Abstract
Sex is beneficial in eukaryotes as it can increase genetic diversity, reshuffle their genomes, and purge deleterious mutations. Yet, its evolution remains a mystery. The eukaryotic clade supergroup Amoebozoa encompasses diverse lineages of polymorphic amoeboid forms, including both free-living and parasitic lineages. The group is generally believed to be asexual, though recent studies show that some of its members are implicated in cryptic forms of sexual cycles. In this study, we conduct a comprehensive inventory and analysis of genes involved in meiosis and related processes, in order to investigate the evolutionary history of sex in the clade. We analyzed genomic and transcriptomic data of 39 amoebozoans representing all major subclades of Amoebozoa. Our results show that Amoebozoa possess most of the genes exclusive to meiosis but lack genes encoding synaptonemal complex (SC). The absence of SC genes is discussed in the context of earlier studies that reported ultrastructural evidence of SC in some amoebae. We also find interclade and intrageneric variation in sex gene distribution, indicating diversity in sexual pathways in the group. Particularly, members of Mycetozoa engage in a novel sexual pathway independent of the universally conserved meiosis initiator gene, SPO11. Our findings strongly suggest that not only do amoebozoans possess sex genes in their genomes, but also, based on the transcriptome evidence, the present sex genes are functional. We conclude that Amoebozoa is ancestrally sexual, contrary to the long held belief that most of its members are asexual. Thus, asexuality in Amoebozoa, if confirmed to be present, is a derived-trait that appeared later in their evolution.
Keywords: sexual reproduction, genome, transcriptome, gene inventory, meiosis, life cycle
Introduction
Understanding the origin and evolution of sex in eukaryotes has proven a formidable task (Bonner 1944; Wenrich 1954; Goodfellow et al. 1974; Bé and Anderson 1976; Schuster 1976; Raikov 1982; Goldstein 1997; Kondrashov 1997; Goldstein 1999b; Parfrey et al. 2008; Garg and Martin 2016). This challenge is exacerbated as microbial eukaryotes have long been excluded from the discussion due to the false assumption that they are primitive (Haeckel 1866) and asexual (Maynard Smith 1978). Despite this assumption, several microbial eukaryotes including some amoebozoans, the focus of this study, are believed to engage in sexual acts (Martin and Alexopoulos 1969; Arnold 1972; Erdos et al. 1973a; Goodfellow et al. 1974; Erdos et al. 1975; Schuster 1976; Dacks and Roger 1999a; Goldstein 1999a; Ramesh et al. 2005; Stanley 2005). However, the exact mechanism of sexual developments in most of these lineages is obscure or deviant from those observed in animals, fungi and plants (Lahr et al. 2011c).
The Amoebozoa encompasses diverse groups of amoebae characterized by complex and diverse life cycles (Erdos et al. 1973b; Goodfellow et al. 1974; Schuster 1976; Raikov 1995; Adl et al. 2012; Tekle et al. 2014), most of which are traditionally considered asexual. The group includes lobose naked (e.g., Amoeba proteus) and testate (e.g., Arcella) amoebae, pelobionts (e.g., E. histolytica), lobose flat-shaped amoebae (e.g., Acanthamoeba), cellular (dictyostelid), and acellular (e.g., myxogastrid) slime molds as well as some less known diverse forms (reticulate, filose and amoeboflagellate). The observed life cycles in the group range from simple binary fission or alternating a/sexual morphotypes to formation of cysts or spores involving fruiting bodies. Asexuality for some members of this group is either used as a defining character (Hurst et al. 1992) or implied due to absence of reports of sexuality (Cavalier-Smith 2002).
The hallmark of sex is meiosis, a well-defined process that allows reduction of parental ploidy by half and genetic exchange through crossing over between homologous chromosomes. The physical and molecular components of meiosis in multicellular eukaryotes are well characterized and have been used as a blueprint to explore sexuality in other microbes (Olive 1963; Raikov 1982; Mignot and Raikov 1992; Ramesh et al. 2005; Poxleitner et al. 2008; Chi, Mahe et al. 2014; Chi, Parrow et al. 2014). Attempts on the observation of meiosis in most microbes have been very challenging, due to a combination of challenges in cultivation combined with complex life cycles. Moreover, canonical meiosis as observed in animals, fungi and plants is rarely observed in amoebae and most other eukaryotic microbes (Erdos et al. 1973b; Raikov 1982; Mihake 1996; Tekle et al. 2014). Thus, lack of physical evidence of sexual stages including meiosis has led some to assume that some microbes are exclusively asexual (Maynard Smith 1978; Tibayrenc et al. 1990). Contrary to this belief, some recent cytological studies using advanced techniques have shown some aspects of sexual stages in a few eukaryotic microbes, including members of Amoebozoa (Poxleitner et al. 2008; Tekle et al. 2014). For example, our recent study shows that Cochliopodium, previously considered asexual, engage in multiple cell fusion followed by karyogamy (nuclear fusion) to form a large polyploid plasmodium, which eventually fragments into uninucleate amoebae (Tekle et al. 2014). This process likely allows the amoeba to undergo genetic exchange through random mixing of chromosomes from multiple individuals.
In general, we can identify four categories of sexual stages where meiosis or nuclear fusion is assumed to occur in Amoebozoa. These include sexual cysts (Erdos et al. 1973b; Goodfellow et al. 1974; Seravin and Goodkov 1987; Mignot and Raikov 1992; Smirnov and Goodkov 1999; Ehrenkaufer et al. 2007), vegetative cellular and/or nuclear fusion (Seravin and Goodkov 1987; Michel and Smirnov 1999; Tekle et al. 2014), a distinct sexual morphotype (Schuster 1976) and putative amoeboid or flagellate gametes (Wrigley de Basanta et al. 2012). The complete stages of canonical meiosis have never been observed in any amoebae studied; meiosis is simply assumed to occur during these various putative sexual stages (Lahr et al. 2011c). It is likely that amoebozoans have several ways of achieving the products of sex, as evidenced by the varied life cycles reported for them (Erdos et al. 1973b; Blanc et al. 1989; Tekle et al. 2014). The evolution of these putative sexual stages within Amoebozoa is poorly understood, as the described life cycles are diverse, and in some instances, amoebae seem to have evolved similar life cycles independently. For example, studies show that amoebozoans such as Endostelium (Olive et al. 1984; Kudryavtsev et al. 2014) are capable of producing fruiting bodies, a character mostly attributed to the distantly related protostelid amoebae (slime molds). Similarly, sexual cysts are reported in some distantly related amoebozoan lineages (Goodfellow et al. 1974; Mignot and Raikov 1992). Lahr et al. (2011) provided a detailed account of amoeboid (a)sexuality, showing that seven of the approximately 14 lineages of Amoebozoa reviewed might be implicated in sex.
Whereas these are compelling reports on sexuality in these amoebae, most of the evidence described needs further investigation due to its incomplete or circumstantial nature. For example, in Cochliopodium, mechanisms of depolyploidization or meiosis still remain to be investigated (Tekle et al. 2014). In some cases, technical complications make a complete study of meiosis in some amoeba difficult. Ultrastructural studies based on transmission electron microscope (TEM) suffer from a lack of sequential sections and other technical difficulties related to fixation that may result in incomplete or artifactual results. Additionally, some amoebae are assumed to undergo meiosis during a cyst stage (Erdos et al. 1973b; Goodfellow et al. 1974; Mignot and Raikov 1992), which creates a technical hurdle for live observation and experimentations; cysts and spores are covered by thick translucent walls, making live observation difficult. Moreover, some published reports have never been reproduced in the laboratory. For example, the life cycle of Trichosphaerium—alternating between two morphs, gamont (sexual) and schizont (asexual)—reported by Schaudinn (1899) has not been observed in more recent laboratory cultures.
Views on the evolution and sexuality of microbial eukaryotes have been changing with the ever-growing molecular genetic data. Particularly, the availability of genomes and RNAseq data for some lineages have allowed us to better understand their evolutionary placement in the tree of life (Hampl et al. 2009; Koonin 2010; Grant and Katz 2014; Tekle et al. 2016). The wealth of genetic data have also opened an opportunity to gain new insights on the molecular basis of sex in microbes. These include discovery of molecular signatures of sex, such as the genes involved in recombination (Ramesh et al. 2005; Malik et al. 2008; Chi et al. 2014a). Several putative asexual eukaryotic microbes such as diplomonads (e.g., Giardia), some ciliates and members of Amoebozoa (e.g., Arcella, Entamoeba) are reported to undergo genetic recombination (Lovlie et al. 1988; Blanc et al. 1989; Deak and Doerder 1998; Caccio and Sprong 2010; Lahr et al. 2011b). Further genome exploration in model organisms (e.g., Dictyostelium) and important human pathogens (e.g., Entamoeba, Giardia) have revealed discovery of homolog genes exclusively used in meiosis (Malik et al. 2008). The discovery of meiosis specific and other sex related genes have been put forward as support for their capability to engage in sexual reproduction, because such genes would likely be pseudogenized if they were no longer used. This leads to an alternative approach to documenting meiosis that has been exploited in recent studies: searching the genomes of amoebae for genetic signs of sex. Consequently, several studies have successfully documented full complements of meiotic genes in diverse putative asexual microbes including some ciliates (e.g., Ichthyophthirius multifiliis) (Chi et al. 2014a), dinoflagellates (Chi et al. 2014b), fungi, diplomonads, and amoebae (Malik et al. 2008; Schurko and Logsdon 2008).
In this study, we conducted a comprehensive sex gene inventory for 39 amoebozoan lineages representing all major subclades of Amoebozoa. We used both genomic and transcriptomic data to investigate the evolution of sex in the group. Our findings show that all analyzed taxa possess several genes unique to meiosis, suggesting conservation of this ancient mechanisms. We conclude that Amoebozoa is ancestrally sexual; thus, asexuality in this group is likely a derived trait that appeared later in their evolution, if indeed they are entirely asexual.
Materials and Methods
Taxa and Genes Studied
We analyzed data for 39 species in Amoebozoa that represent all major subclades (tables 1 and 2). These include taxa belonging to Eudiscosea (17), Mycetozoa (6), Archamoebae (5), Tubulinea (2), Himatismenida (3), Variosea (3), and the incertae sedis taxon Pessonella sp. ATCC® PRA-29. Among these, eight taxa representing three major subclades have completed genomes (table 2), whereas the data for the rest come from different published RNAseq projects (supplementary table S1, Supplementary Material online). Transcriptome data coverage for the latter taxa varied by species (see supplementary table S1, Supplementary Material online).
Table 1.
Phylogenetic Distribution of Sex Genes in Major Subclades of Amoebozoa
| Gene | OG | Archamoeba | Eudiscosea | Himatismenida | Mycetozoa | Tubulinea | Varipodida | Incertae sedis (Pessonella) | |
|---|---|---|---|---|---|---|---|---|---|
| Bouquet Formation | |||||||||
| SAD1 | 129586 | + | + | + | + | + | |||
| Crossover Regulation | |||||||||
| DMC1 | 126834 | + | + | + | |||||
| HOP1 | 128667 | − | − | + | |||||
| HOP2 | 128568 | + | + | + | + | + | |||
| MER3 | 129931 | − | + | − | + | ||||
| MND1 | 127882 | + | + | + | + | + | + | ||
| MSH4 | 130077 | + | + | + | |||||
| MSH5 | 129379 | + | + | + | |||||
| RED1 | 180525 | − | − | − | |||||
| ZIP1 | 171209 | − | − | − | |||||
| DNA Damage Sensing/Response | |||||||||
| MEC1/ATR | 128386 | + | + | + | + | + | + | + | |
| MRE11 | 127969 | + | + | + | + | ||||
| RAD17 | 127538 | − | + | + | + | ||||
| RAD23 | 130351 | + | + | + | + | + | + | ||
| RAD24 | 126706 | + | + | + | + | + | + | + | |
| RAD50 | 127792 | + | + | + | + | + | + | ||
| TEL1/ATM | 128955 | + | + | + | + | + | + | ||
| Double-Strand Break Formation | |||||||||
| SPO11 | 127274 | + | + | − | |||||
| Double-Strand Break Repair (Nonhomologous End Joining) | |||||||||
| KU70 | 129086 | + | + | + | + | + | + | ||
| KU80 | 129372 | − | + | + | + | ||||
| LIG4/DNL1 | 130132 | + | + | + | + | + | + | ||
| XRCC4/LIF1 | 135131 | − | + | + | |||||
| Double-Strand Break Repair and Meiotic Divisions | |||||||||
| REC8 | 150817 | − | − | − | |||||
| Recombinational Repair | |||||||||
| BRCA2 | 131863 | + | + | + | |||||
| DNA2 | 129631 | + | + | + | + | + | + | ||
| EXO1 | 127511 | + | + | + | + | + | |||
| FEN1 | 127472 | + | + | + | + | + | + | ||
| MLH1 | 127201 | + | + | + | + | ||||
| MLH3 | 130552 | + | + | + | + | ||||
| MMS4/EME1(s) | 135664 | − | − | + | |||||
| MPH1/FANCM(a,m) | 128649 | − | + | + | + | + | |||
| MSH2 | 127538 | + | + | + | + | ||||
| MSH3 | 130351 | + | + | + | + | ||||
| MSH6 | 126895 | + | + | + | + | ||||
| MUS81 | 129162 | − | + | + | + | ||||
| PMS1 | 128001 | + | + | + | + | + | + | ||
| RAD51 | 126834 | + | + | + | + | + | + | ||
| RAD52 | 130806 | + | + | + | + | ||||
| RAD54 | 127098 | + | + | + | + | + | + | ||
| RTEL1 | 127294 | + | + | + | + | + | |||
| SGS1 | 126644 | + | + | + | + | + | + | ||
| SLX1 | 128732 | + | + | − | |||||
| SMC5 | 128615 | + | + | + | + | + | + | ||
| SMC6 | 127751 | + | + | + | + | + | + | ||
| Total Detected | 33 | 39 | 17 | 38 | 9 | 28 | 17 | ||
Table 2.
Meiosis Genes Inventoried in Amoebozoa Genomes
| Gene | A. castellanii | D. discoideum | D. purpureum | E. dispar | E. histolytica | E. invadens | E. nuttalli | P. pallidum | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bouquet Formation | ||||||||||||||
| SAD1 | + | + | + | − | + | + | + | + | ||||||
| Crossover Regulation | ||||||||||||||
| DMC1 | + | − | − | + | + | + | + | − | ||||||
| HOP1 | − | − | − | − | − | − | − | − | ||||||
| HOP2 | + | + | + | + | + | + | + | + | ||||||
| MER3 | − | − | − | − | − | − | − | − | ||||||
| MND1 | + | + | + | + | + | + | − | + | ||||||
| MSH4 | + | + | + | + | + | + | + | + | ||||||
| MSH5 | + | + | + | + | + | + | + | + | ||||||
| RED1 | − | − | − | − | − | − | − | − | ||||||
| ZIP1 | − | − | − | − | − | − | − | − | ||||||
| DNA Damage Sensing/Response | ||||||||||||||
| MEC1/ATR | + | + | + | + | + | + | + | + | ||||||
| MRE11 | + | + | + | + | + | + | + | + | ||||||
| RAD17 | + | + | + | − | − | − | − | + | ||||||
| RAD23 | + | + | + | + | + | + | + | + | ||||||
| RAD24 | + | + | + | + | + | + | + | + | ||||||
| RAD50 | + | + | − | + | + | + | + | + | ||||||
| TEL1/ATM | + | + | + | + | + | + | + | + | ||||||
| Double-Strand Break Formation | ||||||||||||||
| SPO11 | + | − | − | + | + | + | + | − | ||||||
| Double-Strand Break Repair (Nonhomologous End-Joining) | ||||||||||||||
| KU70 | + | + | + | − | − | − | − | + | ||||||
| KU80 | + | + | + | − | − | − | − | + | ||||||
| LIG4/DNL1 | + | + | + | + | + | + | + | + | ||||||
| XRCC4/LIF1 | − | + | + | − | − | − | − | + | ||||||
| Double-Strand Break Repair and Meiotic Divisions | ||||||||||||||
| REC8 | − | − | − | − | − | − | − | − | ||||||
| Recombinational Repair | ||||||||||||||
| BRCA2 | + | + | + | − | − | + | − | − | ||||||
| DNA2 | + | + | − | + | + | + | + | + | ||||||
| EXO1 | + | + | + | + | + | + | + | + | ||||||
| FEN1 | + | + | + | + | + | + | + | + | ||||||
| MLH1 | + | + | + | + | + | + | + | + | ||||||
| MLH3 | + | + | + | + | + | + | + | + | ||||||
| MMS4/EME1(s) | − | + | + | − | − | − | − | + | ||||||
| MPH1/FANCM(a,m) | + | + | + | − | − | − | − | + | ||||||
| MSH2 | + | + | + | + | + | + | + | + | ||||||
| MSH3 | + | + | + | − | − | − | − | + | ||||||
| MSH6 | + | + | + | + | + | + | + | + | ||||||
| MUS81 | + | + | + | − | − | − | − | + | ||||||
| PMS1 | + | + | + | + | + | + | + | + | ||||||
| RAD51 | + | + | + | + | + | + | + | + | ||||||
| RAD52 | + | + | + | + | + | + | + | + | ||||||
| RAD54 | + | + | + | + | + | + | + | + | ||||||
| RTEL1 | + | + | + | + | + | + | + | + | ||||||
| SGS1 | + | + | + | + | + | + | + | + | ||||||
| SLX1 | + | − | − | + | + | + | + | − | ||||||
| SMC5 | + | + | + | + | + | + | + | + | ||||||
| SMC6 | + | + | + | + | + | + | + | + | ||||||
We focused on a total of 44 sex-related genes, including 11 meiosis-specific genes, selected based on literature and availability in the OrthoMCL database (http://www.orthomcl.org/orthomcl/). The majority of the genes and taxa analyzed including non-amoebozoan eukaryotes were obtained from a phylogenomic pipeline developed by Grant and Katz (2014). This pipeline includes 13,104 “orthologous groups” (OGs, clusters of homologs organized in OrthoMCL) that are used to capture homologs from newly added taxa. Additional sex related genes and amoebozoan transcriptomes previously not included in the pipeline were later added from OrthoMCL and NCBI databases, respectively. These databases were last accessed June 2016.
Gene Inventory Analysis
Initially, we used the phylogenomics pipeline, a conservative approach, to determine the presence of the sex genes in Amoebozoa. We performed two runs of the pipeline; the first run used a stricter sequence cutoff parameter in Guidance (Penn et al. 2010) of 0.6 and the second used a more relaxed sequence cutoff of 0.3. In both runs, the column cutoff was 0.4. Guidance generates a reference multiple sequence alignment and assigns a score to every residue, which it calculates by building progressive alignments from bootstrap trees (in this case, the number of bootstraps was set to 10) and counting the proportion of alignments that contain the same residue. The average residue score per column and row in the reference alignment can be used to set a column and sequence cutoff, respectively, and any column or row which on average doesn’t meet the cutoff is removed from further analysis.
To ensure that no homologs were missed because of taxon removal during Guidance, we then used local BLAST (Altschul et al. 1990) to retrieve potential homologs from each of the analyzed genomes and transcriptomes, as well as from outgroup genomes across the eukaryotic tree of life and genomes of some bacteria and archaea, where available. For the instances where both the taxon and the gene were taken from the phylogenomics pipeline, we used the BLAST results included in the pipeline to find putative homologs for each gene in each species. In cases where either the gene or the taxon was not found in the pipeline, a new BLASTp search was performed using the gene sequences as queries and the taxon sequences as the subject. In both cases, we collected all hits with an e-value less than 1e−15. As this sometimes resulted in a very large number of hits, custom Python scripts were used to ensure only one sequence per contig or scaffold was retained, and to remove identical sequences from the subsequent list of hits.
Two alternative methods of homology searching were also tested to ensure that no distant homologs were missed. The first was psi-BLAST (Altschul et al. 1997), which uses hits generated from a first BLASTp run to build a protein profile to BLAST against the chosen protein database. This process can be repeated multiple times, with each repeat adding new hits to the profile to better detect distant homologs. The second homology search program we used was HMMer (hmmer.org, version 3.1b2), which uses an input sequence or alignment to build a hidden Markov model including the protein sequence and predicted secondary structure elements, and searches the given protein database for sequences which match the generated model.
To confirm the presence of the genes for each taxon, as well as differentiate paralogous genes from each other, the remaining hits were aligned using MUSCLE (Edgar 2004) via Seaview (Gouy et al. 2010). Columns with more than 75% missing data were masked, and phylogenetic trees were built using RAxML BlackBox (Stamatakis et al. 2008) through the CIPRES portal (Miller et al. 2010). RAxML analyses were run with default parameters, with the exceptions of the Protein Substitution Matrix (RTREV instead of JTT) and the use of empirical base frequencies. For the trees, outgroups from across the eukaryotic tree of life, as well as some bacterial and archaeal taxa when available, were included in the analyses. When the resulting phylogenetic tree showed multiple clusters of sequences (corresponding to gene paralogs), sequences from each cluster were reciprocally BLASTed against NCBI's non-redundant protein database to determine gene identity, and clusters outside the gene of interest were eliminated from the analysis. Presence of each gene in each Amoebozoa taxon was assigned based on remaining taxa in each gene tree; taxa not represented in a given tree were assigned “absent” (with genomes) or “no detection” (with transcriptomes).
Results
Gene Inventory Approaches
We inventoried eight genomes and 31 transcriptomes representing all major subgroups from across Amoebozoa using a phylogenomics pipeline and BLASTp for 11 meiosis-specific and 33 sex-related genes (tables 1 and 2, supplementary table S2 and file S1, Supplementary Material online). Of the genes inventoried, all but three were present in at least one lineage of Amoebozoa, and 15 were present in over half of the taxa analyzed (table 2, supplementary table S2, Supplementary Material online). Similarly, at least one sex related gene was detected in every amoeba analyzed, and 17 of the taxa, including all eight genomes, have over half of the sex genes in this inventory (tables 1 and 2).
Both the strict and relaxed runs of the pipeline returned fewer detections of the sex genes than the BLAST approach, even after confirming BLAST homologs with RAxML trees (data not shown). This is not surprising, as the phylogenomic pipeline was designed for large-scale taxonomic analyses, rather than individual gene identification. Additionally, the results of the psi-BLAST and HMMer searches did not differ significantly from the results of the BLASTp searches (data not shown). For these reasons, as well as to maximize detection of sex genes, we chose to base our further analyses on the hits returned from BLAST and confirmed with RAxML phylogenetic analysis.
Amoebozoa Subclades
Meiosis-related genes were found in every major subclade of Amoebozoa (table 1). Eudiscosea had the largest number of meiosis specific and other sex related genes, with 39 out of 44. Mycetozoa and Archamoebae had similarly high detections, with 38 and 33 genes detected, respectively (table 1). Tubulinea had the lowest rate of detection, with only 9 genes detected. Similarly, subclades Himatismenida and an incertae sedis (ATCC® PRA-29), had the next lowest sex gene detections (table 1). These three lineages (Tubulinea, Himatismenida and ATCC® PRA-29) do not have completed genomes and are represented by lower numbers of transcriptomic data (supplementary table S1, Supplementary Material online). Thus, this low detection may be due to lack of data and representation within these clades, rather than truly lacking many of the meiosis genes.
Eight of the 11 meiosis-specific genes were detected in Amoebozoa. MND1 is detected in every clade except Tubulinea (table 1). HOP2 is in every clade except Tubulinea and Himatismenida (table 1). DMC1, MSH4, and MSH5 are only detected in Archamoebae, Eudiscosea, and Mycetozoa (table 1). MER3 is only detected in Eudiscosea and Varipodida. SPO11 is detected in Archamoebae and Eudiscosea. HOP1 was consistently absent in almost all Amoebozoa analyzed but was oddly found in one member of Mycetozoa, P. polycephalum.
Amoebozoa Genomes
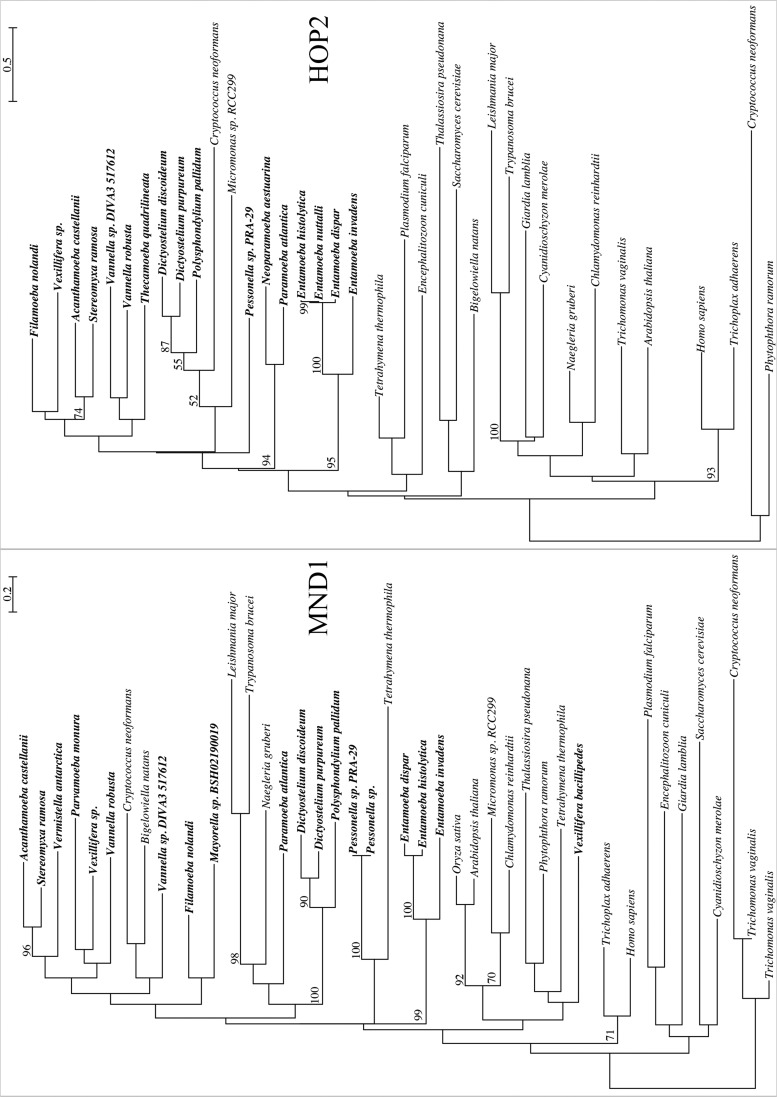
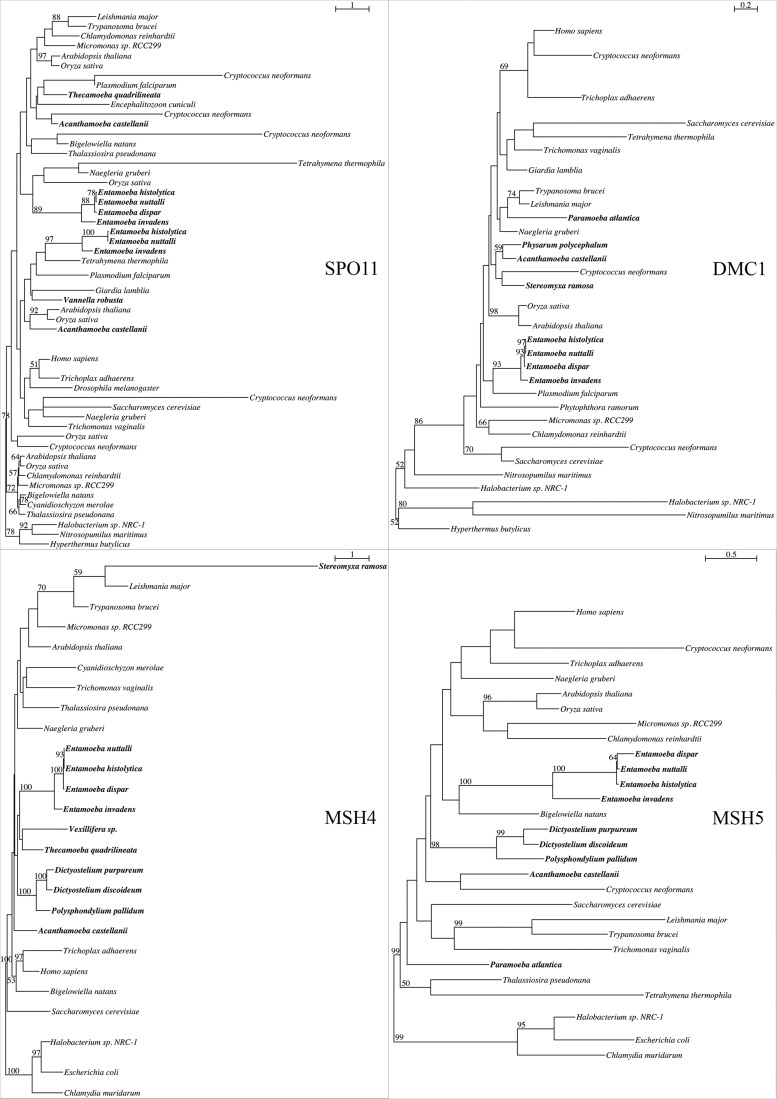
As expected, the eight completed genomes of Amoebozoa showed the highest detection of sex-related and meiosis-specific genes (table 2). Six of the 11 meiosis-specific genes were found within these genomes: SPO11, which initiates recombination by creating double-stranded breaks in DNA; DMC1, which promotes double-stranded break repair using the homologous chromosome; MND1 and HOP2, which form a heterodimer that stabilizes DMC1’s association with DNA and promotes Holliday Junction formation; and MSH4 and MSH5, which form a heterodimer that stabilizes recombination intermediates.
Although three of the six meiosis specific genes (HOP2, MSH4, and MSH5; table 2, figs. 1 and 2) were found in all genomes, we observed some variation in the number of presences in the remaining three genes by clades and individual species. MND1 is found in every genome except Entamoeba nuttalli (table 2, fig. 1). DMC1 and SPO11 are found in all Entamoeba and Acanthamoeba genomes (table 2, fig. 2) but are noticeably absent in the three mycetozoan genomes. Additionally, five meiosis-specific genes—HOP1, ZIP1, RED1, MER3, and REC8—are not found in any of the genomes inventoried. The three genes (HOP1, ZIP1, and RED1), making up the components of the synaptonemal complex (SC), were consistently absent in all genomes (table 2).
Fig. 1.—
Maximum likelihood trees of meiosis-specific genes MND1 and HOP2. Trees rooted at midpoint. Bootstrap support values ≥50% are shown above their bipartitions.
Fig. 2.—
Maximum likelihood trees of meiosis-specific genes SPO11, DMC1, MSH4, and MSH5. Trees rooted based on prokaryotic outgroup position. Bootstrap support values ≥50% shown above or beside their bipartitions.
We also inventoried 33 sex-related genes in these eight genomes (table 2, supplementary file S1, Supplementary Material online). All 33 of these genes were found in at least one Amoebozoa genome; 20 were found in every genome, and an additional four were found in every subclade but not every species within those subclades (table 2). Six were not found in Entamoeba, one was not found in Mycetozoa, and two were not found in Entamoeba or Acanthamoeba (table 2).
Amoebozoa Transcriptomes
In addition to the eight genomes analyzed, we inventoried transcriptome data of 31 species of amoebae for the same set of sex genes (supplementary table S2 and file S1, Supplementary Material online). Detection of sex genes in these amoebae transcriptomes correlated with the size of data analyzed (supplementary fig. S1, Supplementary Material online) and was much lower than in the genomes. Lineages with smaller transcriptome data generally rendered fewer detections (supplementary fig. S1, Supplementary Material online).
Meiosis-specific genes occur in 14 of the transcriptomes analyzed (supplementary table S2, Supplementary Material online). The most common meiosis-specific gene is MND1, occurring in 13 transcriptomes, including three transcriptomes (Vexillifera bacillipedes, Vermistella antarctica, and Parvamoeba monura) for which MND1 is the only detected meiosis-specific gene (fig. 1, supplementary table S2, Supplementary Material online).
The other relatively common meiosis-specific gene is HOP2, occurring in 10 transcriptomes (fig. 2, supplementary table S2 and file S1, Supplementary Material online). MSH4, DMC1, MSH5, SPO11, and MND3 are found in few amoebozoan transcriptomes (figs. 1 and 2, supplementary fig. S2; see supplementary table S2 and file S1 Supplementary Material online). The three genes associated with SC (HOP1, RED1, and ZIP1) and REC8, involved in holding sister chromatids together during meiosis, were not detected in any transcriptome analyzed, except for a single detection of HOP1 in the transcriptome of P. polycephalum (fig. S2 and table S2, Supplementary Material online).
Of the 33 sex-related genes, all 33 were detected in at least one transcriptome, and 12 were detected in half or more of the transcriptomes analyzed (supplementary table S2, Supplementary Material online). The transcriptome with the highest number of detected genes was Vannella sp. DIVA3 517612, with 29 sex-related genes (figs. 1 and 2; supplementary fig. S2 and table S2, Supplementary Material online). Conversely, the transcriptomes with the lowest number of detections were Acanthamoeba healyi, Ovalopodium desertum, and Nolandella abertawensis, each of which only contained RAD24 (supplementary table S2 and fig. S2, Supplementary Material online). The low detection rates in these taxa are likely due to low number of available transcriptome data that might have been caused due to the physiological states of the amoebae during RNA collection or methods of sequencing (supplementary table S1 and fig. S1, Supplementary Material online).
Discussion
Amoebozoa Is Ancestrally Sexual
Evidence for the ancestral origin of sex in eukaryotes is accumulating (Dacks and Roger 1999b; Malik et al. 2008; Lahr et al. 2011b). Our findings reinforce this conclusion by providing comprehensive analyses of sex genes in a mostly putative asexual eukaryotic supergroup, Amoebozoa. The current study provides evidence that amoebozoans possess most of the known molecular genetic toolkit important for sex. This finding lends support to the sexual nature of previously unconfirmed life cycles or sex-like (parasexual) behaviors reported in various groups of amoebozoans [reviewed in Lahr et al. (2011b)]. Amoebozoa not only possess sex genes in their genomes, but these genes are also functional and actively expressed, as confirmed by their detection in our transcriptome data. The presence of these genes in the genomes and most of the transcriptome data representing the major subclades of Amoebozoa demonstrates that amoebozoans are ancestrally sexual. Therefore, our study debunks the long held view that the majority of amoebozoans are purely asexual microbes.
Life Cycle and Mechanism of Sex in Amoebozoa
The members of Amoebozoa are extremely diverse in their life cycles, both asexual and sexual. However, the exact mechanisms of sexual development in Amoebozoa are mostly unknown. Even in those model amoebozoan lineages proposed to undergo meiosis during the cyst stage (Mignot and Raikov 1992) or alternating between haploid and diploid stages (Martin and Alexopoulos 1969; Erdos et al. 1973b). The genetic and ultrastructural basis of meiosis is poorly understood. This is mainly due to lack of observation caused by experimental challenges. Our current study shows that observed variations in life cycle and sexual behavior in Amoebozoa reflect a similar variability at the genetic level.
SC Independent Sex in Amoebozoa?
One of the common genetic features observed among all amoebozoan genomes examined is the consistent absence of three meiosis exclusive genes, HOP1, ZIP1, and RED1, involved in SC formation (Dong and Roeder 2000; Muniyappa et al. 2000). The SC is an ultrastructurally detectable protein structure that forms between two pairs of sister chromatids during meiosis (Heyting 1996, 2005). It is believed to facilitate chromosome pairing, synapsis, and recombination. The SC has been used as one of the reliable indicators for the occurrence of meiosis (Aldrich 1967; Heywood and Magee 1976; Raikov 1995) and is commonly found among eukaryotes that undergo conventional sex (von Wettstein et al. 1984; Heyting 1996) including members of Opisthokonta and Archaeplastida. Interestingly, despite the absence of detectable SC genes, some ultrastructural studies report the physical detection of SC in some members of Amoebozoa, including those inventoried in this study. These include SC observation in cysts of Arcella vulgaris (Mignot and Raikov 1992) and reproductive cysts or spores of several mycetozoans (Carroll and Dykstra 1966; Aldrich 1967; Erdos et al. 1972). Whereas SC independent recombination pathways are known (Lukaszewicz et al. 2013; Chi et al. 2014a) and amoebae are reported to undergo genetic recombination (Lahr et al. 2011a; Singh, et al. 2013), we find the discrepancies between genetic and ultrastructural evidence for SC quite intriguing.
A similar phenomenon is found in a distant eukaryotic lineage, ciliates. There is ample evidence supporting sexuality in ciliates, including some ultrastructural studies that report SC like structures in some species of ciliates (Raikov 1982; Skarlato 1982; Bobyleva 1984). However, similar to amoebae, ciliates lack clear homologs of genes known to encode SC proteins in their genomes (Chi et al. 2014a). Microscopic observations of SC in ciliates are sporadic. For instance, the model organism, Tetrahymena thermophila, is reported to lack SC in meiotic nuclei (Wolfe, et al. 1976). Other ciliates either lack fully mature SC structures or contain only a residual SC structure, resembling those found in fission yeast, Schizosaccharomyces pombe (Chi et al. 2014a). Residual SC, also known as linear elements (LinEs), are simple filamentous structures found in S. pombe (Loidl 2006). The main structural component of LinE is REC10, a distant homolog of RED1 in budding yeast (Lorenz et al. 2004). Both RED1 and REC10 are not detected in amoebae (table 2) or ciliates (Chi et al. 2014a) genomes. The structural and genetic homology of residual SC in ciliates and S. pombe remain to be elucidated.
Studies show that there is morphological and genetic variation in the eukaryotic SC (Bogdanov et al. 2007). Whereas SCs show overall similarity in morphology, SCs in plants and animals display size variation based on genome sizes (Bogdanov et al. 2007). Similarly, members of fungi display species-specific banding patterns of the lateral SC elements (Zickler 1973; von Wettstein et al. 1984). The genetic makeup of SC also varies. The genes encoding the central-space protein of the SC in budding yeast (ZIP1) and mammals (SCP1) share no sequence similarity, but have similar physico-chemical properties (Heyting 1996; Penkina et al. 2002). Therefore, it likely that the microscopically observed SC reported in both amoebae and ciliates might have a different origin or coded by different sets of genes. This might also result due to either rapid evolution of genes or replacement with other gene product. Our current findings necessitate further investigation on the morphologic and genetic origin of SC in both amoebae and ciliates.
A minor crossover (CO) pathway independent of SC is known in plants, vertebrates, and budding yeast (Higgins et al. 2008; Holloway et al. 2008; Lukaszewicz et al. 2013). This type of crossover involves MUS81, a non-meiosis exclusive DNA endonuclease with overlapping function in chromosomal CO pathway. This pathway has been proposed as predominant mechanism of meiotic recombination in lineages that lack SC such as ciliates and budding yeast (Lukaszewicz et al. 2013). Interestingly, amoebae possess a MUS81 homolog (table 2). Given some members of Amoebozoa are reported to engage in meiotic like recombination, amoebae may have independently evolved a mechanism of SC independent meiotic recombination similar to ciliates and budding yeast.
Interaclade and Intrageneric Sexual Pathways Variations
Genome wide exploration of sex genes revealed that variation in sexual pathways might exist in amoebozoans. Previous gene inventory studies that included two amoebozoan genomes (Dictyostelium and Entamoeba) with limited gene sampling show similar results to ours (Malik et al. 2008). However, our study is more thorough, including greater gene inventory sampling from eight completed genomes representing three major subclades of Amoebozoa as well as additional transcriptome data of various amoebae. This comprehensive sampling enabled us to gain some insights into the evolution of sexual pathways in Amoebozoa.
Comparison of gene inventories in the genomes of the three amoebozoan subclades shows that Eudiscosea (Acanthamoeba) and Mycetozoa uniquely share six sex-related genes, which are not detected in Archamoebae (table 2). The functions of these six shared genes include DNA damage sensing/response (RAD17), double-strand break repair (non-homologous end-joining, KU70 and KU80) and recombinational repair (MPH1/FANCM, MSH3 and MUS81, table 2). Archamoebae and Mycetozoa do not share any of the sex genes inventoried that are not also present in Acanthamoeba, whereas Archamoebae shares three sex genes with Acanthamoeba which are not present in Mycetozoa (table 2). It is interesting to note that among the sex genes present in Archamoebae and Acanthamoeba and absent in Mycetozoa are two of the key meiosis exclusive genes, SPO11 and DMC1. SPO11 is one of the central and universally conserved meiosis genes that plays a role in initiation of recombination by forming double-strand breaks in DNA (Keeney et al. 1997). DMC1 encodes the main enzyme in meiosis that promotes recombination between homologous chromosomes by repairing programmed DNA double strand breaks (DSBs) (Bugreev et al. 2011). These differences clearly indicate that there is variation in recombination pathways in Amoebozoa.
The evolutionary relationship of the three subclades (Eudiscosea, Archamoebae, and Mycetozoa) is not well resolved (Tekle et al. 2016). Archamoebae and Mycetozoa are traditionally placed under the more inclusive subclade Conosa (Cavalier-Smith 1998). However, the support for Conosa in molecular studies varies (Tekle et al. 2008; Lahr et al. 2011a; Cavalier-Smith et al. 2014), and it usually is not supported in large-scale analysis (Tekle et al. 2016). The lack of shared sex genes in these subclades is interesting and worth further investigation, though with the current data it is premature to make any evolutionary inferences based on the observed difference of sex genes in these lineages. Besides, sex genes are notorious for convergent evolution, unusual paralogy and relatively accelerated rates of evolution (Malik et al. 2008).
It should be noted that it is common to see independent loss of one or a suite of meiosis-specific genes in sexual eukaryotes. DMC1, along with a suite of other genes (HOP1, HOP2, and MND1), is missing in Drosophila, Anopheles, and Neurospora (Ramesh et al. 2005; Schurko and Logsdon 2008). Similarly, Caenorhabditis lacks HOP2, MND1, and DMC1, while retaining HOP1 (Malik et al. 2008). All these lineages are sexual and the reported variations show a plasticity that exists in the recombination pathways after the first steps of meiosis. However, given the universal role of SPO11, its absence in Mycetozoa is very unusual and poses new evolutionary questions about this subclade. Eukaryotic SPO11 shows interdomain sequence and structure conservation with one of the Archaeal DNA topoisomerase (Topo VI) subunits (Nichols et al. 1999). SPO11 is expressed exclusively during meiosis and plays a critical role in the initiation of sex by catalyzing the formation of DSBs prior to synapsis in prophase I of meiosis (Keeney et al. 1997). SPO11 is ubiquitous; its homologues are found across the tree of life in truly or cryptically sexual eukaryotes (Malik et al. 2008). Whereas independent genes losses and different crossover pathways are known past the first step of meiosis, the initiation of meiosis mediated by SPO11 is well conserved across eukaryotes (Chi et al. 2014a).
The absence of SPO11 in Mycetozoa, as evidenced by its absence in all Mycetozoa genomes as well as transcriptomes, adds a further layer of complexity to the evolutionary conundrum and mechanisms of sex in eukaryotes. Members of Mycetozoa are extensively studied for their sexuality and other cellular processes, and their life cycle involving diploid and haploid stages is well documented (Erdos et al. 1973b, 1975). However, the transition between these stages remains obscure due to the technical challenges described earlier. Members of the Mycetozoa are also known to undergo genetic recombination in a consistent manner as in meiosis (Erdos et al. 1975; Francis 1998). These findings posit a novel mechanism of cryptic sexual processes in this lineage. It further questions the universal role of SPO11 as initiator of meiosis in eukaryotes. A detailed account on the evolutionary role of SPO11 in general and SPO11 independent ploidy reduction in Dictyostelium in particular is described in a recent review article (Bloomfield 2016).
A few SPO11-independent crossover induction pathways have been identified. For example, in SPO11 null mutants of Saccharomyces pombe, knockout of FEN1, an exonuclease critical in Okazaki fragment processing in yeast, substantially increased crossover frequency and viability of spores (Farah et al. 2005). Similarly, expression of vertebrate DNA Deaminases in S. pombe and Caenorhabditis elegans SPO11 null mutants also restored crossing over in both organisms (Pauklin et al. 2009). It is possible that members of Mycetozoa induce crossovers by a similar alternative pathway. Alternatively, crossovers in Mycetozoa may be environmentally induced. Dictyostelium is known to be highly resistant to irradiation (Deering 1968) and has a very low rate of mutation compared with other eukaryotes (Saxer et al. 2012), suggesting a very efficient DNA repair mechanism that may have evolved under increased mutagenesis pressure. If Dictyostelium and other mycetozoans are subjected to increased amounts of DNA damage in their natural environment, this may render additional induction of damage for crossing over redundant and unnecessary (Bloomfield 2016).
In addition to the interclade genomic variation, we also noticed intrageneric genome variations of sex genes in Amoebozoa genomes (table 2). All of observed variations in Mycetozoa are non-meiosis-specific genes, whereas in Entamoeba one of the three intrageneric variations was one of the meiosis exclusive genes, MND1 (table 2); all Entamoeba species have this gene except Entamoeba nuttalli (table 2). This gene is also found in the remaining whole genome lineages and in most of the amoeba transcriptomes examined (tables 1 and 2). MND1 works in close conjunction with HOP2 by forming a heterodimeric complex that interacts with DMC1 to promote meiotic homolog juxtaposition and strand assimilation (Chen et al. 2004). MND1 is one of the indispensable genes for meiotic recombination. It is not clear if its absence is due to genome sequence incompleteness or indicative of another deviant pathway employed in this particular species. As described above, loss and gain of sex genes, particularly those not exclusive for meiosis, are common due to redundancy and function overlaps. However, if intrageneric variation like the above example is authentic and of common occurrence, its investigation will further our understanding of the evolution and mechanisms of such genes in cryptic sexual life cycles.
Our study demonstrates that amoebozoans employ diverse sexual pathway strategies to achieve the products of sex (recombination). It further demonstrates that the mechanism of sexuality is as diverse as the reported life cycles in this major clade of eukaryotes. Given this diversity, further genome wide investigations in Amoebozoa will likely unravel yet more unknown mysteries of sexual like processes and contribute substantially to our understanding of the origin and evolution of sex in general, and evolution and the roles of specific sex genes such as SPO11 in particular.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (1R15GM116103-01 to Y.I.T.) and National Science Foundation (RIA 1409587 to Y.I.T.). The authors thank Prof O. Roger Anderson for his invaluable comments on the early draft of the manuscript. We would like to thank Jessica Grant and Xyrus Maurer-Alcalá for assistance with initial data analysis. Additional support for collaborating groups included National Science Foundation (1436759 to L.A.G.; NSF 1541511 and NIH 1R15GM113177 to L.A.K.).
Literature Cited
- Adl SM, et al. 2012. The revised classification of eukaryotes. J. Eukaryot. Microbiol. 59:429–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldrich HC. 1967. The ultrastructure of meiosis in three species of Physarum . Mycologia 59:127–148. [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410. [DOI] [PubMed] [Google Scholar]
- Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold ZM. 1972. Observations on the Biology of the Protozoan Gromia oviformis Dujardin. Berkley: University of California Press. [Google Scholar]
- Bé AWH, Anderson OR. 1976. Gametogenesis in planktonic foraminifera. Science 192:890–892. [DOI] [PubMed] [Google Scholar]
- Blanc D, Nicholls R, Sargeaunt PG. 1989. Experimental production of new zymodemes of Entamoeba histolytica supports the hypothesis of genetic exchange. Trans. R. Soc. Trop. Med. Hyg. 83:787–790. [DOI] [PubMed] [Google Scholar]
- Bloomfield G. 2016. Atypical ploidy cycles, Spo11, and the evolution of meiosis. Semin. Cell Dev. Biol. 54:158–164. [DOI] [PubMed] [Google Scholar]
- Bobyleva NN. 1984. Synaptonemal complexes and intranuclear pore complexes in the meiotic prophase of the micronucleus of the ciliate Loxodes striatus . Tsitologiya 26:138–142. [Google Scholar]
- Bogdanov YF, Grishaeva TM, Dadashev SY. 2007. Similarity of the domain structure of proteins as a basis for the conservation of meiosis. Int. Rev. Cytol. 257:83–142. [DOI] [PubMed] [Google Scholar]
- Bonner JT. 1944. A descriptive study of the development of the slime mold Dictyostelium discoideum . Am. J. Bot. 31:175–182. [Google Scholar]
- Bugreev DV, et al. 2011. The resistance of DMC1 D-loops to dissociation may account for the DMC1 requirement in meiosis. Nat. Struct. Mol. Biol. 18:56–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caccio SM, Sprong H. 2010. Giardia duodenalis: genetic recombination and its implications for taxonomy and molecular epidemiology. Exp. Parasitol. 124:107–112. [DOI] [PubMed] [Google Scholar]
- Carroll GC, Dykstra R. 1966. Synaptinemal complexes in Didymium iridis . Mycologia 58:166–169. [Google Scholar]
- Cavalier-Smith T. (Cavalier-Smith, T. co-authors). 2002. The phagotrophic origin of eukaryotes and phylogenetic classification of protozoa. Int. J. Syst. Evol. Microbiol. 52:297–354. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. 1998. A revised six-kingdom system of life. Biol. Rev. Cambridge Phil. Soc. 73:203–266. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T, et al. 2014. Multigene phylogeny resolves deep branching of Amoebozoa. Mol. Phylogenet. Evol. 83:293–304. [DOI] [PubMed] [Google Scholar]
- Chen YK, et al. 2004. Heterodimeric complexes of Hop2 and Mnd1 function with Dmc1 to promote meiotic homolog juxtaposition and strand assimilation. Proc. Natl. Acad. Sci. U S A. 101:10572–10577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi J, Mahe F, Loidl J, Logsdon J, Dunthorn M. 2014. Meiosis gene inventory of four ciliates reveals the prevalence of a synaptonemal complex-independent crossover pathway. Mol. Biol. Evol. 31:660–672. [DOI] [PubMed] [Google Scholar]
- Chi J, Parrow MW, Dunthorn M. 2014. Cryptic sex in Symbiodinium (Alveolata, Dinoflagellata) is supported by an inventory of meiotic genes. J. Eukaryot. Microbiol. 61:322–327. [DOI] [PubMed] [Google Scholar]
- Dacks J, Roger AJ. 1999. The first sexual lineage and the relevance of facultative sex. J. Mol. Evol. 48:779–783. [DOI] [PubMed] [Google Scholar]
- Deak JC, Doerder FP. 1998. High frequency intragenic recombination during macronuclear development in Tetrahymena thermophila restores the wild-type SerH1 gene. Genetics 148:1109–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deering RA. 1968. Dictyostelium discoideum: a gamma-ray resistant organism. Science 162:1289–1290. [DOI] [PubMed] [Google Scholar]
- Dong H, Roeder GS. 2000. Organization of the yeast Zip1 protein within the central region of the synaptonemal complex. J. Cell Biol. 148:417–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenkaufer GM, Haque R, Hackney JA, Eichinger DJ, Singh U. 2007. Identification of developmentally regulated genes in Entamoeba histolytica: insights into mechanisms of stage conversion in a protozoan parasite. Cell Microbiol. 9:1426–1444. [DOI] [PubMed] [Google Scholar]
- Erdos GW, Raper KB, Vogen LK. 1972. Fine structure of macrocysts in Polysphondylium violaceum . Cytobiologie 6:351–366. [Google Scholar]
- Erdos GW, Raper KB, Vogen LK. 1973. Mating types and macrocyst formation in Dictyostelium discoideum . Proc. Natl. Acad. Sci. U S A. 70:1828–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdos GW, Raper KB, Vogen LK. 1975. Sexuality in cellular slime-mold Dictyostelium giganteum . Proc. Natl. Acad. Sci. U S A. 72:970–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farah JA, Cromie G, Davis L, Steiner WW, Smith GR. 2005. Activation of an alternative, Rec12 (Spo11)-independent pathway of fission yeast meiotic recombination in the absence of a DNA flap endonuclease. Genetics 171:1499–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis D. 1998. High frequency recombination during the sexual cycle of Dictyostelium discoideum . Genetics 148:1829–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg SG, Martin WF. 2016. Mitochondria, the cell cycle, and the origin of sex via a syncytial eukaryote common ancestor. Genome Biol. Evol. 8:1950–1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein ST. 1999. Foraminifera: a biological overview In: Sen Gupta BK, editor. Modern Foraminifera. Dordrecht: Kluwer; p. 37–56. [Google Scholar]
- Goldstein ST. 1997. Gametogenesis and the antiquity of reproductive pattern in the Foraminiferida. J Foram. Res. 27:319–328. [Google Scholar]
- Goodfellow LP, Belcher JH, Page FC. 1974. A light- and electron-microscopical study of Sappinia diploidea, a sexual amoeba. Protistologica 2:207–216. [Google Scholar]
- Gouy M, Guindon S, Gascuel O. 2010. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27:221–224. [DOI] [PubMed] [Google Scholar]
- Grant JR, Katz LA. 2014. Building a phylogenomic pipeline for the eukaryotic tree of life—addressing deep phylogenies with genome-scale data. PLoS Curr 6:pii: ecurrents.tol.c24b6054aebf3602748ac042ccc8f2e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeckel EHPA. 1866. Generelle morphologie der organismen. Allgemeine grundzüge der organischen formen-wissenschaft, mechanisch begründet durch die von Charles Darwin reformirte descendenztheorie. Berlin: G. Reimer. [Google Scholar]
- Hampl V, et al. 2009. Phylogenomic analyses support the monophyly of Excavata and resolve relationships among eukaryotic “supergroups”. Proc. Natl. Acad. Sci. U S A. 106:3859–3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyting C. 2005. Meiotic transverse filament proteins: essential for crossing over. Transgenic Res. 14:547–550. [DOI] [PubMed] [Google Scholar]
- Heyting C. 1996. Synaptonemal complexes: structure and function. Curr. Opin. Cell Biol. 8:389–396. [DOI] [PubMed] [Google Scholar]
- Heywood P, Magee PT. 1976. Meiosis in protists. Some structural and physiological aspects of meiosis in algae, fungi, and protozoa. Bacteriol. Rev. 40:190–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins JD, Buckling EF, Franklin FC, Jones GH. 2008. Expression and functional analysis of AtMUS81 in Arabidopsis meiosis reveals a role in the second pathway of crossing-over. Plant J. 54:152–162. [DOI] [PubMed] [Google Scholar]
- Holloway JK, Booth J, Edelmann W, McGowan CH, Cohen PE. 2008. MUS81 generates a subset of MLH1-MLH3-independent crossovers in mammalian meiosis. PLoS Genet. 4:e1000186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst LD, Hamilton WD, Ladle RJ. 1992. Covert sex. Trends Ecol. Evol. 7:144–145. [DOI] [PubMed] [Google Scholar]
- Keeney S, Giroux CN, Kleckner N. 1997. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88:375–384. [DOI] [PubMed] [Google Scholar]
- Kondrashov AS. 1997. Evolutionary genetics of life cycles. Ann. Rev. Ecol. Syst. 28:391–435. [Google Scholar]
- Koonin EV. 2010. The origin and early evolution of eukaryotes in the light of phylogenomics. Genome Biol. 11:209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudryavtsev A, Brown MW, Tice A, Spiegel FW, Pawlowski J, Anderson OR. 2014. A revision of the order Pellitida Smirnov et al., 2011 (Amoebozoa, Discosea) based on ultrastructural and molecular evidence, with description of Endostelium crystalliferum n. sp. Protist 165:208–229. [DOI] [PubMed] [Google Scholar]
- Lahr DJ, Grant J, Nguyen T, Lin JH, Katz LA. 2011a. Comprehensive phylogenetic reconstruction of amoebozoa based on concatenated analyses of SSU-rDNA and actin genes. PLoS One 6:e22780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahr DJ, Nguyen TB, Barbero E, Katz LA. 2011b. Evolution of the actin gene family in testate lobose amoebae (Arcellinida) is characterized by two distinct clades of paralogs and recent independent expansions. Mol. Biol. Evol. 28:223–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahr DJ, Parfrey LW, Mitchell EA, Katz LA, Lara E. 2011c. The chastity of amoebae: re-evaluating evidence for sex in amoeboid organisms. Proc. Biol. Sci. 278:2081–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loidl J. 2006. S. pombe linear elements: the modest cousins of synaptonemal complexes. Chromosoma 115:260–271. [DOI] [PubMed] [Google Scholar]
- Lorenz A, et al. 2004. S. pombe meiotic linear elements contain proteins related to synaptonemal complex components. J. Cell Sci. 117:3343–3351. [DOI] [PubMed] [Google Scholar]
- Lovlie A, Haller BL, Orias E. (Orias, E. co-authors). 1988. Molecular evidence for somatic recombination in the ribosomal DNA of Tetrahymena thermophila . Proc. Natl. Acad. Sci. U S A. 85:5156–5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukaszewicz A, Howard-Till RA, Loidl J. 2013. Mus81 nuclease and Sgs1 helicase are essential for meiotic recombination in a protist lacking a synaptonemal complex. Nucleic Acids Res. 41:9296–9309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik SB, Pightling AW, Stefaniak LM, Schurko AM, Logsdon JM., Jr. 2008. An expanded inventory of conserved meiotic genes provides evidence for sex in Trichomonas vaginalis . PLoS One 3:e2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GW, Alexopoulos CJ. 1969. The Myxomycetes. Iowa City: University of Iowa Press. [Google Scholar]
- Maynard Smith J. 1978. The evolution of sex. Cambridge: Cambridge University Press. [Google Scholar]
- Michel R, Smirnov AV. 1999. The genus Flamella Schaeffer, 1926 (Lobosea, Gymnamoebia), with description of two new species. Eur. J. Protistol. 35: 400–410. [Google Scholar]
- Mignot J-P, Raikov IB. 1992. Evidence for meiosis in the testate amoeba Arcella . J. Eurkaryot. Microbiol. 39:287–289. [Google Scholar]
- Mihake A. 1996. Fertilization and sexuality in ciliates In: Hausmann K, Bradbury PC, editors. Ciliates: cells as organisms. Stuttgart: Gustav Fischer; p. 243–290. [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the Gateway Computing Environments Workshop (GCE), p. 1–8.
- Muniyappa K, Anuradha S, Byers B. 2000. Yeast meiosis-specific protein Hop1 binds to G4 DNA and promotes its formation. Mol. Cell Biol. 20:1361–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols MD, DeAngelis K, Keck JL, Berger JM. 1999. Structure and function of an archaeal topoisomerase VI subunit with homology to the meiotic recombination factor Spo11. EMBO J. 18:6177–6188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive LS. 1963. The question of sexuality in cellular slime molds. Bull. Torrey Bot. Club 90:144–147. [Google Scholar]
- Olive LS, Bennett WE, Deasey MC. 1984. The new protostelid genus endostelium. Mycologia 76:884–891. [Google Scholar]
- Parfrey LW, Lahr DJ, Katz LA. 2008. The dynamic nature of eukaryotic genomes. Mol. Biol. Evol. 25:787–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauklin S, et al. 2009. Alternative induction of meiotic recombination from single-base lesions of DNA deaminases. Genetics 182:41–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penkina MV, Karpova OI, Bogdanov Iu F. 2002. Synaptonemal complex proteins: specific proteins of meiotic chromosomes. Mol. Biol. (Mosk) 36:397–407. [PubMed] [Google Scholar]
- Penn O, Privman E, Landan G, Graur D, Pupko T. 2010. An alignment confidence score capturing robustness to guide tree uncertainty. Mol. Biol. Evol. 27:1759–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poxleitner MK, et al. 2008. Evidence for karyogamy and exchange of genetic material in the binucleate intestinal parasite Giardia intestinalis . Science 319:1530–1533. [DOI] [PubMed] [Google Scholar]
- Raikov IB. 1995. Meiosis in protists: recent advances and persisting problems. Eur. J. Protistol. 31:1–7. [Google Scholar]
- Raikov IB. 1982. The Protozoan Nucleus: Morphology and Evolution. Wien: Springer-Verlag. [Google Scholar]
- Ramesh MA, Malik S-B, Logsdon JM. 2005. A phylogenomic inventory of meiotic genes: evidence for sex in Giardia and an early eukaryotic origin of meiosis. 15:185. [DOI] [PubMed] [Google Scholar]
- Saxer G, et al. 2012. Whole genome sequencing of mutation accumulation lines reveals a low mutation rate in the social amoeba Dictyostelium discoideum . PLoS One 7:e46759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurko AM, Logsdon JM., Jr. 2008. Using a meiosis detection toolkit to investigate ancient asexual “scandals” and the evolution of sex. Bioessays 30:579–589. [DOI] [PubMed] [Google Scholar]
- Schaudinn F. 1899. Untersuchungen uber den generationswechsel vonTrichosphaerium sieboldi. Schn. Abh. Konigl. Preuss. Akad. Wiss., Berlin Suppl., pp. 1–93.
- Schuster FL. 1976. Fine Structure of the Schizont Stage of the Testate Marine Ameba, Trichosphaerium sp. Journal of Eurkaryotic Microbiology. 23:86–93. [Google Scholar]
- Seravin LN, Goodkov AV. 1987. Euhyperamoeba fallax Seravin et Goodkov, 1982 (Lobosea, Gymnamoebia)—multinucleate marine limax amoeba—morphology, biological peculiarities and systematic position. Acta Protozool. 26:267–284. [Google Scholar]
- Singh N, Bhattacharya A, Bhattacharya S. 2013. Homologous recombination occurs in Entamoeba and is enhanced during growth stress and stage conversion. PLoS One 8:e74465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarlato SO. 1982. Electron microscope study of the micronuclei of the ciliate Stentor coeruleus during meiosis. Protistologica 18:281–288. [Google Scholar]
- Smirnov AV, Goodkov AV. 1999. An illustrated list of basic morphotypes of Gymnamoebia (Rhizopoda, Lobosea). Protistology 1:20–29. [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML web-servers. Syst. Biol. 57:758–771. [DOI] [PubMed] [Google Scholar]
- Stanley JSL. 2005. The Entamoeba histolytica genome: something old, something new, something borrowed and sex too? Trends Parasitol. 21:451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekle YI, et al. 2016. Phylogenomics of ‘Discosea’: a new molecular phylogenetic perspective on Amoebozoa with flat body forms. Mol. Phylogenet. Evol. 99:144–154. [DOI] [PubMed] [Google Scholar]
- Tekle YI, Anderson OR, Lecky AF. 2014. Evidence of parasexual activity in “asexual amoebae” Cochliopodium spp. (Amoebozoa): extensive cellular and nuclear fusion. Protist 165:676–687. [DOI] [PubMed] [Google Scholar]
- Tekle YI, et al. 2008. Phylogenetic placement of diverse amoebae inferred from multigene analyses and assessment of clade stability within ‘Amoebozoa’ upon removal of varying rate classes of SSU-rDNA. Mol. Phylogenet. Evol. 47:339–352. [DOI] [PubMed] [Google Scholar]
- Tibayrenc M, Kjellberg F, Ayala FJ. 1990. A clonal theory of parasitic protozoa: the population structures of Entamoeba, Giardia, Leishmania, Naegleria, Plasmodium, Trichomonas, and Trypanosoma and their medical and taxonomical consequences. Proc. Natl. Acad. Sci. U S A. 87:2414–2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wettstein D, Rasmussen SW, Holm PB. 1984. The synaptonemal complex in genetic segregation. Annu. Rev. Genet. 18:331–413. [DOI] [PubMed] [Google Scholar]
- Wenrich DH. 1954. Sex in Protozoa: a comparative review. Washington, DC: AAAS. [Google Scholar]
- Wolfe J, Hunter B, Adair WS. 1976. A cytological study of micronuclear elongation during conjugation in Tetrahymena . Chromosoma 55:289–308. [DOI] [PubMed] [Google Scholar]
- Wrigley de Basanta D, Lado C, Estrada-Torres A. 2012. Description and life cycle of a new Physarum (Myxomycetes) from the Atacama Desert in Chile. Mycologia 104:1206–1212. [DOI] [PubMed] [Google Scholar]
- Zickler D. 1973. Fine structure of chromosome pairing in ten Ascomycetes: meiotic and premeiotic (mitotic) synaptonemal complexes. Chromosoma 40:401–416. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.