Abstract
Chaperonins promote protein folding and are known to play a role in the maintenance of cellular stability under stress conditions. The group I bacterial chaperonin complex comprises GroEL, that forms a barrel-like oligomer, and GroES that forms the lid. In most eubacteria the GroES/GroEL chaperonin is encoded by a single-copy bicistronic operon, whereas in cyanobacteria up to three groES/groEL paralogs have been documented. Here we study the evolution and functional diversification of chaperonin paralogs in the heterocystous, multi-seriate filament forming cyanobacterium Chlorogloeopsis fritschii PCC 6912. The genome of C. fritschii encodes two groES/groEL operons (groESL1, groESL1.2) and a monocistronic groEL gene (groEL2). A phylogenetic reconstruction reveals that the groEL2 duplication is as ancient as cyanobacteria, whereas the groESL1.2 duplication occurred at the ancestor of heterocystous cyanobacteria. A comparison of the groEL paralogs transcription levels under different growth conditions shows that they have adapted distinct transcriptional regulation. Our results reveal that groEL1 and groEL1.2 are upregulated during diazotrophic conditions and the localization of their promoter activity points towards a role in heterocyst differentiation. Furthermore, protein–protein interaction assays suggest that paralogs encoded in the two operons assemble into hybrid complexes. The monocistronic encoded GroEL2 is not forming oligomers nor does it interact with the co-chaperonins. Interaction between GroES1.2 and GroEL1.2 could not be documented, suggesting that the groESL1.2 operon does not encode a functional chaperonin complex. Functional complementation experiments in Escherichia coli show that only GroES1/GroEL1 and GroES1/GroEL1.2 can substitute the native operon. In summary, the evolutionary consequences of chaperonin duplication in cyanobacteria include the retention of groESL1 as a housekeeping gene, subfunctionalization of groESL1.2 and neofunctionalization of the monocistronic groEL2 paralog.
Keywords: groEL/groES, Hsp60/Hsp10, heterocyst
Introduction
How new genes emerge and gain functionality is a fundamental question in biology. While the emergence of de novo protein-coding genes from noncoding DNA is extremely rare during evolution, new function often evolves from “molecular tinkering” of preexisting genes or parts of genes that are transformed and give rise to new function (Jacob 1977; Moyers and Zhang 2016). Gene duplication is considered as a major mechanism for the evolution of novel protein-coding genes and the development of phenotypic innovation (Ohno 1970; Lynch 2007). The amplification of gene copies is frequent during microbial evolution and the gene copy number can be highly transient within microbial populations (Romero and Palacios 1997). A temporary gene amplification may result in a dose effect where the protein encoded by the gene is produced in excess (e.g., Fuentes-Hernandez et al. 2015). The long-term retention of duplicated genes can involve, in addition to dose effect, subfunctionalization of the paralogous gene that retains a subset of the original function, or neofunctionalization of the gene duplicate into a novel function (Lynch 2007). Yet, paralogous gene copies are rarely retained during microbial evolution and most genes (89 ± 8%) in prokaryotic genomes are found in a single copy (Bratlie et al. 2010). In addition, the evolution of transcriptional regulation in bacteria may be rapid, leading to high plasticity of the regulatory elements that control the gene transcription level (Oren et al. 2014). Thus, the rapid adaptation of a new protein expression regime can often proceed through a modification of a regulatory region rather than gene amplification (e.g. Amoros-Moya et al. 2010). Hence, paralogous genes encoded in bacterial genomes constitute an exception from the common fate of duplicated genes in the prokaryotic domain.
Here, we study the evolution of chaperonin gene duplication in cyanobacteria. The chaperonin GroEL and its co-chaperone GroES function in unison to promote the folding of client proteins in an ATP-dependent manner (Horwich et al. 1993), while GroEL is also able mediate protein unfolding independently of ATP (Priya et al. 2013). Molecular chaperones are known to mediate stress response in many organisms where their expression is regulated according to various environmental cues including heat, UV radiation, salinity, and light stress (e.g. Webb et al. 1990; Glatz et al. 1997; Yamazawa et al. 1999; Chaurasia and Apte 2009). Moreover, accumulating evidence suggests that the function of chaperones has important consequences for robustness (Queitsch et al. 2002) and adaptation to high mutational load (Sabater-Muñoz et al. 2015). GroESL-mediated folding has been shown to accelerate protein evolution in vitro (Tokuriki and Tawfik 2009). This phenomenon is evident in genomic comparisons of eubacterial organisms as obligatory GroEL substrates were found to evolve faster than casual GroEL interactors (Bogumil and Dagan 2010; Warnecke and Hurst 2010; Williams and Fares 2010).
Chaperonins are encoded in most eubacteria (∼70%) by a single bicistronic groESL operon, while several taxa encode multiple paralogs, including Burkholderiales, Rhizobiales, Mycobacteria, and Cyanobacteria (Lund 2009). Cyanobacteria invented photosynthesis (Mulkidjanian et al. 2006) and they furthermore represent a rare example of genuine cell differentiation within prokaryotes. Cyanobacteria are classified into a monophyletic phylum that includes genera presenting a wide range of phenotypic diversity. Based on their cellular and colony morphology they have been divided into five subsections (Rippka et al. 1979). Under nitrogen-starvation (i.e. diazotrophic conditions), species of the Nostocales (Subsection IV) and Stigonematales (Subsection V) orders differentiate heterocyst cells that supply an oxygen-depleted environment for nitrogen fixation (Flores and Herrero 2010). All cyanobacterial genomes tested so far encode at least one groESL operon and a monocistronic groEL gene (Lund 2009). An exception is Gloeobacter violaceus, which encodes two groESL operons but no monocistronic groEL (supplementary table S1, Supplementary Material online). The recent genome sequencing of stigonematalean cyanobacteria revealed that species in that order encode a second copy of the groESL operon (Dagan et al. 2013). These species are characterized by multi-seriate or true-branching filament formation, the ability to fix nitrogen in heterocysts and differentiate morphologically distinct cell types (Rippka et al. 1979).
The functional role of cyanobacterial groEL paralogs has been studied so far mostly in the unicellular cyanobacterium Synechocystis sp. PCC 6803 and the filamentous, heterocystous cyanobacterium Anabaena sp. L31. In both species, the two groEL paralogs have been shown to be upregulated during heat stress (Glatz et al. 1997). Expression of the monocistronic groEL in Synechocystis is repressed during heat stress when photosynthesis is inhibited (Glatz et al. 1997). Furthermore, complementation experiments in E. coli showed that the monocistronic groEL of Synechocystis sp. and Synechococcus vulcanus cannot complement the native chaperonin (Kovács et al. 2001; Furuki et al. 1996; Tanaka et al. 1997). In Anabaena sp. L31, expression of the monocistronic groEL is repressed when heat stress is applied in diazothrophic conditions (Rajaram and Apte 2008). These studies, as well as studies in Thermosynechococcus elongatus (Sato et al. 2008) and Synechococcus elongatus PCC 7942 (Huq et al. 2010), thus suggest that the monocistronic groEL paralog has undergone a subfunctionalization while the bicistronic groEL paralog maintains a housekeeping function. Here, we study the fate of chaperonin gene duplication in stigonematalean cyanobacteria whose genome includes an additional groESL operon. We reconstruct the evolutionary history of chaperonin gene duplications in that order using phylogenetics. Moreover, we analyze functional divergence of the three chaperonin paralogs in Stigonematales with the cyanobacterium Chlorogloeopsis fritschii PCC 6912 as a representative. By comparing the transcription level and localization under different growth conditions, we show that the groEL paralogs have acquired a distinct transcriptional regulation. Furthermore, we examine the potential of GroES/GroEL paralogs to assemble into a chaperonin complex using protein–protein interaction studies and test for functional diversification of the paralogs by complementation of a groESL depleted E. coli strain. Our study suggests that the retention of groES/groESL duplicates in Stigonematales is accompanied by sub- and neo-functionalization of the paralogs.
Results
Evolution of groEL and groES Paralogs in Cyanobacteria
To study the evolution of groES/groEL paralogs in cyanobacteria we searched for all homologs of the chaperonin subunits in 141 sequenced genomes. Most of the cyanobacteria in our sample (115; 81%) encode a single groESL operon and an additional monocistronic groEL (supplementary table S1, Supplementary Material online). Cyanobacteria species that encode an additional groESL operon are mostly of the filamentous types included in Subsections III, IV, and V. Exceptions among the unicellular cyanobacteria (Subsection I) are Synechococcus sp. PCC 7335 and Synechococcus sp. PCC 7336 that encode two groESL operons. Within Subsection III, all three Pseudoanabaena strains encode two groESL operons, while Leptolyngbya sp. PCC 7375 encodes two groESL operons and two additional monocistronic groEL genes. Many heterocystous cyanobacteria that form linear or branching filaments (Subsections IV and V) encode an additional groESL operon (supplementary table S1, Supplementary Material online).
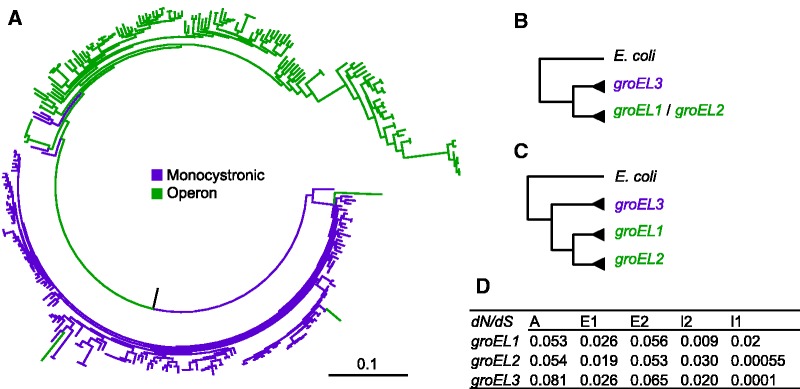
A maximum-likelihood phylogenetic tree reconstructed from all cyanobacterial homologous groEL DNA sequences reveals two main clades that largely correspond to the monocistronic and operon encoded genes (fig. 1A ). Furthermore, most of groEL duplicates encoded in genomes that encode an additional operon (e.g. Subsection V) form a sub-clade that is nested within the operon encoded groEL clade. In the following, we term the genes encoded in the ubiquitous groESL operon by groES1 and groEL1, while the additional operon components are termed groES1.2 and groEL1.2. The monocistronic groEL is termed groEL2. To further study the evolution of groES/groEL paralogs we reconstructed phylogenies that include all strains encoding more than two groEL paralogs and classified the genes into the three named paralogs (supplementary table S1, Supplementary Material online). The tree topology suggests that the split between the operon encoded and monocistronic groEL2 is ancient (fig. 1B ). To test this hypothesis we reconstructed a constrained phylogeny where the split between the monocistronic and operon clades is fixed (fig 1B ) and compared the likelihood of the resulting topology with the maximum likelihood (ML) phylogeny. This comparison revealed that the constrained phylogeny is not significantly different from the ML phylogeny (P = 0.35, using Shimodaira–Hasegawa (SH) test; Shimodaira 2002). Consequently, we conclude that the duplication of the ubiquitous operon groESL1 and the monocistronic groEL2 is ancient and most probably occurred at the base of the cyanobacterial tree.
Fig. 1.—
Phylogenetic reconstruction of groEL paralogs. (A) A ML phylogeny of groEL DNA sequences in 141 cyanobacteria. (B) Constrained topology used to test for an ancient duplication of the monocistronic and operon-encoded groEL. (C) Constrained topology used to test for duplication of groESL1 and groESL1.2. (D) The relative rate of nonsynonymous and synonymous substitutions (dN/dS) in Stigonemalaes groEL domains. A: Apical (amino acids 160-375); E1: 5′ Equatorial (amino acids: 1-133); E2: 3′ Equatorial (amino acids 411-546); I1: 5′ Intermediate (amino acids: 134-189); E2: 3′ Intermediate (amino acids 376-411).
We further tested for monophyly of the operon-encoded groEL duplicates by adding a constrained split between groEL1 and the remaining groEL duplicates (fig. 1C ). The likelihood of that topology is significantly lower than the ML phylogeny (P = 1 × 10− 61, using the SH test). We note, however, that this topology includes also exceptional groEL paralogs that could not be classified into the three orthologous groups such as additional monocistronic groEL paralogs in Leptolyngbya sp. PCC 7375 and N. punctiforme PCC 73102. Testing for a constrained monophyly of groES1 and groES2 revealed that the constrained topology is not significantly different from the ML topology (P = 0.15, using the SH test). To further characterize the evolutionary dynamics of the groES/groEL paralogs in stigonematalean cyanobacteria we constructed a phylogenetic tree that includes all paralogs encoded in the Nostocales (Subsection IV) and Stigonematales (Subsection V) genomes in our sample (supplementary fig. S1, Supplementary Material online). The Nostocales–Stigonematales clade includes all heterocyst forming cyanobacteria and is considered monophyletic (Flores and Herrero 2010; Dagan et al. 2013). The groES phylogeny reveals a clear monophyly of groES1 and groES1.2 that is highly supported by bootstrap (supplementary fig. S1A, Supplementary Material online). Testing for a constrained topology where groEL2 is monophyletic yielded a topology that is not significantly different than the ML phylogeny (P = 0.88, using the SH test). This result confirms that the duplication of groEL1 and groEL2 is ancient. A further test of the split between groEL1 and additional groEL duplicates reveals a topology that is not significantly different than the ML phylogeny (P = 0.15, using SH test). This indicates that the duplication of groESL1 operon occurred before the divergence of Stigonematales. Thus, the heterogeneous distribution of groESL1.2 across the Nostocales and Stigonematales is most probably due to a single duplication event and differential loss of groESL1.2 in the species that lack that operon rather than lateral gene transfer.
Transcriptional Regulation of groEL and groES Paralogs in C. fritschii PCC 6912
Transcriptional regulation of the groESL operon and the monocistronic groEL in cyanobacteria is known to depend on the presence of CIRCE and K-box elements at the 5′-UTR (Kojima and Nakamoto 2007; Sato et al. 2008). Comparative genomics of the cis-regulatory elements upstream of the groES/groEL paralogs in Stigonematales reveals conserved CIRCE and K-box elements upstream of the groESL1 operon (supplementary table S2, Supplementary Material online). The CIRCE element upstream of groESL1.2 is slightly diverged and is absent in Fischerella sp. PCC 73103 and Fischerella thermalis PCC 7521. The groEL2 is preceded in all genomes by a conserved CIRCE element but lacking a K-box element (supplementary table S2, Supplementary Material online). To study differences in the transcriptional regulation of the groEL paralogs, we quantified their transcription level in C. fritschii PCC 6912 using qRT-PCR. In addition, we visualized the promoter activity of the three paralogs in real time using a green fluorescence protein (GFP) marker.
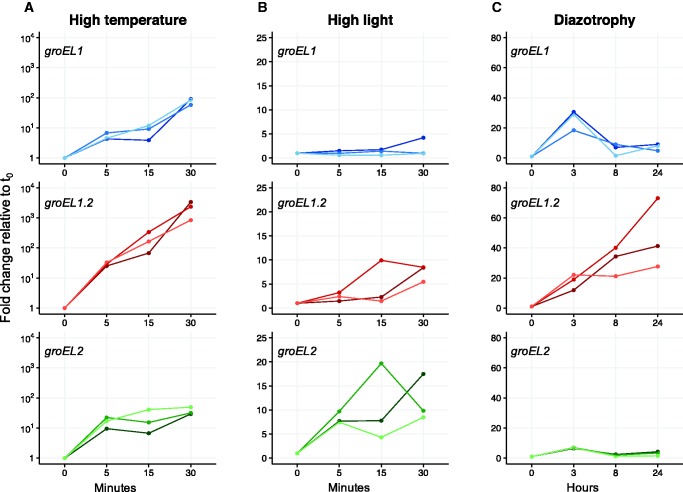
The absolute transcript levels of the three paralogous genes are significantly different under standard growth conditions (P < 0.05, using paired t-test with Bonferroni correction). The groEL1 transcript is the most abundant and groEL2 transcript abundance exceeds that of groEL1.2 (supplementary fig. S2, Supplementary Material online). We then compared the paralogs transcription level under high temperature by incubating C. fritschii at 50 °C. After 5 min of heat exposure (t 5m) the transcription level of groEL1 increased 5-fold on average, while the transcription level of groEL1.2 and groEL2 increased more than 15-fold, on average, in comparison to their transcription level under standard growth conditions (fig. 2A ). After 15 min (t 15m), the transcription level of groEL1 increased 8-fold and that of groEL1.2 increased 189-fold in comparison to the onset of the experiment (t 0). The transcription level of groEL2 remained similar to t 5m. The transcription level of all three paralogs increased further after 30 min of incubation at 50 °C, where the highest increase of about 2,000-fold was observed for groEL1.2. The transcription level of groEL2 increased 37-fold, whereas groEL1 transcription increased 78-fold in comparison to t 0 (fig. 2A ).
Fig. 2.—
Relative transcript abundance of the groEL paralogous genes in C. fritschii PCC 6912 under different growth conditions. Three independent biological replicates are plotted. (A) High temperature (50 °C). (B) High light (70 µE m−2 s−1). (C) Diazotrophy (nitrogen deprivation). Transcript abundance is depicted as fold change relative to expression levels under standard growth condition (37 °C, 24 µE m−2 s−1).
To further validate changes in the transcriptional regulation of the three paralogs under varying growth conditions, we created transcriptional fusions of each groESL/groEL promoter with gfp-mut3.1 (PgroESL1:gfp, PgroESL1.2:gfp, and PgroEL2:gfp). The three transcriptional fusion constructs were introduced independently into C. fritschii. The GFP fluorescence was observed in all transformed strains grown under standard conditions (t 0), indicating that all promoters are active and GFP-fusions are functional. During high temperature stress conditions groESL1 promoter activity increased after 10 min (t 10m), but decreased after 20 min (t 20m) of incubation (fig. 3A ). Elevated promoter activity of groESL1.2 and groEL2 could be clearly observed after 20 min (fig. 3A , supplementary fig. S4A, Supplementary Material online). Thus, promoter activity of the three paralogs as documented by GFP-fusion constructs supports the transcription level changes recorded using qRT-PCR.
Fig. 3.—
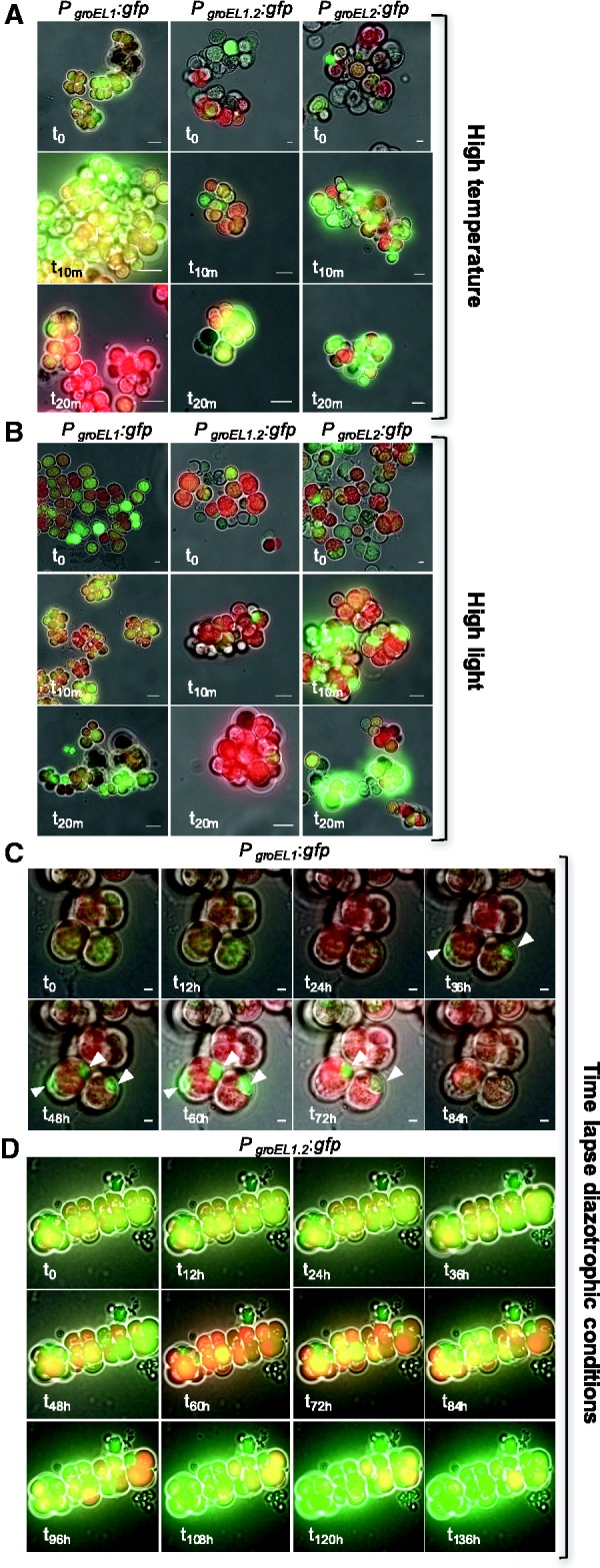
Expression of GFP under control of the different paralogous groEL promoters in C. fritschii PCC 6912. Micrographs of cyanobacterial cells expressing GFP under the transcriptional regulation of the three groEL promoters. For each fusion protein merged pictures of bright-field, GFP- (green), and chlorophyll fluorescence (red) are shown at different time points under various growth conditions (see fig. S4 for separate fluorescence signals). The time is indicated at the bottom left corner of each frame. Scale bars represent 10 µm. The white arrows indicate GFP accumulation in specific cells. (A) High temperature. (B) High light. (C and D) time-lapse of PgroESL1- (C), and PgroESL1.2- (D) driven GFP expression during diazotrophic conditions. Due to the different basal expression of both paralogs, the fluorescent intensity in (C) and (D) was measured with different light intensities and exposure times.
To compare the transcriptional regulation of the paralogs under high light conditions, we incubated C. fritschii PCC 6912 under 3-fold higher light intensity compared with standard conditions (fig. 2B ). The transcription level of groEL1 did not change during the course of the experiment. The groEL1.2 transcript level slightly increased after 30 min of incubation. A continuous increase of groEL2 transcript abundance under high light conditions was observed, up to a maximum of a 12-fold change in comparison to t 0 (fig. 2B and supplementary table S4, Supplementary Material online). The promoter activity of all paralogs, as documented by fluorescence intensity (fig. 3B; supplementary fig. S4B, Supplementary Material online), validates the observed changes in transcription level under high light conditions.
Heterocyst differentiation is a common trait in cyanobacteria with multiple groESL copies. Consequently, we compared the paralogs transcriptional regulation under diazotrophic conditions that induce heterocyst differentiation. Here we used the transcription level of glutamine synthetase (glnA) as a positive indicator for nitrogen deprivation (Tumer et al. 1983). After 3 h incubation under diazotrophic conditions the expression of glnA increased 10-fold, followed by a decrease observed after 8 h (supplementary fig. S31, Supplementary Material online). This was accompanied by an average increase of 26-fold in groEL1 transcript abundance after 3 h followed by a decrease after 8 h (fig. 2C; supplementary table S4, Supplementary Material online). The transcription level changes of groEL2 were similar to that of glnA, but with a maximum of 7-fold change after 3 h. The transcription level of groEL1.2 increased 18-fold after 3 h and continued to increase with a maximum of 47-fold change after 24 h in comparison to t0. The transcription dynamics of groEL1 suggest that it plays a role during the early stages of diazotrophy, while groEL1.2 seems to play a role in later stages of the adaptation to diazotrophic conditions. Consequently, we tested the promoter activity of groESL1 and groESL1.2 under diazotrophic conditions by time-lapse fluorescence microscopy (fig. 3C and D ). Because, C. fritschii grows slower on solid media in comparison to liquid media, we followed the promoter activity for a longer duration than the qRT-PCR experiment. During the first 24 h under diazotrophic conditions no specific promoter activity of groESL1 expression was observed. The fluorescence pattern after 36 h suggests that increased expression of GFP from groESL1 promoter leads to accumulation of GFP in specific cells that seem to differentiate into heterocysts in later stages (fig. 3C ). After 72 h, the groESL1 promoter appeared to be no longer active. The promoter of the second groESL operon (groESL1.2) showed an increased activity after 96 h followed by further elevation in all cells during the time of incubation in diazotrophic conditions (fig. 3D ). These results correspond to the transcription dynamics of groEL1 and groEL1.2 as documented using qRT-PCR, where groEL1 is upregulated in the early stage and groEL1.2 is upregulated in the late response to diazotrophic conditions. Our results further indicate that groESL1 promoter activity during diazotrophy is restricted to specific cells whereas groESL1.2 promoter activity did not show any cell-specific localization. Overall, the differences we observed in the transcriptional regulation of the three paralogs suggest that they have undergone a subfunctionalization.
Chaperonin Assembly
The GroES/GroEL chaperonin complex is composed of multiple GroEL subunits that form a barrel-like structure, whereas subunits of GroES form a lid that binds to the GroEL apical domain (Xu et al. 1997). To further study the functional divergence of the paralogs, we examined the structural differences of the three GroEL proteins by testing their ability to form homo- and heteromeric chaperone complexes. Direct interactions between all GroEL and GroES paralogs were tested using a bacterial two-hybrid system with cyclic adenylate cyclase as a split marker. Pairwise interactions of all GroEL and GroES combinations were tested in vivo in E. coli. All combinations of C- and N-terminally tagged GroES/GroEL pairs were analyzed. Positive interaction was confirmed by β-galactosidase assay (supplementary fig. S5, Supplementary Material online). The results of the interaction assays are summarized in a protein–protein interaction network (fig. 4A ).
Fig. 4.—
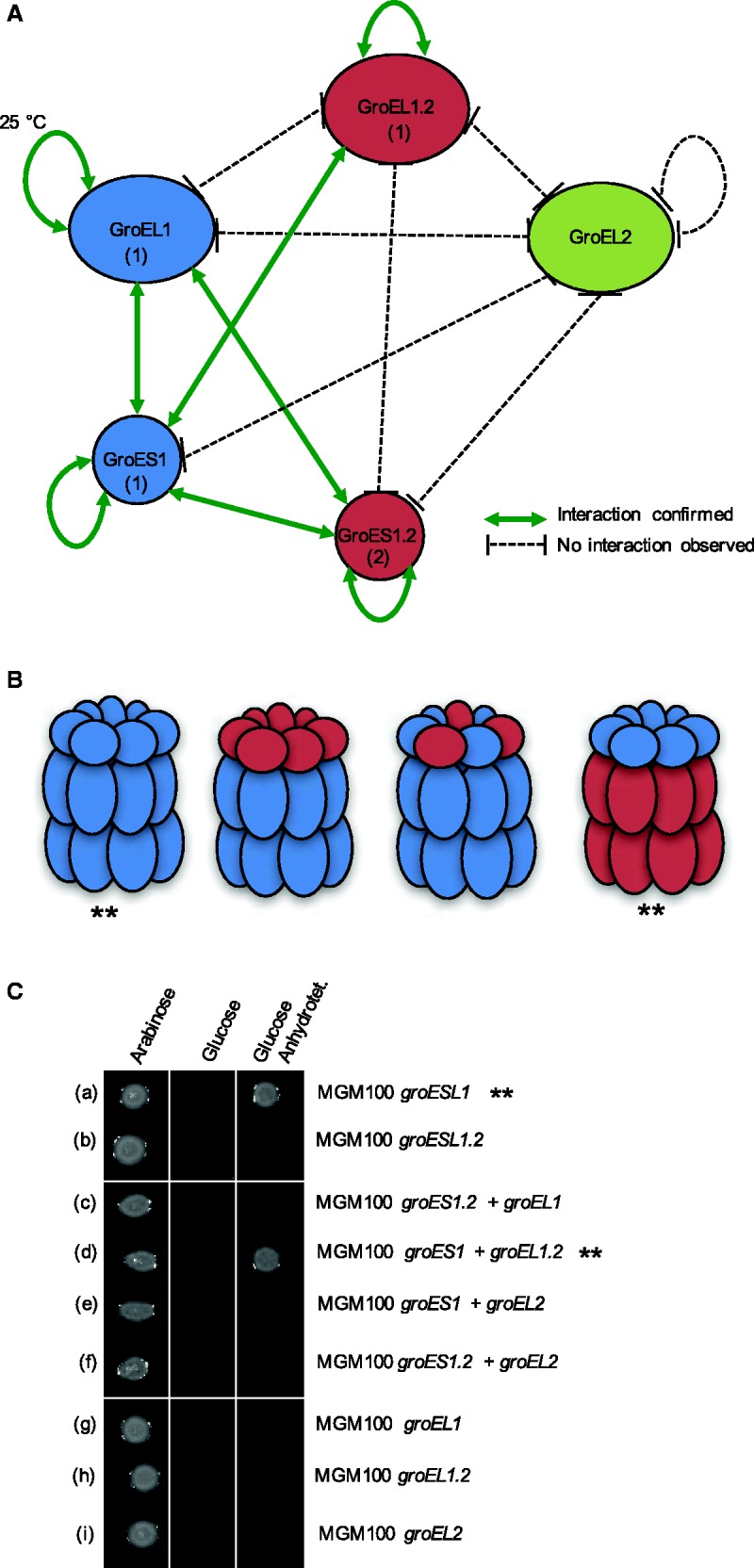
Protein–protein interaction and complementation assay. (A) Protein–protein interaction network of GroEL/GroES paralogs in C. fritschii PCC 6912. Interaction was tested by bacterial two-hybrid assay with either N- or C- terminally tagged proteins. Screening of positive interaction confirmed by a β-galactosidase assay (supplementary fig. S5, Supplementary Material online). The subunits are marked with (1) if the interaction was observed with both C- and N-terminal tagged proteins or (2) if the interaction was observed only with C-terminal tagged proteins. (B) Schematic diagram of putative chaperonin complexes based on the protein–protein interaction network. Chaperonin complexes that perform functional complementation in E. coli MGM100 are marked with **. (C) Complementation assay in the groEL deficient E. coli strain MGM100. Plating on arabinose constitutes a positive control (induction of the native E. coli groESL operon) whereas plating on glucose constitutes the negative control. Anhydrotetracycline induces the expression of the different cyanobacterial groES and groEL paralogs (as indicated on the right side). Combinations that compensate the lack of the native groESL are marked with **.
A homomeric interaction was documented for all chaperonin subunits except for GroEL2. Homomeric interaction of GroEL1 paralogs was only detected at low temperature conditions (25 °C), whereas any GroES1.2 protein interaction was only detected when it was C-terminally tagged. We could not detect an interaction between the different GroEL paralogs. Our results so far indicate that GroEL1 and GroEL1.2 form homogeneous barrel-like structures, while GroEL2 functions probably as a monomer. In contrast, the two small subunits, GroES1 and GroES1.2, were found to interact; hence, it is possible that they can form a heterogeneous lid structure. Moreover, we found GroEL1 and GroES1 to interact; this indicates that the groESL1 encoded proteins can form a chaperonin complex. In contrast, no interaction could be observed between GroEL1.2 and its co-chaperonin GroES1.2, indicating that the groESL1.2 encoded proteins are not forming a chaperonin complex. In addition, the interaction between GroEL1 and GroES1.2, and GroES1 and GroES1.2 suggests that proteins of the two operons can form different types of hybrid GroESL complexes. One type in which GroEL1 forms the barrel and GroES1 and GroES1.2 form a hybrid lid. A second type comprises a GroEL1 barrel and a GroES1.2 lid. The interaction we found between GroEL1.2 and GroES1 suggests that these two paralogs can form an additional hybrid chaperone complex (fig. 4B ). The absence of interaction between GroEL2 and any other tested proteins, suggests that GroEL2 does not form a typical chaperonin complex.
GroEL/GroES Complementation in E. coli
To further characterize the functional diversification of the GroES/GroEL paralogs, we performed a complementation assay in E. coli MGM100, in which the expression of the native groES/groEL operon is controlled by an arabinose-inducible promoter (McLennan and Masters 1998). To this end, we cloned groES/groEL combinations into compatible expression vectors, under the control of the anhydrotetracycline-inducible promoter (Ptet), and introduced them into E. coli MGM100. The ability of the introduced groES/groEL combination to complement the native E. coli groESL operon was scored positive if colonies were able to form upon induction with anhydrotetracycline. Using this approach we tested the complementation potential of the native C. fritschii operons as well as all GroES/GroEL combinations. In addition, because GroEL was shown to refold thermally denatured substrates independently of GroES in Anabaena sp. L31 (Potnis et al. 2016), the complementation potential of all GroEL paralogs was tested in the absence of GroES.
Our results revealed that GroES1/GroEL1 can successfully complement the native E. coli chaperonin, whereas GroES1.2/GroEL1.2 cannot (fig. 4C ). This result is in agreement with the lack of interaction between the GroES1.2 and GroEL1.2. From the observed hybrid chaperone complexes only the GroES1/GroEL1.2 combination was found to complement the native E. coli chaperonin (fig. 4C ). This result is consistent with the observed interaction between GroES1 and GroEL2 but not with the observed interaction between GroES1.2 and GroEL1. The expression of GroEL2 in all combinations could not complement the E. coli chaperonin. This supports the notion that GroEL2 does not function as a typical chaperonin. None of the C. fritschii GroEL paralogs is able to complement the native E. coli groESL operon in the absence of a GroES co-chaperonin (fig. 4C ). A recent study revealed that groEL1 encoded in Anabaena sp. L-31 is able to compensate for the lack of groESL in E. coli cultured in 42 °C (Potnis et al. 2016). We note, however, that Anabaena sp. L-31 encodes only groESL1 and groEL2, hence our result could be explained by the functional diversification of groESL1 and groESL1.2 in Stigonematales.
Discussion
The phylogenetic reconstruction of groEL and groES evolutionary history indicates that the ancestor of heterocystous cyanobacteria (Nostocales and Stigonematales) possessed a duplicated groESL operon. Furthermore, the comparison of transcriptional regulation, chaperonin assembly and function among groESL1 and groESL1.2 paralogs in the stigonematalean cyanobacterium C. fritschii PCC 6912 demonstrates that they are functionally diverged. In those species where groEL diversification has been studied, frequently one groEL copy has maintained the housekeeping function while other paralogs have undergone sub- or neo-functionalization. For example, the genome of Sinorhizobium meliloti, an alphaproteobacterium that is often found in symbiosis with legumes, encodes a total of five groEL paralogs. Only one paralog (groEL1) serves the housekeeping function and is also essential for the symbiotic interaction with plants and the establishment of nodules in the root (Bittner et al. 2007). A second paralog (groEL2) can substitute the housekeeping gene but its lower expression level is insufficient for the establishment of symbiosis in S. meliloti (Bittner et al. 2007). Earlier studies of groEL paralogs in various cyanobacteria indicate that GroESL1 comprise the housekeeping chaperonin (Glatz et al. 1997; Tanaka et al. 1997; Kovacs et al. 2001). Our results demonstrate that the housekeeping function of the chaperonin encoded by groESL1 is also maintained in C. fritschii PCC 6912.
The transcription level of groESL1.2 varies from that of groESL1 under standard and different growth conditions. A comparison of the 5′-UTR of the two operons revealed variation in the position and sequence of the regulatory elements (supplementary table S2, Supplementary Material online). Yet, the amino acid sequence similarity between the two chaperonins is high (GroEL1 vs. GroEL1.2: 87.2%; GroES1 vs. GroES1.2: 73.4% identical amino acids), hence, the mode of action is probably not different. This is supported by the ability of GroEL1.2 to complement a groESL deficient E. coli strain. Nevertheless, the lack of evidence for interaction between GroES1.2 and GroEL1.2 suggests that the groESL1.2 operon is dysfunctional, in a sense that its members do not interact, although transcribed together (ΔCT Ratio 1.20; supplementary table S4, Supplementary Material online). We note that groESL1.2 evolves under purifying selection (fig. 1D ) hence nonfunctionalization of that operon can be ruled out. One reason for the lack of GroES1.2–GroEL1.2 interaction may be due to amino acid replacements at the GroES–GroEL binding sites. However, the known GroES–GroEL interface amino acids (Xu et al. 1997) are conserved among the paralogs in C. fritschii. Furthermore, both subunits of the second operon can interact with GroES1, and even complement the E. coli chaperonin as shown in the case of GroES1–GroEL1.2. We note that oligomerization of GroEL1 was observed only at 25 °C while GroEL1.2 oligomers could be validated at 30 °C. Additionally, groEL1.2 transcription is upregulated under high temperature to a higher extent in comparison to groEL1. Hence, it is likely that heterologous GroEL1.2–GroES1 chaperonin complexes can be formed in C. fritschii and contributes to the high temperature tolerance. The upregulation of groEL1.2 transcription and the strong promoter activity of both groESL1 and groESL1.2 during diazotrophic condition suggest a functional role of the two groESL paralogs during heterocyst differentiation, albeit in different stages thereof.
Heterologous chaperonin complexes may also have divergent substrate sets. For example, a study of groESL paralogs in the deltaproteobacterium Myxococcus xanthus DK1622 reported that the functional diversification of the two duplicates involved a diversification of their substrate sets. Only 35% of the GroEL clients interact with both paralogs in that organism (Wang et al. 2013). The deletion of either one of the paralogs resulted in significantly different development, predation and heat-shock response phenotypes. Hence, the division of client sets between the two paralogs in M. xanthus has prominent functional consequences (Wang et al. 2013). Thus, it is possible that the heterologous chaperonin complexes in C. fritschii might also have divergent substrate sets. We note that GroES1.2 in combination with either GroEL1.2 or GroEL1 could not complement the E. coli chaperonin, which could be due to a different substrate set of that chaperonin complex. Yet, an interaction between GroES1.2 with GroES1 and GroES1.2 with GroEL1 is observed. This raises the possibility that the retention of groES1.2 has a dose effect by elevating the number of GroES subunits required to form the chaperonin lid. Overall, our study suggests that the retention of groESL1.2 in Stigonematales is accompanied by a subfunctionalization of that operon.
Our phylogenetic analysis suggests that the duplication of the monocistronic groEL2 is ancient and most probably was already present in the ancestor of cyanobacteria. The ubiquitous distribution of groEL2 orthologs in cyanobacteria and the differential regulation under light stress in C. fritschii and other cyanobacteria (Glatz et al. 1997) suggest that its function is related to photosynthesis. Our study furthermore shows that GroEL2 cannot form an oligomer (i.e., the typical chaperonin barrel structure) and cannot complement the E. coli chaperonin. Thus the GroEL2 mode of action is distinct from that of GroEL1 and GroEL1.2. We conclude that groEL2 has undergone a neofunctionalization in cyanobacteria.
In summary, our study adds evidence to the hypothesis that groES and groEL duplicates can be retained during prokaryote evolution and evolve new or modified functions in the cell. Prominent examples of groESL subfunctionalization are found in bacterial organisms whose lifestyle includes several developmental stages such as M. xanthus (Wang et al. 2013) or lifestyles as in S. meliloti (Bittner et al. 2007) and Mycobacterium smegmatis (Rao and Lund 2010). Here, we show that evolution of heterocyst differentiation in cyanobacteria was accompanied by groESL duplication and diversification of the paralogs.
Materials and Methods
Phylogenetic Analysis
The search for GroES/GroEL homologs was performed with BLAST+ (Camacho et al. 2009) using the amino acid sequence of GroES and GroEL encoded in Escherichia coli K12 MG1655 as a query. The search database included all sequenced cyanobacterial genomes in NCBI RefSeq (ver. July 2014) (Tatusova et al. 2014) and Joint Genome Institute (JGI; Grigoriev et al. 2012). An e-value <10− 10 was used as a sequence similarity threshold. Annotations of the BLAST hits were determined manually according to RefSeq database. Hits that lacked an annotation were validated by reBLASTing against NCBI. Multiple sequence alignments of the GroES/GroEL orthologs were computed with MAFFT ver. 7.027b (Katoh and Standley 2013). Phylogenetic trees were reconstructed using PhyML ver. 3.1 with the Le-Gascuel (LG) substitution model (Le and Gascuel 2008) for amino acid sequences and general time-reversible (GTR) substitution model (Lanave et al. 1984) for nucleotide sequences and SPR search algorithm (Guindon et al. 2010). Testing hypotheses regarding the tree topology was performed by reconstructing a maximum-likelihood tree with a user tree constraint. The comparison of ML phylogeny and the constrained phylogeny was performed using CONSEL ver. 1.2 (Shimodaira and Hasegawa 2001). The calculation of synonymous (dS) and nonsynonymous (dN) substitution rates was performed with CodeML (Yang 2007). Codon alignments for the dN/dS calculation were prepared with PAL2NAL (Suyama et al. 2006). Phylogenetic trees were plotted with FigTree (http://tree.bio.ed.ac.uk/software/figtree/).
Strains, Culture Conditions and Standard Techniques
Chlorogloeopsis fritschii PCC 6912 was obtained from the Pasteur Culture Collection (PCC) of cyanobacteria. Stock cultures were grown photoautotrophically in liquid BG11 medium supplemented with or without (BG11o) combined nitrogen at 37 °C and a light intensity of 24 µE m− 2 s− 1 (Rippka et al. 1979). Transformation of C. fritschii was performed by triparental mating (Stucken et al. 2012) with selection on 1% agarose BG11 plates supplemented with neomycin (Nm) 30 µg ml− 1. E. coli strains XL1 blue and HB101 were used for cloning and conjugation, E. coli MGM100 (McLennan and Masters 1998) for complementation and E. coli BHT101 for two-hybrid assays. Escherichia coli MGM100 was transformed by electroporation. If not otherwise stated, all E. coli strains were grown in LB or LB agar. Kanamycin (Km) 15 and 50 µg ml− 1, ampicillin (Amp) 100 µg ml− 1, or spectinomycin (Sp) 50 µg ml− 1 were used for selection.
DNA Techniques
Genomic DNA was isolated from C. fritschii PCC 6912 grown at 37 °C in BG11 medium with orbital shaking (100 rpm). A 30-ml of mid exponentially growing C. fritschii cells were harvested by centrifugation and DNA-Isolation was performed as described in (Franche and Damerval 1988), followed by removal of residual RNA with RNase A. PCRs were carried out with Phusion™ polymerase (Thermo Scientific, Germany) following the guidelines of the manufacturer. All primers used in this study are listed in supplementary table S3, Supplementary Material online.
RNA Isolation and Quantitative Real-Time PCR (qRT-PCR)
Samples for qRT-PCR were harvested by filtration before and 5, 15, and 30 min after induction of high light (70 µE m− 2 s− 1) and high temperature (50 °C) growth conditions. Samples from diazotrophic conditions were harvested 3, 8, and 24 h after nitrogen deprivation. Upon harvesting samples were frozen in liquid nitrogen. Total RNA was isolated using Concert Plant RNA Reagent (Invitrogen, Germany) according to the manufacturer’s instruction and treated with RNase-free DNaseI™ (Ambion, Germany). One microgram of total RNA was used for single-strand cDNA synthesis with iScript™ (Biorad, Germany). Quantitative RT-PCR was performed with primers listed in supplementary table S3, Supplementary Material online in a StepOne Real-Time PCR System (Biorad) with Power SYBR Green PCR master mix (Applied Biosystems, Germany) and 50 ng cDNA as template. All samples were run in biological and technical triplicates. The transcript levels of groEL paralogous genes were normalized using housekeeping gene rnpB as a reference. Relative transcript levels were calculated using the ΔΔCt method (Livak and Schmittgen 2001). Absolute transcript levels were calculated as described in (Lu et al. 2012).
Fluorescence Microscopy and Transcriptional Fusions
Samples of the cyanobacterial cultures for fluorescence microscopy were taken before and after 10 and 20 min after the cultures were transferred to high-light conditions (140 µE m− 2 s− 1) and high temperature conditions (50 °C), respectively. Samples from diazotrophic conditions were documented before and every 12 h after nitrogen deprivation over a period of 5 days. To fuse the promoter of groEL and groESL operons (PgroE) to gfp:mut3.1, PstI and BamHI restriction sites were introduced to 5′ and 3′ ends of gfp-mut3.1 (amplified from pRL153-GFP; Tolonen et al. 2006) and cloned in pBluescript SK(+). EcoRI and PstI sites were introduced to each PgroE fragment and these were fused upstream to gfp-mut3.1. The PgroE:gfp-mut3.1 fusions were excised from pBSK+ with EcoRI and BamHI and cloned in the cyanobacterial shuttle vector pRL25C (Wolk et al. 1988).
For time-lapse imaging, gene frame cover slips (Thermo Scientific) were attached to glass slides and the resulting chamber filled with 150 µl of BG11o medium containing agarose (0.5%) and supplemented with neomycin (30 µg ml− 1). To ensure aeration, 0.5 mm slices of solid media were removed from each side of the chamber. Twenty microliters of cells were then placed on top of the agarose slice and the chamber was sealed with a glass coverslip and visualized with an epifluorescence microscope (Zeiss Axio Imager 2, Plan-Apochromat 63×/1.40 Oil DIC M27 objective).
Protein–Protein Interaction Network
To characterize the physical association between components of the C. fritschii GroESL chaperonin complex, proteins were tested systematically for pairwise interaction by using the Bacterial Adenylate Cyclase Two-Hybrid System Kit (BACTH System Kit; Karimova et al. 1998). Both C- and N-termini tags were tested for all paralogs. PstI and BamHI sites were introduced by PCR to all groES and groEL genes. DNA fragments were digested with PstI and BamHI and cloned into the corresponding sites of the different BACTH plasmids. Screening for the ability to interact was performed according to the protocol on LB agar plates supplemented with X-gal (40 µg ml− 1), IPTG (0.5 mM), Amp and Km. Blue colonies indicated positive interaction and the expression of the reporter gene was subsequently confirmed by β-galactosidase assay (supplementary fig. S4, Supplementary Material online).
Complementation Assays
Complementation assays were performed with E. coli MGM100 expressing groESL under the regulation of the arabinose (0.2%) inducible pBAD promoter (McLennan and Masters 1998). The two operons (groESL1 and groESL1.2) and all chaperon subunit encoding genes (groEL1, groEL1.2, groEL2, groES1, and groES1.2) of C. fritschii PCC 6912 were amplified using the primers listed in supplementary table S3, Supplementary Material online. DNA fragments were cloned into pASK-IBA3plus (iba-lifesciences, Göttingen, Germany) or pASK-IBA3sp, an aadA (SpR) cassette marker containing derivative of pASK-IBA3plus, using BsaI cutsites. Expression of the proteins was regulated by anhydrotetracycline treatment (0.2 µg ml− 1, 3 h). For MGM100 co-expressing C. fritschii groEL and groES paralogs from different plasmids, both plasmids were co-transformed by electroporation. Transformed MGM100 was selected on LB plates containing Km, arabinose, Amp or Km, arabinose, Amp, Sp (for co-expression), and cells were grown in liquid media over night. Serially diluted cells were spotted onto selective solidified LB and grown at 30 °C for 18 hours. Growth of E. coli MGM100 on plates supplemented with glucose (and anhydrotetracycline) occurred only if the expressed groES/groEL from C. fritschii can complement the E. coli MGM100 strain.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Anne Kupczok, Giddy Landan, Claudia Walda, Tanita Wein and Samer Kadibalban for fruitful discussions and critical comments on the manuscript. We thank Peter Lund for kindly providing us with E. coli MGM100 strain. The study was supported by the European Research Council (Grant No. 281357 to T.D.) and the German science foundation (Grant No. STU513/2-1 to K.S.).
Literature Cited
- Amoros-Moya D, Bedhomme S, Hermann M, Bravo IG. 2010. Evolution in regulatory regions rapidly compensates the cost of nonoptimal codon usage. Mol Biol Evol. 27:2141–2151. [DOI] [PubMed] [Google Scholar]
- Bittner AN, Foltz A, Oke V. 2007. Only one of five groEL genes is required for viability and successful symbiosis in Sinorhizobium meliloti . J Bacteriol. 189:1884–1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogumil D, Dagan T. 2010. Chaperonin-dependent accelerated substitution rates in prokaryotes. Genome Biol Evol. 2:602–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratlie MS, et al. 2010. Gene duplications in prokaryotes can be associated with environmental adaptation. BMC Genomics 11:588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, et al. 2009. BLAST+: architecture and applications. BMC Bioinform. 10:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaurasia AK, Apte SK. 2009. Overexpression of the groESL operon enhances the heat and salinity stress tolerance of the nitrogen-fixing cyanobacterium Anabaena sp. strain PCC7120. Appl Environ Microbiol. 75:6008–6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagan T, et al. 2013. Genomes of Stigonematalean Cyanobacteria (Subsection V) and the Evolution of Oxygenic Photosynthesis from prokaryotes to plastids. Genome Biol Evol. 5:31–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores E, Herrero A. 2010. Compartmentalized function through cell differentiation in filamentous cyanobacteria. Nat Rev Microbiol. 8:39–50. [DOI] [PubMed] [Google Scholar]
- Franche C, Damerval T. 1988. Test on nif probes and DNA hybridizations. Methods Enzymol. 167:803–808. [Google Scholar]
- Fuentes-Hernandez A, et al. 2015. Using a sequential regimen to eliminate bacteria at sublethal antibiotic dosages. PLoS Biol. 13:e1002104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuki M, Tanaka N, Hiyama T, Nakamoto H. 1996. Cloning, characterization and functional analysis of groEL-like gene from thermophilic cyanobacterium Synechococcus vulcanus, which does not form an operon with groES . Biochim Biophys Acta. 1294:106–110. [DOI] [PubMed] [Google Scholar]
- Glatz A, et al. 1997. Chaperonin genes of the Synechocystis PCC 6803 are differentially regulated under light-dark transition during heat stress. Biochem Biophys Res Commun. 239:291–297. [DOI] [PubMed] [Google Scholar]
- Grigoriev IV, et al. 2012. The genome portal of the Department of Energy Joint Genome Institute. Nucleic Acids Res. 40:D26–D32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, et al. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59:307–321. [DOI] [PubMed] [Google Scholar]
- Horwich AL, Low KB, Fenton WA, Hirshfield IN, Furtak K. 1993. Folding in vivo of bacterial cytoplasmic proteins: role of GroEL. Cell 74:909–917. [DOI] [PubMed] [Google Scholar]
- Huq S, Sueoka K, Narumi S, Arisaka F, Nakamoto H. 2010. Comparative biochemical characterization of two GroEL homologs from the Cyanobacterium Synechococcus elongatus PCC 7942. Biosci Biotechnol Biochem. 74:2273–2280. [DOI] [PubMed] [Google Scholar]
- Jacob F. 1977. Evolution and tinkering. Science 196:1161–1166. [DOI] [PubMed] [Google Scholar]
- Karimova G, Pidoux J, Ullmann A, Ladant D. 1998. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci U S A. 95:5752–5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima K, Nakamoto H. 2007. A novel light- and heat-responsive regulation of the groE transcription in the absence of HrcA or CIRCE in cyanobacteria. FEBS Lett. 581:1871–1880. [DOI] [PubMed] [Google Scholar]
- Kovács E, et al. 2001. The chaperonins of Synechocystis PCC 6803 differ in heat inducibility and chaperone activity. Biochem Biophys Res Commun. 289:908–915. [DOI] [PubMed] [Google Scholar]
- Lanave C, Preparata G, Saccone C, Serio G. 1984. A new method for calculating evolutionary substitution rates. J Mol Evol. 20:86–93. [DOI] [PubMed] [Google Scholar]
- Le SQ, Gascuel O. 2008. An improved general amino acid replacement matrix. Mol Biol Evol. 25:1307–1320. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of Relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. [DOI] [PubMed] [Google Scholar]
- Lu Y, Xie L, Chen J. 2012. A novel procedure for absolute real-time quantification of gene expression patterns. Plant Methods 8:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund PA. 2009. Multiple chaperonins in bacteria – why so many? FEMS Microbiol Rev. 33:785–800. [DOI] [PubMed] [Google Scholar]
- Lynch M. 2007. The Origins of Genome Architecture. Sunderland (MA): Sinauer Associates. [Google Scholar]
- McLennan N, Masters M. 1998. GroE is vital for cell-wall synthesis. Nature 392:139–139. [DOI] [PubMed] [Google Scholar]
- Moyers BA, Zhang J. 2016. Evaluating phylostratigraphic evidence for widespread de novo gene birth in genome evolution. Mol Biol Evol. 33:1245–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulkidjanian AY, et al. 2006. The cyanobacterial genome core and the origin of photosynthesis. Proc Natl Acad Sci U S A. 103:13126–13131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. 1970. Evolution by Gene Duplication. Berlin, Heidelberg: Springer Science & Business Media; [Google Scholar]
- Oren Y, et al. 2014. Transfer of noncoding DNA drives regulatory rewiring in bacteria. Proc Natl Acad Sci U S A. 111:16112–16117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potnis AA, Rajaram H, Apte SK. 2016. GroEL of the nitrogen-fixing cyanobacterium Anabaena sp. strain L-31 exhibits GroES and ATP-independent refolding activity. J Biochem. 159:295–304. [DOI] [PubMed] [Google Scholar]
- Priya S, et al. 2013. GroEL and CCT are catalytic unfoldases mediating out-of-cage polypeptide refolding without ATP. Proc Natl Acad Sci U S A. 110:7199–7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queitsch C, Sangster TA, Lindquist S. 2002. Hsp90 as a capacitor of phenotypic variation. Nature 417:618–624. [DOI] [PubMed] [Google Scholar]
- Rajaram H, Apte SK. 2008. Nitrogen status and heat-stress-dependent differential expression of the cpn60 chaperonin gene influences thermotolerance in the cyanobacterium Anabaena . Microbiology 154:317–325. [DOI] [PubMed] [Google Scholar]
- Rao T, Lund PA. 2010. Differential expression of the multiple chaperonins of Mycobacterium smegmatis . FEMS Microbiol Lett. 310:24–31. [DOI] [PubMed] [Google Scholar]
- Rippka R, Stanier RY, Deruelles J, Herdman M, Waterbury JB. 1979. Generic assignments, strain histories and properties of pure cultures of Cyanobacteria. J Gen Microbiol. 111:1–61. [Google Scholar]
- Romero D, Palacios R. 1997. Gene amplification and genomic plasticity in prokaryotes. Annu Rev Genet. 31:91–111. [DOI] [PubMed] [Google Scholar]
- Sabater-Muñoz B, et al. 2015. Fitness trade-offs determine the role of the molecular chaperonin GroEL in buffering mutations. Mol Biol Evol. 32:2681–2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S, Ikeuchi M, Nakamoto H. 2008. Expression and function of a groEL paralog in the thermophilic cyanobacterium Thermosynechococcus elongatus under heat and cold stress. FEBS Lett. 582:3389–3395. [DOI] [PubMed] [Google Scholar]
- Shimodaira H, Hasegawa M. 2001. CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics 17:1246–1247. [DOI] [PubMed] [Google Scholar]
- Shimodaira H. 2002. An approximately unbiased test of phylogenetic tree selection. Syst Biol. 51:492–508. [DOI] [PubMed] [Google Scholar]
- Stucken K, Ilhan J, Roettger M, Dagan T, Martin WF. 2012. Transformation and conjugal transfer of foreign genes into the filamentous multicellular Cyanobacteria (Subsection V) Fischerella and Chlorogloeopsis . Curr Microbiol. 65:552–560. [DOI] [PubMed] [Google Scholar]
- Suyama M, Torrents D, Bork P. 2006. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34:W609–W612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka N, Hiyama T, Nakamoto H. 1997. Cloning, characterization and functional analysis of groESL operon from thermophilic cyanobacterium Synechococcus vulcanus . Biochim Biophys Acta. 1343:335–348. [DOI] [PubMed] [Google Scholar]
- Tatusova T, Ciufo S, Fedorov B, O'Neill K, Tolstoy I. 2014. RefSeq microbial genomes database: new representation and annotation strategy. Nucleic Acids Res. 42:D553–D559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuriki N, Tawfik DS. 2009. Chaperonin overexpression promotes genetic variation and enzyme evolution. Nature 459:668–673. [DOI] [PubMed] [Google Scholar]
- Tolonen AC, Liszt GB, Hess WR. 2006. Genetic manipulation of Prochlorococcus strain MIT9313: green fluorescent protein expression from an RSF1010 plasmid and Tn5 transposition. Appl Environ Microbiol. 72:7607–7613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumer NE, Robinson SJ, Haselkorn R. 1983. Different promoters for the Anabaena glutamine synthetase gene during growth using molecular or fixed nitrogen. Nature 306:337–342. [Google Scholar]
- Wang Y, et al. 2013. Mechanisms involved in the functional divergence of duplicated GroEL chaperonins in Myxococcus xanthus DK1622. PLoS Genet. 9:e1003306. e1003311. [DOI] [PMC free article] [PubMed]
- Warnecke T, Hurst LD. 2010. GroEL dependency affects codon usage—support for a critical role of misfolding in gene evolution. Mol Syst Biol. 6:340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb R, Reddy KJ, Sherman LA. 1990. Regulation and sequence of the Synechococcus sp. strain PCC 7942 groESL operon, encoding a cyanobacterial chaperonin. J Bacteriol. 172:5079–5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams TA, Fares MA. 2010. The Effect of chaperonin buffering on protein evolution. Genome Biol Evol. 2:609–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolk CP, et al. 1988. Isolation and complementation of mutants of Anabaena sp. strain PCC 7120 unable to grow aerobically on dinitrogen. J Bacteriol. 170:1239–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Horwich AL, Sigler PB. 1997. The crystal structure of the asymmetric GroEL-GroES-(ADP)7 chaperonin complex. Nature 388:741–750. [DOI] [PubMed] [Google Scholar]
- Yamazawa A, Takeyama H, Takeda D, Matsunaga T. 1999. UV-A-induced expression of GroEL in the UV-A-resistant marine cyanobacterium Oscillatoria sp. NKBG 091600. Microbiology 145(4):949–954. [DOI] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24:1586–1591. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.