Abstract
Recent work in human glioblastoma (GBM) has documented recurrent mutations in the histone chaperone protein ATRX. We developed an animal model of ATRX-deficient GBM and show that loss of ATRX reduces median survival and increases genetic instability. Further, analysis of genome-wide data for human gliomas showed that ATRX mutation is associated with increased mutation rate at the single nucleotide variant (SNV) level. In mouse tumors, ATRX deficiency impairs non-homologous end joining (NHEJ) and increases sensitivity to DNA-damaging agents that induce double-stranded DNA breaks. We propose that ATRX loss results in a genetically unstable tumor, which is more aggressive when left untreated, but is more responsive to double-stranded DNA-damaging agents, resulting in improved overall survival.
Introduction
Glioblastoma (GBM) is a lethal primary brain tumor with a median survival of less than two years. Recent work in human gliomas has documented recurrent mutations in the histone chaperone protein ATRX. ATRX mutation in glioma is primarily seen in adolescents and young adults (age 10–30) (1). In pediatric patients, ATRX was reported to be mutated in 31% of patients with primary GBM (WHO grade IV glioma), often with concurrent mutation of TP53 and point mutation of the gene encoding the histone H3.3 variant, H3F3A (1, 2). In adults (age >30), ATRX is mutated less frequently in primary GBM, but is frequently found in lower grade (WHO grade II/III) and secondary glioblastoma (2–4). Recent profiling of adult grade II and III gliomas revealed that a majority (~75%) of the sub-type of low-grade gliomas that carry TP53 and IDH1 mutations also harbor ATRX mutations, thus underscoring their fundamental role in gliomagenesis (4).
ATRX mutation is seen in at least 15 types of human cancers, including neuroblastoma, osteosarcoma, and pancreatic neuro-endocrine (PanNET) tumors (5, 6). However, the role of ATRX in tumorigenesis remains largely unknown. The ATRX protein likely plays an important epigenetic role, depositing histones at heterochromatin and telomeric DNA (7, 8). Previous characterization of human GBM and PanNET tumors has shown an association between ATRX loss and the maintenance of telomere length by alternative lengthening of telomere (ALT), or non-telomerase-based, pathways (1, 8). Transgenic loss of ATRX in a mouse model is embryonic-lethal, and post-natal conditional loss of ATRX alone impairs cortex development without causing tumor formation (7). Mutations in ATRX result in loss of ATRX protein by immunostaining and are thought to mediate loss of function (1).
The development of genetically engineered mouse (GEM) models allows for the systematic evaluation of the contribution of specific genetic lesions to glial tumor development. The Sleeping Beauty (SB) transposase system is particularly well-suited to providing a platform for the rapid validation of proposed driver mutations (9). The SB system uses a synthetic plasmid DNA encoding a transposase gene that inserts a desired transposon DNA element stably into genomic DNA. Using various combinations of human oncogenes and inhibitors of tumor suppressor function, tumors resembling human GBM can be reliably generated (10, 11).
We used the SB transposase system to create an animal model of ATRX-deficient GBM. Loss of ATRX accelerated tumor growth rate and reduced median survival, uncovering the impact of ATRX loss on glioma tumor proliferation. We show that ATRX loss causes genetic instability in mouse GBM, including both microsatellite instability and impaired telomere maintenance. In accordance with this, analysis of publically-available human glioma genome-wide data integrated from multiple sequencing platforms showed that ATRX mutations are associated with increased mutation rate at the single nucleotide variant (SNV) level, but not at the chromosomal/copy number level. We also show that loss of ATRX results in impairment of non-homologous end joining (NHEJ) activity and is strongly correlated with loss of activated (phospho-) DNA-protein kinase core (pDNA-PKcs) subunit staining. By uncovering the connection of ATRX mutation and impaired NHEJ, we provide a mechanism for genetic instability and an actionable therapeutic target for ATRX-deficient GBM. Taken together, these results provide insights into the role of ATRX mutations in human glioma.
Results
Generation of ATRX-deficient mouse GBM using Sleeping Beauty model
To assess the impact of ATRX loss on GBM, we developed an endogenous mouse model using the Sleeping Beauty (SB) transposase system (10). We cloned an ATRX knockdown sequence (shATRX) into a plasmid with flanking sequences recognized by the SB transposase for insertion into host genomic DNA (fig. 1A and S1). We induced glioblastoma in mice by injecting plasmids encoding: (1) SB transposase/firefly luciferase, (2) shp53, and (3) NRAS, with or without (4) shATRX, into the lateral ventricle of neonatal mice. Transfection efficiency and tumor growth were monitored by in vivo imaging of luminescence (fig. 1B). ATRX loss was tested in the context of over-expression of the oncogene NRAS and a short hairpin against p53, because ATRX mutation by itself is not associated with cancer development in humans (12), and glial tumors with ATRX mutations almost always include TP53 mutations (1). The receptor tyrosine kinase-RAS-PI3 kinase (RTK-RAS-PI3K) pathway is mutated in a large percentage of adult and pediatric high-grade gliomas (1, 13, 14). Thus, many genetically engineered animal models of GBM have taken advantage of the glioma-promoting effects of NRAS, either directly by activating mutations or through loss of NF1expresion which results in NRAS up-regulation (Table S1).
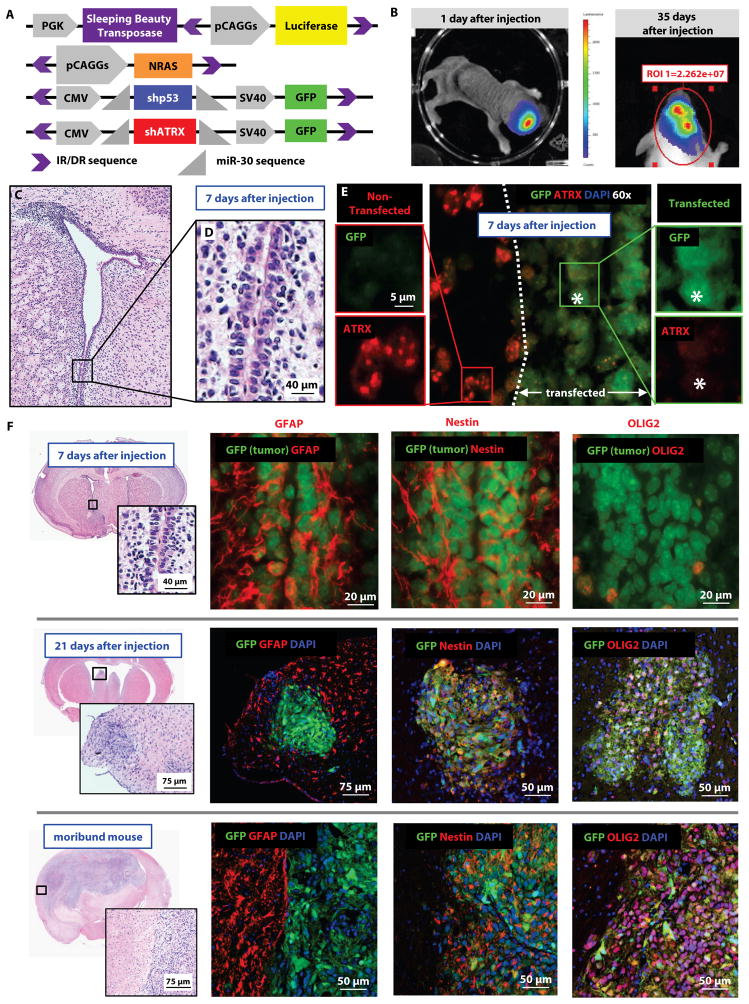
Fig. 1. Sleeping Beauty mouse model represents ATRX-deficient GBM.
(A) Constructs of Sleeping Beauty (SB) plasmids, including the plasmid with short hairpin against ATRX. Purple brackets represent IR/DR (inverted repeat/direct repeat) sequences; the area between these IR/DR sequences is recognized by the SB transposase for insertion into the host genomic DNA. miR-30 sequences represent the 5′ and 3′ flanking sequences of the 300 nucleotide primary microRNA molecule. (B) Plasmid insertion and tumor growth are monitored by in vivo luminescence. (C) H&E staining of 8 day old mouse brain (7 days after injection), including inset of subventricular zone (SVZ) lining the lateral ventricle (D). (E) Immuno-fluorescence of the region depicted in panel D, with transfected cells (GFP-positive, to the right of the dotted line) showing ATRX reduction at 7 days after injection. (F) Immunohistochemistry staining of transfected cells/tumors for markers associated with neural stem cells, including GFAP, OLIG2, and Nestin in mice injected with shp53/NRAS/shATRX.
After injection of plasmids (shp53, NRAS, and shATRX), mice were euthanized at early time points to characterize the model. At 7 days after injection, transfected cells (GFP positive) were found within 50–100 μm of the lateral ventricles and showed early loss of ATRX expression by immunohistochemistry (IHC) (fig. 1C–E). To determine if all three plasmids (shp53, NRAS, and shATRX) were being expressed in the same cells, we injected mice with plasmids expressing single fluorescent markers: (1) shp53-GFP (green); (2) NRAS-katushka (red), and (3) shATRX-noGFP (detected by IHC with immuno-fluorescent blue secondary antibody (405 nm)). At 15 days after injection, cells showed evidence of co-transfection with all plasmids (fig. S2).
In both experimental conditions (shp53/NRAS with or without shATRX), GFP-positive transfected cells co-expressed GFAP and Nestin, markers of neural stem cells (fig. 1F) (15), and not myosin VIIa, a marker of ependymal cells (16), (fig. S3). By day 15, we found multiple clusters of proliferative cells, which were GFAP negative by this time point, but remained OLIG2 and Nestin positive. This expression pattern persisted through late-stage tumors from moribund animals (fig. 1F and fig. S4–S7). Tumors in moribund animals showed strong pERK staining as well (fig. S6), which was downstream of RAS and expressed in human GBM (17).
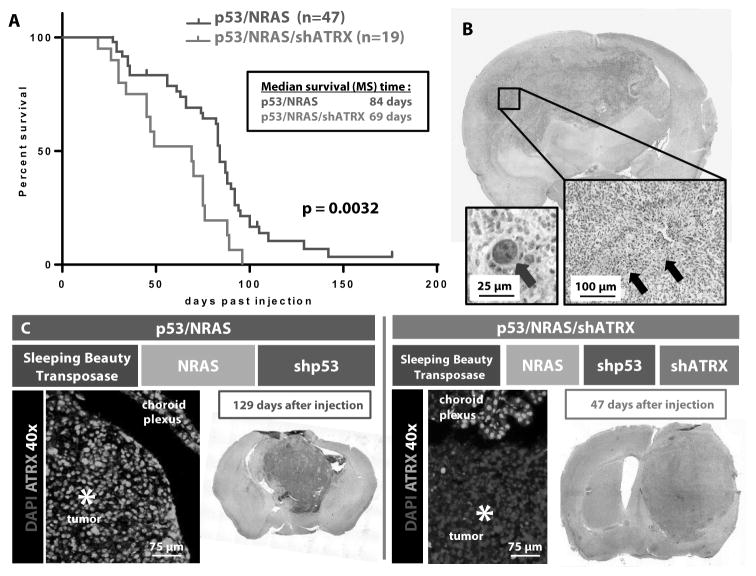
In the process of optimizing and characterizing this model, we established a large cohort of mice injected with shp53/NRAS plasmids (n=47). We then compared their survival with our experimental group of mice injected with shp53/NRAS/shATRX plasmids (n=19). The median survival of mice injected with shp53/NRAS/shATRX was significantly decreased (69 days) compared to that of mice injected with shp53/NRAS (84 days, p=0.0032) (fig. 2A). All tumors (with or without shATRX) showed histological hallmarks of human GBM, including pseudopalisading necrosis (fig. 2B, black arrows). Loss of ATRX was localized only within tumors generated by injection of the shATRX-expressing plasmid and not in the adjacent normal cortex or choroid plexus (fig. 2C). ATRX loss was evident throughout the entire tumor, and mice injected with shp53/NRAS/shATRX had larger tumors at earlier time points (fig. 2C and table S2). These data show that ATRX loss accelerates GBM tumor growth and reduces survival in mice bearing GBM.
Fig. 2. ATRX loss decreases median survival in mice bearing SB-generated GBMs.
(A) Kaplan-Meier survival curves of C57BL/6 mice bearing SB-induced tumors (p=0.0032, Mantel log-rank test). (B) Representative brain tumors with histologic hallmarks of glioblastoma (GBM): pseudopalisading necrosis and nuclear atypia (black arrows and blue arrow, respectively). (C) Tumors with addition of shATRX plasmid are larger at earlier time points than tumors with shp53/NRAS alone and show ATRX loss throughout the tumor.
Microsatellite instability in ATRX-deficient GBM
ATRX encodes a protein with a DNA-binding finger and a SWI2/SNF2-like ATPase motif, making it a member of a family of ATP-dependent chromatin-associated proteins (18). Other members of this class participate in DNA damage repair pathways, in particular nucleotide excision repair and double-stranded break repair (19). Previous work has shown that ATRX is recruited to sites of DNA damage (20). Thus, we hypothesized that ATRX may play a role in maintaining genetic stability in glioblastoma.
One type of genetic instability that is seen in human GBM is microsatellite instability (MSI). Microsatellites are repeated mono-nucleotide and di-nucleotide sequences that are prone to error during DNA replication. Loss of DNA mismatch repair results in variation of the length of microsatellite sequences in some human tumors, including GBM (21, 22). Both ATRX mutation (1) and microsatellite instability (MSI) (21) are seen more frequently in younger patients with GBM. We used established primer sets for mouse microsatellite sequences (23, 24) to assess our mouse GBMs and determine if ATRX loss resulted in the presence of MSI.
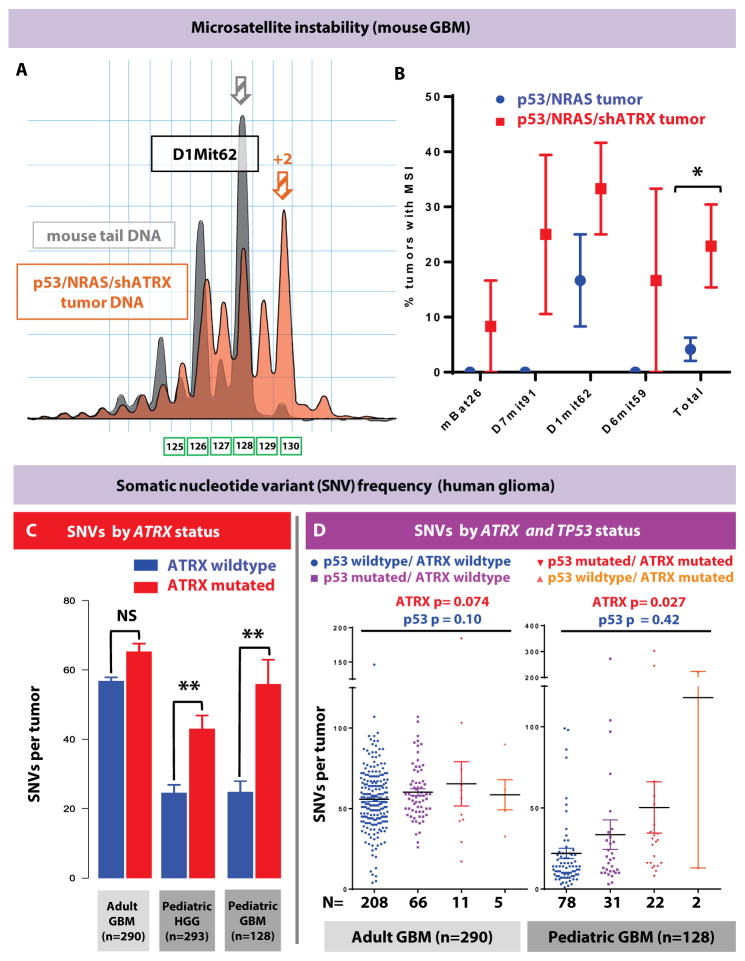
Using established MSI software parameters, tumors were scored for differences in predominant microsatellite (PCR fragment) length between tumor and control tail DNA. Tumors with ATRX loss showed greater rates of MSI using four distinct microsatellite markers (fig. 3A–B and table S3). Overall, ATRX loss increased the rate of instability five-fold (p = 0.014, n = 48 total tumor vs. control DNA comparisons). The rate of MSI in ATRX-deficient mouse tumors (21%) was comparable to the rate of MSI in human pediatric GBM (19%) (21).
Fig. 3. ATRX loss increases MSI in mouse GBM, and SNV frequency in human glioma.
(A) Representative microsatellite lengths from a tumor with MSI-high positivity (log2 ratio of >4 for predominant tumor sample allele versus control mouse tail DNA) using marker D1Mit62. Plot shows a shift in populations of allele lengths in p53/NRAS/shATRX tumor (orange) compared to control mouse tail DNA (gray). Predominant allele size in this example is two nucleotides longer (orange arrow) than control mouse tail DNA allele (gray arrow). (B) Comparison of the percentage of tumors with MSI positivity using four independent MSI primer sets (mBat26, D7mit91, D1mit62, and D6mit59) (n=48 tumor vs. control DNA comparisons). Data shown as mean ± SEM; *P < 0.05 using two-sided Chi-square analysis. (C) Analysis of matched human tumor/germline integrated sequencing datasets showing SNV frequency in tumors by ATRX mutational status (GBM, glioblastoma, WHO Grade IV; pediatric GBM excludes diffuse intrinsic pontine glioma; HGG, high-grade glioma, WHO Grade III and IV). **P < 0.005 using unpaired Mann-Whitney test. (D) Analysis of significance of contribution to SNV rate by ATRX and TP53 mutational status, using a two-way ANOVA model in adult GBM (n=290) and pediatric GBM (n=128). Each data point represents an individual human tumor; line represents mean ± SEM.
Increased somatic variant rate in human glial tumors with ATRX mutations
The finding of MSI in ATRX-deficient mouse GBM pointed to ATRX playing a role in maintaining genetic stability at the sequence level. To validate this finding in human glioma, we evaluated whether ATRX mutation was associated with increased somatic nucleotide variant (SNV) rate in genome-wide data from multiple datasets. We retrieved multiple publicly available genome-wide datasets available in the European Genome Archive (EGA). We then integrated these multiple sequencing platforms, along with additional pediatric high-grade glioma samples (deposited at EGA accession number (EGAS00001001436), to produce full somatic sequence and copy number information on 293 pediatric high-grade glioma (HGG) samples (up to age 30), of which 38/293 (13%) samples contained ATRX mutations. We also retrieved and analyzed 290 GBM samples (age >30) available from The Cancer Genome Atlas (TCGA (10, 25). After analyzing our integrated pediatric and adult datasets, we did not observe any clustering of mutations in the SNF2-DNA binding region of the gene (fig. S8), as has been described by others (1). A higher average somatic nucleotide variant (SNV) rate was seen in ATRX-mutated pediatric high-grade glioma in all anatomical locations (p = 0.0038), and particularly in pediatric GBM (p = 0.0005), but not in adult GBM (p =0.71) (fig. 3C).
ATRX mutation is frequently found with concurrent TP53 mutation in human glioma. Because loss of p53 function may be permissive of genetic instability in tumor cells (26), we evaluated whether our finding of increased somatic variant rate in ATRX-mutated tumors was confounded by TP53 mutation in human GBM. TP53 mutation was not a predictor of variant rate in either dataset, whereas ATRX was a predictor in pediatric non-brainstem GBM (ANOVA p value 0.027), but not in adult GBM (fig. 3D and table S4). IDH1 mutation was also not a predictor of variant rate in pediatric or adult datasets, with ANOVA p values of 0.14 (adult GBM) and 0.19 (pediatric GBM) (table S4).
Association between ATRX loss and copy number alterations
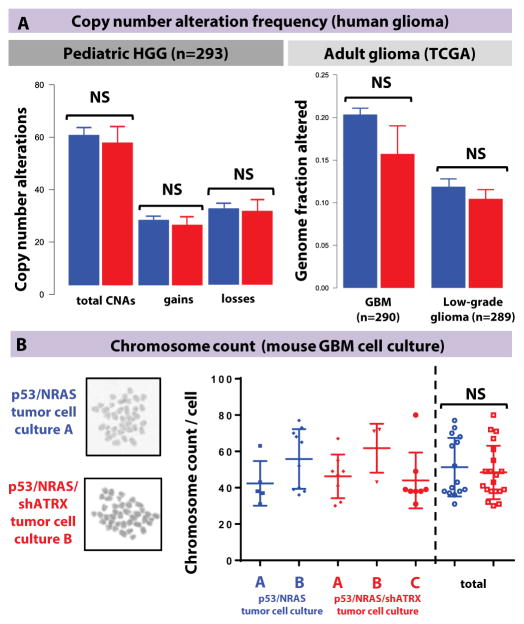
Previous analysis of pediatric GBM has shown an association between tumors with concurrent mutations in H3F3A, TP53, and ATRX and copy number alterations (1). In our dataset, ATRX mutations were not associated with total copy number alterations, nor gains or losses considered separately, in the pediatric dataset, or with percentage genome fraction altered in the adult glioma dataset (fig. 4A).
Fig. 4. ATRX loss does not cause structural/chromosomal alterations or change chromosome count.
(A) Analysis of chromosomal alterations, including copy number alterations, in the integrated pediatric GBM datasets, and percentage of genomic alterations in the adult TCGA dataset shows no difference by ATRX mutational status. Line represents mean ± SEM; NS = > 0.05 using unpaired Mann-Whitney test. (B) Chromosome count by metaphase preparation of independent GBM neurosphere cultures. Line represents mean ± SEM; NS = > 0.05 using unpaired Mann-Whitney test. Both groups had similar coefficients of variation [31.4% in p53/NRAS tumor cells (n=15) and 30.7% in p53/NRAS/shATRX tumor cells (n=20)].
To further examine the impact of ATRX on stability at the chromosomal level, we investigated whether ATRX loss was associated with karyotypic changes in our mouse GBMs. We generated primary cell cultures from SB-generated mouse GBMs and demonstrated that tumor neurosphere cultures generated with p53/NRAS/shATRX tumors showed stable reduction of ATRX expression compared to p53/NRAS cells (fig. S9). Using a previously established method (27), karyotypes of multiple independent GBM cell cultures showed similar mean chromosomal counts and coefficients of variation between neurospheres with and without shATRX (fig. 4B).
Assessment for Alternative Lengthening of Telomeres (ALT) in mouse GBM
Previous characterization of human GBM has shown an association between mutations of ATRX and the maintenance of telomere length by alternative lengthening of telomeres (ALT), or non-telomerase-based, pathways (1, 8). To establish a causal link, we assessed our mouse GBMs for evidence of impact on telomere lengthening. We isolated DNA from GFP-positive mouse GBM tumor tissue (with or without shATRX) and noted no difference in telomere lengths in tumors with ATRX loss by established telomere qPCR primers (28) (fig. S10 and table S5). We then surveyed DNA extracted from tumors and neurospheres for c-circle amplification, which has been shown to be a specific assay for ALT (29). C-circles were found in a subset of tumor and neurosphere samples with ATRX loss, but not in DNA extracted from normal mouse brains (fig. S11).
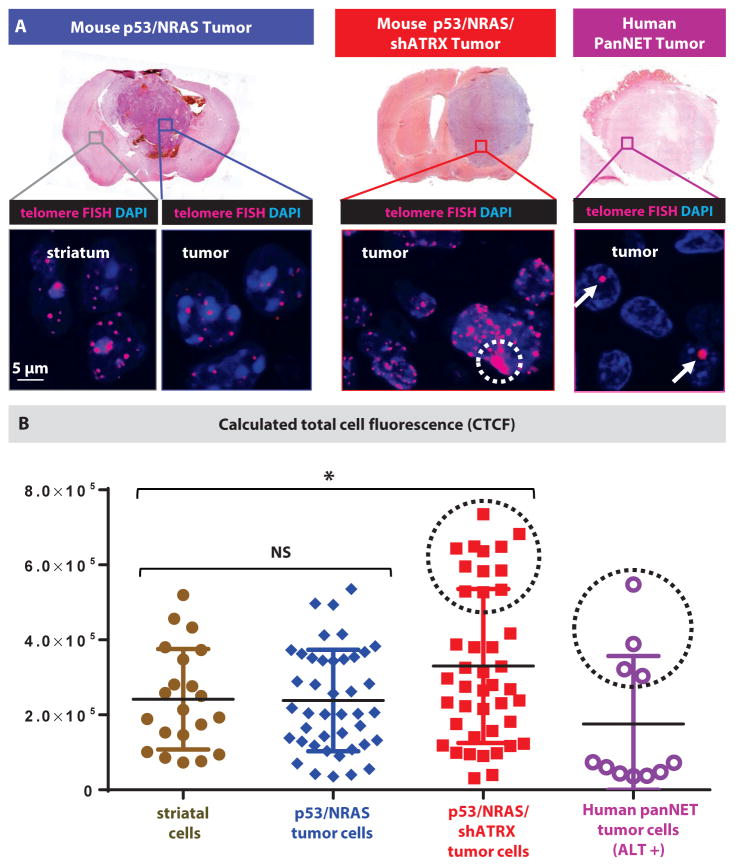
Recently, ultra-bright spots seen on human tumor tissue by telomeric FISH have been demonstrated as a sensitive indicator for the presence of ALT (8). The majority of human pancreatic neuroendocrine tumors (PanNETs) have been found to harbor both ATRX mutations and ALT by telomeric FISH (8). We therefore hybridized FISH probes against human PanNET tumors to be used as positive controls, and noted distinct bright spots (fig. 5A, white arrow) in a subset of cells in PanNET tumors, which are consistent with tumor ALT positivity.
Fig. 5. ATRX-deficient mouse GBMs display ALT.
(A) Detection of ALT using telomeric FISH assay showing characteristic ultra-bright spots in human PanNET tumors (white arrows, positive control). A distinct population of cells with increased telomere signal is seen in ATRX-deficient tumors (white dotted circle). (B) Calculated total cell fluorescence (CTCF) in arbitrary units for telomeric FISH signal (data points represent individual cells from three tumors in each condition); black dotted circles denote cells qualitatively showing ultra-bright spots, consistent with ALT. Line represents mean ± SD; * P < 0.05 using unpaired Mann-Whitney test.
In our mouse tumors, we noted a population of tumor cells with higher fluorescent signal in ATRX-deficient mouse GBM (fig. 5A, white dotted circle) that was not seen in control tumors or striatal cells. To quantify the difference, we calculated total cell fluorescence (CTCF) in tumor cells randomly chosen from multiple tumors in each experimental condition. Mean CTCF from p53/NRAS tumors was similar to that of mouse striatal cells, but was increased in p53/NRAS/shATRX tumor cells (fig. 5B). Again, we saw a distinct set of cells with higher CTCF in both p53/NRAS/shATRX tumors and human PanNET tumors (fig. 5B, black dotted circles), consistent with ALT physiology.
ATRX loss and impaired non-homologous end joining
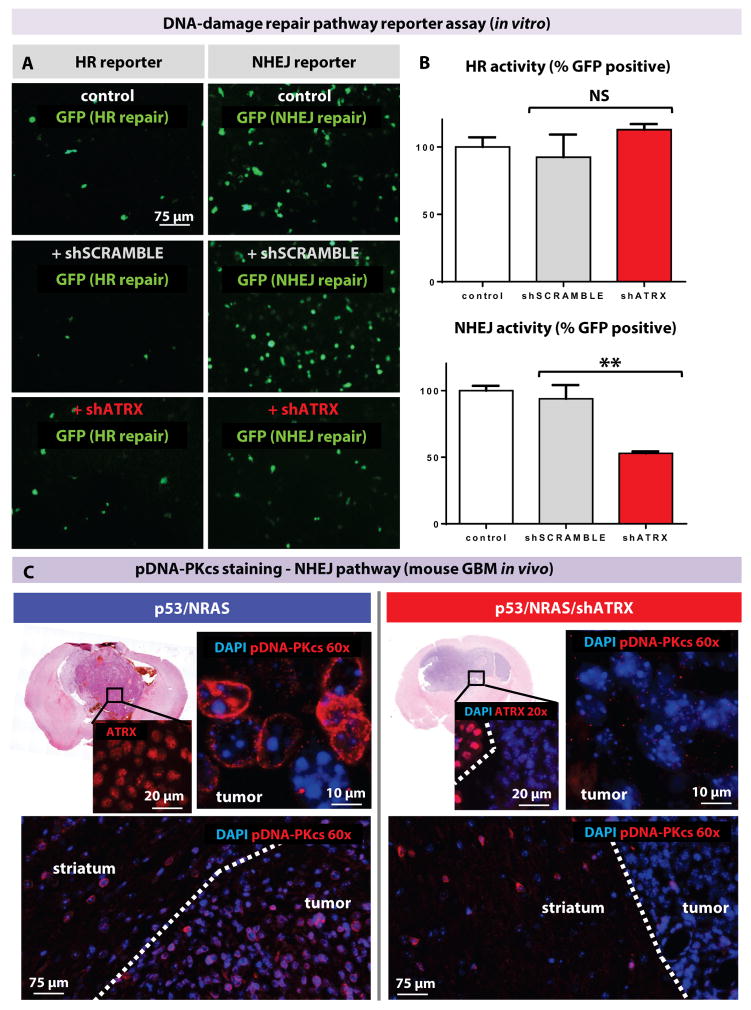
To determine the mechanism by which ATRX loss impacts genomic and telomeric stability, we assessed its impact on DNA-damage repair (DDR) pathways. We used reporter plasmids previously designed to quantify homologous recombination (HR) and non-homologous end joining (NHEJ) efficacy (30). Hepa 1–6 cells were transfected with linearized reporters harboring a DNA-damaged GFP that can be restored by NHEJ or HR, depending on the plasmid system used. Co-transfection with shATRX reduced NHEJ function by 50% (p=0.0024) but HR activity remained unaffected (fig. 6A) when quantified by flow cytometry (percentage of GFP-positive cells after transfection with shATRX or shSCRAMBLE normalized to mean control; fig. 6B and fig. S12).
Fig. 6. ATRX loss reduces NHEJ repair.
(A) Reporter assay with GFP expression that is restored by NHEJ or HR in the appropriate plasmids. Addition of shATRX impairs NHEJ (assay performed in triplicate). (B) Flow cytometric quantification of HR and NHEJ activity as assessed by percentage of GFP-positive cells normalized to control (differences in HR activity are non-significant). Line represents mean ± SD; ** P < 0.005 using unpaired t-test. (C) Loss of ATRX in mouse GBM decreases pDNA-PKcs immunostaining (showing representative results of 3 mouse tumors per condition). Dotted line represents distinction between tumor and non-tumor brain.
To confirm that the NHEJ pathway was impaired in vivo in mouse GBM with ATRX loss, we investigated whether key NHEJ pathway proteins were impacted. Previous studies have shown that the proteins, namely Ku70 and Ku80, first bind to double-stranded DNA breaks and then recruit the catalytic subunit of DNA-dependent protein kinase catalytic subunit (DNA-PKcs). DNA-PKcs can then recruit and phosphorylate other NHEJ pathway proteins, as well as phosphorylate itself to promote its activity (pDNA-PKcs). These activated pathway proteins then bridge broken ends to facilitate re-joining (30).
We stained mouse GBMs (with and without shATRX) and found that ATRX loss was strongly correlated with loss of pDNA-PKcs by immunofluorescence staining (fig. 6C and table S6). Tumors with p53/NRAS (n=3) alone showed robust pDNA-PKcs staining throughout the tumor, but it was almost absent in all tumors with p53/NRAS/shATRX (n=3). We observed staining for pDNA-PKcs in the non-tumor striatum of all animals in both groups (fig. 6C). Immunohistochemistry for the NHEJ DNA repair enzymes Ku70 and XRCC4 revealed no overt changes in expression (fig. S13). Similar results were obtained for HR repair pathway enzymes RAD51 and BRCA1 (fig. S14) and base excision repair protein PARP1 (table S6). On the basis of our finding of increased MSI in ATRX-deficient mouse GBM, we also surveyed for mismatch repair proteins known to impact MSI (MLH1, MSH6, and PMS2) and found similar expression patterns in tumors with and without ATRX loss (fig. S15).
Sensitivity of ATRX-deficient GBM tumor cells to double-stranded DNA-damaging treatment
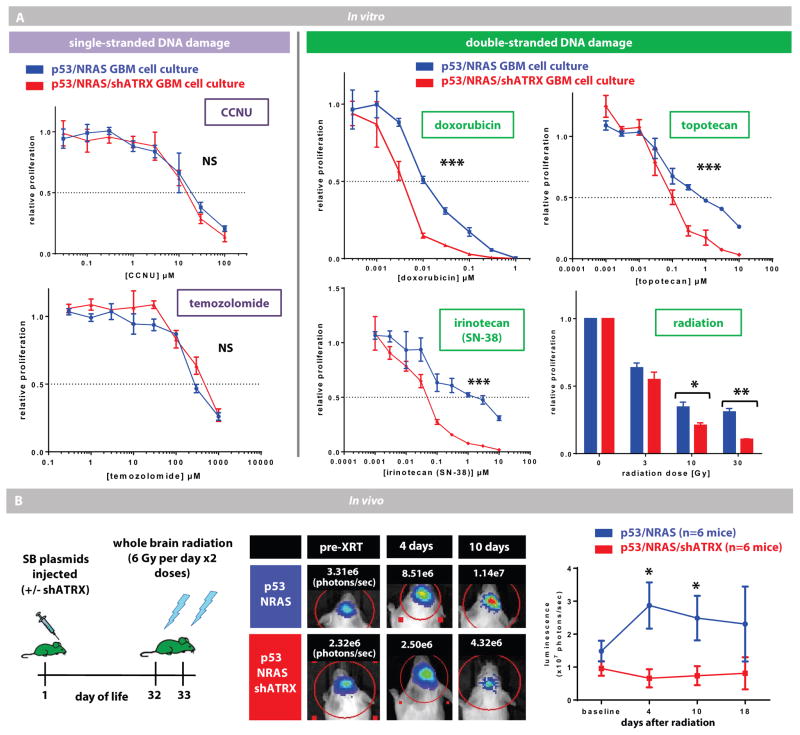
Inhibition of NHEJ in cancer cells induces an accumulation of double-stranded DNA-breaks, which increases susceptibility to radiation (31). Because our data show that ATRX loss impairs NHEJ, we hypothesized that ATRX-deficient GBM cells would have increased sensitivity to DNA-damaging agents that primarily induce double-stranded breaks. Indeed, ATRX-deficient GBM cells were more sensitive in vitro to treatment with radiation and previously published doses of doxorubicin, irinotecan (SN-38), and topotecan (fig. 7A) (32–35). In contrast, treatment of GBM cells with agents that primarily induce single-stranded defects (CCNU and temozolomide (32)) was not affected by ATRX status. To confirm our findings in vivo, we assessed bioluminescence in mice with GBMs treated with whole brain radiation. ATRX-deficient mice showed reduced growth at days 4 and 10 after radiation compared to controls (mice with shp53/NRAS) (fig. 7B: treated and fig. S16: untreated).
Fig. 7. ATRX-deficient GBM cells are sensitive to double-stranded DNA-damaging treatments.
(A) In vitro data showing proliferation of mouse GBM cell cultures (with or without shATRX) after exposure to escalating doses of cytotoxic agents. ATRX-deficient tumor cells have reduced proliferation only after exposure to agents that induce double-stranded breaks. Line represents mean ± SEM;* P < 0.05 using F-test of logIC50. (B) Schematic of whole brain radiation for mice with GBM (with or without shATRX). Tumor growth assessed by in vivo luminescence is reduced in ATRX-deficient tumors (representative mice and their luminescence values are shown). Plot on right shows average tumor luminescence for all mice at early time points after radiation (n=6 mice in each group). Line represents mean ± SEM;* P < 0.05 using unpaired t-test.
We found that ATRX-deficient tumors showed reduction in pDNA-PKcs at multiple time points after radiation (fig. S17), whereas control tumors and non-tumor brain showed both pDNA-PKcs and ATRX expression. In vitro, we found that ATRX-deficient tumor cells showed an increase in γH2A.X expression, a sensitive marker for double-stranded DNA breaks, 24 hours after treatment with doxorubicin (35), compared to control tumor cells (fig. S18A). In vivo, we saw increased γH2A.X expression in ATRX-deficient tumors 24 hours after radiation (single dose of 6 Gy) (fig. S18B).
ATRX mutation and improved survival in treated human GBM
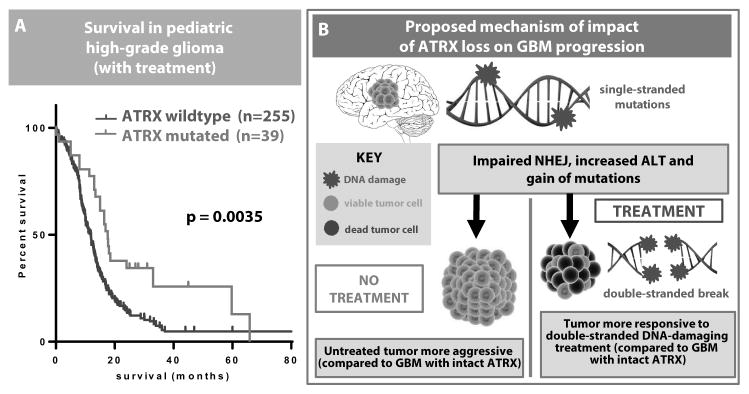
Recent data have shown that adults with treated ATRX-mutated GBM have a survival advantage (3, 4). We used our integrated human glioma genome-wide dataset to confirm that ATRX mutation provides a survival advantage in pediatric high-grade glioma patients as well (p = 0.0035, fig. 8A).
Fig. 8. Pediatric patients with high-grade glioma and ATRX mutation survive longer.
(A) Kaplan-Meier curve based on genome-wide data from multiple pediatric datasets (n=293) showing the survival benefit of ATRX mutation in treated patients. (B) Schematic of impact of ATRX loss on GBM.
In contrast, our data indicate that ATRX loss in untreated mice confers a survival disadvantage (fig. 2). Increased sensitivity of ATRX-deficient GBM to radiation and/or chemotherapy that induces double-stranded breaks could explain this apparent discrepancy. Thus, we propose that ATRX loss causes impaired NHEJ and genetic instability in glioma, which is more aggressive when untreated but is more responsive to double-stranded DNA-damaging therapy, ultimately resulting in improved overall survival (fig. 8B).
Discussion
Recurrent mutations in ATRX point to its critical role in tumor progression in multiple human cancers (5, 6). Despite this, little is known about the molecular mechanism by which it impacts tumor development and growth. We used an animal model of ATRX-deficient GBM to uncover the impact of ATRX loss on glioma tumor proliferation and loss of genetic stability. We show that ATRX reduction impairs non-homologous end joining (NHEJ) and pDNA-PKcs recruitment, thus providing a mechanism for genetic instability and a molecular target. These results provide insights into the impact of ATRX mutations in human glioma, and possibly other human tumors.
Our data link ATRX to regulation of NHEJ. Other chromatin remodelers are known to play important roles in providing proteins access to damaged DNA and telomeres (36). We hypothesize that reduced ATRX causes conformational changes in heterochromatin restricting the access of NHEJ proteins such as DNA-PKcs to damaged DNA. Our finding of impaired NHEJ is consistent with previous research, which has demonstrated that ALT is dependent on homologous recombination of sister telomere chromatids (37). Our results suggest that a relative increase in HR in relation to NHEJ in ATRX-deficient glioma may be supportive of ALT (proposed schematic, Fig. S19).
Our experimental data reproduce ATRX loss resulting in ALT in an animal model. Human (non-GBM) immortalized cell lines with ALT have been associated with genetic instability and altered DNA-damage response (38). Our data substantiate the connection between ATRX, ALT, and the DNA-damage response. Our animal model showed ALT in ATRX-deficient tumors by telomere FISH and in a subset of our ATRX-deficient tumors and tumor neurospheres by c-circle assay, but did not display elongated telomeres by telomere qPCR. Overall, our results suggest that ATRX loss in GBM is capable of producing ALT, but that additional factors and/or species differences may also play a role. Mouse telomeres are much longer than human telomeres (28); thus we hypothesize that this species difference could influence ALT assays and ALT physiology.
According to the mutator hypothesis of oncogenesis, early mutations in “caretaker genes” can drive further tumor development (39). Our data show that ATRX plays this role in glioma because it is required for NHEJ DNA repair. It is possible that the genetic instability in ATRX-deficient GBM drives proliferation by affecting cell cycle control or differentiation, as has been shown in other genetically-unstable tumor models (40, 41). Additionally, impaired apoptotic signaling through defective DNA-PKcs phosphorylation (42) and/or concurrent TP53 mutations could provide an additional proliferative advantage to ATRX-mutated tumors.
We show that ATRX is implicated in maintaining stability at the sequence level in our animal model and in our pediatric human datasets. Of note, in our adult human datasets, we did not see a difference in SNV rate between ATRX-mutated and non-mutated tumors, possibly making this finding most applicable to younger patients. The choice of DDR pathway influences the quality of the repair and the introduction of new somatic mutations and chromosomal re-arrangements (43). Recent work has highlighted that NHEJ is actually a combination of two related but distinct processes: (1) canonical NHEJ (C-NHEJ), which is the traditional pathway involving repair of double-stranded breaks using DNA-PKcs, and (2) alternative NHEJ (A-NHEJ), which is driven by PARP1, uses microhomology-mediated end-joining, and is associated with deletions at repair junctions (44). Because our tumors (with or without ATRX loss) showed robust PARP1 staining (table S6), it is possible that the reduction in canonical NHEJ in ATRX-deficient glioma results in mutagenesis from use of the more error-prone alternative-NHEJ pathway (A-NHEJ). Increase in A-NHEJ in ATRX-deficient glioma could potentially explain the increase in point mutations that result in both increased somatic mutation rate and MSI (which is also the accumulation of single-stranded mutations), thus explaining the phenotypic features seen in our animal and human data.
An important feature of endogenous animal models of glioma is the choice of genetic drivers. One possible limitation of our model is the use of NRAS mutation, which is only rarely mutated in human GBM. Nevertheless, RAS provides a useful driver of GBM formation, which retains relevant histologic features of the human disease while activating the RTK/RAS/PI(3)K pathway, for which a majority of GBMs contain signal alterations (13, 14). Although ATRX mutation co-occurs with IDH-R132H mutation in adult glioma, it almost never does in pediatric GBM and instead frequently co-occurs with histone H3.3 mutations. In both cases, it almost always co-occurs with TP53 mutation (1–4). This overlap highlights the fact that ATRX mutation can impact tumor development with multiple other glioma drivers, as long as TP53 signaling is impaired, a feature our model also encapsulates.
Our results have translational relevance in that we modeled ATRX loss in mice to identify its role in furthering GBM progression and therapy response. We believe that this seemingly paradoxical role will assist in the design of future therapy for ATRX-mutated glioma. For example, detection of ATRX mutation and/or loss of ATRX by immunostaining may be evidence of a treatment-responsive subtype of glioma that would encourage the use of radiation and/or chemotherapy agents that induce double-stranded breaks. Our data showing both a reduction in NHEJ activity and an increase in response to double-stranded DNA-damaging agents in tumor cells with ATRX loss are consistent with previous research showing that NHEJ inhibition reduces double-stranded break repair and increases sensitivity to radiation (31). Further pre-clinical studies could highlight regimens that more selectively target the defects in NHEJ and double-stranded break repair in ATRX-deficient glioma. Our data raise the possibility that topo-isomerase inhibitors (topotecan or irinotecan) might be clinically useful to exploit this defect.
These results lay a foundation for uncovering the molecular impact of ATRX mutations in the pathogenesis and response to therapeutics in human GBM. Additionally, this mouse model provides a platform for the development of targeted therapy for GBM patients harboring ATRX mutations.
Supplementary Material
Fig. S1. Sleeping Beauty responsive shATRX plasmids are cloned to explore the impact of ATRX on GBM development.
Fig. S2. Cells co-transfected with shp53, shATRX, and NRAS plasmids are characterized at 15 days after injection.
Fig. S3. SB-mediated transfected cells are distinct from ependymal cells in mice.
Fig. S4. Characterization of p53/NRAS/shATRX mice 7 days after injection shows tumor cells expressing GFAP and Nestin..
Fig. S5. p53/NRAS/shATRX mice at 21 days after injection show tumor cells expressing OLIG2 and Nestin.
Fig. S6. Moribund p53/NRAS/shATRX mice show tumor cells expressing OLIG2, Nestin, and pERK.
Fig. S7. p53/NRAS/shATRX mice show loss of GFAP expression by 15 days after injection.
Fig. S8. ATRX mutations do not cluster in SNF2/helicase domain in human glioma.
Fig. S9. Primary GBM cell cultures are generated from SB-induced mouse tumors.
Fig. S10. No difference is seen in telomere length in tumors with or without ATRX by qPCR.
Fig. S11. Assessment of ALT by c-circle assay with DNA from mouse tumors and neurospheres.
Fig. S12 Cells transfected with shATRX plasmid show reduction in NHEJ by flow cytometry.
Fig. S13. Non-homologous end joining (NHEJ) pathway is characterized by IHC.
Fig. S14. Homologous recombination (HR) pathway analysis is characterized by IHC.
Fig. S15. Mismatch repair (MMR) pathway analysis is characterized by IHC.
Fig. S16. SB mouse tumor growth (without radiation treatment) is characterized by luminescence.
Fig. S17. Tumors with ATRX loss show reduction in phospho-DNA-PKcs before and after radiation treatment.
Fig. S18. Mouse tumors with ATRX loss show increased expression of γH2A.X in vitro and in vivo.
Fig. S19. Detailed schematic shows proposed impact of ATRX loss on GBM progression.
Table S1. Animal models of glioma all have RTK/RAS/PI3K alterations.
Table S2. Mice injected with p53/NRAS/shATRX have faster growing tumors.
Table S3. MSI rate is increased in p53/NRAS/shATRX tumors.
Table S4. IDH1 and TP53 mutations do not alter SNV rate calculated by two-way ANOVA model.
Table S5. Telomere qPCR ATLR worksheet and standard curve show similar results in all experimental groups.
Table S6. Immunofluorescence analysis shows reduced pDNA-PKcs expression in ATRX-deficient mouse GBM.
Table S7. Antibodies used for tissue analysis are summarized in table form.
Acknowledgments
We thank Dr. John Ohlfest (University of Minnesota, deceased), for his generous support of our implementation of the Sleeping Beauty model. Dr. Koschmann wishes to thank Drs. Patricia Robertson and Hugh Garton for their academic support. We gratefully acknowledge Mr. Philip Jenkins and the Dept. of Neurosurgery at the University of Michigan Medical School for their support of our work. We are also grateful to Dr. Karin Muraszko for her academic leadership, and D. Tomford, S. Napolitan, M. Dahlgren, and C. Shaw for superb administrative support. This study makes use of data generated by the St Jude Children’s Research Hospital – Washington University Pediatric Cancer Genome Project; the Principal Investigator Dr. Cynthia Hawkins and the Hospital for Sick Children; the McGill University Health Center and the DKFZ-University of Heidelberg Pediatric Brain Tumour Consortium; and the Institute of Cancer Research – Institut Gustav Roussy – Hospital San Joan de Deu collaborative group.
Funding: This work was supported by National Institutes of Health/National Institute of Neurological Disorders & Stroke (NIH/NINDS) grants 1RO1-NS 054193, 1RO1-NS 061107, and 1RO1-NS082311 to PRL; grants 1UO1-NS052465, 1RO1-NS 057711, and 1RO1-NS074387 to MGC, and grant NIH/National Cancer Institute R01CA172380 to A Meeker. CK was supported by the St. Baldrick’s Foundation Fellowship, and the Alex’s Lemonade Stand Foundation /Northwestern Mutual Young Investigator Grant. CJ, A Mackay and JFS acknowledge NHS funding to the NIHR Biomedical Research Centre at The Royal Marsden and the ICR, and the INSTINCT network funded by The Brain Tumour Charity, Great Ormond Street Children’s Charity and Children with Cancer UK.
Footnotes
Author contributions: CK carried out the animal and in vitro studies and drafted the manuscript. AAC, FJN, DT, FM, NK, MD, LM, JK, RL, YL and SR participated in the animal and in vitro studies. LZ performed statistical analysis. A. Meeker and JBC assisted with design and interpretation of ALT studies (FISH and C-circle). HA provided human PanNET samples. DF assisted with design and interpretation of metaphase preparation/chromosome counting. A. Mackay, JFS and CJ performed analysis of human pediatric glioma datasets. VG assisted with design and interpretation of DNA damage repair plasmid assays. MGC and PRL conceived and supervised the study, participated in its design and coordination, and helped to draft and edit the manuscript. All authors read and approved the final manuscript.
Competing interests: The authors report that they have no competing interests.
Materials and data availability: We have deposited previously unpublished sequence data for additional pediatric high-grade glioma samples (C. Jones, EGAS00001001436).
References and Notes
- 1.Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, Sturm D, Fontebasso AM, Quang DA, Tonjes M, Hovestadt V, Albrecht S, Kool M, Nantel A, Konermann C, Lindroth A, Jager N, Rausch T, Ryzhova M, Korbel JO, Hielscher T, Hauser P, Garami M, Klekner A, Bognar L, Ebinger M, Schuhmann MU, Scheurlen W, Pekrun A, Fruhwald MC, Roggendorf W, Kramm C, Durken M, Atkinson J, Lepage P, Montpetit A, Zakrzewska M, Zakrzewski K, Liberski PP, Dong Z, Siegel P, Kulozik AE, Zapatka M, Guha A, Malkin D, Felsberg J, Reifenberger G, von Deimling A, Ichimura K, Collins VP, Witt H, Milde T, Witt O, Zhang C, Castelo-Branco P, Lichter P, Faury D, Tabori U, Plass C, Majewski J, Pfister SM, Jabado N. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 2.Jiao Y, Killela PJ, Reitman ZJ, Rasheed AB, Heaphy CM, de Wilde RF, Rodriguez FJ, Rosemberg S, Oba-Shinjo SM, Nagahashi Marie SK, Bettegowda C, Agrawal N, Lipp E, Pirozzi C, Lopez G, He Y, Friedman H, Friedman AH, Riggins GJ, Holdhoff M, Burger P, McLendon R, Bigner DD, Vogelstein B, Meeker AK, Kinzler KW, Papadopoulos N, Diaz LA, Yan H. Frequent ATRX, CIC, FUBP1 and IDH1 mutations refine the classification of malignant gliomas. Oncotarget. 2012;3:709–722. doi: 10.18632/oncotarget.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suzuki H, Aoki K, Chiba K, Sato Y, Shiozawa Y, Shiraishi Y, Shimamura T, Niida A, Motomura K, Ohka F, Yamamoto T, Tanahashi K, Ranjit M, Wakabayashi T, Yoshizato T, Kataoka K, Yoshida K, Nagata Y, Sato-Otsubo A, Tanaka H, Sanada M, Kondo Y, Nakamura H, Mizoguchi M, Abe T, Muragaki Y, Watanabe R, Ito I, Miyano S, Natsume A, Ogawa S. Mutational landscape and clonal architecture in grade II and III gliomas. Nat Genet. 2015;47:458–468. doi: 10.1038/ng.3273. [DOI] [PubMed] [Google Scholar]
- 4.Eckel-Passow JE, Lachance DH, Molinaro AM, Walsh KM, Decker PA, Sicotte H, Pekmezci M, Rice T, Kosel ML, Smirnov IV, Sarkar G, Caron AA, Kollmeyer TM, Praska CE, Chada AR, Halder C, Hansen HM, McCoy LS, Bracci PM, Marshall R, Zheng S, Reis GF, Pico AR, O’Neill BP, Buckner JC, Giannini C, Huse JT, Perry A, Tihan T, Berger MS, Chang SM, Prados MD, Wiemels J, Wiencke JK, Wrensch MR, Jenkins RB. Glioma Groups Based on 1p/19q, IDH, and TERT Promoter Mutations in Tumors. The New England journal of medicine. 2015;372:2499–2508. doi: 10.1056/NEJMoa1407279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer discovery. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science signaling. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berube NG, Mangelsdorf M, Jagla M, Vanderluit J, Garrick D, Gibbons RJ, Higgs DR, Slack RS, Picketts DJ. The chromatin-remodeling protein ATRX is critical for neuronal survival during corticogenesis. J Clin Invest. 2005;115:258–267. doi: 10.1172/JCI22329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heaphy CM, de Wilde RF, Jiao Y, Klein AP, Edil BH, Shi C, Bettegowda C, Rodriguez FJ, Eberhart CG, Hebbar S, Offerhaus GJ, McLendon R, Rasheed BA, He Y, Yan H, Bigner DD, Oba-Shinjo SM, Marie SK, Riggins GJ, Kinzler KW, Vogelstein B, Hruban RH, Maitra A, Papadopoulos N, Meeker AK. Altered telomeres in tumors with ATRX and DAXX mutations. Science. 2011;333:425. doi: 10.1126/science.1207313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rankin SL, Zhu G, Baker SJ. Review: insights gained from modelling high-grade glioma in the mouse. Neuropathology and applied neurobiology. 2012;38:254–270. doi: 10.1111/j.1365-2990.2011.01231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wiesner SM, Decker SA, Larson JD, Ericson K, Forster C, Gallardo JL, Long C, Demorest ZL, Zamora EA, Low WC, SantaCruz K, Largaespada DA, Ohlfest JR. De novo induction of genetically engineered brain tumors in mice using plasmid DNA. Cancer research. 2009;69:431–439. doi: 10.1158/0008-5472.CAN-08-1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calinescu AA, Nunez FJ, Koschmann C, Kolb BL, Lowenstein PR, Castro MG. Transposon mediated integration of plasmid DNA into the subventricular zone of neonatal mice to generate novel models of glioblastoma. Journal of visualized experiments : JoVE. 2015 doi: 10.3791/52443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gibbons RJ, Higgs DR. Molecular-clinical spectrum of the ATR-X syndrome. Am J Med Genet. 2000;97:204–212. doi: 10.1002/1096-8628(200023)97:3<204::AID-AJMG1038>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 13.Paugh BS, Qu C, Jones C, Liu Z, Adamowicz-Brice M, Zhang J, Bax DA, Coyle B, Barrow J, Hargrave D, Lowe J, Gajjar A, Zhao W, Broniscer A, Ellison DW, Grundy RG, Baker SJ. Integrated molecular genetic profiling of pediatric high-grade gliomas reveals key differences with the adult disease. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2010;28:3061–3068. doi: 10.1200/JCO.2009.26.7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.N. Cancer Genome Atlas Research. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alcantara Llaguno S, Chen J, Kwon CH, Jackson EL, Li Y, Burns DK, Alvarez-Buylla A, Parada LF. Malignant astrocytomas originate from neural stem/progenitor cells in a somatic tumor suppressor mouse model. Cancer cell. 2009;15:45–56. doi: 10.1016/j.ccr.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wei D, Levic S, Nie L, Gao WQ, Petit C, Jones EG, Yamoah EN. Cells of adult brain germinal zone have properties akin to hair cells and can be used to replace inner ear sensory cells after damage. Proc Natl Acad Sci U S A. 2008;105:21000–21005. doi: 10.1073/pnas.0808044105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sheng Z, Li L, Zhu LJ, Smith TW, Demers A, Ross AH, Moser RP, Green MR. A genome-wide RNA interference screen reveals an essential CREB3L2-ATF5-MCL1 survival pathway in malignant glioma with therapeutic implications. Nature medicine. 2010;16:671–677. doi: 10.1038/nm.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xue Y, Gibbons R, Yan Z, Yang D, McDowell TL, Sechi S, Qin J, Zhou S, Higgs D, Wang W. The ATRX syndrome protein forms a chromatin-remodeling complex with Daxx and localizes in promyelocytic leukemia nuclear bodies. Proc Natl Acad Sci U S A. 2003;100:10635–10640. doi: 10.1073/pnas.1937626100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Osley MA, Tsukuda T, Nickoloff JA. ATP-dependent chromatin remodeling factors and DNA damage repair. Mutation research. 2007;618:65–80. doi: 10.1016/j.mrfmmm.2006.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leung JW, Ghosal G, Wang W, Shen X, Wang J, Li L, Chen J. Alpha thalassemia/mental retardation syndrome X-linked gene product ATRX is required for proper replication restart and cellular resistance to replication stress. J Biol Chem. 2013;288:6342–6350. doi: 10.1074/jbc.M112.411603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Viana-Pereira M, Lee A, Popov S, Bax DA, Al-Sarraj S, Bridges LR, Stavale JN, Hargrave D, Jones C, Reis RM. Microsatellite instability in pediatric high grade glioma is associated with genomic profile and differential target gene inactivation. PLoS One. 2011;6:e20588. doi: 10.1371/journal.pone.0020588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haines J, Bacher J, Coster M, Huiskamp R, Meijne E, Mancuso M, Pazzaglia S, Bouffler S. Microsatellite instability in radiation-induced murine tumours; influence of tumour type and radiation quality. International journal of radiation biology. 2010;86:555–568. doi: 10.3109/09553001003734600. [DOI] [PubMed] [Google Scholar]
- 23.Bacher JW, Abdel Megid WM, Kent-First MG, Halberg RB. Use of mononucleotide repeat markers for detection of microsatellite instability in mouse tumors. Molecular carcinogenesis. 2005;44:285–292. doi: 10.1002/mc.20146. [DOI] [PubMed] [Google Scholar]
- 24.Vilar E, Bartnik CM, Stenzel SL, Raskin L, Ahn J, Moreno V, Mukherjee B, Iniesta MD, Morgan MA, Rennert G, Gruber SB. MRE11 deficiency increases sensitivity to poly(ADP-ribose) polymerase inhibition in microsatellite unstable colorectal cancers. Cancer research. 2011;71:2632–2642. doi: 10.1158/0008-5472.CAN-10-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.N. Cancer Genome Atlas Research, Comprehensive. Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. The New England journal of medicine. 2015;372:2481–2498. doi: 10.1056/NEJMoa1402121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y, Yang J, Zheng H, Tomasek GJ, Zhang P, McKeever PE, Lee EY, Zhu Y. Expression of mutant p53 proteins implicates a lineage relationship between neural stem cells and malignant astrocytic glioma in a murine model. Cancer cell. 2009;15:514–526. doi: 10.1016/j.ccr.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferguson DO, Sekiguchi JM, Chang S, Frank KM, Gao Y, DePinho RA, Alt FW. The nonhomologous end-joining pathway of DNA repair is required for genomic stability and the suppression of translocations. Proc Natl Acad Sci U S A. 2000;97:6630–6633. doi: 10.1073/pnas.110152897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Callicott RJ, Womack JE. Real-time PCR assay for measurement of mouse telomeres. Comparative medicine. 2006;56:17–22. [PubMed] [Google Scholar]
- 29.Henson JD, Cao Y, Huschtscha LI, Chang AC, Au AY, Pickett HA, Reddel RR. DNA C-circles are specific and quantifiable markers of alternative-lengthening-of-telomeres activity. Nature biotechnology. 2009;27:1181–1185. doi: 10.1038/nbt.1587. [DOI] [PubMed] [Google Scholar]
- 30.Mao Z, Jiang Y, Liu X, Seluanov A, Gorbunova V. DNA repair by homologous recombination, but not by nonhomologous end joining, is elevated in breast cancer cells. Neoplasia. 2009;11:683–691. doi: 10.1593/neo.09312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Azad A, Jackson S, Cullinane C, Natoli A, Neilsen PM, Callen DF, Maira SM, Hackl W, McArthur GA, Solomon B. Inhibition of DNA-dependent protein kinase induces accelerated senescence in irradiated human cancer cells. Molecular cancer research : MCR. 2011;9:1696–1707. doi: 10.1158/1541-7786.MCR-11-0312. [DOI] [PubMed] [Google Scholar]
- 32.Plowman J, Waud WR, Koutsoukos AD, Rubinstein LV, Moore TD, Grever MR. Preclinical antitumor activity of temozolomide in mice: efficacy against human brain tumor xenografts and synergism with 1,3-bis(2-chloroethyl)-1-nitrosourea. Cancer research. 1994;54:3793–3799. [PubMed] [Google Scholar]
- 33.Pollack IF, Erff M, Bom D, Burke TG, Strode JT, Curran DP. Potent topoisomerase I inhibition by novel silatecans eliminates glioma proliferation in vitro and in vivo. Cancer research. 1999;59:4898–4905. [PubMed] [Google Scholar]
- 34.Sarkaria JN, Carlson BL, Schroeder MA, Grogan P, Brown PD, Giannini C, Ballman KV, Kitange GJ, Guha A, Pandita A, James CD. Use of an orthotopic xenograft model for assessing the effect of epidermal growth factor receptor amplification on glioblastoma radiation response. Clin Cancer Res. 2006;12:2264–2271. doi: 10.1158/1078-0432.CCR-05-2510. [DOI] [PubMed] [Google Scholar]
- 35.Wolff JE, Trilling T, Molenkamp G, Egeler RM, Jurgens H. Chemosensitivity of glioma cells in vitro: a meta analysis. Journal of cancer research and clinical oncology. 1999;125:481–486. doi: 10.1007/s004320050305. [DOI] [PubMed] [Google Scholar]
- 36.Lord CJ, Ashworth A. The DNA damage response and cancer therapy. Nature. 2012;481:287–294. doi: 10.1038/nature10760. [DOI] [PubMed] [Google Scholar]
- 37.Ramamoorthy M, Smith S. Loss of ATRX Suppresses Resolution of Telomere Cohesion to Control Recombination in ALT Cancer Cells. Cancer cell. 2015;28:357–369. doi: 10.1016/j.ccell.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lovejoy CA, Li W, Reisenweber S, Thongthip S, Bruno J, de Lange T, De S, Petrini JH, Sung PA, Jasin M, Rosenbluh J, Zwang Y, Weir BA, Hatton C, Ivanova E, Macconaill L, Hanna M, Hahn WC, Lue NF, Reddel RR, Jiao Y, Kinzler K, Vogelstein B, Papadopoulos N, Meeker AK A. L. T. S. C. Consortium. Loss of ATRX, genome instability, and an altered DNA damage response are hallmarks of the alternative lengthening of telomeres pathway. PLoS genetics. 2012;8:e1002772. doi: 10.1371/journal.pgen.1002772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability--an evolving hallmark of cancer. Nature reviews Molecular cell biology. 2010;11:220–228. doi: 10.1038/nrm2858. [DOI] [PubMed] [Google Scholar]
- 40.Sherman MH, Bassing CH, Teitell MA. Regulation of cell differentiation by the DNA damage response. Trends in cell biology. 2011;21:312–319. doi: 10.1016/j.tcb.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schneider L, Pellegatta S, Favaro R, Pisati F, Roncaglia P, Testa G, Nicolis SK, Finocchiaro G, d’Adda di Fagagna F. DNA damage in mammalian neural stem cells leads to astrocytic differentiation mediated by BMP2 signaling through JAK-STAT. Stem cell reports. 2013;1:123–138. doi: 10.1016/j.stemcr.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burma S, Chen DJ. Role of DNA-PK in the cellular response to DNA double-strand breaks. DNA repair. 2004;3:909–918. doi: 10.1016/j.dnarep.2004.03.021. [DOI] [PubMed] [Google Scholar]
- 43.Roos WP, Kaina B. DNA damage-induced cell death by apoptosis. Trends in molecular medicine. 2006;12:440–450. doi: 10.1016/j.molmed.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 44.Betermier M, Bertrand P, Lopez BS. Is non-homologous end-joining really an inherently error-prone process? PLoS genetics. 2014;10:e1004086. doi: 10.1371/journal.pgen.1004086. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Sleeping Beauty responsive shATRX plasmids are cloned to explore the impact of ATRX on GBM development.
Fig. S2. Cells co-transfected with shp53, shATRX, and NRAS plasmids are characterized at 15 days after injection.
Fig. S3. SB-mediated transfected cells are distinct from ependymal cells in mice.
Fig. S4. Characterization of p53/NRAS/shATRX mice 7 days after injection shows tumor cells expressing GFAP and Nestin..
Fig. S5. p53/NRAS/shATRX mice at 21 days after injection show tumor cells expressing OLIG2 and Nestin.
Fig. S6. Moribund p53/NRAS/shATRX mice show tumor cells expressing OLIG2, Nestin, and pERK.
Fig. S7. p53/NRAS/shATRX mice show loss of GFAP expression by 15 days after injection.
Fig. S8. ATRX mutations do not cluster in SNF2/helicase domain in human glioma.
Fig. S9. Primary GBM cell cultures are generated from SB-induced mouse tumors.
Fig. S10. No difference is seen in telomere length in tumors with or without ATRX by qPCR.
Fig. S11. Assessment of ALT by c-circle assay with DNA from mouse tumors and neurospheres.
Fig. S12 Cells transfected with shATRX plasmid show reduction in NHEJ by flow cytometry.
Fig. S13. Non-homologous end joining (NHEJ) pathway is characterized by IHC.
Fig. S14. Homologous recombination (HR) pathway analysis is characterized by IHC.
Fig. S15. Mismatch repair (MMR) pathway analysis is characterized by IHC.
Fig. S16. SB mouse tumor growth (without radiation treatment) is characterized by luminescence.
Fig. S17. Tumors with ATRX loss show reduction in phospho-DNA-PKcs before and after radiation treatment.
Fig. S18. Mouse tumors with ATRX loss show increased expression of γH2A.X in vitro and in vivo.
Fig. S19. Detailed schematic shows proposed impact of ATRX loss on GBM progression.
Table S1. Animal models of glioma all have RTK/RAS/PI3K alterations.
Table S2. Mice injected with p53/NRAS/shATRX have faster growing tumors.
Table S3. MSI rate is increased in p53/NRAS/shATRX tumors.
Table S4. IDH1 and TP53 mutations do not alter SNV rate calculated by two-way ANOVA model.
Table S5. Telomere qPCR ATLR worksheet and standard curve show similar results in all experimental groups.
Table S6. Immunofluorescence analysis shows reduced pDNA-PKcs expression in ATRX-deficient mouse GBM.
Table S7. Antibodies used for tissue analysis are summarized in table form.