Abstract
Temperature, perhaps more than any other environmental factor, is likely to influence the evolution of all organisms. It is also a very interesting factor to understand how genomes are shaped by selection over evolutionary timescales, as it potentially affects the whole genome. Among thermophilic prokaryotes, temperature affects both codon usage and protein composition to increase the stability of the transcriptional/translational machinery, and the resulting proteins need to be functional at high temperatures. Among eukaryotes less is known about genome evolution, and the tube-dwelling worms of the family Alvinellidae represent an excellent opportunity to test hypotheses about the emergence of thermophily in ectothermic metazoans. The Alvinellidae are a group of worms that experience varying thermal regimes, presumably having evolved into these niches over evolutionary times. Here we analyzed 423 putative orthologous loci derived from 6 alvinellid species including the thermophilic Alvinella pompejana and Paralvinella sulfincola. This comparative approach allowed us to assess amino acid composition, codon usage, divergence, direction of residue changes and the strength of selection along the alvinellid phylogeny, and to design a new eukaryotic thermophilic criterion based on significant differences in the residue composition of proteins. Contrary to expectations, the alvinellid ancestor of all present-day species seems to have been thermophilic, a trait subsequently maintained by purifying selection in lineages that still inhabit higher temperature environments. In contrast, lineages currently living in colder habitats likely evolved under selective relaxation, with some degree of positive selection for low-temperature adaptation at the protein level.
Keywords: hydrothermal vents, thermal adaptation, RNAseq, protein composition, selection
Introduction
Genomic investigations of thermophilic prokaryotes have shown that no general mechanism of thermal adaptation exists, with the noticeable exception of the charged residues increase, the depletion of thermolabile residues, structural RNA GC content, and the mRNA purine load (Hurst and Merchant 2001; Hickey and Singer 2004; Paz et al. 2004). On the contrary, several “evolutionary strategies” may be implemented or combined over evolutionary timescales to confer thermal tolerance (Jaenicke and Böhm 1998; Hickey and Singer 2004), depending on the genome-wide mutational bias and phylogenetic load of the investigated species (Chen et al. 2004). Among prokaryotes, this is probably the result of parallel evolution in LUCA descendants in the early history of life (Boussau et al. 2008).
Despite the lack of “universal” mechanisms of adaptation, thermal selection has, however, a profound influence on the residue composition of proteins in prokaryotes (Hickey and Singer 2004; Berezovsky and Shakhnovich 2005; Glyakina et al. 2007). The structure of proteins is stabilized by decreasing their flexibility while maintaining their functional state (i.e. their activity) with increasing temperature. Adaptation to high temperature usually includes 1) protein protection within the cell environment by chaperone molecules or the accumulation of osmolytes (Baross and Holden 1996; Kandror et al. 2002), 2) more plastic membranes (Cossins and Macdonald 1989), 3) targeted intrinsic protein modification that stabilizes the structure through the establishment of noncovalent interactions (including Van der Waals interactions, hydrogen bonds, electrostatic, or salt bridges; Querol et al. 1996; Vogt et al. 1997) or the induction of oligomerization (Fraser et al. 2016), and 4) on a more global scale, amino acid replacements favoring protein compactness through the increase of hydrophobic/aromatic residues (Zeldovich et al. 2007). This latter process has been reported in some prokaryotes (mostly archaea) that have presumably evolved under hot conditions for a long time. Because of such a variety of adaptive strategies, the numerous proposed thermophilic indices are mainly based on the frequency of either some of the most biased charged residues. Most of these indices are usually strongly positively correlated with the optimal growth temperature of the microorganisms. Amongst them, the ERK bias (Glyakina et al. 2007), the EK/QH ratio (de Farias and Bonato 2003), the CvP_bias (Charged versus polar residues: Suhre and Claverie 2003), or the IVYWREL index (Zeldovich et al. 2007) have often been used to discriminate prokaryotes according to their thermal preference.
Such a “structural” adaptation hypothesis, which postulates that elevated thermal regimes likely shape the amino acid composition of proteins regardless of function, has received little attention in eukaryotes. While it is known that a diversity of animals can live at temperatures up to 50° C, notably the scorpions Buthidae, the desert ants Cataglyphis, ostracods from geothermal settings, and alvinellids at hydrothermal vents (Wickstrom and Castenholz 1973; Gehring and Wehner 1995; Girguis and Lee 2006; Ravaux et al. 2013), we do not know whether these organisms exhibit specific residue patterns when compared with closely related mesophilic or psychrophilic species. Wang and Lercher (2010) attempted to explore conditions for thermal adaptation in warm-blooded animals and vertebrates, and found several adaptive analogies with thermophilic prokaryotes. A recent study based on thermophilic yeast genomes also shed some light on the thermal fingerprint on their proteomes (van Noort et al. 2013; Bock et al. 2014), depicting strategies adopted by these strains when facing high temperature (i.e. trehalose accumulation: Kandror et al. 2002). In this yeast study, comparisons of amino acid composition and proteomic/transcriptomic data clearly indicated that thermophily is only governed by changes in the primary structure of proteins and not by the differential expression of thermally inducible genes (Bock et al. 2014). Specifically, replacements of lysine to arginine were common (van Noort et al. 2013). The thermophilic mold Chaetomium thermophilum also displays some amino acid peculiarities that were shared with other thermophilic fungi of the family Eurotiomycetidae, including a high proportion of IVYWREL due to an increase of aromatic residues and isoleucine and a depletion of glycine. According to these authors, such thermal adaptations may have been gained by convergent evolution.
As mentioned, thermophilic metazoans are also found on the chimney walls of hydrothermal vents. Amongst them, the polychaete family Alvinellidae, which comprises two of the most extreme eukaryotic thermophiles on Earth (Chevaldonné et al. 1992; Cary et al. 1998; Girguis and Lee 2006): The Pompeii worm Alvinella pompejana (Desbruyères et al. 1998) on the East Pacific Rise (EPR) and its ecological homolog: The sulfide worm Paralvinella sulfincola in the northern Pacific (Tunnicliffe et al. 1993). Alvinellid worms live close to vent fluids coming out of hydrothermal chimneys and thus, requires specific functional adaptations, such as protein thermostability (Jollivet et al. 1995; Sicot et al. 2000; Shin et al. 2009; Kashiwagi et al. 2010), adaptations to hypoxia with greater gill surface area and higher affinity respiratory pigments (Hourdez and Lallier 2007), detoxification of sulfide (Powell and Somero 1986), renaturation of proteins through chaperones and heat shock proteins (Baross and Holden 1996) and an efficient reactive oxygen species elimination arsenal (Marie et al. 2006; Dilly et al. 2012). The bases of their adaptation to high temperatures are, however, poorly understood because of the difficulties associated with sampling them (it is hard to know their precise thermal history), studying them in the lab (they require specialized high-pressure aquaria) and the complexity of these thermophilic traits. Overall “structural” effects due to the ecological history of the species have not been yet carefully examined, although the first analyses of the transcriptome/proteome of A. pompejana led to adaptive trends analogous to both the thermophilic prokaryotes and the mold C. thermophilum (Jollivet et al. 2012; Holder et al. 2013). In particular, the ribosomal proteins of A. pompejana displayed several thermophilic features similar to those of ultrathermophilic bacteria such as a high proportion of positively charged and of large side-branched hydrophobic residues (Ile, Leu, Tyr: Jollivet et al. 2012), a marked CvP_bias similar to C. thermophilum (Holder et al. 2013) and, more surprisingly, a strong increase in the number of alanine, a characteristics that is only shared with the thermophilic fungi Thielavias (van Noort et al. 2013).
The annelid family Alvinellidae therefore represents an exciting biological model to focus on long-term adaptation of ectothermic species to high temperatures. This family indeed includes closely related species that live at similar depths (hydrostatic pressure: 150–250 bars), but inhabit contrasted thermal environments spanning nearly all temperatures possibly observed for metazoans (Desbruyères and Laubier 1986; Jollivet et al. 1995). Indeed, even if alvinellid worms such as A. pompejana or P. sulfincola inhabit chimney walls and can tolerate punctual bursts of temperatures up to 100 °C (Chevaldonné et al. 1992), the family also includes species (e.g. P. grasslei and Paralvinella pandorae) that live in the coolest (2–5 °C) part of the hydrothermal environment. These in situ observations have been confirmed by laboratory experiments that determined the thermal preferendum of P. sulfincola and A. pompejana in pressurized aquaria, (between 40 and 55 °C). Species from colder habitats such as P. grasslei or Paralvinella palmiformis are, however, more plastic than previously thought and in situ survival temperatures range from 2 to 30 °C (Dilly et al. 2012, personal observation). This indicates that many alvinellids are thermotolerant, with a thermal range shift to higher temperatures in species, which live in the hottest part of the environment. One plausible explanation for the observed pattern is that the last common ancestor to all alvinellids was thermophilic (fossil evidence suggests that alvinellid-like worms inhabited vent chimneys as early as the Devonian; Haymon et al. 1984; Little et al. 1999). Alternatively, it is also possible that the thermophilic character was acquired secondarily and independently in different lineages through a parallel and convergent evolution. However, if the thermophilic trait has been recently derived (i.e. a few millions years ago), one could expect that positively selected mutations at the base of protein thermostability should be different between the thermophilic species and easily detectable.
In the present work, we propose to test such evolutionary hypotheses in a constrained phylogenetic framework to reconstruct the thermal evolution of these worms using proteins as a proxy. Such a strategy allows for the control of phylogenetic constraints, trait similarities by descent, and more importantly stabilizing ecological parameters affecting all the species tested [such as hydrostatic pressure (Somero 1992) or strong limitation in oxygen (Webster 2003)] that could direct genome evolution. The aim of the study was therefore to 1) examine and compare nucleotide composition, and codon and amino acid usage between a set of alvinellid species living in contrasted thermal environments to detect putative structural signatures of thermal adaptation and, 2) to estimate species evolutionary rates and the nature of the mutations recently accumulated in these species (derived amino acid replacements). We produced a phylogenomic tree based on the set of orthologous genes to provide the evolutionary framework in which to conduct all analyses. Testing such hypotheses builds on the fact that recent parallel evolution to increased temperatures should produce a higher rate of replacements producing a signature of positive selection, as structural adaptation requires a large number of beneficial mutations all over the genome. On the contrary, if thermophily represents an ancestral trait in this worm family, present-day species that live under “hot” conditions should have evolved under more constrained conditions with slower rates of replacements. Both evolutionary scenarios were therefore also investigated by looking at signatures of selection using d N/d S ratio-based approaches. These ratios were determined in an original pipeline processing of high-throughput data that deals with both the lack of close well-annotated genomes and the recently highlighted limitation of codon models in the research of selection footprints.
Materials and Methods
Animal Collection, mRNA Extraction, Sequencing, and Assembly
Alvinella pompejana (Ap), Alvinella caudata (Ac), Paralvinella grasslei (Pg), and Paralvinella pandorae irlandei (Pp) were collected from hydrothermal vents on the EPR at a depth of 2,550 m during the Mescal oceanographic cruise in April/May 2010. Specimens were collected using the submersible Nautile, and upon recovery on board, flash-frozen in liquid nitrogen. Paralvinella fijiensis was sampled from hydrothermal vent chimneys of the Lau back-arc basin (Tui’Malina, 1,980 m) during the Lau Basin 2009 USA expedition with the ROV Jason and the R/V Thompson and directly preserved in liquid nitrogen. Once back in the laboratory, total RNA extraction was performed with Trizol, and the isolated RNA were re-precipitated after PVPP treatment to eliminate polysaccharides.
An EST library for the thermophilic Paralvinella sulfincola (Girguis and Lee 2006) was acquired as part of an earlier Joint Genome Initiative project to P.R. Girguis and S. Hourdez. The dataset consists of 24,702 transcripts assembled using Newbler (Margulies et al. 2005) based on reads obtained by 454 Roche technology. The specimens were sampled on the Juan de Fuca Ridge during the JdFR 2008 oceanographic cruise in August/September on board of the R/V Atlantis with the submersible Alvin. Transcriptomes for the remaining 5 species included in this study were assembled after Illumina sequencing, with 108-bp paired end reads. For Paralvinella fijiensis, the sequencing effort was 1 full lane (160 million reads), and for the 4 other species, the sequencing effort was a quarter of a lane (40 million reads) per species on a HiSeq 2000 at Genome Québec (Metzker 2009). For the five Illumina sequenced species, de novo assemblies were produced with the Velvet/Oases software (Zerbino and Birney 2008) following a bioinformatic procedure detailed in supplementary material S1, Supplementary Material online.
Search for Orthologs
A reciprocal best tblastx top hit search was performed between all possible transcriptomes pairs (Tatusov et al. 1997; Savard et al. 2006) using a stringent e-value cutoff of 10− 20 and a “medium soft filtering” for low similarity regions (Moreno-Hagelsieb and Latimer 2008). In this very conservative approach, any cluster of sequences with more than one copy per species was discarded from the dataset to avoid potential paralogy. Putative orthologous groups (POGs) were locally aligned on the corresponding complete transcript sequences using the BlastAlign algorithm (with a proportion of gaps per sequence <50%), which accounts for long indels in a conservative way (Belshaw and Katzourakis 2005). Additionally, because of the subsequent mutigene dN/dS-based study, a script was developed to detect ORFs (with and without the initial methionine and a minimal coding sequence length of 50 a.a.) to produce a series of loci alignments in the right coding frame without gaps in order to avoid false signal in case of misalignment in the areas flanking the indel (Gu and Li 1994 but see Fletcher and Yang 2010 and the discussion of the method in supplementary material S1, Supplementary Material online).
Base Composition
The transcript composition (GC content at the two first positions, GC12, and GC content at the third position, GC3) was first measured on the set of the 423 concatenated orthologs and, then on each gene separately to account for the intergenes variance. In the latter case, the standard error and the median value of each variable was calculated for each species. The purine load index (PLI) was also computed following the Forsdyke’s (2011) formula.
To evaluate whether the relative synonymous codon usage of each species vary between species and the thermal preference, we computed a standardized matrix of codon frequencies in all species from the concatenated set of coding sequences using the amino acid frequencies of the translated proteins. This matrix was then used as an input to perform a codon analysis to identify the preferred (Fop) codons following their relative contributions and coordinates on the two main axes of the correspondence analysis using the seqinR package, which controls for the amino acid composition of the proteome (Charif and Lobry 2007).
Amino Acid Composition
The relative amino acid composition of the proteome of each species was calculated based on the counts of residues found in the translated sequences of the orthologous transcripts from both the concatenated set of the 423 genes and each gene separately using an in-house Python script. Based on these counts, several thermophilic criteria previously described in the literature to discriminate between hyperthermophilic and mesophilic prokaryotes (de Farias and Bonato 2003; Suhre and Claverie 2003; Zeldovich et al. 2007; Wang and Lercher 2010) were calculated. According to Jollivet et al. (2012) and Holder et al. (2013), three thermophilic criteria that better characterize adaptation to hot temperatures in metazoans were more specifically tested: 1) the CvP (charged: DEKR vs polar: GHNPQST) bias criteria, 2) the EK/QH ratio, and 3) the IVYWREL criterion. Finally, the amino acid proxies for the wide-genome GC bias, that is, the relative amino acid counts of GC-rich codons (GARP) and of AT-rich codons (FYMINK) were also computed, as these indices did not conform to the theoretical GC-content expectations in the thermophilic worm A. pompejana genome (Jollivet et al. 2012). All these amino acid counts and criteria were then used in statistical analyses run under the R package for cross-species comparisons. These included across-genes median and variance estimates within each species and, analyses of variance and comparison of means using multiple Wilcoxon tests between species.
We also explored the relative effect of each of the 20 amino acids over the protein adaptation of the alvinellid species to construct a new metazoan criterion of thermophily. A matrix of amino acid counts at orthologous sites was produced based on the POGs alignments, in which rows represented species, and columns the amino acid frequencies. The factorial component analysis (FCA) (Teil 1975) was conducted following a count normalization (transformation and scaling of the dataset) using the FactomineR package (Hê et al. 2008) to explore how the amino acid composition (i.e. the overall variance) is partitioned between species. The amino acids that contribute the most in separating the “hot” versus “cold” species were then used to establish a new criterion.
Disordered Regions in Translated Sequences
Increasing the number and extent of disordered regions (i.e. unfolded polypeptide regions of low complexity and high flexibility, mainly involved in molecular binding/recognition) in proteins has been shown to represent a good proxy for protein adaptation to cold (Tantos et al. 2009). We estimated the proportion of disordered regions by calculating for each residue the probability that it takes a disordered state using our translated set of sequences and the predictive method of disorder-order transition adapted from Isin’s (1925) model by Lobanov and Galzitskaya (2011). A detailed description of our analyses with Isin’s model is provided in supplementary material S1, Supplementary Material online.
Phylogenomic Reconstruction
Previous phylogenetic analyses done with either ribosomal DNA (Féral et al. 1994; Rousset et al. 2003) or the mitochondrial Cox1 gene (Vrijenhoek 2013) led to alternative scenarios about the monophyly or the polyphyly of the genus Paralvinella. Depending on the gene used, the dichotomy between the two morphologically distinct genera (i.e. Alvinella vs Paralvinella) is not fully supported (see supplementary fig. S1, Supplementary Material online). Because of such a discrepancy, an unrooted phylogenomic tree of the six alvinellid species was reconstructed with RAxML-VI-HPC (Stamatakis 2006) and the GTR + Gamma nucleotide substitution model, in which the substitution rate variation among sites is modeled by the gamma distribution with four rate categories. Tree parameters and substitution models were optimized on the concatenated dataset of coding sequences from the aligned and filtered POGs using a codon partition model regardless of the substitution model used. Branch bootstrap values were obtained from the dataset resampling of 1,000 replicates. Alternative substitution models or partitions (concatenation without gene partitions or concatenation with codon-position partition) were also tested.
Reconstruction of the Residue Composition for Ancestral Nodes
Ancestral states of the residue composition of alvinellid proteins were reconstructed over the whole set of concatenated genes from the amino acid frequency of the sequences inferred from the Bayesian reconstruction of ancestral nodes in the “aaml” package in PAML v4.0 software (Yang 2007) using the classical Jones’ matrix of amino acid substitutions and a P-value threshold of 0.90 on amino acid replacements. To test whether the thermophilic trait of the worms is ancestral or recently derived in parallel in a few species, the new metazoan criterion (as designed from the result of the factorial correspondence analysis and the ecological a priori binomial tests) was computed from the frequency of residues of the reconstructed protein sequences at the four ancestral nodes of the alvinellid tree and on sequences of the present species.
Derived Mutation Rates in Recent Lineages
Derived amino acid substitution rates in recent lineages (i.e. terminal branches) were also estimated from both the POGs dataset (concatenated and 423 individually). In this case, the derived substitution rate strictly refers to “private” substitutions, that is, that only occur in one lineage with all other lineages sharing the same alternative residue—a parsimonious ancestral state allowing us to orientate the mutation. The idea here is to solely target the relatively recent substitution events possibly prone to positive selection. This allows us to test whether thermophily is an ancestral or derived trait and alternative scenarios of parallel evolution in either chimney species (e.g. Ap, Ac, Ps, Pf) or “cold” species (Pp and Pg). To test whether the derived mutation rate can be higher in some species, the number of derived replacements was weighted by the overall divergence accumulated in terminal branches to account for the unequal time elapsed since the speciation between species. This was achieved by the construction of an ultrametric tree corresponding to the ML tree previously obtained using the “mid-rooting position” approach implemented in Seaview 4.0 (Gouy et al. 2010), and the “ultrametrize” option of “Chronopl” from the Ape package in R (Paradis et al. 2004). The null hypothesis of constant rate of accumulation of derived mutations was tested with a χ 2 statistics.
We also tested whether the direction of substitution could lead to a specific enrichment in several classes of residues based on their electro-chemical properties. The selected classes were: Positively charged, negatively charged, polar, aliphatic, and aromatic plus the separate counts of un-categorizable residues cysteine, proline and glycine. We then computed the relative differences in amino acid classes/criteria observed between lineages (e.g. the increase of positively charged amino acids out of the derived substitutions in Ap compared with the other residue categories), and perform a factorial correspondence analysis on the derived substitutions (aaancestral → aaderived) to characterize the nature of amino acid substitutions in a given habitat from the interspecies substitution variance observed.
Strength and Mode of Selection over Lineages
The nonsynonymous/synonymous substitution ratio ω was also computed for each branch of the alvinellid phylogenomic tree using the free-ratio branch model (M1) implemented in the package CodeML of the PAML v4.0 software (Yang 2007). The small number of species used in the tree should reduce the risk of over-parametrization. Using the tree topology obtained with RaxML ((Ap,Ac),(Pp,(Pg,(Pf,Ps)))) as a reference, we compared this model with the one-ratio model (M0) using a Likelihood Ratio Test (LRT) with a df of n-1 branches for either the concatenated set of sequences or each of 423 POGs independently. The ω values were estimated for each terminal branch and subsequently used to calculate an intergene median per branch and its standard error after filtering (see supplementary material S1, Supplementary Material online ). In addition, putative codon sites under positive selection were investigated using branch-site codon models, which account for the ω variation among codons in a foreground lineage when compared with the other branches. In this approach, a LRT was used to compare the selection branch-site model (M2a) with three ω classes (ω 0 < 1, ω 1 = 1, and ω2 > 1) to the nearly neutral model (M1a) with two ω classes (ω 0 < 1, ω 1 = 1) or its alternative M2a model in which ω 2 is fixed to 1. The significance of the LRT was tested against a mixture distribution 50:50 between mass point 0 and χ2 with one degree of freedom at a threshold of 10%, 5%, and 1%. Under the acceptance of the “selective” model, putative codons under positive selection were identified among foreground branches using a Bayesian empirical Bayes (BEB) approach (see supplementary material S1, Supplementary Material online for more details).
Binomial Tests with Ecological Priors
Because of the very high intergene variance of most of our variables (i.e. the nucleotide composition, residue composition, thermostability criteria and dN/dS ratio) within each species, we developed a Binomial sign-test analysis with ecological priors, that is, the a priori knowledge of the thermal habitat of the worms: “COLD” for Pp and Pg and “HOT” for the thermophilic Ap and Ps. For each gene, species were ranked according to the variable tested. For each species, we then run a sign test, which compares the number of times the value of a variable (e.g. residue frequency, ω) is greater or lower for the species under scrutiny when compared with its opposite ecological trend (i.e. COLD or HOT) where n + represents the number of times the species displays a greater residue frequency than its opposite ecological background, n − = number of times the species displays a lower residue frequency than its opposite ecological background, and n T is the total number of trials (n + + n −). A binomial test is then run to test whether n + is significantly different from n- given the total number of cases: n T. This test therefore contrasts the most extreme classes of an ecological distribution, and examines whether the number of positive and negative cases are significantly different at a 5% threshold (see supplementary material S1, Supplementary Material online for details).
Results
Assembly and Gene Orthology
The total number of assembled transcripts obtained from the Velvet/Oases procedure (with a kmer = 21) ranged from 20,018 in P. pandorae to 40,843 in P. grasslei, with the noticeable exception of P fijiensis (one Illumina lane), for which a maximum of 80,939 transcripts were successfully assembled (see supplementary table S1, Supplementary Material online for assembly statistics). Based on these sequence datasets, two sets of candidate orthologous loci were generated: One without missing species (423 POGs) and the other allowing one missing species for each POG (1340 POGs).
Nucleotide Composition of Transcripts
The GC content, purine load and codon frequencies were calculated for each transcript, separately and over the whole concatenated set of genes for the six alvinellid species (table 1). The overall GC content was rather low (43.7–44.9%, table 1) but not markedly different between alvinellid species when compared with their intergene variation (30–65%, data not shown). The 18S rDNA GC content was much greater than coding sequences with values ranged between 50.1% and 52.2% but not correlated with the worm’s habitat and nearly identical between P. grasslei (cold-adapted) and P. fijiensis (hot-adapted). The GC3 was lower than GC12 but, as opposed to GC12, not correlated with the thermal habitat of the species (see supplementary material S1, Supplementary Material online for more details). The purine load of alvinellid worms was positive (around 104 bases per kilobase) and slightly greater than to the average value estimated for living organisms. Purine load was, however, not different between cold- (+104) and hot-adapted (+103.4) species (table 1).
Table 1.
Nucleotidic and Amino Acid Compositions (Percentages Based on Direct Counts) of Alvinellid Coding Sequences and Translated Proteins Obtained from the Six Transcriptome Assemblies
| Species | A. pompejana# | A. caudata# | P. sulfincola# | P. fijiensis# | P. grasslei § | P. pandorae § |
|---|---|---|---|---|---|---|
| Nucleic acid content | ||||||
| Total GC% | 44.42 | 44.58 | 43.75 | 44.85 | 44.26 | 44.17 |
| GC12% | 44.90 | 44.80 | 44.90 | 45.00 | 44.70 | 44.60 |
| GC3% | 43.55 | 44.05 | 41.51 | 44.64 | 43.47 | 43.32 |
| Purine Load | 101.82 | 102.52 | 105.20 | 103.98 | 104.23 | 103.90 |
| rDNA-18S | 52.22 | 52.20 | 51.92 | 51.76 | 51.71 | 51.10 |
| Charged content | 27.13 *** | 27.04 * | 27.08 * | 27.36 *** | 26.85** | 26.70** |
| D (Asp) | 6.08 | 6.11 | 5.97*** | 6.06*** | 6.62 ** | 6.87 ** |
| E (Glu) | 7.25 | 7.07 | 7.43 ** | 7.27 *** | 6.69** | 6.71 |
| H (His) | 2.46 | 2.34 | 2.22 | 2.32 | 2.37** | 2.25 |
| K (Lys) | 6.62 | 6.85* | 6.45 | 6.51 | 6.55 | 6.43 |
| R (Arg) | 4.72 | 4.67 | 5.01 *** | 5.20 *** | 4.62 | 4.44 |
| Polar content | 29.99* | 29.84* | 30.18* | 29.55* | 31.17 ** | 31.13 ** |
| Q (Gln) | 4.08 | 4.09 | 4.19 | 4.04 | 4.24 | 4.09 |
| G (Gly) | 3.28 | 3.28 | 3.10 | 2.97* | 3.37 | 3.57 |
| N (Asn) | 5.08 | 5.07 | 5.31 | 5.28 | 5.29 | 5.17 |
| P (Pro) | 2.55 ** | 2.62 ** | 2.48 ** | 2.56 ** | 2.24 | 2.12 |
| S (Ser) | 8.08** | 8.13** | 8.27** | 8.05* | 9.19 *** | 9.48 *** |
| T (Thr) | 6.92* | 6.65 | 6.83 | 6.65 | 6.84 | 6.70 |
| Aliphatic content | 36.96 ** | 37.17 *** | 36.65 * | 36.93 ** | 36.21* | 36.39* |
| A (Ala) | 7.86 *** | 8.11 *** | 7.90 *** | 8.42 *** | 6.99** | 7.15* |
| C (Cys) | 1.42* | 1.35 | 1.36 | 1.29 | 1.37 | 1.17 |
| I (Ile) | 8.40 | 8.56 | 8.00 | 8.38 | 8.38 | 8.85* |
| L (Leu) | 6.98** | 6.90** | 6.96* | 6.69* | 6.63 | 6.68 |
| M (Met) | 3.42 | 3.50 | 3.20 | 3.22 | 3.59 *** | 3.98 *** |
| V (Val) | 8.88 | 8.75 | 9.23 | 8.93 | 9.25 | 8.56 |
| Aromatic content | 5.92 | 5.97 | 6.13 * | 6.15 ** | 5.76 | 5.81 |
| F (Phe) | 3.12 | 3.08 | 3.25 | 3.29 | 3.19 | 3.32 |
| W (Trp) | 0.30 | 0.31 | 0.33 | 0.32 * | 0.27 | 0.29 |
| Y (Tyr) | 2.50 | 2.58 * | 2.55 | 2.54 * | 2.30* | 2.20* |
Note.—All frequencies of each species were compared against either “cold” species (§) or “hot” species (#) according to an a priori ecological sign-test. Bold results discriminated hot from cold lineages.
Level of significance associated with the a priori ecological sign-test,
P < 0.05,
P < 0.01,
P < 0.001.
A factorial correspondence analysis of the codon frequencies did not indicate any variation of codon usage between the six species, which seems to be phylogenetically inherited in alvinellid worms (supplementary fig. S2, Supplementary Material online).
Amino Acid Composition of Translated Sequences
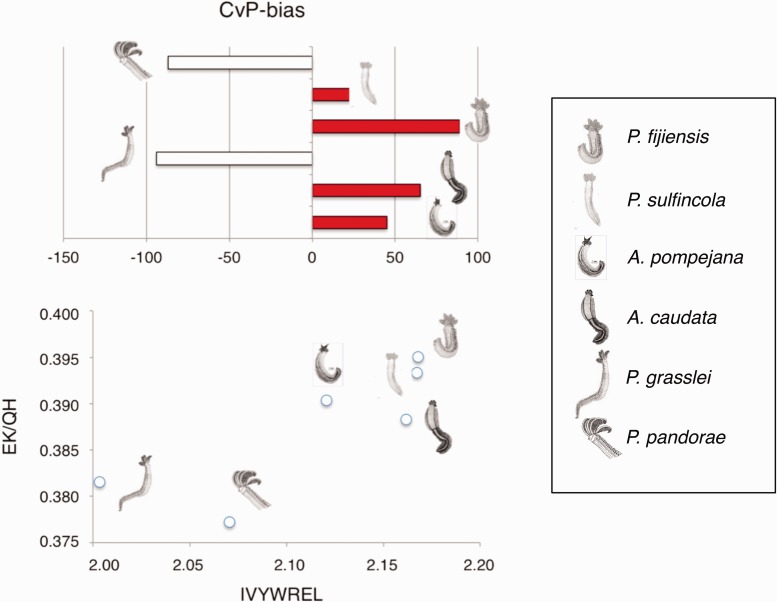
Cumulative ranked curves of three thermophilic criteria, CvP_bias, EK/QH and GARP-FYMINK, were plotted for the 423 translated ORFs and, clearly show that CvP_bias is the only criterion separating “hot” and “cold”-adapted species (see supplementary material S1, Supplementary Material online and fig. 1). However, the IVYWREL versus EK/QH biplot also successfully segregates species according to habitat (fig. 1). Looking at amino acids associated with GC-rich codons, ranked values of GARP markedly correlated with the ranked GC12 values, while no negative correlation was observed with the FYMINK index. The cumulative deviation of these two variables showed three distinct trends: An increase of GARP in the thermophilic Paralvinella species, a very weak deviation of GARP-FYMINK toward zero in the two Alvinella species and a clear increase of FYMINK in the cold-adapted Paralvinella species (supplementary material S1, Supplementary Material online).
Fig. 1.—
Main prokaryotic thermophilic amino acid indices (IVYWREL, EK/QH, and CvP_bias criteria) in “hot” and “cold” alvinellid species using translated sequences of 423 orthologous transcripts. (a) Overall CvP_bias between species, (b) overall EK/QH vs. IVYWREL biplot between species.
The AMOVA with nonparametric Kruskal–Wallis and paired Wilcoxon tests revealed no significant difference between alvinellid species of the average amino acid composition on the 423 translated proteins or their computed thermostability criteria (data not shown). However, these tests have limited power due primarily to the non-normality of the amino acid distributions within each species and also to the large intergene variance associated with these distributions. Sign-tests with ecological priors, however, provided strong evidence for the structural adaptation of alvinellid worms at the scale of their proteome. Results indicated that A, P, and L were significantly less frequent and M, G, S, and D significantly more frequent in P. pandorae and P. grasslei when compared with the “hot” adapted species (table 1). R, E, and Y were also more frequent in “hot” species but not always significant in all chimney species (table 1). Sign-test comparisons on thermophilic criteria and amino acid classes confirmed the occurrence of highly significant differences in amino acid patterns between “hot” and cold-adapted species. A greater number of genes were indeed ranked top for both the FYMINK (AT-ended codons) and STNQ (polar) criteria in the cold-adapted species whereas a greater number of genes exhibited maximal values for the GARP (GC-ended codons), IVYWREL, aromatic (FYW), and charged (RHKDE) residues in the “hot”-adapted species.
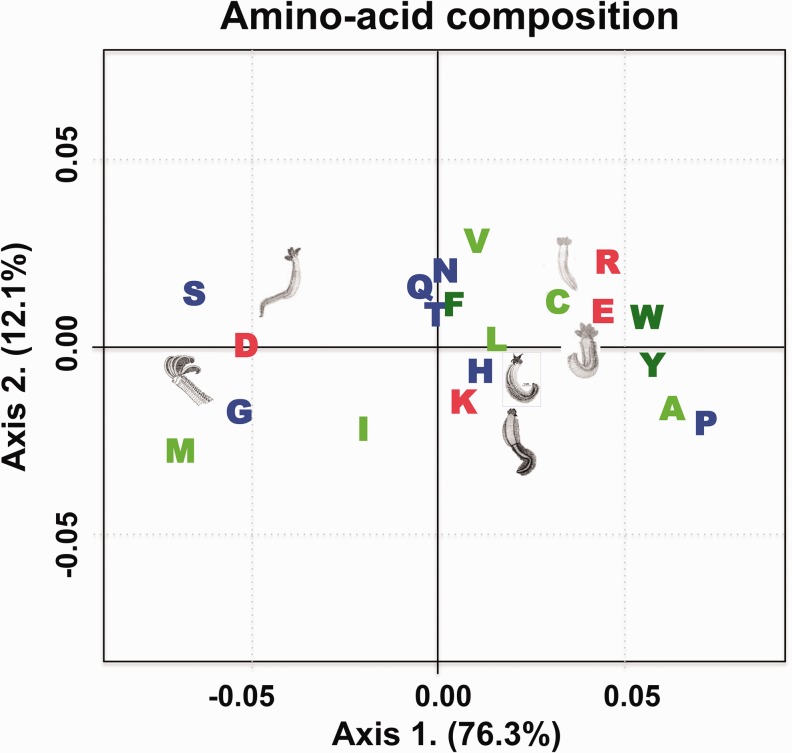
A FCA was performed on the residue frequencies of the 6 alvinellid species to understand the relationship between temperature and amino acid composition (fig. 2). The first axis explains 76.3% of the overall variance of the proteins in terms of amino acid composition and clearly separates the two cold-adapted species (P. grasslei and P. pandorae) from the four other species that live on the chimney walls. The proteins of the two cold-adapted species were enriched in serine, aspartic acid, glycine and methionine (DGMS) whereas the chimney-species proteins exhibited more alanine, leucine, glutamic acid, proline, and tyrosine (PAYLE). The detailed inspection of the amino acid composition of alvinellid proteins therefore suggests that species are subdivided into two groups according to their thermal preference. These groups can be discriminated by a higher frequency of four to five amino acid residues, which can be summarized by the indices PAYLE for the “hot” species and DGMS for the “cold” species. This new criterion corresponds to amino acids that also significantly differed between the two ecological groups on the basis of the sign test with ecological priors. Additionally, the arrangement of species on the first factorial axis was not the direct consequence of the GC genome-wide bias, as there is no correlation between the GC content and the coordinates of amino acid on the first FCA axis. Finally, cold-adapted species also displayed specific signatures of disordered residues using the Ising prediction software of Lobanov and Galzitskaya (2011). Although not very conclusive, results showed that P. pandorae displays a greater proportion of disordered residues (about 42%) when compared with the other species (33–37%; see supplementary fig. S3, Supplementary Material online).
Fig. 2.—
Factorial correspondence analysis of the amino acid frequencies between protein orthologous sequences of six alvinellid species.
Phylogenomic Tree
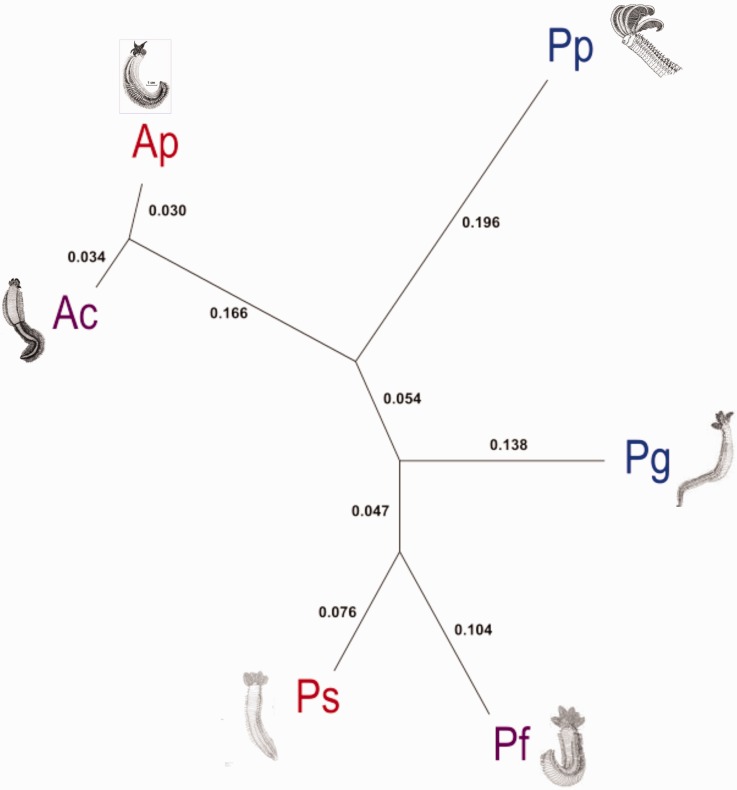
Although restricted to 6 species, the phylogenomic ML reconstruction of Alvinellidae ancestry with 423 POGs led to 3 putative species lineages (fig. 3, i.e. concatenation of 1,340 POGs without indel filtering and one missing species led to the same topology), and confirmed the separate evolution of P. pandorae, previously placed in the subgenus Nautalvinella (Desbruyères and Laubier 1993) on the basis of its gill morphology. This latter species is clearly basal to the group formed by the three other paralvinellid species (all belonging to the subgenus of Paralvinella). Most nuclear loci supported the dichotomy between the two formal genera (distances greater between Alvinella and Paralvinella than between P. pandorae and the other species) and placed the root between the Alvinella and Paralvinella species under the mid-point rooting option. Both rooted and unrooted trees confirmed the very close proximity of P. sulfincola and P. fijiensis, although living in very distinct geographic areas (North-East Pacific versus Western Pacific). As a consequence, the two cold-adapted species (P. pandorae and P. grasslei) displayed the greatest patristic divergence while branching at a more basal position in the Alvinellidae tree, under the mid-point rooting hypothesis separating the two genera. The hypothesis of global molecular clock was not significantly better than the no-clock model implemented in baseML (Yang 2007). However, branch lengths associated with Alvinella species were much shorter than those estimated for the Paralvinella species, suggesting that the species formation in Alvinella was either more recent or that these sister species evolved much more slowly than the other genus.
Fig. 3.—
Unrooted phylogenomic ML tree of alvinellids with patristic distances (in bold). All nodes were fully supported by 100% bootstrap values.
Rate and Nature of Derived Amino Acid Replacements
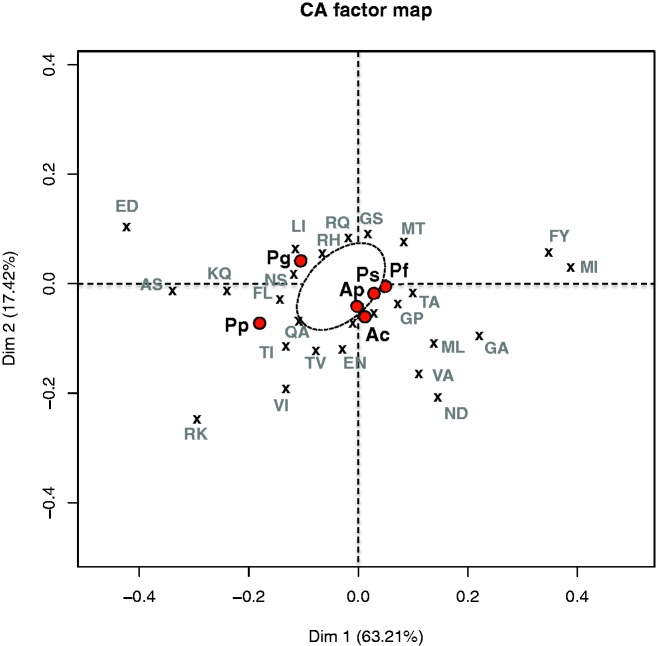
Derived mutations (i.e. orientated replacements specific to only one species) were counted and sorted into biochemical classes (e.g. charged, polar, aliphatic, aromatic) to estimate the mutational asymmetry of residue changes (e.g. VL–LV) between each species and its direct ancestor in a given lineage. The results showed a remarkable decrease in charged residues for the two cold-adapted species (table 2). Proline also decreased remarkably (−16 for P. grasslei and −27 for P. pandorae). Such a general decrease in charged residues and proline was entirely compensated by the gain of polar residues in P. grasslei (+73). In P. pandorae, the compensation also involved the gain of a large number of aliphatic residues (+36) in addition to polar ones (+65). A FCA was then performed on the matrix of mutational asymmetries between the 20 amino acids (fig. 4). The first two factorial axes explained about 80% of the total variance of the derived mutations among species (axis 1: 63.2% and axis 2: 17.4%). Along the first axis, the four chimney species were tightly grouped and well separated from the two cold-adapted species. This indicates that the chimney species have a very similar pattern of replacements. Relative to chimney species, the differences in substitutional patterns of cold-adapted species are expressed by their position on axis 2. A more detailed analysis of the relative increase/decrease of specific residues between biochemical groups (fig. 4), indicated that derived mutations accumulated very differently in the two cold-adapted species (E → D, K → Q, or A → S preferred for P. grasslei, against R → K, V → I, and T → I for P. pandorae).
Table 2.
Gain and Loss of Residue Classes in the Present-day Species Obtained from the Derived Substitutions Accumulated Within Each Terminal Branch of the Phylogenomic ML Tree
| Species | Polar | Aliphatic | Aromatic | Aa + | Aa − | Cys | Pro | Gly |
|---|---|---|---|---|---|---|---|---|
| A. pompejana | +11 | −4 | +1 | −4 | −3 | +1 | −5 | +3 |
| A. caudata | −4 | +13 | +9 | −1 | −1 | −5 | −4 | −7 |
| P. sulfincola | +12 | +4 | 0 | −1 | −5 | +1 | −4 | −7 |
| P. fijiensis | +4 | +10 | +9 | +8 | −21 | +3 | −4 | −9 |
| P. grasslei | +73 | −14 | −6 | −18 | −13 | +2 | −16 | −8 |
| P. pandorae | +65 | +36 | +4 | −45 | −33 | −9 | −27 | +9 |
Note.—Aa+: positively charged amino acids, Aa−: negatively charged amino acids.
Fig. 4.—
Factorial correspondence analysis (FCA) of orientated amino acid substitution frequencies between orthologous protein sequences of alvinellid species. The dashed envelope represents the cloud of remaining substitution points.
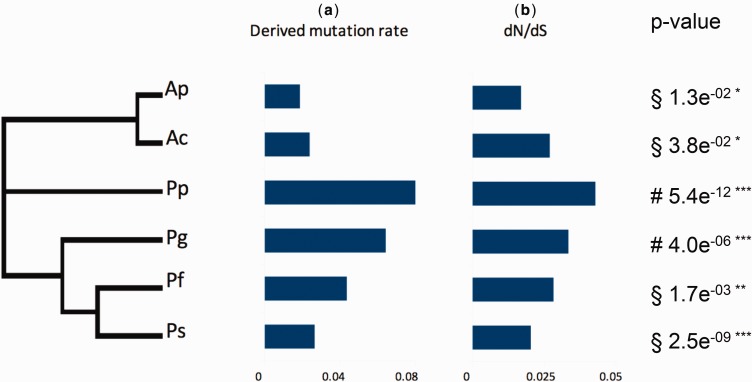
Finally, each count of derived mutations was divided by the ultrametrized synonymous length of its corresponding terminal branch under the mid-point rooting hypothesis to test whether nonsynonymous substitutions have accumulated at the same rate (fig. 5a ). The hypothesis of a constant accumulation of amino acid replacements in terminal branches was rejected (χ 2 = 1326, df = 5, P-value < 0.001), indicating that amino acid replacements were more pronounced in the terminal branches of the two cold-adapted species. However, these results, although weighted by the branch length in the absence of a molecular clock constraint, were very sensitive to the substitution model and to the gene partition during the phylogenomic reconstruction.
Fig. 5.—
Derived amino acid replacement rates and median intergenes dN/dS ratios observed on the terminal branches of the tree. Each rate was weighted with the length (t) of the corresponding terminal branch obtained after tree ultrametrization. Absolute timing inferred under a clock hypothesis on the mt Cox1 gene (see supplementary fig. S1, Supplementary Material online) and its evolutionary rate estimated from Chevaldonné et al. (2002) produced exactly the same results. The null hypothesis of constant rate of derived replacements was rejected using a χ 2 test (χ 2 = 1326, dfl = 5). (a) Derived mutation rates, (b) dN/dS ratios in terminal branches, (c) P-values represent the level of dN/dS significance following an ecological a priori Binomial test (i.e. values of a given species significantly greater or lower than either “cold” (§) or “hot” (#) species).
Relaxation of Selection on “Cold” Adapted Genes
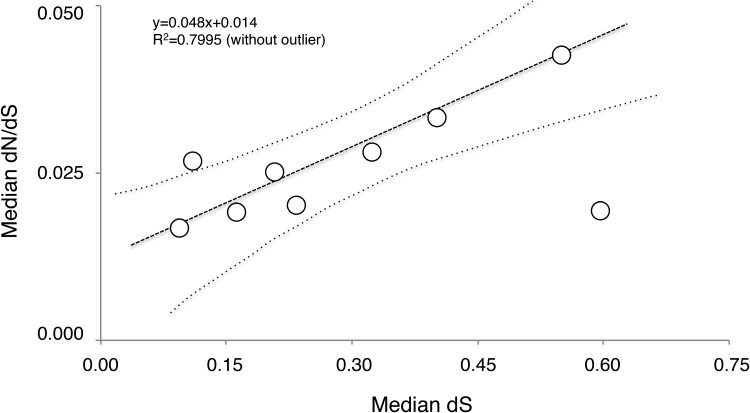
The nonsynonymous versus synonymous mutation ratios (ω) were estimated for 423 POGs over the branches with a significant LRT values against the null M0 (one ω for the whole tree) model of the alvinellid RaxML tree (fig. 3) using the free-ratio (M1) selective model of CodeML. According to the analysis, alvinellids evolved under great purifying selective constraints with ω never exceeding 0.05 over the 423 POGs (fig. 5b ). These latter estimates were on average very close to the median values estimated from the concatenated set of genes for the three internal branches (0.019–0.025). In contrast, values for genes of the two cold-adapted species were higher (0.043 for P. pandorae and 0.033 for P. grasslei) and suggested that the genes of these two species could have experienced some relaxation of selection. These observations are unlikely to be the consequence of a systematic lack of power in the estimation of dS because the distribution of ω values does not seem to be constrained by its lower extent (i.e. dS always above zero) and because the ω values linearly increased with species divergence (i.e. dS; see Fig. 6). However, interestingly, the internal branch leading to Alvinella spp. is very constrained by purifying selection (very low dN value), yielding a dN/dS that represents an outlier in the other correlated values of dN/dS vs dS plot (Fig. 6). The observed differences in the median estimates of ω between the hot-adapted and cold-adapted species are confirmed by the Binomial sign-test, with ecological priors accounting more informatively for the intergenes variability (P-values in fig. 5c ). Both P. pandorae and P. grasslei displayed significantly more genes with a higher ω value than those found in the thermophilic representatives, whereas all the four species inhabiting chimneys significantly displayed more genes with a lower ω value than those of the two cold representatives.
Fig. 6.—
Biplot of dN/dS versus dS medians obtained over branches of the phylogenomic ML tree of the six alvinellid species with the free-ratio branch-model M1 implemented in PaML 4.0 (Yang 2007). Branch values are significantly (P < 0.01: Pearson’s r test) linearly regressed without using the branch leading to Alvinella spp. (outside of the confidence envelope at 95% for the whole dataset).
For each terminal branch, a specific search for codon sites under positive selection was performed by comparing the selection model (M2A) of codon substitution with the nearly neutral model (M1A). Branches leading to cold-adapted species produced a greater number of genes with a significantly better fit to M2A. The likelihood ratio tests indicate that 21 genes displayed an additional class of ω > 1 (positive selection) in cold-adapted species while this number was only 6 in the thermophilic species. Among this subset of genes under positive selection (i.e. with a significant LRT), 12 genes contained sites under positive selection with a BEB probability greater than 0.95 in the two cold-adapted species and only two genes in A. pompejana and P. sulfincola. However, only one replacement under positive selection led to a substantial change in the amino acid property of the protein (charged to polar) while all other mutations occurred within the same biochemical class of residues.
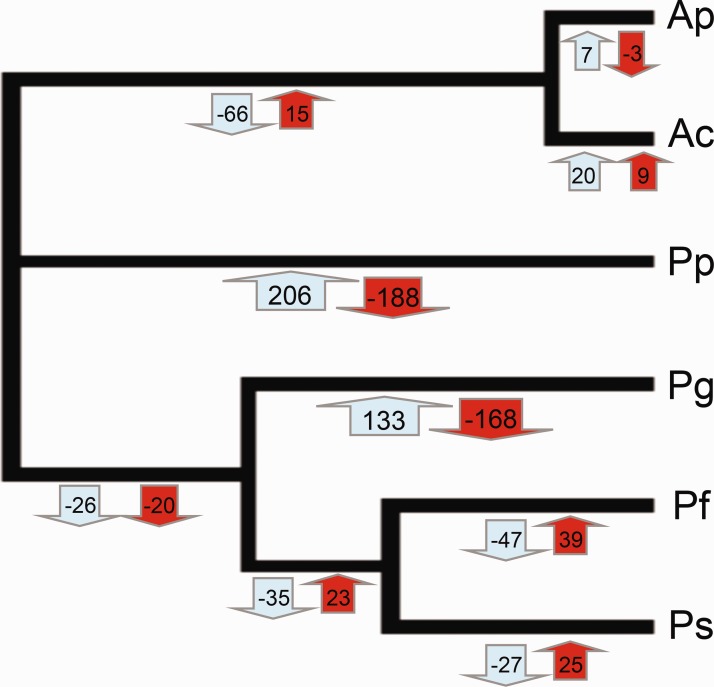
Evolution of Thermophilic Criteria from the Ancestor to Present Taxa
The criteria specific to alvinellids, PAYLE vs. DGMS, which opposed hot-adapted to cold-adapted species (see above), were used to reconstruct the history of thermal adaptation from ancestral sequences. To this extent, we measured the relative increase or decrease of the two amino acid groups from the most recent common ancestor (MRCA) of alvinellid worms (obtained from the mid-rooting point between the genera Alvinella and Paralvinella) to the present-day taxa. Most ancestral protein sequences were reconstructed with a high Bayesian probability (>90%) and, thus gave us a good level of confidence to evaluate the evolution of our criteria along the branches of the phylogenomic tree of alvinellid worms (fig. 7). The cold-adapted species displayed a great increase of the DGMS proportion of residues in their proteins relative to their ancestral state (net increase of 206 and 133 residues in P. pandorae and P. grasslei, respectively) and this increase was directly linked to the decrease of PAYLE for the same species (188 and 168 residues). On the contrary, PAYLE increased significantly in hot-adapted Paralvinella species and in the internal branch leading to Alvinella spp. (i.e. a very small increase in charged and aromatic residues but a loss of polar residues in Alvinella).
Fig. 7.—
Ancestral reconstruction of amino acid changes using the alvinellid criteria PAYLE vs. DGMS along the phylogenomic tree of alvinellid worms following the Bayesian reconstruction of ancestral sequences implemented in aaML of PaML (Yang 2007).
Discussion
Thermophily is Likely Driven by a Rather Old and Global Structural Effect on Proteins
Alvinellid polychaetes represent a family of worms that successfully colonized all the vent habitats of the Pacific Ocean (Desbruyères and Laubier 1986, 1993). Out of the twelve species described so far, at least five live on the walls of “hot” vent chimneys. Two of these worms, A. pompejana and P. sulfincola, have been experimentally shown to be the most thermophilic metazoans known on Earth (Girguis and Lee 2006; Ravaux et al. 2013). The polyphyletic nature of thermophily (i.e. a specific trait shared by the two well-separated genera of Alvinellidae) raises the question of whether thermophily has evolved independently several times in the recent history of the family or resulted from a very long history of thermal adaptation, initiated several tens of million years ago. Here we performed a comprehensive analysis of proteomes using 6 alvinellid species living in contrasted thermal habitats to answer this question in a well-constrained phylogenetic framework.
Out of 423 orthologous putative ORFs, the cold- and hot-adapted species displayed contrasted patterns of protein composition. The direct comparison of overall residue frequencies between species indicated a smaller number of charged residues, and especially positively charged residues in P. pandorae and P. grasslei. Positively charged residues were mainly replaced by neutral polar and small aliphatic residues in these latter species. The FCA on protein composition also provided a strong support for adaptive patterns while separating the two cold-adapted species (and especially P. pandorae) from the four other species living on chimneys (76% of the whole variance associated with the first axis). Residues contributing the most to this difference were Pro, Ala, Tyr, Leu, Glu (PAYLE), and Arg, which allowed the clustering of the four chimney species, and Asp, Gly, Met, and Ser (DGMS) that grouped the two cold-adapted species. Although restricted to a small number of species that could limit the power of significance, these diagnostic residues segregated the hot and cold species on the first FCA axis. This was supported by a significant a priori ecological sign-test unbalance between the two groups of species (and especially significant differential patterns favoring Ala, Leu, Pro, and Tyr in the hot-adapted species). This pattern was not phylogenetically driven: Alvinella species tightly clustered with the two thermophilic Paralvinella species, whereas P. grasslei, which is phylogenetically and morphologically very close to P. fijiensis and P. sulfincola (Desbruyères and Laubier 1993) grouped with the most divergent species P. pandorae. The absence of relationship in the FCA plot between the species coordinates and their GC contents, and the lack of marked codon usage differences suggested that these observed amino acid patterns were not due to a genome-wide GC-bias effect across species (contrary to what is usually observed for microorganisms; Chen et al. 2004). The FCA analysis coupled with the Binomial test with ecological priors unambiguously confirmed previous results obtained by Jollivet et al. (2012) based on a pair of “hot” and “cold”-adapted alvinellid species (A. pompejana and P. grasslei). According to gene ontogeny (GO analysis, see supplementary material S1, Supplementary Material online), the study also indicated that these amino acid changes are likely to affect all categories of proteins and thus are mostly the reflection of a global structural effect on proteins, reminiscent of what has been described for thermophilic and hyperthermophilic microorganisms (Kreil and Ouzounis 2001; Hickey and Singer 2004; Zeldovich et al. 2007). As such, and despite the smaller thermal amplitude encountered by the worms (<60 °C) when compared with the thermophilic prokaryotes, our hot- and cold-adapted species appear to be well discriminated by the thermophilic criteria documented for prokaryotes such as the CvP_bias (Suhre and Claverie 2003), the de Farias and Bonato’s (2003) EK/QH or the IVYWREL (Zeldovich et al. 2007), even if not perfectly adapted to our metazoan models (see further discussion on the PAYLE vs DGMS criteria in supplementary material S1, Supplementary Material online).
Thermophily is an Ancestral Trait with a Long History of Parallel Evolution
The main purpose of the study was to shed light on the ancestry of the thermophily trait in the family of these peculiar worms. As previously noticed from both the recovered fossils in “old” ophiolites (i.e. fossil vent rocks) and phylogenetic analyses (Little et al. 1999; Vrijenhoek 2013), alvinellid worms may have experienced a long history of speciation events possibly starting at the beginning of the Cretaceous in this hostile and highly selective environment. The first resulting observation of the present study is that a very large proportion of protein positions were not free to vary in both thermophilic and cold-adapted alvinellid species with median ω values ranged from 0.01 (A. pompejana) to about 0.04 (P. pandorae). Even if synonymous rates are mostly driven by effective population size and generation time, which in turn may lower dN/dS in species with high fecundity and rapid turnover, these ratios are far smaller than those recorded for mammalians (0.20–0.25: Ohta 1995) or marine species such as oysters or tunicates (0.10–0.20: Gayral et al. 2013). The second main result is that alvinellid species that live on chimneys (i.e. thermophilic species) are all under stronger purifying selection (small ω ratios) than their cold-adapted counterparts even if the small number of species studied here could impact the significance of these comparisons. All thermophilic species indeed display very low rates of nonsynonymous mutations in their recent history (i.e. terminal branches). In contrast, the cold-adapted lineages (also under purifying selection) have derived nonsynonymous rates two to three times greater than their thermophilic counterparts. Such a difference is, however, not due to a phylogenetic effect because 1) all alvinellid worms display the same codon usage and 2) the two cold-adapted species are not closely related, and thus display phylogenetically independent accelerated derived mutation patterns in comparison to the other four species. This difference does not seem to be the outcome of great differences in the effective population sizes and/or generation times either, as most species exhibit the same reproductive biology and nearly the same census size of populations (Chevaldonné and Jollivet 1993; Zal et al. 1995; Jouin et al. 2002; Copley et al. 2003; Faure et al. 2007) and have a very long history of evolution because their last speciation event (about 28 Ma for the most closely related and sibling species (see Chevaldonné et al. 2002).
Nevertheless, because of the very small nonsynonymous rates and the lack of positive selection in thermophilic species, the enrichments in Arg, Glu, Ala, Ile, and Tyr at ancestral nodes and their global effect on proteomes, we suggest that the alvinellid primitive proteome was preadapted to “hot” conditions, giving support to the hypothesis that thermophily is an ancestral state. Although the question of whether LUCA was thermophilic is still under great debate (Galtier et al. 1999; Di Giulio 2003; Boussau et al. 2008), most studies based on protein resurrection suggest that ancestors to bacteria and archaeal were probably thermophilic (Gaucher et al. 2003; Akanuma et al. 2013). This situation also seems to have occurred in alvinellid worms, probably because of their emergence before or at the beginning of Cretaceous (Haymon et al. 1984; Little et al. 1999). Long term-history of purifying selection on thermostable proteins, and especially those of thermophilic alvinellids may have resulted in an evolutionary slow-down leading to the assumption that thermophiles were more likely to resemble the ancestral state. To this extent, protein surface interactions, and especially the superficial increase of ionic and hydrogen bonds, and the physical constraints associated with protein folding may influence (and even “freeze”) the evolutionary rate of a protein-encoding gene as previously stated by Tóth-Petróczy and Tawfik (2011) and Romero-Romero et al. (2016). In the case of alvinellids, the hypothesis of ancestry is also well supported by the fact that only very few or no loci were shown to be under positive selection on the terminal branches leading to the thermophilic species. If thermophily was recently gained in some of the alvinellid lineages, one would indeed expect to see more innovation in proteins of the thermophilic species.
Even if conditioned by the number of species used, the ancestral state of alvinellid adaptation to high temperature was also supported by the relative change of PAYLE and DGMS over the course of alvinellid evolution. These results suggest that the two cold-adapted species have accumulated much more DGMS residues against PAYLE residues than their chimney relatives. Comparatively, the amount of observed replacements from the ancestral nodes to the thermophilic species was much more moderate (with a much lower variance of amino acid counts over branches). For example, A. pompejana exhibited virtually no difference in its protein composition compared with its direct ancestor and a limited number of differences with its sister species A. caudata. Adding differences from the root trifurcation (MRCA node) to A. pompejana led to the accumulation of only 18 PAYLE residues, while this number decreased by 204 and 166 on the terminal branches leading to P. pandorae and P. grasslei, respectively.
Based on these results, the ability to live in highly variable and extreme temperatures was likely gained in the early history of alvinellid evolution. However, looking at the derived mutation rates indicates that chimney species also accumulated charged and aromatic mutations independently, as derived replacements are unique to each taxon. For the four chimney species, the nature of derived replacements was very similar, with an overall increase of charged and aromatic residues but these substitutions were not always on the same proteins orthologs. This indicates that, even if alvinellids were pre-adapted to high temperatures, they also co-evolved separately for millions years through a long process of parallel evolution to nearly the same extreme environment. Indeed, even if changes in protein thermostability (i.e. at least stabilizing or increasing their T m) may occur linearly in time as a consequence of directional selection, proteins are likely to explore a wide space of conformational pathways through the process of “thermodynamic system shift” (Hart et al. 2014). As a consequence, the longer species live in similarly constrained environments, the more they are likely to diverge by this process despite strong purifying selection and habitat convergence. Fixation of similar adaptive mutations in replicate populations or closely related species seems to be a common issue with a high probability of occurrence (Orr 2005) and often involves the same set of genes (Nadeau and Jiggins 2010). The maintenance of equal phenotypic performances in different species is therefore a very long process usually governed by epistatic interactions, which imposes many compensatory replacements to either maximize the benefit or minimize the deleterious effect of a single mutation in different genetic backgrounds (Berezovsky et al. 2007; Lunzer et al. 2010; Serohijos and Shakhnovich 2014). This contrasts sharply with the two cold-adapted species for which the rate and the nature of derived mutations were significantly different leading to the preferential replacements of charged residues by polar ones in P. grasslei and aliphatic ones in P. pandorae. These results agree well with a diversifying selective process (as opposed to the “thermodynamic system shift” process) as supported by previous experimental studies on bacterial evolution which provide examples of parallel molecular evolution in replicates evolving in similar (thermal) environments while adapting specifically to the environment often leading to a fitness loss in other environments due to antagonistic gene pleiotropy effects (Elena and Lenski 2003).
Cold Adaptation Emerged Several Times during the Alvinellid Radiation
Nonsynonymous to synonymous ratios on terminal branches of the alvinellid tree indicate that the protein evolution of the two cold-adapted lineages was less constrained than for the four chimney species. As selective relaxation should affect equally protein residues that are free to change (i.e. covarions, Zuckerkandl and Pauling 1965), one would expect that most amino acid replacements under relaxed selection should be randomly distributed into the biochemical classes of residues. Here, the accumulation of amino acid changes impacts more specifically some classes of residues, with striking differences in the two cold-adapted species that does not seem to be attributable to genetic drift alone. Although the radiation of alvinellid worms seems to be quite ancient (>100 Ma), this differential accumulation of residues in the two species was not consistent with the universal trend of amino acid losses and gains through the course of evolution reported by Jordan et al. (2005) in the three domains of life, but rather suggests a convergent effect specifically directed on “cold” species during their independent evolutionary trajectories. Interestingly, the proteins of the ancestors and chimney species are enriched in amino acids known to decline in modern taxa and thus suggesting that proteins of the four “thermophilic” species are more structurally related to the ancestor. On the contrary, DGMS enrichments in proteins of cold-adapted species do not fit the expectations associated with the accumulation of late-coming amino acids in the evolution of life. Differences in the nature of derived mutations (aliphatic vs. polar) between the two cold-adapted species suggest therefore that species experienced specific phylogenetic constraints or adaptive specificities for living in cold environments. Both types of enrichments are, however, observed in species living in polar environments and can be viewed as a mean to increase protein flexibility at low temperatures to preserve function (Saunders et al. 2003). Colder species and especially P. pandorae also exhibited a greater proportion of disordered polypeptidic regions, which is characteristic mostly of psychrophilic species to compensate the limited conformational movements of the protein at environmental temperatures (Russell 2000; Saunders et al. 2003; Siddiqui and Cavicchioli 2006). In psychrophilic archaea, adaptation to cold led to the increase of glycine, glutamine and threonine (providing a greater conformational mobility of the protein backbone), the decrease of proline in loop regions and arginine in the exposed parts of the molecule, and the reduction of large (aromatic) hydrophobic residues in the core of the protein (Saunders et al. 2003; Reed et al. 2013). The increase of nearly all these residues in the cold-adapted alvinellid proteomes at a greater rate and in a nonrandom manner between lineages suggests that part of the protein evolution was driven by positive selection. This view is supported by a greater number of codons under positive selection in the two cold-adapted lineages. In addition, the distribution of GO functions in loci displaying dN/dS values significantly greater in cold-adapted species indicates that selective relaxation and/or positive selection was not targeting specific pathways, suggesting a more global structural effect (see supplementary material S1, Supplementary Material online). Although not significant, some metabolic processes, such as homeostasis, and nucleic acid production and repairs, may have, however, been more commonly affected.
An alternative explanation to the differences observed between the two cold-adapted species may come from the timing of the colonization of the cold hydrothermal vent habitats during the evolutionary history of alvinellid species. Indeed P. pandorae belongs to the subgenus Nautalvinella, which according to Vrijenhoek (2013) was the first to split and diverge inside the genus Paralvinella (i.e. about 60 Ma). Because the three species of this clade are all cold-adapted species, they are likely to represent the first colonizers of colder vent environments and display specific morphological characteristics, including penned gills, more adapted to survive in the well-oxygenated deep-sea surroundings (Desbruyères and Laubier 1993). In contrast, P. grasslei belongs to the Paralvinella subgenus, from which, at least three species (P. sulfincola, P. fijiensis and P. hessleri) are known to live exclusively on vent chimneys walls. The radiation of Paralvinella into subgenera and two ecological groups (three thermophilic and four mesophilic species) therefore date from very different times. This could have played a role by affecting the time spent exploring the combination of residues to obtain the best conformational issue of the proteins. As these two groups evolved separately for a very long period of time, both lineages could have experimented different adaptive strategies and a much longer period of time to adapt to colder environments (i.e. to replace charged residues by hydrophobic ones, possibly via transient polar states) in the case of the Nautalvinella subgenus group.
Conclusion
The reconstruction of ancestral states with confidence strongly depends on the number of species in a phylogeny and on the divergence between the sequences. Some caution in the interpretation of inferred changes should therefore be taken. However, our results indicate that present-day alvinellid lineages have experienced habitat shifts during their evolution. Contrary to expectations that thermophily should be a derived character in alvinellids, the rates and nature of derived substitutions, as well as the strength of selection occurring on terminal and internal branches of their phylogenetic tree, rather suggest that adaptation to high temperatures has occurred early in the evolutionary history of these worms. Thermophily was then maintained through strong purifying selection and potentially reinforced in some hot-adapted lineages. This suggests that the long-term parallel evolution of the worms since their radiation may have played a non-negligible role in shaping the worm proteomes. Adaptation to colder temperatures seems to have happened secondarily but independently and probably not at the same time, and lead to slightly different strategies, overall replacing charged residues by polar or by aliphatic ones.
The classical view of local adaptation considers that selective sweeps should have had a very narrow, gene-specific, impact spectrum while demographic effects are expected to have a broader impact on the whole genome, as they bring nearly neutral and slightly deleterious mutations randomly to fixation more rapidly (Ohta 1992; Barton and Mallet 1996). However, this view may be too simplistic, and we should consider that adaptation to extreme environments, within which temperature and hydrostatic pressure conditions directly affect the entropic stability of the whole proteome (and thus, the relationship between the cost and function of proteins), produces nearly similar genome imprints, at least in the coding regions.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We are particularly grateful to the chief scientists that lead the oceanographic cruises Biospeedo 2004 (D. Jollivet, doi:10.17600/4010050), JdFR 2008 Expedition (OCE06-23554, P.R. Girguis, R.W. Lee), Lau Basin 2009 Expedition (C.R. Fisher) and Mescal 2010 (N. Le Bris and F.H. Lallier, doi:10.17600/12010020) and to the “Nautile”, “Alvin”, and “Jason II” crews for their technical support and efforts during the expeditions for the animal collection. We thank the Roscoff ABIMS platform of bioinformatics, and more particularly S. Caron for his help in the regular use of computing cluster facilities and to M. Monsoor for developing the pipeline suite (AdaptSearch) for the Galaxy environment. This research was supported by a joint research postdoc grant funded by both the Institut Ecologie et Environnements (InEE) du CNRS and the Conseil Général du Finistère (CG29) and by the ANR (project 05-BLAN-0407), the Total Foundation and, the programme “Molecular and Cellular Biology” (01201353567) for O.V.G. and M.Y.L. We also thank Nick Goldman for his advices on the ML approach for detecting positive selection on coding sequences.
Literature Cited
- Akanuma S, et al. 2013. Experimental evidence for the thermophilicity of ancestral life. Proc Nat Acad Sci U S A. 110:11067–11072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baross JA, Holden JF. 1996. Overview of hyperthermophiles and their heat-shock proteins In: Adams MWW, editor. Advances in protein chemistry. Vol. 48 San Diego: Academic Press, Inc; p. 1–34. [DOI] [PubMed] [Google Scholar]
- Barton NH, Mallet J. 1996. Natural selection and random genetic drift as causes of evolution on islands (and discussion). Phys Trans Roy Soc B. 351:785–795. [DOI] [PubMed] [Google Scholar]
- Belshaw R, Katzourakis A. 2005. BlastAlign: a program that uses blast to align problematic nucleotide sequences. Bioinformatics 21:122–123. [DOI] [PubMed] [Google Scholar]
- Berezovsky IN, Shakhnovich EI. 2005. Physics and evolution of thermophilic adaptation. Proc Natl Acad Sci U S A. 102:12742–12747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezovsky IN, Zeldovich KB, Shakhnovich EI. 2007. Positive and negative design in stability and thermal adaptation of natural proteins. PLoS Comput Biol. 3(3):e52–498. 507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock T, et al. 2014. An integrated approach for genome annotation of the eukaryotic thermophile Chaetomium thermophilum . Nuc Acids Res. doi:10.1093/nar/gku1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boussau B, Blanquart S, Necsulea A, Lartillot N, Gouy M. 2008. Parallel adaptations to high temperatures in the Archaean eon. Nature 456:942–946. [DOI] [PubMed] [Google Scholar]
- Cary SC, Shank T, Stein J. 1998. Worms bask in extreme temperatures. Nature 391:545–546. [Google Scholar]
- Charif D, Lobry JR. 2007. SeqinR 1.0-3: a contributed package to the R project for statistical computing devoted to biological sequences retrieval and analysis. In: Structural approaches to sequence evolution. Berlin: Springer. [Google Scholar]
- Chevaldonné P, Desbruyères D, Childress JJ. 1992. And some even hotter…. Nature 359:593–594. [Google Scholar]
- Chevaldonné P, Jollivet D. 1993. Videoscopic study of deep-sea hydrothermal vent alvinellid polychaete populations: estimation of biomass and behaviour. Mar Ecol Prog Ser. 95:251–262. [Google Scholar]
- Chevaldonné P, Jollivet D, Desbruyères D, Lutz RA, Vrijenhoek RC. 2002. Sister-species of eastern Pacific hydrothermal vent worms (Ampharetidae, Alvinellidae, Vestimentifera) provide new mitochondrial COI clock calibration. Cah Biol Mar. 43:367–370. [Google Scholar]
- Chen SL, Lee W, Hottes AK, Shapiro L, McAdams HH. 2004. Codon usage between genomes is constrained by genome-wide mutational processes. Proc Nat Acad Sci U S A. 101:3480–3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copley JTP, Tyler PA, Van Dover CL, Philp SJ. 2003. Spatial variation in the reproductive biology of Paravinella palmiformis (Polychaeta: Alvinellidae) from a vent field on the Juan de Fuca Ridge. Mar Ecol Prog Ser. 255:171–181. [Google Scholar]
- Cossins AR, Macdonald AG. 1989. The adaptation of biological membranes to temperature and pressure: fish from deep and cold. J Bioenerg Biomembr. 21:115–135. [DOI] [PubMed] [Google Scholar]
- De Farias ST, Bonato MC. 2003. Preferred amino acids and thermostability. Genet Mol Res. 2:383–393. [PubMed] [Google Scholar]
- Desbruyères D, et al. 1998. Biology and ecology of the ‘Pompeii worm’ (Alvinella pompejana Desbruyères & Laubier), a normal dweller of an extreme deep-sea environment: a synthesis of current knowledge and recent developments. Deep-Sea Res II 45:383–422. [Google Scholar]
- Desbruyères D, Laubier L. 1986. Les Alvinellidae, une famille nouvelle d’annélides polychètes inféodées aux sources hydrothermales sous-marines: systématique, biologie et écologie. Can J Zool. 64:2227–2245. [Google Scholar]
- Desbruyères D, Laubier L. 1993. New species of Alvinellidae (Polychaeta) from the North Fiji back-arc basin hydrothermal vents (southwestern Pacific). Proc Biol Soc Wash 106:225–236. [Google Scholar]
- Dilly GF, Young CR, Lane WS, Pangilinan J, Girguis PA. 2012. Exploring the limit of metazoan thermal tolerance via comparative proteomics: thermally induced changes in protein abundance by two hydrothermal vent polychaetes. Proc Roy Soc Lond B. 279:3347–3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giulio M. 2003. The universal ancestor was a thermophile or a hyperthermophile: tests and further evidence. J Theor Biol. 221:425–436. [DOI] [PubMed] [Google Scholar]
- Elena SF, Lenski RE. 2003. Evolution experiments with microorganisms: the dynamics and genetic bases of adaptation. Nature Rev. 4:457–469. [DOI] [PubMed] [Google Scholar]
- Faure B, Chevaldonné P, Pradillon F, Thiébaut E, Jollivet D. 2007. Spatial and temporal dynamics of reproduction and settlement in the Pompeii worm (Polychaeta: Alvinellidae). Mar Ecol Prog Ser. 348:197–211. [Google Scholar]
- Féral JP, Philippe H, Desbruyéres D, Laubier L, Derelle E, Chenuil A. 1994. Phylogénie moléculaire de polychètes Alvinellidae des sources hydrothermales actives de l'océan Pacifique. C R Acad Sci Paris, Sciences de la vie, Ser. 3(317):771–779. [Google Scholar]
- Fletcher W, Yang Z. 2010. The effect of insertions, deletions, and alignment errors on the branch-site test of positive selection. Mol Biol Evol. 27:2257–2267. [DOI] [PubMed] [Google Scholar]
- Forsdyke DR. 2011. Mutation In: Evolutionary bioinformatics. Berlin: Springer; p. 131–151. [Google Scholar]
- Fraser NJ, et al. 2016. Evolution of protein quaternary structure in response to selective pressure for increased thermostability. J Mol Biol. doi: 10.1016/j.jmb.2016.03.014. [DOI] [PubMed] [Google Scholar]
- Gagnière N, et al. 2010. Insights into metazoan evolution from Alvinella pompejana cDNAs. BMC Genomics 11:634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galtier N, Tourasse N, Gouy M. 1999. A nonhyperthermophilic common ancestor to extant life forms. Science 283:220–221. [DOI] [PubMed] [Google Scholar]
- Gaucher EA, Thomson JM, Burgan MF, Benner SA. 2003. Inferring the palaeoenvironment of ancient bacteria on the basis of resurrected proteins. Nature 425:285–288. [DOI] [PubMed] [Google Scholar]
- Gayral P, et al. 2013. Reference-free population genomics from next-generation transcriptome data and the vertebrate-invertebrate gap. PLoS Genet. 9:e1003457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring WJ, Wehner R. 1995. Heat shock protein synthesis and thermotolerance in Cataglyphis, an ant from the Sahara desert. Proc Natl Acad Sci U S A. 92:2994–2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girguis PR, Lee RW. 2006. Thermal preference and tolerance of alvinellids. Science 312:231. [DOI] [PubMed] [Google Scholar]
- Glyakina AV, Garbuzynskiy SO, Lobanov MY, Galzitskaya OV. 2007. Different packing of external residues can explain differences in the thermostability of proteins from thermophilic and mesophilic organisms. Bioinformatics 23:2231–2238. [DOI] [PubMed] [Google Scholar]
- Gouy M, Guindon S, Gascuel O. 2010. Seaview version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 27:221–224. [DOI] [PubMed] [Google Scholar]
- Gu X, Li WH. 1994. A model for the correlation of mutation rate with GC content and the origin of GC-rich isochores. J Mol Evol. 38:468–475. [DOI] [PubMed] [Google Scholar]
- Hart KM, et al. 2014. Thermodynamic system drift in protein evolution. PLoS Biol. 12:e1001994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haymon RM, Koski RA, Sinclair C. 1984. Fossils of hydrothermal vent worms from Cretaceous sulfite ores of the Samail ophiolites, Oman. Science 223:1407–1409. [DOI] [PubMed] [Google Scholar]
- Hê S, Josse J, Husson F. 2008. FactoMineR: an R package for multivariate analysis. J Stat Soft. 25:1–18. [Google Scholar]
- Hickey DA, Singer GAC. 2004. Genomic and proteomic adaptations to growth at high temperatures. Genome Biol. 5:117–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holder T, et al. 2013. Deep transcriptome-sequencing and proteome analysis of the hydrothermal vent annelid Alvinella pompejana identifies the CvP-bias as a robust measure of eukaryotic thermostability. Biol Direct. 8:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hourdez S, Lallier FH. 2007. Adaptations to hypoxia in hydrothermal-vent and cold-seep invertebrates. Rev Environ Sci Biotechnol. 6:143–159. [Google Scholar]
- Hurst LD, Merchant AR. 2001. High guanine-cytosine content is not an adaptation to high temperature: a comparative analysis amongst prokaryotes. Proc R Soc Lond B. 268:493–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenicke R, Böhm G. 1998. The stability of proteins in extreme environments. Curr Opin Struct Biol. 8:738–748. [DOI] [PubMed] [Google Scholar]
- Jollivet D, Desbruyères D, Ladrat C, Laubier L. 1995. Evidence for differences in allozyme thermostability in deep-sea hydrothermal vent polychaetes Alvinellidae: a possible selection by habitat. Mar Ecol Prog Ser. 123:125–136. [Google Scholar]
- Jollivet D, et al. 2012. Proteome adaptation to high temperatures in the ectothermic hydrothermal vent Pompeii worm . PLoS One 7:e31150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan IK, et al. 2005. A universal trend of amino acid gain and loss in protein evolution. Nature 433:633–637. [DOI] [PubMed] [Google Scholar]
- Jouin C, Mozzo M, Hourdez S. 2002. Ultrastructure of spermatozoa in four species of Alvinellidae (Annelida: Polychaeta). Cah Biol Mar. 43:391–394. [Google Scholar]
- Kandror O, DeLeon A, Goldberg AL. 2002. Trehalose synthesis is induced upon exposure of Escherichia coli to cold and is essential for viability at low temperatures. Proc Nat Acad Sci U S A. 99:9727–9732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwagi S, et al. 2010. Characterization of a Y-Family DNA polymerase eta from the eukaryotic thermophile Alvinella pompejana . J Nuc Acids 10. doi:10.4061/2010/701472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreil DP, Ouzounis CA. 2001. Identification of thermophilic species by the amino acid compositions deduced from their genomes. Nucleic Acids Res. 29:1608–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little CTS, Maslennikov VV, Morris NJ, Gubanov AP. 1999. Two Palaeozoic hydrothermal vent communities from the southern Ural mountains, Russia. Palaeontology 42:1043–1078. [Google Scholar]
- Lobanov MY, Galzitskaya OV. 2011. The Ising model for prediction of disordered residues from protein sequence alone. Phys Biol. 8. doi:10.1088/1478-3975/8/3/035004. [DOI] [PubMed] [Google Scholar]
- Lunzer M, Golding GB, Dean AM. 2010. Pervasive cryptic epistasis in molecular evolution. PLoS Genet. 6:e1001162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies M, et al. 2005. Genome sequencing in microfrabricated high-density picolitre reactors. Nature 437:376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie B, Genard B, Rees J-F, Zal F. 2006. Effect of ambient oxygen concentration on activities of enzymatic antioxidant defences and aerobic metabolism in the hydrothermal vent worm, Paralvinella grasslei . Mar Biol. 150:273–284. [Google Scholar]
- Metzker ML. 2009. Sequencing technologies - the next generation. Nat Rev Genet. 11:31–46. [DOI] [PubMed] [Google Scholar]
- Moreno-Hagelsieb G, Latimer K. 2008. Choosing BLAST options for better detection of orthologs as reciprocal best hits. Bioinformatics 24:319–324. [DOI] [PubMed] [Google Scholar]
- Nadeau NJ, Jiggins CD. 2010. A golden age for evolutionary genetics? Genomic studies of adaptation in natural populations. Trends Genet. 26:484–492. [DOI] [PubMed] [Google Scholar]
- Ohta T. 1992. The nearly neutral theory of molecular evolution. Ann Rev Ecol Syst. 23:263–286. [Google Scholar]
- Ohta T. 1995. Synonymous and nonsynonymous substitutions in mammalian genes and the nearly neutral theory. J Mol Evol. 40:56–63. [DOI] [PubMed] [Google Scholar]
- Orr HA. 2005. The genetic theory of adaptation: a brief history. Nat Rev Genet. 6:119–127. [DOI] [PubMed] [Google Scholar]
- Paradis E, Claude J, Strimmer K. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20:289–290. [DOI] [PubMed] [Google Scholar]
- Paz A, Mester D, Baca I, Nevo E, Korol A. 2004. Adaptive role of increased frequency of polypurine tracts in mRNA sequences of thermophilic prokaryotes. Proc Nat Acad Sci U S A. 101:2951–2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell MA, Somero GN. 1986. Adaptation to sulfide by hydrothermal vent animals: sites and mechanisms of detoxification and metabolism. Biol Bull. 171:274–290. [Google Scholar]
- Querol E, Perez-Pons JA, Mozo-Villarias A. 1996. Analysis of protein conformational characteristitics related to thermostability. Protein Eng. 9(3):265–271. [DOI] [PubMed] [Google Scholar]
- Ravaux J, et al. 2013. Thermal limit for metazoan life in question: in vivo heat tolerance of the Pompeii worm. PLoS One 8(5):e64074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed CJ, Lewis H, Trejo E, Winston V, Evilia C. 2013. Protein adaptations in archaeal extremophiles. Archaea, Hindawi Publishing Corp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Romero ML, et al. 2016. Selection for protein kinetic stability connects denaturation temperatures to organismal temperatures and provides clues to archaean life. PLoS One. doi:10.1371/journal.pone.0156657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousset V, Rouse GW, Féral JP, Desbruyères D, Pleijel F. 2003. Molecular and morphological evidence of Alvinellidae relationships (Terebelliformia, Polychaeta, Annelida). Zool Scripta. 32:185–197. [Google Scholar]
- Russell NJ. 2000. Toward a molecular understanding of cold activity of enzymes from psychrophiles. Extremophiles 4:83–90. [DOI] [PubMed] [Google Scholar]
- Saunders NFW, et al. 2003. Mechanisms of thermal adaptation revealed from the genomes of the Antarctic archaea Methanogenium frigidum and Methanococcoides burtonii . Genome Res. 13:1580–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serohijos AWR, Shakhnovich EI. 2014. Merging molecular mechanism and evolution: theory and computation at the interface of biophysics and evolutionary population genetics. Curr Opin Struct Biol. 26:84–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savard JI, et al. 2006. Phylogenomic analysis reveals bees and wasps (Hymenoptera) at the base of the radiation of Holometabolous insects. Genome Res. 16:1334–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin DS, et al. 2009. Superoxide dismutase from the eukaryotic thermophile Alvinella pompejana: structures, stability, mechanism, and insights into amyotrophic lateral sclerosis. J Mol Biol. 385:1534–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sicot FX, et al. 2000. Molecular adaptation to an extreme environment: origin of the stability of the Pompeii worm collagen. J Mol Biol. 302:811–820. [DOI] [PubMed] [Google Scholar]
- Siddiqui KS, Cavicchioli R. 2006. Cold-adapted enzymes. Ann Rev Biochem. 75:403–433. [DOI] [PubMed] [Google Scholar]
- Somero GN. 1992. Adaptations to high hydrostatic pressure. Ann Rev Physiol. 54:557–577. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. [DOI] [PubMed] [Google Scholar]
- Suhre K, Claverie J-M. 2003. Genomic correlates of hyperthermostability, an update. J Biol Chem. 278:17198–17202. [DOI] [PubMed] [Google Scholar]
- Tantos A, Friedrich P, Tompa P. 2009. Cold stability of intrinsically disordered proteins. FEBS Lett. 583:465–469. [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Koonin EV, Lipman DJ. 1997. A genomic perspective on protein families. Science 278:631–637. [DOI] [PubMed] [Google Scholar]
- Teil H. 1975. Correspondence factor analysis: an outline of its method. Math Geol. 7:3–12. [Google Scholar]
- Tóth-Petróczy Á, Tawfik DS. 2011. Slow protein evolutionary rates are dictated by surface-core association. Proc Nat Acad Sci U S A. 108:11151–11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunnicliffe V, Desbruyères D, Jollivet D, Laubier L. 1993. Systematic and ecological characteristics of Paralvinella sulfincola Desbruyères and Laubier, a new Polychaete (Family Alvinellidae) from Northeast Pacific hydrothermal vents. Can J Zool. 71:286–297. [Google Scholar]
- Van Noort V, et al. 2013. Consistent mutational paths predict eukaryotic thermostability. BMC Evol Biol. 13:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt G, Woell S, Argos P. 1997. Protein thermal stability, hydrogen bonds, and ion pairs. J Mol Biol. 269(4):631–643. [DOI] [PubMed] [Google Scholar]
- Vrijenhoek RC. 2013. On the instability and evolutionary age of deep-sea chemosynthetic communities. Deep-Sea Res. II 92:189–200. [Google Scholar]
- Wang G-Z, Lercher MJ. 2010. Amino-acid composition in endothermic vertebrates is biased in the same direction as in thermophilic prokaryotes. BMC Evol Biol. 10:263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster KA. 2003. Evolution of the coordinate regulation of glycolytic enzyme genes by hypoxia. J Exp Biol. 206:2911–2922. [DOI] [PubMed] [Google Scholar]
- Wickstrom CE, Castenholz RW. 1973. Thermophilic ostracods: aquatic metazoan with the highest known temperature tolerance. Science 181:1063–1064. [DOI] [PubMed] [Google Scholar]
- Yang Z. 2007. PaML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24:1586–1591. [DOI] [PubMed] [Google Scholar]
- Zal F, Jollivet D, Chevaldonné P, Desbruyères D. 1995. Reproductive strategy and population structure of the deep-sea hydrothermal vent worm Paralvinella grasslei (Polychaeta: Alvinellidae) at 13°N on the East Pacific Rise. Mar Biol. 122:637–648. [Google Scholar]
- Zeldovich KB, Berezovsky IN, Shakhnovich EI. 2007. Protein and DNA sequence determinants of thermophilic adaptation. PLoS Comput Biol. 3:62–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuckerkandl E, Pauling L. 1965. Evolutionary divergence and convergence in proteins In: Bryson V, Vogel HJ, editors. Evolving genes and proteins. Academic Press, New York, p. 97–166. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.