data support a posterior probability of 95% to observe an excess <3% of ASE genes among the selected ones compared with the other genes.
Fig. 2.—
Pie chart distribution of ASE and non-ASE contigs among all the analyzed contigs (left panel) and among the selected contigs (central panel) and P value of the enrichment test of ASE contig among the selected contigs (one-sided Fisher’s exact test) for the two wild pearl millet hybrids: P9Ax8106A (top row) and 8106AxP9A (bottom row).
Intersecting the set of contig tested for ASE in the present study with the set of contig tested for association genetics in Ousseini et al. (2016), we were able to retain 89 contigs tested both for association with phenotypic traits of interest and for ASE for the P9Ax8106A hybrid. Among them, 35 were associated with the phenotypic traits studied here, 14 were found as ASE and 5 were both associated and ASE. We found a lower proportion of ASE contigs among the associated ones compared with all the tested contigs leading to a nonsignificant P value of 0.709 when testing for enrichment (one-sided Fisher’s exact test). The same tendency (P value = 0.686) was found for the 8106AxP9A hybrid with 94 tested contigs among which only three were found to be both associated and ASE. Using simulations, we found a low probability of 9% (for P9Ax8106A) and 16% (for 8106AxP9A) to detect an excess of 10% of ASE genes among the selected one compared with the other genes from our data. This probability achieved almost 100% for an excess of 40% of ASE genes. Inferring the posterior distribution of ASE genes proportion among the selected genes fitting our data, we found a probability of 95% to observe an excess <7% of ASE genes among the associated ones compared with the other genes.
Discussion
Cis-Regulatory Divergence Quantification in Wild-Pearl Millet Accessions
In this study, we quantified allele-specific variation using whole transcriptome sequencing to investigate the role of cis-regulatory variation in adaptation at the intraspecific level. We found ∼15% genes showing unequal expression of the two parental alleles within F1 hybrids due to cis-regulatory differences between the two wild pearl millet accessions sampled from contrasting environmental habitats. This rate is at the lower-bound of ASE rate values ranging from ∼15% (He et al. 2012) to 80% (Skelly et al. 2011) in studies investigating ASE genome-wide for other organisms.
Several factors could explain the relatively low rate of ASE genes found in our study and the high variability of the proportion of ASE found between experiments. First, as the ASE rate depends on genetic variability among the two genotypes being studied for cis-regulatory divergence, this ratio is expected to depend on the divergence time separating the two genotypes. Thus, interspecific crosses should have a higher ASE rate than intraspecific crosses. This was observed in experiments investigating the two cases with the same pipeline of analysis. For instance, He et al. (2012) found 13.5% of the genes with unbalanced expression of the two parental alleles from an intraspecific cross of Arabidopsis thaliana. The ratio reached 15% for an interspecific cross between Arabidopsis thaliana and Arabidopsis lyrata. The same tendency was observed by Steige et al. (2017 and 2015) in the Capsella genus with 35% ASE genes at the intraspecific level compared with the 40% ASE genes at the interspecific level. He et al. (2012) reported that the difference between the two specific levels is not as high. They suggested that the extent of functional divergence should be constrained. It should also be noted that the methodological identification of ASE genes relies on crosses between divergent genotypes and should therefore constrain ASE experiments to species that have not diverged too far.
Second, the observed ASE ratio is probably strongly influence by the stringency of bioinformatics pipeline and the statistical analysis of the data of each specific study. The classification of ASE using RNA-seq data basically relies on read mapping to extract allele-specific read counts at heterozygous positions followed by statistical analysis of the expression data. For now, these two technical issues are still the subject of debate and vary greatly among ASE analyses with NGS (reviewed in Castel et al. 2015) and should therefore partly explain the high variability of the ASE rate between the different studies. Degner et al. (2009) showed that the reliability in ASE estimation depends to a great extent on the ability to control for read-mapping bias. As the reference genome contains only one allele at each genomic position, reads carrying the reference allele have more chance of being mapped at the right position than reads carrying the alternative allele, thus presenting at least one mismatch with the reference genome. Not taking this issue into account is expected to lead to overestimation of the ASE. Several approaches have been proposed to circumvent the mapping bias issue such as SNP-masked genome at known heterozygous positions (Degner et al. 2009), including known polymorphism data in an enhanced reference genome (Vijaya Satya et al. 2012) or mapping reads on the two parental genomes (McManus et al. 2010; Rozowsky et al. 2014). As the read mapping bias is also probably influenced by unidentified polymorphisms, the two first methods failed to fully remove the mapping bias (Degner et al. 2009; Castel et al. 2015). The latter strategy has been found to be more reliable but relies on the haplotype reconstruction of the two parental genomes. This approach could be challenging for a nonmodel organism with a large genome and heterozygous parental genotypes collected directly in the field. In our study, we chose to adopt a stringent filtering strategy of heterozygous positions by removing too polymorphic regions (Wood et al. 2015) and limiting the read counts to sections with only one identified SNP within a 100-bp window corresponding to the maximum size of a read. This strategy allowed us to efficiently control for mapping bias and to attain a reference ratio of 0.518, which is slightly above the expected nonbiased 0.5 reference ratio. Furthermore, the remaining mapping bias was taken into account in the statistical test being performed for ASE classification. Finally, the proportion of ASE should greatly vary depending on the statistical test being applied to classify genes as ASE. The binomial test is the simplest way of testing for ASE and is widely used (Fontanillas et al. 2010; Fraser et al. 2011; Cubillos et al. 2014). Other tests based on a beta-binomial modeling of allele-specific read counts have been developed (Skelly et al. 2011; León-Novelo et al. 2014; van de Geijn et al. 2015). As many methods and pipelines have been recently published, further tests are now needed to compare the available approaches for ASE inference.
Contribution of C is-Regulatory Variation to Adaptive Evolution at the Intraspecific Level
The ASE analysis performed here allowed the identification of genes exhibiting cis-regulatory variation between accessions evolving in contrasted environments. The parental genotypes used in our study were collected directly in the field and originated from very divergent habitats particularly with respect to humidity and temperature variables. Phenotypic characterization in common garden experiments highlight the high phenotypic variability among populations originating from the two parental genotypes (Ousseini et al. 2016). Northern populations, originated from a drier area, present smaller plants with fewer spikes that flower earlier than southern populations when evaluated in a common environment. These repeated observations along two environmental gradients of collection (in Niger as in the present study and in Mali) suggest that these changes are adaptive.
From GO annotations of pearl millet genes, we found that the genes with cis-regulatory divergence were significantly enriched in some GO terms that were specific to each hybrid. This result depending on the direction of the cross could be explained either by the difference in depth of sequencing between the two hybrids or by false positive inherent to GO term enrichment approaches (Pavlidis et al. 2012). We further found that the genes with cis-regulatory divergence between the two parental accessions were significantly enriched in GO terms related to “defense response” (GO:0006952) in the two hybrids. Thus, cis-regulatory variable genes are probably involved in the adaptive phenotypic divergence observed among populations. However, when we intersected our ASE gene set with the set of genes potentially involved in adaptation identified in another study based on the same populations (Berthouly-Salazar et al. 2016), we found that cis-regulatory variation contribute to adaptation of our populations but they are not over-represented among the selected genes nor among genes potentially involved in adaptive trait variation. Altogether, our results suggest that adaptation do not rely more on cis-regulatory variation in our experiment than expected by chance. It should be noticed that the method used to detect the genes under selection is more effective for strong signature of selection (Berthouly-Salazar et al. 2016). Such approach might be less effective to identify selection from standing variation or with small phenotypic effect inducing soft signature of selection (Yeaman 2015). Therefore, from our experimental design we were not able to exclude the possibility of higher contribution of cis-regulatory variation to the latter type of selection. However, our finding is consistent with other studies claiming that the implication of cis-regulation in phenotypic evolution varies depending on the divergence time between genotypes. The idea first emerged from a compilation of studies showing a greater proportion of null coding mutations causing phenotypic variation in intraspecific comparisons that contrast with the greater proportion of cis-regulatory mutations in interspecific comparisons (Stern and Orgogozo 2008, 2009). Some experimental validations then appeared in comparative studies investigating both cis and trans-regulatory divergence from intra and inter-crosses within the drosophila genus (Wittkopp et al. 2008; Coolon et al. 2014) and in yeast (Emerson et al. 2010). The main argument supporting discordance depending on the divergence time is that trans-acting mutations are more frequent whereas cis-acting mutations arise less often but are more likely to be fixed within a population over time due to the reduced pleiotropic effect of those mutations (Wittkopp et al. 2008) and a higher effect on gene expression (Gruber et al. 2012; Metzger et al. 2016). From our experimental design, we were not able to quantify trans-regulatory divergence in our plant material since we used highly heterozygous parental accessions directly collected in the field which prevented us from precisely identifying the parental origin of the alleles in the hybrids. However, our study is the first to investigate the intraspecific contribution of cis-regulatory divergence to adaptive evolution using accessions from natural populations instead of inbred parental lines from laboratories usually used in such studies.
Conclusion
In our study, we succeeded in quantifying ASE in wild pearl millet, a nonmodel organism, from parental genotypes collected directly in the field. With a careful and stringent analysis pipeline, we were able to confidently identify genes with cis-regulatory divergence between genotypes at the intraspecific level. The low ASE rate compared with other studies was probably due to the short divergence time and to the stringency of the pipeline we used. Finally, our results are consistent with the expectation that at the intraspecific level cis-regulatory variation are not predominantly involved in adaptive evolution. While our study helped investigate the contribution of cis-regulatory divergence to adaptive traits, technical advances will enable us to advance our understanding of the genetic basis of phenotypic evolution, for example by investigating the role of isoforms in adaptive evolution (Gao et al. 2015; Mallarino et al. 2016) and offer an integrative view of the interactions between regulatory mechanisms and alternative transcription.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Authors Contributions
Y.V. designed the study; M.C. and C.M. generated the data; C.B.S. and I.S.O. contributed data; B.R. performed the analyses; and B.R. and Y.V. wrote the draft, comments from authors were included.
Supplementary Material
Acknowledgments
This work was supported by the Agence Nationale de la Recherche [ANR 12-ADAP-0002] to Y.V. We thank the GeT-genotoul platform in Toulouse and the MGX platform in Montpellier for RNA and DNA sequencing.
Literature Cited
- Alexa A, Rahnenfuhrer J, Lengauer T. 2006. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 22:1600–1607. [DOI] [PubMed] [Google Scholar]
- Alonso CR, Wilkins AS. 2005. Opinion: the molecular elements that underlie developmental evolution. Nat Rev Genet. 6:709–715. [DOI] [PubMed] [Google Scholar]
- Arunkumar R, Maddison TI, Barrett SCH, Wright SI. 2016. Recent mating-system evolution in Eichhornia is accompanied by cis-regulatory divergence. New Phytol. 211:697–707. [DOI] [PubMed] [Google Scholar]
- Berthouly-Salazar C, et al. 2016. Genome scan reveals selection acting on genes linked to stress response in wild pearl millet. Mol Ecol. 25:5500–5512. [DOI] [PubMed] [Google Scholar]
- Castel SE, Levy-Moonshine A, Mohammadi P, Banks E, Lappalainen T. 2015. Tools and best practices for data processing in allelic expression analysis. Gen Biol. 16:195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coolon JD, McManus CJ, Stevenson KR, Graveley BR, Wittkopp PJ. 2014. Tempo and mode of regulatory evolution in Drosophila . Genome Res. 24:797–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubillos FA, et al. 2014. Extensive cis-regulatory variation robust to environmental perturbation in Arabidopsis . Plant Cell Online 26:4298–4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degner JF, et al. 2009. Effect of read-mapping biases on detecting allele-specific expression from RNA-sequencing data. Bioinformatics 25:3207–3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, et al. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dussert Y, et al. 2013. Polymorphism pattern at a miniature inverted-repeat transposable element locus downstream of the domestication gene Teosinte-branched1 in wild and domesticated pearl millet. Mol Ecol. 22:327–340. [DOI] [PubMed] [Google Scholar]
- Emerson JJ, et al. 2010. Natural selection on cis and trans regulation in yeasts. Genome Res. 20:826–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engström PG, et al. 2013. Systematic evaluation of spliced alignment programs for RNA-seq data. Nat Methods 10:1185–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontanillas P, et al. 2010. Key considerations for measuring allelic expression on a genomic scale using high-throughput sequencing. Mol Ecol. 19:212–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser HB, et al. 2011. Systematic detection of polygenic cis-regulatory evolution. PLoS Genet. 7:e1002023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q, Sun W, Ballegeer M, Libert C, Chen W. 2015. Predominant contribution of cis-regulatory divergence in the evolution of mouse alternative splicing. Mol Syst Biol. 11:816816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Alcalde F, et al. 2012. Qualimap: evaluating next-generation sequencing alignment data. Bioinformatics 28:2678–2679. [DOI] [PubMed] [Google Scholar]
- Gruber JD, Vogel K, Kalay G, Wittkopp PJ. 2012. Contrasting properties of gene-specific regulatory, coding, and copy number mutations in Saccharomyces cerevisiae: frequency, effects, and dominance. PLoS Genet. 8:e1002497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, et al. 2012. Genome-wide analysis of cis-regulatory divergence between species in the Arabidopsis genus. Mol Biol Evol. 29:3385–3395. [DOI] [PubMed] [Google Scholar]
- He F, et al. 2016. The footprint of polygenic adaptation on stress-responsive cis-regulatory divergence in the Arabidopsis genus. Mol Biol Evol. 33:2088–2101. [DOI] [PubMed] [Google Scholar]
- Hoekstra HE, Coyne JA. 2007. the locus of evolution: evo devo and the genetics of adaptation. Evolution 61:995–1016. [DOI] [PubMed] [Google Scholar]
- Josephs EB, Lee YW, Stinchcombe JR, Wright SI. 2015. Association mapping reveals the role of purifying selection in the maintenance of genomic variation in gene expression. Proc Natl Acad Sci. 112:15390–15395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MC, Wilson AC. 1975. Evolution at two levels in humans and chimpanzees. Science 188:107–116. [DOI] [PubMed] [Google Scholar]
- Lemmon ZH, Bukowski R, Sun Q, Doebley JF. 2014. The role of cis regulatory evolution in maize domestication. PLoS Genet. 10:e1004745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- León-Novelo LG, McIntyre LM, Fear JM, Graze RM. 2014. A flexible Bayesian method for detecting allelic imbalance in RNA-seq data. BMC Genomics 15:920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch VJ, Wagner GP. 2008. Resurrecting the role of transcription factor change in developmental evolution. Evolution 62:2131–2154. [DOI] [PubMed] [Google Scholar]
- Mack KL, Campbell P, Nachman MW. 2016. Gene regulation and speciation in house mice. Genome Res. 26:451–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallarino R, Linden TA, Linnen CR, Hoekstra HE. 2016. The role of isoforms in the evolution of cryptic coloration in Peromyscus mice. Mol. Ecol. 26:245–258 [DOI] [PubMed] [Google Scholar]
- Mariac C, et al. 2011. Genetic basis of pearl millet adaptation along an environmental gradient investigated by a combination of genome scan and association mapping. Mol Ecol. 20:80–91. [DOI] [PubMed] [Google Scholar]
- Mariac C, et al. 2014. Cost-effective enrichment hybridization capture of chloroplast genomes at deep multiplexing levels for population genetics and phylogeography studies. Mol Ecol Resources 14:1103–1113. [DOI] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17:10. [Google Scholar]
- McKenna A, et al. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus CJ, et al. 2010. Regulatory divergence in Drosophila revealed by mRNA-seq. Genome Res. 20:816–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger BPH, et al. 2016. Contrasting frequencies and effects of cis- and trans-regulatory mutations affecting gene expression. Mol Biol Evol. 33:1131–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oumar I, Mariac C, Pham J-L, Vigouroux Y. 2008. Phylogeny and origin of pearl millet (Pennisetum glaucum [L.] R. Br) as revealed by microsatellite loci. Theor Appl Genet. 117:489–497. [DOI] [PubMed] [Google Scholar]
- Ousseini IS, et al. 2016. Myosin XI is associated with fitness and adaptation to aridity in wild pearl millet. Heredity. http://dx.doi.org/10.5061/dryad.mn3g7. [DOI] [PMC free article] [PubMed]
- Pavlidis P, Jensen JD, Stephan W, Stamatakis A. 2012. A critical assessment of storytelling: gene ontology categories and the importance of validating genomic scans. Mol Biol Evol. 29:3237–3248. [DOI] [PubMed] [Google Scholar]
- Prud’homme B, Gompel N, Carroll SB. 2007. Emerging principles of regulatory evolution. Proc Natl Acad Sci. 104:8605–8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanel A, Lago S, Prandi D, Sboner A, Demichelis F. 2015. ASEQ: fast allele-specific studies from next-generation sequencing data. BMC Med Genomics 8:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozowsky J, et al. 2014. AlleleSeq: analysis of allele-specific expression and binding in a network framework. Mol Syst Biol. 7:522–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skelly DA, Johansson M, Madeoy J, Wakefield J, Akey JM. 2011. A powerful and flexible statistical framework for testing hypotheses of allele-specific gene expression from RNA-seq data. Genome Res. 21:1728–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steige KA, Laenen B, Reimegård J, Scofield D, Slotte T. 2017. Genomic analysis reveals major determinants of cis-regulatory variation in Capsella grandiflora [cited 2016 Nov 23]. Available from: http://biorxiv.org/lookup/doi/10.1101/034025. [DOI] [PMC free article] [PubMed]
- Steige KA, Reimegård J, Koenig D, Scofield DG, Slotte T. 2015. Cis-regulatory changes associated with a recent mating system shift and floral adaptation in Capsella . Mol Biol Evol. 32:2501–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern DL, Orgogozo V. 2008. The loci of evolution: how predictable is genetic evolution?. Evolution 62:2155–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern DL, Orgogozo V. 2009. Is genetic evolution predictable?. Science 323:746–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Geijn B, McVicker G, Gilad Y, Pritchard JK. 2015. WASP: allele-specific software for robust molecular quantitative trait locus discovery. Nat Methods 12:1061–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijaya Satya R, Zavaljevski N, Reifman J. 2012. A new strategy to reduce allelic bias in RNA-Seq readmapping. Nucleic Acids Res. 40:e127–e127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner GP, Lynch VJ. 2008. The gene regulatory logic of transcription factor evolution. Trends Ecol Evol. 23:377–385. [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, Haerum BK, Clark AG. 2008. Regulatory changes underlying expression differences within and between Drosophila species. Nat Genet. 40:346–350. [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, Kalay G. 2011. Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nat. Rev. Genet. 13:59–49 [DOI] [PubMed] [Google Scholar]
- Wood DLA, et al. 2015. Recommendations for accurate resolution of gene and isoform allele-specific expression in RNA-Seq data. PLoS One 10:e0126911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray GA. 2007. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet. 8:206–216. [DOI] [PubMed] [Google Scholar]
- Yeaman S. 2015. Local adaptation by alleles of small effect. Am Nat. 186:S74–S89. [DOI] [PubMed] [Google Scholar]
- Zhang X, Borevitz JO. 2009. Global analysis of allele-specific expression in Arabidopsis thaliana . Genetics 182:943–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.