Abstract
Nuclear receptors (NRs) regulate numerous aspects of the endocrine system. They mediate endogenous and exogenous cues, ensuring a homeostatic control of development and metabolism. Gene duplication, loss and mutation have shaped the repertoire and function of NRs in metazoans. Here, we examine the evolution of a pivotal orchestrator of cholesterol metabolism in vertebrates, the liver X receptors (LXRs). Previous studies suggested that LXRα and LXRβ genes emerged in the mammalian ancestor. However, we show through genome analysis and functional assay that bona fide LXRα and LXRβ orthologues are present in reptiles, coelacanth and chondrichthyans but not in cyclostomes. These findings show that LXR duplicated before gnathostome radiation, followed by asymmetric paralogue loss in some lineages. We suggest that a tighter control of cholesterol levels in vertebrates was achieved through the exploitation of a wider range of oxysterols, an ability contingent on ligand-binding pocket remodeling.
Keywords: nuclear receptors, cholesterol, liver X receptor, chordates
Introduction
The appearance of complex endocrine systems, coordinating distinct biological functions such as development, metabolism or reproduction, represents a hallmark of bilaterian evolution (Bertrand et al. 2004). This increased complexity required a homeostatic coalescence of tissue-specific metabolic pathways and signaling cascades, with nuclear receptors (NRs) as major mediators of endocrine processes. These metazoan-specific transcription factors are mostly triggered by ligands of diverse origin (hormonal, nutritional, environmental), selectively modulating transcription upon recognition of specific DNA responsive elements, in the promoter region of target genes (Laudet and Gronemeyer 2002).
Evolutionary events like gene duplication, loss or mutation significantly contributed to functionally diversify NRs with likely impacts on organism physiology (e.g., Bertrand et al. 2004; Escriva et al. 2006; Bridgham et al. 2008, 2010; Carroll et al. 2008; Ogino et al. 2016). Gene duplication was particularly relevant in the case of vertebrates (e.g., Gutierrez-Mazariegos et al. 2016). As with many other gene families, the vertebrate NR repertoire augmented as a consequence of whole-genome duplications (WGD) in early vertebrate evolution (Thornton 2001; Bertrand et al. 2004; Escriva et al. 2006; Lecroisey et al. 2012). This is thought to underscore the complexity and elaboration of the vertebrate endocrine system.
Here, we scrutinize a particularly intriguing case involving a NR group, the liver X receptor (LXR, NR1H). LXR plays a critical role in cholesterol homeostasis, regulating the expression of genes involved in the efflux, transport, and excretion (Kalaany and Mangelsdorf 2006; Laurencikiene and Ryden 2012). Originally classified as “ligand orphan” (Willy et al. 1995), it was later discovered that oxysterols (cholesterol oxidized derivatives) such as 24(S)-hydroxycholesterol, 22(R)-hydroxycholesterol and 24(S),25-epoxycholesterol are their bona fide ligands at physiological concentrations (Janowski et al. 1996; Lehmann et al. 1997). The repertoire of LXRs genes is surprisingly unequal in the investigated taxa. In mammals, two genes LXRα and LXRβ that share similar binding properties have been identified. Despite these similarities they control specific as well as overlapping physiological processes (Peet et al. 1998; Alberti et al. 2001; Juvet et al. 2003; Steffensen et al. 2003; Gerin et al. 2005; Korach-Andre et al. 2010). In contrast to mammals, a single gene was identified in birds (e.g., Gallus gallus), teleosts (e.g., Danio rerio) and amphibians (e.g., Xenopus tropicalis and X. laevis) and tunicates (Maglich et al. 2003; Reschly et al. 2008; Krasowski et al. 2011). This phylogenetic distribution was interpreted as a result from a duplication of a single LXR gene in mammalian ancestry (Maglich et al. 2003; Reschly et al. 2008; Krasowski et al. 2011). Upon duplication in the mammalian ancestor one paralogue retained a more ubiquitous expression, while the second evolved specific roles in cholesterol metabolism (Reschly et al. 2008). However, an alternative hypothesis involving secondary loss of one LXR independently in multiple lineages would also account for the observed evolutionary pattern.
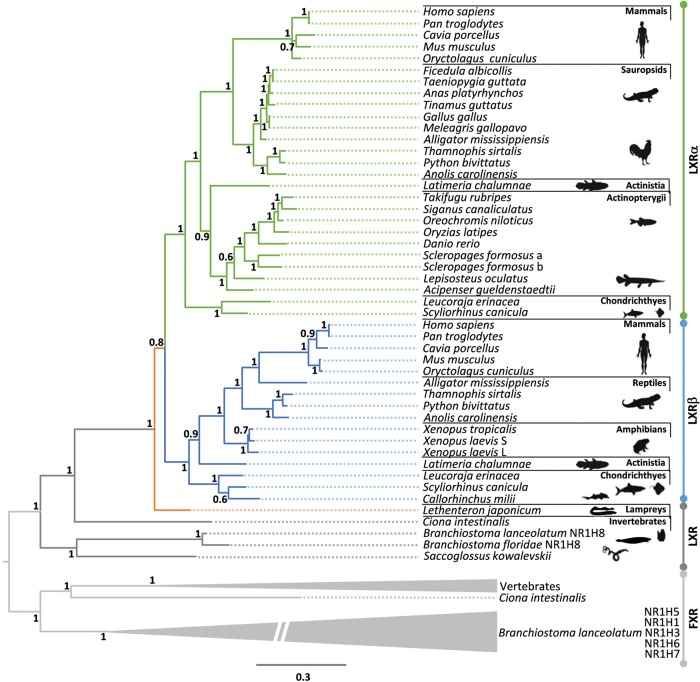
To discriminate between these evolutionary scenarios we investigated a broad range of chordate clades, including the chondrichthyans, cyclostomes and cephalochordates. To examine the LXR gene repertoire in vertebrate species, we searched the genome and transcriptome sequences of selected species from mammals, birds, reptiles, amphibians, coelacanth, teleosts, lepisosteiformes, chondrichthyans and cyclostomes. Confirming previous findings, LXRα and LXRβ were identified in mammals (Reschly et al. 2008; Krasowski et al. 2011). In contrast, single-copy LXR genes were retrieved from spotted gar, most of teleosts and birds, consistent with previous observations (Reschly et al. 2008; Krasowski et al. 2011). However, our extensive searches uncovered some gnathostome lineages with two LXR sequences: the Asian arowana (osteoglossomorpha), the coelacanth and the anole lizard. Further scrutiny of the available transcriptomes of the elephant shark, revealed a complete sequence and two nonoverlapping LXR sequence fragments (supplementary fig. S1, Supplementary Material online). Searches of additional genome and transcriptome sequences of cartilaginous fishes, the little skate Leucoraja erinacea and the small spotted catshark Scyliorhinus canicula, yielded several overlapping partial sequences with similarity to either LXRα or LXRβ from bony vertebrates. Through a combination of PCR strategies we were able to recover the full or near-full coding sequence of two LXR genes in the little skate and the small spotted catshark. In contrast, a single LXR-like sequence was identified in the genome and transcriptome datasets of the Japanese lamprey, Lethenteron japonicum. Searches to the genome assembly and transcriptomes of the sea lamprey, Petromyzon marinus, allowed also the identification of a single LXR-like gene although too short for phylogenetic analysis (not shown). The investigation of the genome sequences from two cephalochordate species allowed the recovery of 10 LXR/FXR-like sequences (NR1H1-10), similar to those found in previous studies (Bertrand et al. 2011; Lecroisey et al. 2012). However, a clear identity was only verified in seven, as three B. floridae sequences have been discontinued, namely NR1H2 (XP_002224320.1) NR1H4 (XP_002224321.1) and NR1H10 (XP_002246474.1). Additionally, another sequence (NR1H9) shows a truncated DBD (not shown). Thus, a total of six cephalochordate sequences were considered for the main phylogenetic analysis.
To assign orthology/paralogy of the recovered sequences we next carried out phylogenetic analysis (fig. 1 and supplementary fig. S2, Supplementary Material online). Two monophyletic clades containing, respectively, LXRα and LXRβ are observed, out-grouped by single-copy LXR sequences from the Japanese lamprey, cephalochordates, sea squirt and hemichordate (fig. 1 and supplementary fig. S2, Supplementary Material online). Thus, data derived from genome, transcriptome and phylogenetics indicate that LXRα orthologues are present in all the examined gnathostome species (except amphibians), while LXRβ is found in mammals, reptiles, amphibians, coelacanth and cartilaginous fish. The little skate and the small spotted catshark LXRs genes robustly groups with the LXRα and LXRβ clades, respectively (fig. 1), providing unequivocal support for their orthology. The Asian arowana sequences are both of the LXRα type, a probable consequence of the teleost-specific genome duplication (3R) or a lineage-specific duplication. In summary, phylogenetic analysis suggests a much earlier origin of LXRα and LXRβ than the timing of mammalian radiation (Reschly et al. 2008), predating gnathostome divergence but after splitting from cyclostomes (fig. 1). Our analysis also confirms that the unusual NR1H gene number in cephalochordates is the result of a lineage specific expansion of the FXR clade as previously suggested (fig. 1 and supplementary fig. S2, Supplementary Material online) (Bertrand et al. 2011; Lecroisey et al. 2012).
Fig. 1.—
Phylogenetic analysis of NR1H nuclear receptors (LXRs/FXRs). Bayesian phylogenetic tree of LXR and FXR amino acid sequences; numbers at nodes indicate posterior probabilities.
To discriminate between true gene loss and absence of sequencing data we next investigated the synteny of LXR genes in selected species with available genome data (fig. 2). We find strong synteny conservation in the examined LXRα loci. In both Xenopus species the loss of LXRα is confirmed since the locus is conserved with no LXR-like intervening sequence. Orthologous flanking genes in the LXRβ loci are not as evident but still statistically supported, in reptiles and amphibians (fig. 2). This locus is entirely absent in birds as previously noted (Lovell et al. 2014). The LXRβ of the elephant shark maps to a single gene scaffold impeding synteny comparisons (not shown). The cephalochordate LXR locus shows some degree of conservation when compared with vertebrates (fig. 2). One flanking gene, ACP-like, presents the corresponding human orthologues, ACP2 and ACPT, located in close proximity to LXRα (Chr11 p11.2 47.20 Mb) and LXRβ (Chr19 q13.33 50.8 Mb), respectively. A second flanking gene in the cephalochordate LXR locus, PARV-like, has the human orthologues locating to the LXRα locus, PARVA (Chr11 p15.3 12.40 Mb) and PARVB in human chromosome 22 (Chr22 q13.31 44 Mb) (fig. 2). In contrast, the cephalochordate FXR-like genes localize to separate and nonsyntenic genomic locations with respect to the LXR loci (supplementary fig. S3, Supplementary Material online).
Fig. 2.—
Synteny maps of LXRα and LXRβ loci. (A) Detail of the LXRα locus and LXRβ locus in the selected vertebrate and cephalochordate species; Chr and Sca indicate chromosome and scaffold, respectively. (B) Statistical support of synteny analysis; P values indicate the probability of identifying nonhomologous chromosomal segments, and S indicates the size of the chromosomal segment identified.
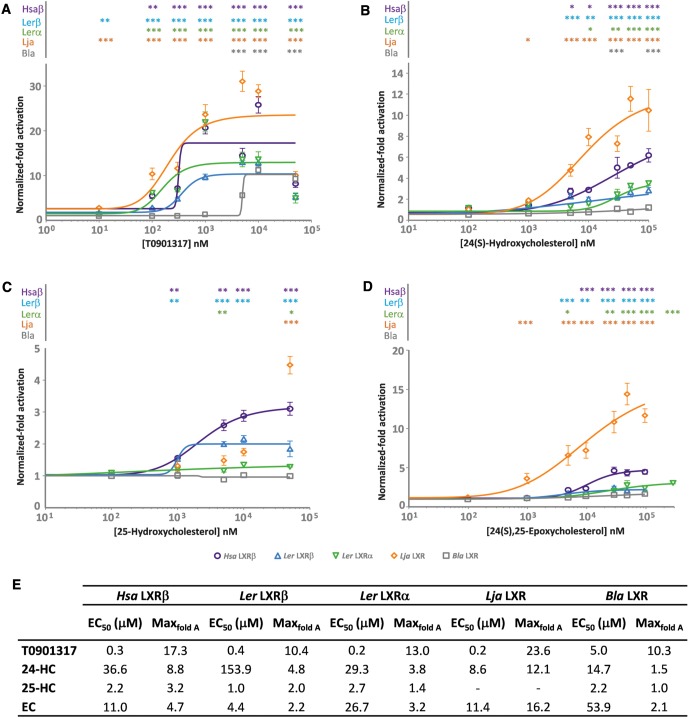
To determine the ligand binding properties of the little skate LXRα and LXRβ, and the single proteins of Japanese lamprey and European amphioxus, we investigated their capacity to bind to physiological and synthetic LXR ligands. The synthetic compound T0901317 is known to be a LXR agonist in several species. The agonistic response was observed for the two little skate receptors and for the lamprey receptor with significant activations (P < 0.001), in all of the tested concentrations (fig. 3A ), suggesting that this synthetic compound is also a potent agonist for these three receptors (L. erinacea LXRα EC50 = 0.2 μM, L. erinacea LXRβ EC50 = 0.4 μM, L. japonicum LXR EC50 = 0.2 μM). Interestingly, amphioxus LXR is less sensitive to this agonist, displaying statistical significant activation (P < 0.001) only at higher concentrations (EC50 = 5.0 μM). We next assayed three oxysterols: 24(S)-hydroxycholesterol (24-HC), 25-hydroxycholesterol (25-HC) and 24(S),25-epoxycholesterol (24,25-EC) (fig. 3B–D ). Previous studies including human and mouse LXRs reported that 24-HC and 24,25-EC robustly activated both LXRα and LXRβ; 25-HC, on the other hand, induced lower transcriptional responses at the tested concentrations, with a more prominent decrease observed for LXRα (Reschly et al. 2008). A similar pattern was observed for the little skate α and β isoforms. The lamprey LXR was also strongly induced by 24-HC and 24,25-EC, producing the highest maximal activations. Upon exposure to 25-HC, lamprey LXR transcriptional response was also prominent and statistically significant (P < 0.001), yet only at higher concentrations. Contrary to the vertebrate isoforms, the amphioxus LXR was residually, but significantly, activated by 24-HC alone. Overall, the obtained EC50 values for the tested LXR/oxysterol pairs are much higher, in the micromolar range than those of T0901317 (fig. 3E ). Likewise, the respective maximal activations were lower than for T0901317, similar to mammalian LXRs (Reschly et al. 2008). Regarding the Japanese lamprey LXR exposed to 25-HC, due to the absence of a dose–response plateau, within the tested concentration range, the estimation of EC50 and maximal activation values was not performed.
Fig. 3.—
Functional analysis of L. erinacea, L. japonicum and B. lanceolatum LXRs LBD. Dose–response curves for LXRs activation by T0901317 (A), 24(S)-hydroxycholesterol (B), 25-hydroxycholesterol (C) and 24(S),25-epoxycholesterol (D) for H. sapiens LXRβ (○), L. erinacea LXRβ (Δ) and LXRα (∇), L. japonicum (◊) and B. lanceolatum (□); EC50 and maximum normalized-fold activation (Maxfold A) values for HsaLXRβ, LerLXRβ and LerLXRα (E). The activation of LXR was normalized to the control condition (DMSO without ligand) represented by 10 − 2 M. Hsa stands for H. sapiens, Ler stands for L. erinacea, Lja stands for L. japonicum and Bla stands for B. lanceolatum. The values represented are the means with ±SE from three separate experiments. *P < 0.05, **P < 0.01, ***P < 0.001.
By performing a comprehensive search of the LXR gene repertoire in chordate genomes, we unveil the accurate evolutionary functional diversification of this NR group (fig. 4). A single LXR member was previously found in basal chordates such as tunicates (Reschly et al. 2008), similar to what we describe here in cephalochordates and cyclostomes. In contrast, we also report that LXRα and LXRβ are gnathostome-specific paralogues found in a wide array of lineages including the chondrichthyans, with independent gene loss of either paralogue in several lineages (supplementary fig. S4, Supplementary Material online). The overall timing of LXR gene duplication as determined from our phylogenetic analysis is coincident with that of whole genome duplications in vertebrate ancestry (1R/2R) (Putnam et al. 2008; Smith and Keinath 2015), although the absence of synteny data from lamprey and chondrichthyans impedes a detailed discussion.
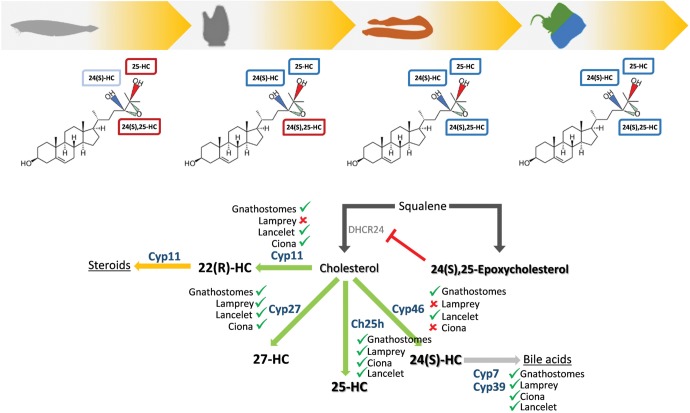
Fig. 4.—
Elaboration of the metabolic and signaling oxysterols cascades in chordate evolution. Top—Binding specificities of LXRs to oxysterols in cephalochordates, tunicates, cyclostomes and chondrichthyans: red and blue boxes represent no activation and activation of LXR-dependent transcription, respectively, and light blue boxes represent residual LXR activation. Bottom—Schematic representation of cholesterol synthesis and oxidation pathways: green ✓ and red X stand for presence or absence of the corresponding cytochrome P450s oxygenase (CYP) gene, respectively (Nelson et al. 2013).
Unlike vertebrates, the amphioxus orthologue displays low to null capacity to activate transcription upon exposure to any of the tested oxysterols. The tunicate LXR, we should recall, exhibits an intermediate pharmacology: not activated by synthetic LXR agonists, yet induced by some oxysterols and other steroidal compounds, such as androstenol and androstanol. Besides steroids, the tunicate LXR was strongly activated by 6-Formylindolo(3,2-b)carbazole, a tryptophan photoproduct and proposed endogenous aryl hydrocarbon receptor ligand (Reschly et al. 2008). Certainly, if steroids, or other polycyclic compounds, are the tunicate LXR physiological ligands, they differ from those of vertebrates (Reschly et al. 2008). Yet, vertebrate LXR ligand capacity seems contingent on an emerging ability to accommodate oxysteroid backbones: originally limited to a narrower set of oxysterols in the ancestor of tunicates and vertebrates. The broader variety of oxysterol ligands, capable of specifically activating LXRs, first appeared in the ancestor of vertebrates, as depicted in our results with the cyclostome and chondrichthyan LXR receptors. In agreement, the comparison of the LXR ligand binding pockets (LBP) from invertebrate and vertebrate species indicates that a significant remodeling occurred, after the separation of tunicates from vertebrates, with multiple substitutions entrenched in the vertebrate lineage (supplementary fig. S5, Supplementary Material online). Curiously, this parallels the assembly of bile acid synthesis pathways, of which oxysterols serve as intermediaries. Classic and alternate synthesis pathways are triggered by cytochrome P450s (CYPs) oxygenase-dependent cholesterol oxidation (fig. 4). Similarly to the NR repertoire, the original, tandemly repeated, CYP clans were expanded by duplication (Nelson et al. 2013). According to the ligand exploitation paradigm, selection for novel biosynthetic pathways generates a collection of intermediaries and prompts new combinations of ligand/receptor pairs (Thornton 2001). Thus, it is conceivable that both metabolic (oxysterols) and signaling (LXRs) pathways evolved, in parallel and opportunistically, towards the integrated network observed in gnathostomes.
The coexistence of different complements of LXR genes in gnathostome lineages is enigmatic. In effect, the retention of LXRα and LXRβ in many lineages indicates that both NRs evolved separate roles in the aftermath of gene duplication. Paradoxical though, the vertebrate LXR paralogues examined to date exhibit some redundancy in ligand specificity. Interestingly, the zebrafish and the amphibian LXRα and LXRβ, respectively, also show conserved ligand specificity compared with both mammalian LXRs (Reschly et al. 2008). Thus, the coexistence of one (birds, amphibians and teleosts) or two (chondrichthyans, coelacanths, reptiles and mammals) LXR paralogues in diverse gnathostome lineages does not imply distinguishable differences in LXR ligand preference. Given the apparent functional redundancy of both receptors at ligand preference, it is possible that there could be differences in the transcriptional regulation of the genes coding for the receptors per se. Data from mouse supports this hypothesis. In fact, Peroxisome Proliferator-Activated Receptors (PPARs), major regulators of lipid metabolism, were suggested to induce the transcription of LXRα, but not LXRβ, indirectly regulating cholesterol metabolism (Tobin et al. 2000; Chawla et al. 2001). Nevertheless, differential activation mechanics, such as nuclear targeting, co-repressor or co-activator binding, phosphorylation, could favor specific, isoform-specific, metabolic pathways.
Materials and Methods
Amino acid sequences were retrieved through BLAST searches in the publically available genome databases (Ensembl, GenBank, Skatebase; http://skatebase.org/), using as reference annotated human LXRα and LXRβ sequences. Sequence sampling included representatives of major vertebrate lineages: mammals (Homo sapiens, Pan troglodytes, Mus musculus, Cavia porcellus, Dasypus novemcinctus, Oryctolagus cuniculus), reptiles (Thamnophis sirtalis, Python bivittatus, Anolis carolinensis, Alligator mississippiensis, Alligator sinensis) birds (G. gallus, Meleagris gallopavo, Ficedula albicollis, Taeniopygia guttata, Anas platyrhynchos), amphibians (X.laevis, X. tropicalis), sarcopterygii (Latimeria chalumnae), euteleostei (Takifugu rubripes, Siganus canaliculatus, Oryzias latipes, Oreochromis niloticus, D. rerio) osteoglossomorpha (Scleropages formosus), holostei (Lepisosteus oculatus), chondrichthyans (L. erinacea, S. canicula, Callorhinchus milii), cyclostomes (L. japonicum and P. marinus) and four invertebrate deuterostomes (Ciona intestinalis, Branchiostoma lanceolatum, B. floridae and Saccoglossus kowalevskii). Retrieved sequences and corresponding accession numbers are listed in the Supplementary Material online. All sequences were aligned with MAFFT alignment software (Katoh and Toh 2010) using the E-INS-i model. Sequence alignment was visualized and edited in Geneious® v7.1.7 (available upon request). The columns containing 90% of gaps were stripped. The final sequence alignment contained 86 sequences and 547 positions and was used to perform phylogenetic analysis. Bayesian phylogenetic calculation was performed with MrBayes v 3.2.3 sited in the CIPRES Science Gateway V3.3 (Miller et al. 2015). Calculation parameters were as follows, generation number = 1000000, rate matrix for aa = mixed (Jones), nruns = 2, nchains = 4, temp = 0.2, sampling set to 500 and burnin to 0.25.
LXRα and LXRβ genes were localized onto the human chromosomes Chr11 and Chr19, respectively, corresponding LXR gene and the neighboring genes were collected from Ensembl and GenBank databases. Human loci (GRCh38) were further used as a reference to assemble the synteny maps of the remaining species: G. gallus (Galgal4), A. carolinensis (AnoCar2.0), X. tropicalis (GCF_000004195.3), X. laevis (GCF_001663975.1), L. chalumnae (LatCha1), D. rerio (GRCz10), L. oculatus (LepOcu1), B. floridae (GCF_000003815.1) and B. lanceolatum (BraLan2). Synteny statistics was calculated using CHSminer v1.1 (Wang et al. 2009) search parameters were maintained as default: maximal gap ≤ 30 and size ≥ 2, with the exception of the D. rerio Ch7 versus H. sapiens Chr11, were maximal gap was set to 80 to accommodate the highly rearranged locus in D. rerio.
Leucoraja erinacea were collected from the coast of Woods Hole, MA. All tissues were collected and preserved in RNAlater and stored at −20 °C. Total RNA was isolated using an Illustra RNAspin Mini RNA Isolation Kit (GE Healthcare, UK) according to the manufacturer’s recommendations, including the on-column treatment of isolated RNA with RNase-free DNase I. Using 500ng of liver RNA as input cDNA was synthetized with the iScript cDNA Synthesis Kit (Bio-Rad), according to the manufacturers’ recommendations. Using two partial LXRα-like segments retrieved through BLAST searches in Skatebase, a set of primers were designed to isolate the partial open reading frame (ORF) of LerLXR with Phusion Flash master mix (Thermo Fisher Scientific). The isolated partial ORF was further extended through rapid amplification of cDNA ends (RACE) technique. For this 5´ RACE ready cDNA was prepared from previously isolated RNA using the SMARTER RACE cDNA amplification kit (Clontech) according to manufactures recommendations. The full ORF of LerLXRα was amplified using Phusion Flash master mix (Thermo Fisher Scientific). For LXR isolation in amphioxus (KY094511), B. lanceolatum, adult specimens were collected from Ria Formosa, Portugal. Total RNA and cDNA synthesis were performed as described earlier. A combination of PCR strategies (e.g., degenerate primers, RACE PCR and genome database search) was employed to isolate the full ORF of the LXR orthologue.
The ligand binding domain (LDB) including the hinge region of H. sapiens LXRβ (U07132.1), L. erinacea LXRα (Ler LXRα) and L. erinacea LXRβ (LerLXRβ) (transcriptome Contig89816 from Skatebase), L. japonicum LXR (LjaLXR) and B. lanceolatum LXR (BlaLXR) were isolated by PCR with Phusion Flash master mix (Thermo Fisher Scientific) using the specific primers (HsaLXRβ PF-ACTGGGATCCTAGATCCGGAAGAAGAAGATTCGG and PR-ATATCTAGATCACTCGTGGACGTCCCAGAT; LerLXRα PF-ACTGGGATCCGGAA GAAAATGAAGAAGCTGGAG and PR-ATATCTAGAAGTCATTCCTGCATGTCCCAG; LerLXRβ PF-ACTGGGATCCAGAAGAAGCAGAGGAAGCGGGAG and PR-ATATCTAG ACCCTCCGTCACTCATGCAC; LjaLXR PF-CCCTCTAGACGTCGGAAAAACGACGA ACC and PR-AAAGGTACCTCACTCGTGAACGTCCCAGA; BlaLXR PF-GCATCTAGA CTCCGCGACAGAGCACC and PR-CCGGGTACCCTACTGTGGAACGTCCCATAT). PCR reaction comprised an initial denaturation step at 98°C for 10 s followed by 40 cycles of denaturation at 98°C for 1 s annealing at 62°C (LerLXRs) or 60°C (LjaLXR and BlaLXR) for 5 s and extension at 72°C for 15 s, with an final extension step for 60 s. The resulting PCR products and pBIND (AF264722; Promega) were digested with BamHI and XbaI (LerLXRs) or XbaI and KpnI (LjaLXR and BlaLXR) restriction enzymes (Promega) and ligated withT4 ligase (Promega) to produce GAL4-LBD “chimeric” receptor. COS-1 cells were seeded into a 24-well plate at a concentration of 2×105 cells/ml 24 h prior to transfection. Cells were transfected with lipofectamine 2000 reagent (Invitrogen) and the transfection medium OptiMEM (Life Technologies) according manufacturer’s indications, using 500 ng of pBIND constructions and 1,000 ng of pGL4.31[luc2P/GAL4UAS/Hygro] vector (DQ487213; Promega). After 5 h of incubation, transfection media was replaced with phenol red-free DMEM (PAN-Biotech) supplemented with 10% of charcoal stripped fetal bovine serum (PAA Laboratories) and cells were treated with varying concentration of oxysterols (ranging from 101 to 105.5 nM) in DMSO. Cells were lysed 24 h after transfection and assayed for luciferase activity with Dual-Luciferase Reporter Assay System (Promega), according to the manufacturer’s instructions. All transfections were performed in triplicate. Data was presented as means± standard error (SE) from three separate experiments. SigmaPlot 11.0 software was used to calculate the EC50 values from the sigmoidal dose–response curves and the differences between groups variation were analysed with one-way ANOVA. Holm–Sidak was used to identify significant differences in the normalized-fold activation of the LXR receptors with the several compounds tested. The level of significance (P value) was set to 0.05.
The synthetic LXR agonist T0901317 and 24(S)-hydroxycholesterol (24-HC) were obtained from Enzo, 25-hydroxycholesterol (25-HC) and 24(S),25-epoxycholesterol (24,25-EC) were obtained from Santa Cruz Biotechnology. All test compounds were diluted in DMSO in order to obtain the desired concentrations.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Neelakanteswar Aluru and Pedro Morais for the samples of little skate and amphioxus, respectively. This work was supported by Norte2020 and FEDER (Coral—Sustainable Ocean Exploitation—Norte-01-0145-FEDER-000036), EXPL/MAR-EST/1540/2012. SFRH/BD/79305/2011 to E.F., SFRH/BD/84238/2012 to M.L.-M. and SFRH/BPD/72519/2010 to R.R. L.F.C.C. and M.M.S. are supported by the European Regional Development Fund (ERDF) through the COMPETE—Operational Competitiveness Programme and POPH—Operational Human Potential Programme and national funds through FCT—Foundation for Science and Technology (UID/Multi/04423/2013). This work was supported by the Branchiostoma lanceolatum genome consortium that provided access to the Branchiostoma lanceolatum genome sequence. We thank three anonymous reviewers for their comments and suggestions.
Literature Cited
- Alberti S, et al. 2001. Hepatic cholesterol metabolism and resistance to dietary cholesterol in LXRbeta-deficient mice. J Clin Invest. 107:565–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand S, et al. 2004. Evolutionary genomics of nuclear receptors: from twenty-five ancestral genes to derived endocrine systems. Mol Biol Evol. 21:1923–1937. [DOI] [PubMed] [Google Scholar]
- Bertrand S, Belgacem MR, Escriva H. 2011. Nuclear hormone receptors in chordates. Mol Cell Endocrinol. 334:67–75. [DOI] [PubMed] [Google Scholar]
- Bridgham JT, Brown JE, Rodriguez-Mari A, Catchen JM, Thornton JW. 2008. Evolution of a new function by degenerative mutation in cephalochordate steroid receptors. PLoS Genet. 4:e1000191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridgham JT, et al. 2010. Protein evolution by molecular tinkering: diversification of the nuclear receptor superfamily from a ligand-dependent ancestor. PLoS Biol. 8:e1000497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SM, Bridgham JT, Thornton JW. 2008. Evolution of hormone signaling in elasmobranchs by exploitation of promiscuous receptors. Mol Biol Evol. 25:2643–2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla A, et al. 2001. A PPAR gamma-LXR-ABCA1 pathway in macrophages is involved in cholesterol efflux and atherogenesis. Mol Cell 7:161–171. [DOI] [PubMed] [Google Scholar]
- Escriva H, et al. 2006. Neofunctionalization in vertebrates: the example of retinoic acid receptors. PLoS Genet. 2(7):e102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerin I, et al. 2005. LXRbeta is required for adipocyte growth, glucose homeostasis, and beta cell function. J Biol Chem. 280:23024–23031. [DOI] [PubMed] [Google Scholar]
- Gutierrez-Mazariegos J, et al. 2016. Evolutionary diversification of retinoic acid receptor ligand-binding pocket structure by molecular tinkering. R Soc Open Sci. 3:150484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janowski BA, Willy PJ, Devi TR, Falck JR, Mangelsdorf DJ. 1996. An oxysterol signalling pathway mediated by the nuclear receptor LXR alpha . Nature 383:728–731. [DOI] [PubMed] [Google Scholar]
- Juvet LK, et al. 2003. On the role of liver X receptors in lipid accumulation in adipocytes. Mol Endocrinol. 17:172–182. [DOI] [PubMed] [Google Scholar]
- Kalaany NY, Mangelsdorf DJ. 2006. LXRS and FXR: the yin and yang of cholesterol and fat metabolism. Annu Rev Physiol. 68:159–191. [DOI] [PubMed] [Google Scholar]
- Katoh K, Toh H. 2010. Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics 26:1899–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korach-Andre M, et al. 2010. Separate and overlapping metabolic functions of LXRalpha and LXRbeta in C57Bl/6 female mice. Am J Physiol Endocrinol Metab. 298:E167–E178. [DOI] [PubMed] [Google Scholar]
- Krasowski MD, Ni A, Hagey LR, Ekins S. 2011. Evolution of promiscuous nuclear hormone receptors: LXR, FXR, VDR, PXR, and CAR . Mol Cell Endocrinol. 334:39–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laudet V, Gronemeyer H. 2002. The nuclear receptors factsbook. London: Academic press. [Google Scholar]
- Laurencikiene J, Ryden M. 2012. Liver X receptors and fat cell metabolism. Int J Obes (Lond). 36:1494–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecroisey C, Laudet V, Schubert M. 2012. The cephalochordate amphioxus: a key to reveal the secrets of nuclear receptor evolution. Brief Funct Genomics 11:156–166. [DOI] [PubMed] [Google Scholar]
- Lehmann JM, et al. 1997. Activation of the nuclear receptor LXR by oxysterols defines a new hormone response pathway. J Biol Chem. 272:3137–3140. [DOI] [PubMed] [Google Scholar]
- Lovell PV, et al. 2014. Conserved syntenic clusters of protein coding genes are missing in birds. Genome Biol. 15:565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maglich JM, et al. 2003. The first completed genome sequence from a teleost fish (Fugu rubripes) adds significant diversity to the nuclear receptor superfamily. Nucleic Acids Res. 31:4051–4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MA, et al. 2015. A RESTful API for access to phylogenetic tools via the CIPRES science gateway. Evol Bioinform Online 11:43–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DR, Goldstone JV, Stegeman JJ. 2013. The cytochrome P450 genesis locus: the origin and evolution of animal cytochrome P450s. Philos Trans R Soc Lond B Biol Sci. 368:20120474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino Y, et al. 2016. Neofunctionalization of androgen receptor by gain-of-function mutations in teleost fish lineage. Mol Biol Evol. 33:228–244. [DOI] [PubMed] [Google Scholar]
- Peet DJ, et al. 1998. Cholesterol and bile acid metabolism are impaired in mice lacking the nuclear oxysterol receptor LXR alpha . Cell 93:693–704. [DOI] [PubMed] [Google Scholar]
- Putnam NH, et al. 2008. The amphioxus genome and the evolution of the chordate karyotype. Nature 453:1064–1071. [DOI] [PubMed] [Google Scholar]
- Reschly EJ, et al. 2008. Ligand specificity and evolution of liver X receptors. J Steroid Biochem Mol Biol. 110:83–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JJ, Keinath MC. 2015. The sea lamprey meiotic map improves resolution of ancient vertebrate genome duplications. Genome Res. 25:1081–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffensen KR, et al. 2003. Gene expression profiling in adipose tissue indicates different transcriptional mechanisms of liver X receptors alpha and beta, respectively. Biochem Biophys Res Commun. 310:589–593. [DOI] [PubMed] [Google Scholar]
- Thornton JW. 2001. Evolution of vertebrate steroid receptors from an ancestral estrogen receptor by ligand exploitation and serial genome expansions. Proc Natl Acad Sci U S A. 98:5671–5676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobin KA, et al. 2000. Cross-talk between fatty acid and cholesterol metabolism mediated by liver X receptor-alpha. Mol Endocrinol. 14:741–752. [DOI] [PubMed] [Google Scholar]
- Wang Z, Ding G, Yu Z, Liu L, Li Y. 2009. CHSMiner: a GUI tool to identify chromosomal homologous segments. Algorithms Mol Biol. 4:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willy PJ, et al. 1995. LXR, a nuclear receptor that defines a distinct retinoid response pathway. Genes Dev. 9:1033–1045. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.