Abstract
Intrinsic signal optical imaging (ISI) is a rapid and noninvasive method for observing brain activity in vivo over a large area of the cortex. Here we describe our protocol for mapping retinotopy to identify mouse visual cortical areas using ISI. First, surgery is performed to attach a head frame to the mouse skull (~1 h). The next day, intrinsic activity across the visual cortex is recorded during the presentation of a full-field drifting bar in the horizontal and vertical directions (~2 h). Horizontal and vertical retinotopic maps are generated by analyzing the response of each pixel during the period of the stimulus. Last, an algorithm uses these retinotopic maps to compute the visual field sign and coverage, and automatically construct visual borders without human input. Compared with conventional retinotopic mapping with episodic presentation of adjacent stimuli, a continuous, periodic stimulus is more resistant to biological artifacts. Furthermore, unlike manual hand-drawn approaches, we present a method for automatically segmenting visual areas, even in the small mouse cortex. This relatively simple procedure and accompanying open-source code can be implemented with minimal surgical and computational experience, and is useful to any laboratory wishing to target visual cortical areas in this increasingly valuable model system.
INTRODUCTION
Since its first demonstration in the 1980s1, ISI of the cortex has become a fundamental technique in systems neuroscience. By imaging reflected light at longer wavelengths, it is possible to measure the hemodynamic response elicited by the heightened metabolic demand of active neurons2–5. In the past two decades, ISI has become a common way to visualize organizing principles of cortex, such as orientation6, ocular dominance7, and retinotopy8 maps. Retinotopic mapping has been widely used in humans and primates to identify visual areas based on the locations of distinct visual field maps, a defining feature of a visual area9–11. Neighboring reversals of horizontal and vertical retinotopic gradients is a well-described pattern across the visual cortex12–14. Identifying these reversals is a useful way to draw borders between visual areas, and additional criteria, such as the coverage of the visual field, can then be applied to assure accurate parsing of visual areas.
Functional mapping provides useful advantages over anatomical reference frames, particularly increased precision of areal targeting in live animals. This is especially important for experiments in animals with small brains, such as mice, which are rapidly becoming an important model for visual neuroscience15–17. We therefore sought to develop a protocol that would robustly and automatically map mouse visual cortex and identify area boundaries. By doing so, we can reliably identify visual areas in the mouse in order to perform targeted studies to understand their connectivity and function8,18,19.
Applications of method
The development of powerful techniques for cell-specific observation and manipulation in transgenic animals, combined with the experimental tractability of the visual system17, has enabled the mouse visual cortex to become a fertile ground for systems neuroscience15. Numerous systems neuroscience laboratories would benefit from reliably defining areas of the mouse cortex for studies of feedforward and feedback processes20, multisensory processing21, and perceptual decision-making22. As several of these regions, such as the anterolateral and anteromedial areas (AL and AM, respectively), are <0.25 mm2 and can vary in exact size and position across animals4, it is necessary to define these areas functionally rather than with stereotaxic coordinates. Therefore, automatically defining the borders between the small, yet functionally distinct, visual areas in the mouse is a valuable and necessary step for researchers wishing to enter this field. As we have previously demonstrated, it is possible to obtain retinotopic maps of mouse visual cortex, automatically generate borders between visual areas, and use these maps to target locations of interest for anatomical or functional studies8,18,19. We have used this ISI protocol to show that mice have 11 retinotopically defined visual areas8 with distinct spatial and temporal frequency preferences18, as well as nonlinear selectivity for pattern motion19.
As our method does not require removal of the skull, mice can be imaged over multiple days or weeks for the purposes of developmental or pathological studies. Furthermore, ISI does not require a functional activity indicator (e.g., GCaMP), and it can therefore be performed in various strains of unaltered mice. Still, it should be noted that intrinsic signals are much weaker than changes measured through the skull with calcium indicators such as GCaMP6 (ref. 23). In addition, before implanting the head frame, marks can be left on the skull for stereotaxic targeting of other brain regions. For example, if the investigator would like to target viral injections to both the thalamus and a higher visual area, stereotaxic coordinates and a marker can be used to designate the spot of pipette insertion on the skull, and this mark will remain over the course of imaging until the time of injection. Last, the simplicity and flexibility of this technique are well suited for the robust identification of area borders in mice, and should work well in other species, including rats, ferrets, and macaques.
Limitations
As compared with imaging with chemical or genetically encoded indicators, intrinsic signals have lower spatial resolution and can be more difficult to detect. Due to optical blurring (e.g., from scatter and a finite depth of field) and the imprecise localization of a hemodynamic signal, the point-spread function of optical imaging is estimated to be 200–250 μm full width at half maximum24,25, ~20 times larger than the point-spread function of spiking activity26. However, it is not necessary to have cellular resolution in order to identify visual areas, as the area sizes, even in the mouse, are substantially larger. The temporal resolution is limited by the hemodynamic signal, which is on the order of seconds. If investigators are interested in single-cell properties, or phenomena occurring on faster time scales, intrinsic imaging is not ideal. Finally, automatic segmentation of visual areas is contingent upon maps with smooth retinotopy, few artifacts (e.g., those caused by blood vessels, breathing), and fairly strong responses. It may be necessary to increase the number of repeats of the stimulus in order to achieve maps with sufficient information for automatic border generation with our code. In addition to ISI data, our visual area segmentation code will work with other types of data (e.g., wide-field calcium imaging, flavoprotein autofluorescence), provided the investigators have already calculated the retinotopic maps from their data.
Overview of procedure
To identify visual areas, intrinsic signals measured from the cortical surface of anesthetized mice are acquired during the presentation of visual stimuli, and these measures are used to derive retinotopic maps that are the basis for areal segmentation. First, a surgery is performed to attach a head frame to the mouse. Although we provide a protocol below, there are various examples in the literature of devices for head fixation that may be used27–30. After allowing the mouse to recover (preferably the next day), a clean region of the skull over the visual cortex is exposed. Alternatively, investigators may wish to perform a craniotomy and implant a window for subsequent chronic two-photon or wide-field imaging28,29, although this is not needed for successful intrinsic imaging. Mice are given a sedative (chlorprothixene), allowing us to use a lower level of isoflurane anesthesia during imaging. This is important, as isoflurane is known to disrupt visual responses in the cortex31,32. Although the amplitude of the intrinsic signal is lower in sedated mice33, ISI in the awake mouse is associated with larger artifacts from heart rate and breathing. If investigators choose to use this protocol in an awake mouse, they should thoroughly habituate the mouse to the setup before imaging. The mouse is then moved under a camera with large dynamic range and a stable light source, adjacent to a large monitor. During periodic visual stimulation with vertically and horizontally moving stimuli, intrinsic hemodynamic activity is recorded with luminance from (long-pass) orange light. The reflected light reaching the camera goes through a near-infrared bandpass filter. Responses from each pixel are then analyzed at the frequency of the stimulus to identify the location on the screen that drove a peak response to produce a retinotopic map34. Finally, an automated algorithm9 identifies visual area borders based on retinotopic reversals (maps in adjacent areas are mirror images) and overlays these borders with blood vessel maps to enable accurate targeting of higher visual areas. Below, we explore the equipment and setup involved in these steps, and then outline a procedure to implement them.
Experimental design
Lights, camera, optics
The hemodynamic response has several components that can be optimized with different wavelengths of light2. Near the site of activity, there is an initial increase in deoxygenated hemoglobin (HbR), which is followed by a dip in HbR as oxygenated hemoglobin enters the region35. Many intrinsic imaging configurations isolate the peak in this oxymetry component by using an orange light source (595–605 nm)4. As HbR has a higher absorption coefficient (>600 nm) than oxygenated blood, more-active regions can be distinguished from less-active regions by using wavelengths above 600 nm. Another component of the response, an increase in blood volume due to filling of capillaries, is slower and more widespread, but it can be isolated with lower-wavelength light (~570 nm)4. The last component of the hemodynamic response, caused by a change of light scattering in active tissue, is also observed using red light (>605 nm)4.
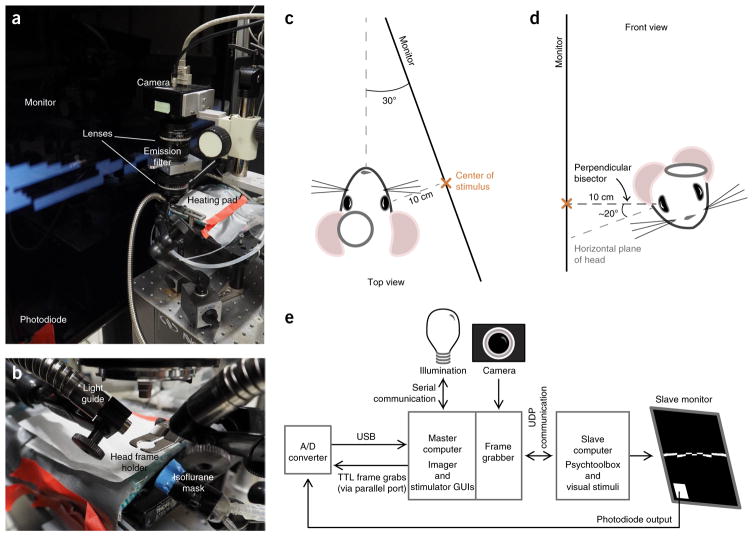
The best cameras for intrinsic imaging are able to capture a large number of photons before saturation, as signal-to-noise ratio increases with photon count. In turn, a camera that has a high well capacity across the entire sensor is desired. For instance, compute the total number of pixels and multiply by the well capacity of each pixel for a normalized metric across cameras. Here, we use the Dalsa Pantera 1m60, which has a well depth of 500,000 electrons and about 1 million total pixels. Quantum efficiency is less important because it is easy to simply shine more light with reflectance imaging. The lenses described here have a higher magnification (pixels per millimeter) than those described in our previous publications to increase the resolution of the image using lenses versus the digital zoom8,18,36. The primary goal of the lens configuration is to fill the image sensor with your region of interest, with minimal distortion. A tandem configuration can be used to achieve higher magnification in addition to a reasonable working distance for in vivo imaging (Fig. 1a)37. In addition, adding bandpass filters in front of the lens will reduce unwanted light from other sources such as the monitor or room. Light guides are used to effectively direct the light over the exposed skull or brain (Fig. 1b).
Figure 1.
Intrinsic imaging setup. (a) Intrinsic imaging rig overview. (b) Close-up of mouse head frame holder, isoflurane mask, and light guides. (c) Mouse position in relation to the monitor, viewed from the top of the setup (INTRODUCTION). The orange ‘x’ marks the center of the stimulus. (d) Mouse position viewed from the front (orange ‘x’ marks the center of the stimulus). The mouse is positioned 10 cm away from the monitor, and the perpendicular bisector of the eye is approximately perpendicular to the monitor. (e) Diagram of connectivity between master and slave computers and associated hardware. Details on each piece of equipment can be found in the ‘EQUIPMENT’ and ‘EQUIPMENT SETUP’ sections.
Choice of imaging preparation
Depending on what the investigators wish to do before or after mapping visual areas, they may wish to thin the skull or implant a chronic window. Thinning the skull can improve imaging, but it should be done only if the skull integrity does not matter for beyond a week after the experiment—for example, if injections or a craniotomy will be done after intrinsic imaging. Injections can be performed immediately after imaging, or after the animal recovers. In other experimental designs, it may be best to completely remove the skull. For example, if the investigator would like to perform chronic two-photon imaging over the visual cortex after mapping the higher visual areas, we would recommend implanting a window. This surgery could be done at the same time as the head frame surgery, and extra time should be allowed for the mouse and imaging window to recover, especially in case of blood loss. We refer the reader to several previously published protocols for implanting a chronic imaging window28,29,38.
Episodic versus continuous stimulation
Standard intrinsic imaging protocols present randomized, episodic stimuli and average responses to the stimuli39. However, these approaches are easily corrupted by biological artifacts such as those from heartbeat and breathing, and many repeats of the stimulus are required to overcome these artifacts. To address this problem, various groups have developed methods using periodic stimulation and analysis of the response at the frequency of the stimulus, reducing both biological artifacts and stimulus time by systematically placing the stimulus frequency outside of the frequency bands of the biological noise10,34,40,41. We have built on this approach by using a reverse checkerboard drifting bar stimulus that periodically drifts slowly across the entire visual field and allows for automatic segmentation of small visual areas in the mouse (see Supplementary Video S1 and S2 in Marshel et al.18).
Accurate stimulation of the entire visual field
The position and orientation of the eye relative to the screen is paramount in a retinotopic mapping experiment. At rest, the mouse’s eyes deviate 22° above the plane parallel to the ground, and 60° lateral to the midline42. The screen should be placed perpendicular to the optic axis of the eye. To account for the horizontal deviation of the eye from the midline, the screen should be tilted inward toward the nose by 30° (Fig. 1c). The mouse’s nose should be pointing directly at the left edge of the screen to ensure that the entire left hemifield is stimulated (Fig. 1c). If the mouse’s head is held in stereotaxic coordinates, tilt the top of the screen inward toward the mouse by ~20° to account for the deviation of the eye from the horizon. In our setup, the typical angle of the head frame placed over the visual cortex is ~20° lateral to the horizontal, and thus when the mouse is head-fixed with the head frame parallel to the ground the optic axis of the eye is shifted downward by ~20°, placing it approximately perpendicular to the horizon (Fig. 1d). In this case, the screen does not need to be tilted vertically. If the head frame is placed at an angle >20° to the horizontal plane of the mouse’s head, tilt the screen to account for the difference.
Generation of retinotopic maps
To effectively drive the cortex, a contrast-reversing checkerboard bar stimulus is used (see Supplementary Video S1 and S2 in Marshel et al.18). The checkerboard bar extends across the screen and drifts orthogonally in either a vertical or horizontal manner, to produce maps of altitude and azimuth, respectively. The altitude stimulus is adjusted from previous studies such that the size (degrees) and speed (degrees/s) of the bar are constant relative to the mouse18. If the experimenter needs to identify only the centers of the major visual areas (e.g., the primary visual cortex (V1) or the lateromedial area (LM)), it is not necessary to stimulate the entire visual field, and instead a smaller stimulus can be used.
From the raw movies, we take the response time course for each pixel and compute the phase of the Fourier transform at the stimulus frequency34. The bar is drifted in opposite directions in order to subtract the delay in the intrinsic signal relative to neural activity34. The resulting phase maps are converted to retinotopic coordinates, in degrees (e.g., Fig. 2), using known information about the geometry of the experimental setup (e.g., screen distance from the eye).
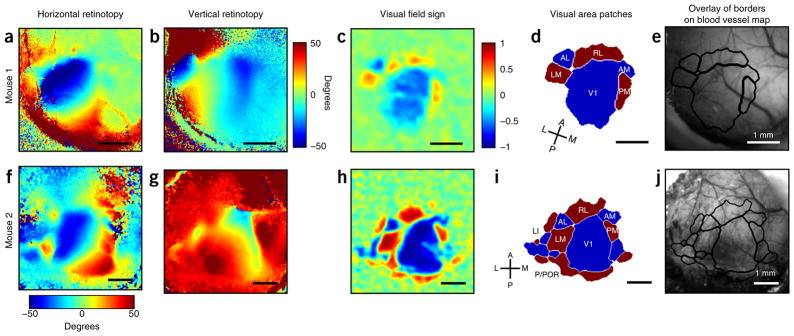
Figure 2.
Example data from ISI experiments. Retinotopic maps from two different mice with a thinned skull and five (Mouse 1; panels a–e) or 10 (Mouse 2, panels f–j) repeats of the stimulus. Mouse 2 data (f–j) have previously been published8. (a) Map of horizontal retinotopy; color scale is in degrees of visual space. (b) Map of vertical retinotopy; color scale is in degrees of visual space. (c) Map of visual field sign; scale is −1 to 1. The visual field sign is computed by taking the sine of the difference between the vertical and horizontal retinotopic gradients at each pixel9. (d) Thresholded visual field sign patches, revealing key visual areas. Areas with mirror representations are blue, and areas with nonmirror representations are red. Regions with values close to zero do not have a clear topographic structure. (e) Visual area borders overlaid on blood vessel map (acquired with CCD camera and green light). (f–h) Same as a–c but with a different mouse and more repeats. (i) Thresholded visual field sign patches. As demonstrated here, more repeats of the stimulus are necessary to confidently map the smaller, more lateral visual areas. (j) Visual area borders overlaid on blood vessel map. For additional examples, see Garrett et al.8 and the sample data hosted on Github at https://snlc.github.io/ISI/. Scale bars, 1 mm. All work involving live animals was conducted according to a procedure approved by the Salk Institute IACUC. A, anterior; AL, anterolateral area; AM, anteromedial area; L, lateral; LI, laterointermediate area; LM, lateromedial area; M, medial; P, posterior; PM, posteromedial area; POR, postrhinal; RL, rostrolateral area; V1, primary visual cortex. Image adapted with permission of the Journal of Neuroscience, from ‘Topography and areal organization of mouse visual cortex’, Garrett, M.E., Nauhaus, I., Marshel, J.H. & Callaway, E.M., 34, 12587–12600 (2014); permission conveyed through Copyright Clearance Center.
Automatic identification of area borders
After imaging, the retinotopic maps can be analyzed to automatically segment visual areas using several criteria (see Table 1 in Garrett et al.8), primarily based on the principle that reversals in retinotopic gradients correspond to boundaries between visual areas. When viewed from the surface, each region of the cortex can be characterized by how well its retinotopy matches the representation of visual space in the retina; in some regions, the visual field is faithfully projected onto the cortex (nonmirror representation), whereas in some regions this projection is flipped (mirror representation). We can define local regions of the cortex as mirror or nonmirror by calculating the point of visual space represented by each point of cortex and determining the gradient of this map for each local region9. After low-pass filtering of the retinotopic maps, we identify a vector for each pixel that represents the direction of the retinotopic gradient in either the altitude or azimuth directions. By taking the sine of the angle between these two gradients for each pixel, we can compute a map of visual field sign9, which represents the direction of the retinotopic gradients as a single value between 1 and −1 (Fig. 2c,h). Regions in the map where the visual field sign reverses from positive to negative values indicate a reflection in the retinotopic gradient at the border between two areas. Adjacent pixels with similar visual field signs can be combined into patches, which represent putative visual areas. Additional criteria are imposed to ensure that each identified patch corresponds to a single map of visual space; for example, patches are fused if they represent different regions of visual space, or split if there are redundant representations8 (Fig. 2d). We can then overlay these maps on blood vessel images, which provide a guide for the targeting of areas for injections or recording (Fig. 2e).
MATERIALS
REAGENTS
Mice and surgery
Animals: 1- to 3-month-old C57BL/6 mice (Jackson Laboratory, cat. no. 000664) are imaged here, although we have also successfully imaged other strains of mice. Older mice may be used, but the skull thickens over the course of development, making skull thinning more difficult ! CAUTION All experiments should be performed in accordance with institutional guidelines and local and national regulations. All work presented here was conducted according to a procedure approved by the Salk Institute Institutional Animal Care and Use Committee (IACUC).
Mini Dissecting Scissors, 8.5 cm, straight, sharp tips (WPI, cat. no. 503667)
Tweezer, no. 5 (Kent Scientific, cat. no. INS600098)
Isoflurane (Isothesia; Allivet, cat. no. 50562)
Chlorprothixene hydrochloride (Sigma-Aldrich, cat. no. C1671)
Agarose Type III-A (Sigma-Aldrich, cat. no. A9793)
Carpofen (Rimadyl Injectable; Allivet, cat. no. 27180)
C&B Metabond Radiopaque L-Powder (Parkell, cat. no. S396)
C&B Metabond ‘B’ Quick Base (Parkell, cat. no. S398)
C&B Metabond ‘C’ Universal TBB Catalyst (Parkell, cat. no. S371)
Adjustable Precision Applicator Brushes (Parkell, cat. no. S379)
Nair Hair Removal (Amazon, cat. no. ASIN B001G7PTWU)
Vetbond (Santa Cruz Biotechnology, cat. no. sc-361931)
Eye ointment (Rugby, cat. no. 370435)
Silicon oil (OFNA Racing, cat. no. 10236)
Artificial cerebral spinal fluid (ACSF; prepared according to Cold Spring Harbor Protocol found at http://cshprotocols.cshlp.org/content/2011/9/pdb.rec065730.full?)
Kwik-Cast Sealant (World Precision Instruments, cat. no. KWIK-CAST)
Lidocaine (VWR International, cat. no. 95033-980)
Ibuprofen Oral Suspension (Perrigo, cat. no. 45802-952-43)
1-ml syringes (BD, cat. no. 309659)
Sterilized saline (0.9% sodium chloride for injection; Hospira, cat. no. 0409-4888-10)
Sterilized water for injection (Hospira, cat. no. 0409-4887-10)
Kimwipes (Kimtech, cat. no. 34120)
EQUIPMENT
Mice and surgery
Oxygen cylinders (Airgas, cat. no. OX USP300)
Mouse anesthesia system with isoflurane box (Parkland Scientific, cat. no. V3000PK)
Small rodent stereotax fitted with anesthesia mask (Narishige, cat. no. SG-4N)
Dental drill (Osada, model no. EXL-M40)
0.9-mm burrs for micro drill (Fine Science Tools, cat. no. 19007-09)
8-mm coverslips, no. 1 thickness (Warner Instruments, cat. no. 64-0701)
Cotton applicators (Fisher Scientific, cat. no. 19-062-616)
Microwave
Heat block or water bath set to 70 °C (Thomas Scientific, cat. no. D1100)
T/Pump Warm Water Recirculator (Kent Scientific, cat. no. TP-700) with warming pad (Kent Scientific, cat. no. TPZ)
Mouse head frame and head frame holder (Supplementary Data 1; alternative designs available via Janelia Open Science (https://www.janelia.org/open-science))
Breadboard (Thor Labs, cat. no. MB810)
Television, computer, optics, and camera
Television/monitor (Samsung, 55-inch 1080p LED TV, > 20-inch is sufficient)
Photodiode (Thor Labs, cat. no. SM05PD1A)
Light source with serial port communication (Illumination Technologies, cat. no. 3900)
Light guides (Illumination Technologies, cat. no. 9240HT)
Excitation filters (510–590 nm and 610 nm; Illumination Technologies or Chroma)
Emission filter, 525-nm bandpass filter (Edmund Optics, cat. no. 87-801)
Emission filter, 700-nm bandpass filter (Edmund Optics, cat. no. 88-018)
Charge-coupled device (CCD) or complementary metal oxide silicon (CMOS) camera with camera link (we use a Dalsa Pantera TF 1M60)
Camera mount (ideally easily adjustable; Edmund Optics, cat. no. 54-123)
Nikon AI-S FX Nikkor 50 mm f/1.2 manual focus lens
Nikon Ai 85 mm f/2 manual focus lens
2 NogaFlex Holders (Noga, cat. no. NF61003)
Frame-grabbing card (Matrox)
Master computer with at least 8 GB RAM
Slave computer that meets the specifications for Psychtoolbox (http://psychtoolbox.org/requirements/)
REAGENT SETUP
Chlorprothixene
Put 2 mg of chlorprothixine in 10 ml of sterilized water; mix the solution and administer 0.1 ml/20 g per mouse. It is best to make a fresh solution of chlorprothixene each day. If you are performing multiple experiments in 1 d, the chlorprothixene solution should be stored at 4 °C between experiments. The chlorprothixene solution should be made fresh each day.
Agarose
To make 1.5% (wt/vol) agarose, add 75 mg of agarose to 5 ml of ACSF, microwave on high for ~15 s, and then put it in a heat block or water bath at 70 °C. The agarose should be made fresh on the day of the experiment and discarded after use.
Carpofen
Add 0.1 ml of carpofen (50 mg/ml) to 10 ml of saline. This solution can be stored at 4 °C for 1 week.
EQUIPMENT SETUP
Camera, lens, and optics
Configure the camera on a stable camera mount with a manual focus knob, which moves the entire configuration up and down. Ensure that the optic axis of the camera is perpendicular to the plane of imaging through the brain. You may choose to attach the lenses to the camera in a ‘tandem’ configuration, as a way of magnifying the image with minimal distortion (Fig. 1a). The top lens aperture should initially be set to f/5.6 and the bottom aperture should be set to f/2.8, but you may want to adjust the bottom lens aperture, depending on the strength of your light source (see PROCEDURE Step 42). Under the camera, position a head frame holder, heating pad, and light guides from the light source (Fig. 1b). ▲ CRITICAL The entire imaging setup, including monitor, camera, and optics, should be either in an enclosed space or in a room in which the lights can be turned off during experiments.
Monitor and computers
Mount the TV used for visual stimulation on a custom, fully adjustable floor stand. The TV here is mounted vertically to fully stimulate the upper visual field. Configure two computers, a frame grabber, and a DAQ interface as shown in Figure 1e.
Stimulus Presentation and Image Acquisition Code
Download the ‘Imager.zip’ and ‘Stimulator.zip’ files from GitHub (https://snlc.github.io/ISI/). Unzip these files into a folder, and add this folder to your MATLAB path. The ‘Stimulator’ folder also includes a sample parameter (.param) file. In the ‘Imager.zip’ folder, open ‘Intrinsic Imaging Rig Instructions.pdf.’ Follow the installation instructions in this document in order to configure two computers to acquire imaging data and run a stimulus. In addition, you will need to install the Matrox Imaging Library (the ‘light’ version is suitable). An extensive guide to using the imager and Stimulator programs, including additional capabilities of these programs, can be found at https://sites.google.com/site/iannauhaus/home/matlab-code. Here, we present a streamlined version for use in this ISI protocol. ▲ CRITICAL The stimulus and acquisition code provided is for use in MATLAB 2008 (Mathworks) on a machine with Windows XP. Check GitHub for modifications to this code for use with newer versions of MATLAB and Windows, as well as releases for Python.
Automatic Area Segmentation Code
For automatic segmentation of maps in MATLAB, use the ‘Visual Area Segmentation’ repository available from https://snlc.github.io/ISI/; for Python, use the ‘retinotopic_mapping’ package from https://github.com/zhuangjun1981/retinotopic_mapping.
Mouse surgery setup
Attach an isoflurane regulator to an oxygen tank, and run the tubing to a small mask for the mouse’s nose (Fig. 3a). Position a head frame holder on a breadboard with a heating pad (Fig. 3a).
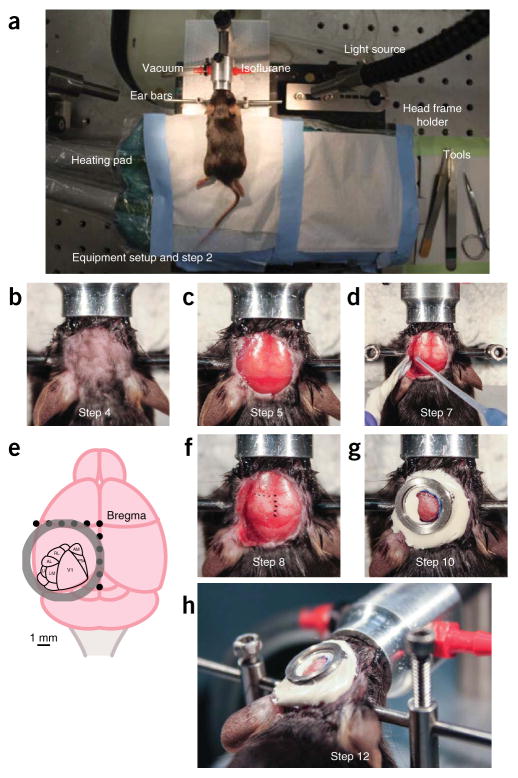
Figure 3.
Mouse head frame surgery to prepare for intrinsic imaging (Steps 1–12). (a) Overview of surgery setup (left) and head frame holder setup (right). (b–g) Surgery steps to attach head frame. (b) Hair is trimmed and lidocaine ointment is applied. (c) Skin is removed from the entire surface of the head. (d) After removing side muscles, Vetbond is carefully applied to glue the skin. (e) Proper position of the head frame on the mouse skull to cover the visual cortex. Dots on skull are 1 mm apart. (f) Dots (1 mm apart) are drawn on the skull to demonstrate proper placement of the head frame. (g) Head frame is attached with cement. (h) Side view of attached head frame, ensuring that there is no cement in the grooves. All work involving live animals was conducted according to a procedure approved by the Salk Institute IACUC. AL, anterolateral area; AM, anteromedial area; LI, laterointermediate area; LM, lateromedial area; PM, posteromedial area; RL, rostrolateral area; V1, primary visual cortex.
PROCEDURE
Mouse head frame surgery ● TIMING ~1 h
-
1|
Anesthetize the animal with isoflurane (2% in oxygen for induction, 1.5% during surgery).
▲ CRITICAL STEP All work involving live animals was conducted according to a procedure approved by the Salk Institute IACUC. Before conducting animal research, obtain the necessary approval from your institution.
-
2|
Remove the animal from isoflurane box, apply eye lubricant to the mouse’s eyes, and secure the animal in stereotax on a heat pad (Fig. 3a).
-
3|
Administer 0.1 ml carpofen/10 g mouse (5 mg/kg) as an analgesic.
-
4|
Using scissors, clippers, and/or Nair, remove all hair from behind the eyes to the back of the neck and apply lidocaine ointment to the scalp (Fig. 3b). Alternatively, liquid lidocaine may be injected under the skin. If you are using scissors, it may help to wet the hair and scalp with ethanol before hair removal.
-
5|
Make an incision down the midline with scalpel or scissors, from just behind the eyes (~1 mm above the bregma suture) to behind the lambda suture. Cut out a circle of skin with scissors to expose the entire left hemisphere and part of the right hemisphere (Fig. 3c).
▲ CRITICAL STEP The more skull that is exposed for the dental cement to adhere to, the stronger the bond between the skull and the head frame. However, exposing too much may irritate the animal. Be particularly careful near the mouse’s eyes.
-
6|
Clean the skull using a cotton applicator and use tweezers to pull away the fascia from the midline outward. Carefully remove the muscle on the side and back of the skull with a scalpel or tweezers in order to expose lateral areas. Clean away any blood or loose tissue with a Kimwipe or cotton applicator.
-
7|
Apply Vetbond to the edges of the incision to secure the skin to the skull (Fig. 3d).
▲ CRITICAL STEP Avoid getting glue near the eyes, and be careful not to glue the ears to the bars or the scalp to the anesthesia mask.
-
8|
Determine the head frame placement. If desired, approximate V1 bregma coordinates (−3.2 to −4.8 anterior/ posterior (AP), −2.7 medial/lateral (ML)) by drawing dots on the skull every millimeter from the bregma using a marker and a ruler (Fig. 3e,f).
▲ CRITICAL STEP While the mouse is in the stereotax, the investigator may want to mark additional points on the skull for the purposes of future injections, electrode placements, and so on. These marks can be made with a permanent marker, and the investigator should avoid covering them with cement.
-
9|
Mix two scoops of Metabond powder, seven drops of base, and two drops of catalyst in the mixing tray with the applicator brush. Using the brush, trace the head frame location and apply cement around the head frame location to the edges of the incision.
-
10|
Once you have built up a pedestal of cement, lay the head frame in place (Fig. 3g).
▲ CRITICAL STEP Perform this step as quickly and as carefully as possible. Taking too long to build up cement will cause a film of dried cement to form over the top, preventing good adhesion to the headframe.
▲ CRITICAL STEP Use a cooled ceramic tray to mix the cement, making it pliable for a longer period.
-
11|
Use the brush to add extra cement to fill in any holes and over the wings of the head frame.
-
12|
Before the cement is completely dry (but after it is relatively firm), use tweezers to remove any dental cement that has come up into the outer grooves of the frame (Fig. 3h). Be careful not to move the frame. At this stage, a cranial window may also be implanted, after the cement has dried completely28,29.
-
13|
Put the mouse on a heating pad/warmer in the cage until it wakes up and is active.
-
14|
Give 3 ml of ibuprofen in a water bottle for postoperation analgesia.
Preparation of the mouse for imaging ● TIMING ~20–30 min
▲ CRITICAL The surgery to affix the head frame should be completed at a minimum of 1 h before imaging, allowing the animal to fully recover. Ideally, this surgery should be done 1 d in advance, but not more than 5 d in advance, to ensure that the head frame is still securely attached to the skull, as this attachment may degrade over time.
-
15|
After the mouse has fully recovered (the mouse should be active in the cage without being hunched over or demonstrating any sign of pain; this takes at least 1 h), place the mouse in the isoflurane chamber at a 2% flow rate.
-
16|
Once the mouse is fully anesthetized (1 breath per s), position the mouse in the head frame holder (Fig. 4a). If necessary, drill away any cement blocking the head frame from fully fitting into the holder.
-
17|
Administer 0.1 ml/20 mg of chlorprothixene (1 mg/kg) via intramuscular injection. After injection, reduce isoflurane to 1.5%, and continue to reduce as time passes, turning down 0.1–0.5% every 5–10 min.
▲ CRITICAL STEP Keep the amount of anesthesia to a minimum (<0.5%) to ensure that the mouse is in the best state for imaging. It should not take longer than 20 min to prepare the mouse for imaging.
-
18|
Clear away the cement using the dental drill at low speed. After clearing the cement, the skull can be thinned for acute experiments (option A) or left completely intact (option B). Figure 4b shows an intact skull before thinning, and Figure 4c shows a skull that has been thinned:
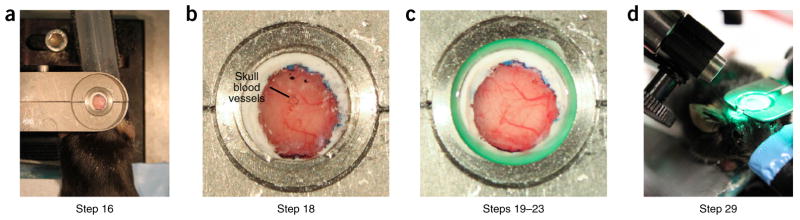
Figure 4.
Preparation of mouse for imaging (Steps 16–29). (a) Top view of mouse head frame holder. Cement and Vetbond have been cleared from the skull. (b) View of skull before thinning. Saline is on the skull to allow visualization of blood vessels. Blood vessels in the skull appear darker and more clumped than the brain-surface blood vessels. (c) View of skull after thinning, with agarose, a coverslip, and KwikCast to seal the gap between the coverslip and the head frame. Surface blood vessels are much more visible. (d) Mouse in head frame holder under camera with green light to visualize blood vessels. Note: room lights are on to allow a clear view of the setup; all imaging should be done in an enclosed space or dark room. All work involving live animals was conducted according to a procedure approved by the Salk Institute IACUC.
(A) Preparation for an intact skull (if blood vessels are sufficiently visible)
-
Drill off any remaining Vetbond on the skull so that the surface is smooth (Fig. 4b).
▲ CRITICAL STEP Be sure to drill only at low speeds and with a light touch, as the drill can become hot and cause damage or bleeding.
(B) Skull thinning to visualize blood vessels
-
Carefully and lightly drill the skull until it is flexible and just past the point of spongy, flaky bone (Fig. 4b). Blood vessels should be clearly visible when saline or ACSF is applied to the brain (Fig. 4c)30.
▲ CRITICAL STEP The skull can become overheated during thinning. Be sure to stop drilling periodically, and apply ACSF to cool the skull.
▲ CRITICAL STEP A thinned skull may become soft and covered with scar tissue after 1 week. Skull thinning should be used only for acute experiments.
? TROUBLESHOOTING
-
19|
Apply saline or ACSF to the skull and allow it to soak for several minutes to help make the bone more transparent. If you cannot see any blood vessels, the skull needs to be thinned further as in Step 17B (compare Fig. 4b,c). Repeat Steps 18 and 19 as necessary.
-
20|
Remove the ACSF from the skull surface with a Kimwipe.
-
21|
Set aside an 8-mm coverslip and then drop warm (~50–60°) agarose into the frame so that it just fills the frame.
-
22|
Quickly place an 8-mm glass coverslip over the agarose and center in the frame.
-
23|
Seal the coverslip by applying Kwik-Cast to the edges (Fig. 4c).
▲ CRITICAL STEP Make sure that the coverslip is sealed, and that there are no bubbles in the agarose. These can expand and disrupt imaging. Ensure that the coverslip is clear of debris and that the Kwik-Cast does not leak into the center of the coverslip. If it does, redo the agarose and coverslip entirely.
Setup of the mouse, light source, and camera ● TIMING ~10 min
▲ CRITICAL It may be useful to image several mice in succession, using the time of imaging to prepare the next mouse.
▲ CRITICAL Importantly, the chlorprothixene will begin to wear off after 5 h. If images are not acquired within this time, it is best to either administer a booster shot of chlorprothixene (.05 ml) or allow the mouse to recover and reimage on a subsequent day.
-
24|
When the Kwik-Cast has solidified, move the mouse to the ISI setup and position the mouse in the headframe holder so that the nose is at a 30-degree angle to the screen (Fig. 1c). The screen should be 10 cm from the perpendicular bisector of the eye (Fig. 1d).
-
25|
Lower the isoflurane concentration to 0.5–1%. The mouse should be breathing at a rate of ~1–1.5 breaths per s.
-
26|
Remove eye lubricant with a cotton applicator and apply silicone oil to the right eye by putting a generous drop of silicone oil onto the end of a fresh cotton applicator and gently rolling this over the mouse’s eye.
-
27|
Turn on the light source, camera, and television, and turn off the lights in the room, or otherwise block light from entering the setup.
-
28|
Make sure that the photodiode is properly positioned on the monitor, in the bottom left corner. Light should not be able to leak into the photodiode from places other than the screen.
-
29|
Make sure that the light source has a green filter in place to view the vasculature, and position the light guides to optimally illuminate the brain (Fig. 4d).
Acquisition of intrinsic signal images during visual stimulation ● TIMING ~1–2 h
-
30|
In MATLAB, open the acquisition and stimulation programs by running the following commands:
≫ imager <enter> ≫ Stimulator <enter>
-
31|
In the ‘Main Window’, enter your animal identification details (Supplementary Fig. 1).
-
32|
In the ‘Imager’ window, make sure that the ‘ExperimentID’ drop-down shows the correct animal number.
-
33|
In the top dropdown menu, change ‘ExperimentID’ to ‘Control Panel’. In the dropdown menu below, change ‘Stopped’ to ‘Grab Continuous’. If necessary, move the camera so that the brain is in center of the image.
? TROUBLESHOOTING
-
34|
Change zoom to +2 or +3, and find the brain in the image so that you can see the surface vasculature. The best digital zoom will depend on your lens configuration.
-
35|
Adjust the illumination intensity as necessary.
-
36|
Turning the manual focus knob, move the camera up and down until the image is clear, and then focus slightly lower so that it is just barely blurry. Focusing on the blood vessels may work all right, but focusing the camera below the blood vessels will help to smooth any blood vessel artifacts (see below).
▲ CRITICAL STEP If your camera is mounted on a micromanipulator, you can systematically determine the best depth for imaging in your setup. It is ideal to focus ~400–500 μm below the brain surface34.
-
37|
Change ‘Grab Continuous’ to ‘Stopped’.
▲ CRITICAL STEP Be careful not to touch or move the camera or change the positioning of the mouse in the head frame. This placement should be consistent across all experiments so that functional data can be aligned with images of surface vasculature taken at the end of the experiment. If movement of the camera over the course of imaging is a concern, take a snapshot by choosing ‘Grab Single,’ and saving this under your animal’s directory.
-
38|
Switch the filter in the light source to a red filter, and change the filter under the camera to a red 700-nm filter.
-
39|
Adjust the illumination light intensity for the red light. In the ‘Imager’ window, change ‘Stopped’ to ‘Adjust Illumination’ (Supplementary Fig. 1).
-
40|
Click ‘enable box’ followed by the ‘illumination ROI’ button, and then click the center of the brain on the right image. The box should be centered on the brain.
-
41|
Adjust the light intensity with the arrows on the bar until the brain is saturated (white dots in the image are saturated pixels), and then go two clicks to the left. There should be no white pixels on the brain, and the distribution should be as far to the right as possible, and there should be no pixels in the farthest right bin.
-
42|
If the illumination is irregular across the brain or insufficiently bright, adjust the light guides or the aperture on the bottom lens to optimally illuminate the brain.
-
43|
Check the illumination intensity in the ‘adjust illumination’ menu periodically after every few experiments to ensure that it is not saturated—the light source may get brighter over time and this can cause pixel saturation and ruin the signal at these points.
-
44|
Go back to ‘Stopped’.
-
45|
Make sure that there is nothing obstructing the mouse’s field of view and that the anesthesia mask is secure.
-
46|
In the ‘ParamSelect’ window, load the sample .param file. Make sure that ‘stim_time’ is 15 s and then hit ‘Send’; wait for the stimulus to be generated, and then hit ‘Play’. Watch the stimulus and make sure that it is centered on the eye and that the altitude lines become flat (parallel to the ground) at the perpendicular bisector of the mouse’s eye.
? TROUBLESHOOTING
-
47|
Click ‘select ROI’ (region of interest) and make a box around the brain inside the frame. Double-click inside the box.
If you move anything about the animal or the camera before the experiment starts, you will need to reset the ROI. If you restart MATLAB for any reason, use the ‘last ROI’ button. This is important to be able to use experiments before/after MATLAB restarts.
-
48|
In the ‘Main window’, check the eye that you will be imaging (this protocol is set up for the ‘right eye’) and, if desired, check ‘random’. If checked, the ‘random’ option will loop through the stimuli randomly; we generally leave this unchecked.
-
49|
In the ‘ParamSelect’ window, change the stimulus time to 183 s for 10 sweeps of the bar. Make sure that ‘# Repeats’ in the ‘Looper’ window is set to 1. The first row of ‘Symbol’ should say “ori”, and the ‘Value Vector’ should be [90 270] (Supplementary Fig. 1). If your TV/monitor is oriented horizontally, these values will be [0 180]. When all of these values are set, click ‘Run’ in the ‘Main Window’. click ‘Run’ in the ‘Main Window’.
-
50|
When the experiment has been completed, click ‘save’ in the window that pops up.
-
51|
Check the processed maps that appear for evidence of any structure. You should see clear evidence of V1, in the form of a small, triangular rainbow. In addition, you should see smaller rainbows lateral to V1 (similar to those in the maps in Fig. 2).
? TROUBLESHOOTING
-
52|
Advance the experiment in the ‘Main window’ by clicking the experiment number once.
? TROUBLESHOOTING
-
53|
Change the ‘altazimuth’ parameter in the ‘ParamSelect’ window to the opposite (for example, change to azimuth if you just ran an altitude experiment). Adjust the number of repeats if desired.
▲ CRITICAL STEP It is best to begin with a test experiment using one repeat to assess experimental conditions and brain state. Continue running test experiments with one repeat until coarse maps are visible. If maps are not visible after several test experiments, lower the isoflurane concentration and check that the eye is open, clear, and not obscured. Once coarse maps are visible, run a final experiment with 3–10 repeats. See discussion of repeat count in ‘Anticipated Results’.
-
54|
Hit ‘Run’ in the ‘Main window’ once you have changed the settings.
-
55|
Once you have completed data collection, take a vasculature image. On the light source, replace the red filter with the green filter. Under the camera, carefully remove the 700-nm filter and replace it with the green emission filter.
-
56|
In ‘Imager’, go from ‘Stopped’ to ‘Grab continuous’.
-
57|
Adjust the illumination intensity using the slider bar until you can see the vasculature clearly.
-
58|
Hit ‘Grab Single’ to save an image of the blood vessel anatomy using the same ROI that was used for data collection. Hit ‘yes’ and ‘ok’ for the dialog boxes that appear. It may also be useful to take a full-field image by choosing ‘full field ROI’ and then pressing ‘Grab Single’. Save this image in the main animal folder.
-
59|
Remove the coverslip and agarose and replace with Vetbond, Kwik-Cast, or Metabond to protect the skull.
-
60|
Return the mouse to a recovery cage on a heat pad. Allow ~1–2 h for recovery from sedation.
▲ CRITICAL STEP If you would like to reimage the mouse, allow 24 h for recovery from the sedative before imaging again. The agarose will dry over time, and therefore it should be removed after the experiment. A thinned skull should be checked daily for its health if the investigator wishes to conduct further experiments beyond 3–5 d.
Processing of images ● TIMING ~5 min
-
61|
The Stimulator program will automatically save raw images in a folder as .mat files, one for each frame of the stimulus. There are 1,870 frames per condition (e.g., altitude or azimuth) and repeat of the stimulus. Within the .mat file, the variable ‘im’ is the raw data image. The .analyzer file contains information about each stimulus condition, as well as sync information for each condition (Supplementary Fig. 2). These are labeled ‘syncInfo1’, ‘syncInfo2’, and so on. The ‘syncInfo’ structure contains time stamps for each frame. The code will also generate phase maps and a processed .mat file for each experiment. These processed files can be used in the following analysis.
-
62|
These processed files can be used in two different ways, using either MATLAB (option A) or Python (option B) code.
(A) For MATLAB
-
After your experiment is complete, enter the following command:
≫ getAreaBorders(‘anim’, ‘azimuth expt’, ‘altitude expt’)
This script will generate a variety of maps that are useful, including response magnitude maps, segmentation of area borders, and overlays of retinotopy, blood vessel images, and area borders. Figure 2 shows examples of maps generated with this code.
(B) For Python
-
The visual areas can also be segmented by a retinotopic_mapping python package from precalculated altitude and azimuth maps (ideally in .tif format). A detailed description of how to use this package can be found on Github (https://github.com/zhuangjun1981/retinotopic_mapping/blob/master/retinotopic_mapping/examples/retinotopic_mapping_example.ipynb).
? TROUBLESHOOTING
? TROUBLESHOOTING
Troubleshooting advice can be found in Table 1.
TABLE 1.
Troubleshooting.
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 18 | Skull bleeding | A blood vessel was hit in the skull, or blood is coming from underneath the skull | Wipe away blood and wait; in most cases, it will stop bleeding. Bleeding under the skull can take an hour, or potentially much longer, to clear. In most cases, the investigator can proceed with imaging if there is restricted bleeding. This blood will not interfere with the intrinsic signal, but it will impact clarity in the blood vessel image. To prevent bleeding underneath the skull, it is best to drill very lightly, stopping periodically to prevent overheating |
| 33 | Nothing is visible in ‘Imager’ | Imaging equipment is not set up properly | Ensure that the camera is on, the imaging path is clear, and the camera is connected to the computer |
| 46 | Stimulus is not running | Poor communication between the master and slave computers | Enter ‘≫configDisplayCom’ on the master computer. Double-check that IP addresses are correct on both the master and slave computer ‘configDisplayCom’ files. Make sure that the monitor/TV is on |
| 51 | Image on screen but no clear structure to maps | The mouse is not in the proper state | Reduce isoflurane flow, and wait for the mouse to wake up slightly such that breathing is >1 breath per s and whiskers are alert. If maps do not improve after 1 h, consider waking the mouse completely, re-administering chlorprothixene, and restarting, either on the same day or a subsequent day |
| 52 | Blood vessel artifacts | Poor camera focus | Focus the camera deeper below the skull and reimage |
| 62 (B)(i) | Segmentation does not work well | Weak or distorted maps | Increase the low-pass filtering value in the code Check plots for signal intensity to make sure that there is a clear response, at least for V1 |
● TIMING
The experimenter should allow several days to set up and troubleshoot the hardware and software components of stimulus presentation and image acquisition. It is best to assure that the system is working properly (the camera is working, stimulus plays, and so on) before beginning experiments.
Steps 1–14, surgery to affix the head frame: ~1 h (should be completed a minimum of 1 h before imaging, allowing the animal to fully recover)
Steps 15–23, preparation of the mouse for imaging: ~20–30 min
Steps 24–60, imaging: ~1–2 h for each mouse (depending on how many repeats of the stimulus are needed)
Steps 61 and 62, processing of images: ~5 min
ANTICIPATED RESULTS
With this procedure, one can acquire complete retinotopic maps of the mouse visual cortex (Fig. 2a,b,f,g). Figure 2a–e illustrates results from a typical imaging experiment, with five repeats of the altitude and azimuth stimuli. Using these maps and the segmentation code, visual area borders can be automatically segmented (Fig. 2d,e), although the analysis failed to properly segment the anteromedial area (Fig. 2d). Although it is quite easy to visualize V1 and LM, areas such as the posterior, postrhinal, and laterolateral anterior areas require many repeats and a carefully thinned skull (Fig. 2f–j). Although five repeats of the stimulus are sufficient for most visual areas, 10 or more repeats may be necessary for more difficult-to-segment areas (Fig. 2f–j).
With these maps, it is also possible to calculate additional values such as eccentricity and cortical magnification8. Successfully segmented borders overlaid on blood vessel images (Fig. 2e,j) are useful in subsequent studies in order to target higher visual areas for virus injection, calcium imaging, or electrode placement.
Supplementary Material
Acknowledgments
We thank the Callaway laboratory, especially P. Li, T. Ito, and E. Kim, for their help in developing this setup and protocol; D. Ringach (University of California, Los Angeles) for writing the initial version of the image acquisition code; and O. Odoemene (Cold Spring Harbor Laboratory) for encouraging the inception of this article. We would also like to thank the Allen Institute for Brain Science founders, Paul G. Allen and Jody Allen, for their vision, encouragement, and support. This work was supported by National Institutes of Health grants EY022577, MH063912, and EY019005, as well as grants from the Gatsby Charitable Foundation and the Rose Hills Foundation to M.E.G. A.L.J. was supported by the National Science Foundation and the Martinet Foundation.
Footnotes
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS A.L.J., I.N., M.E.G., and E.M.C. designed the experiments, developed the protocol, and wrote the manuscript. A.L.J. and M.E.G. collected and analyzed intrinsic signal imaging data. I.N., M.E.G., and A.L.J. wrote MATLAB code. J.Z. wrote Python code.
COMPETING FINANCIAL INTERESTS
The authors declare that they have no competing financial interests.
References
- 1.Grinvald A, Lieke E, Frostig RD, Gilbert CD, Wiesel TN. Functional architecture of cortex revealed by optical imaging of intrinsic signals. Nature. 1986;324:361–364. doi: 10.1038/324361a0. [DOI] [PubMed] [Google Scholar]
- 2.Malonek D, et al. Vascular imprints of neuronal activity: relationships between the dynamics of cortical blood flow, oxygenation, and volume changes following sensory stimulation. Proc Natl Acad Sci USA. 1997;94:14826–14831. doi: 10.1073/pnas.94.26.14826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Attwell D, Laughlin SB. An energy budget for signaling in the grey matter of the brain. J Cereb Blood Flow Metab. 2001;21:1133–1145. doi: 10.1097/00004647-200110000-00001. [DOI] [PubMed] [Google Scholar]
- 4.Frostig RD, Lieke EE, Ts’o DY, Grinvald A. Cortical functional architecture and local coupling between neuronal activity and the microcirculation revealed by in vivo high-resolution optical imaging of intrinsic signals. Proc Natl Acad Sci USA. 1990;87:6082–6086. doi: 10.1073/pnas.87.16.6082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shmuel A, Grinvald A. Functional organization for direction of motion and its relationship to orientation maps in cat area 18. J Neurosci. 1996;16:6945–6964. doi: 10.1523/JNEUROSCI.16-21-06945.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonhoeffer T, Grinvald A. Iso-orientation domains in cat visual cortex are arranged in pinwheel-like patterns. Nature. 1991;353:429–431. doi: 10.1038/353429a0. [DOI] [PubMed] [Google Scholar]
- 7.Ts’o DY, Frostig RD, Lieke EE, Grinvald A. Functional organization of primate visual cortex revealed by high resolution optical imaging. Science. 1990;249:417–420. doi: 10.1126/science.2165630. [DOI] [PubMed] [Google Scholar]
- 8.Garrett ME, Nauhaus I, Marshel JH, Callaway EM. Topography and areal organization of mouse visual cortex. J Neurosci. 2014;34:12587–12600. doi: 10.1523/JNEUROSCI.1124-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sereno MI, McDonald CT, Allman JM. Analysis of retinotopic maps in extrastriate cortex. Cereb Cortex. 1994;4:601–620. doi: 10.1093/cercor/4.6.601. [DOI] [PubMed] [Google Scholar]
- 10.Sereno MI, et al. Borders of multiple visual areas in humans revealed by functional magnetic resonance imaging. Science. 1995;268:889–893. doi: 10.1126/science.7754376. [DOI] [PubMed] [Google Scholar]
- 11.Wandell BA, Winawer J. Imaging retinotopic maps in the human brain. Vision Res. 2011;51:718–737. doi: 10.1016/j.visres.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sereno MI, Allman JM. The Neural Basis of Visual Function. Macmillan; 1991. pp. 160–172. [Google Scholar]
- 13.Allman JM, Kaas JH. A representation of the visual field in the caudal third of the middle temporal gyrus of the owl monkey (Aotus trivirgatus) Brain Res. 1971;31:85–105. doi: 10.1016/0006-8993(71)90635-4. [DOI] [PubMed] [Google Scholar]
- 14.Kaas JH. Extra-Striate Cortex in Primates. Vol. 12. Springer; 1997. pp. 91–119. [Google Scholar]
- 15.Huberman AD, Niell CM. What can mice tell us about how vision works? Trends Neurosci. 2011;34:464–473. doi: 10.1016/j.tins.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niell CM. Exploring the next frontier of mouse vision. Neuron. 2011;72:889–892. doi: 10.1016/j.neuron.2011.12.011. [DOI] [PubMed] [Google Scholar]
- 17.Luo L, Callaway EM, Svoboda K. Genetic dissection of neural circuits. Neuron. 2008;57:634–660. doi: 10.1016/j.neuron.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marshel JH, Garrett ME, Nauhaus I, Callaway EM. Functional specialization of seven mouse visual cortical areas. Neuron. 2011;72:1040–1054. doi: 10.1016/j.neuron.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Juavinett AL, Callaway EM. Pattern and component motion responses in mouse visual cortical areas. Curr Biol. 2015;25:1759–1764. doi: 10.1016/j.cub.2015.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glickfeld LL, Andermann ML, Bonin V, Reid RC. Cortico-cortical projections in mouse visual cortex are functionally target specific. Nat Neurosci. 2013;16:219–226. doi: 10.1038/nn.3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olcese U, Iurilli G, Medini P. Cellular and synaptic architecture of multisensory integration in the mouse neocortex. Neuron. 2013:1–15. doi: 10.1016/j.neuron.2013.06.010. [DOI] [PubMed] [Google Scholar]
- 22.Raposo D, Kaufman MT, Churchland AK. A category-free neural population supports evolving demands during decision-making. Nat Neurosci. 2014;17:1784–1792. doi: 10.1038/nn.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vanni MP, Murphy TH. Mesoscale transcranial spontaneous activity mapping in GCaMP3 transgenic mice reveals extensive reciprocal connections between areas of somatomotor cortex. J Neurosci. 2014;34:15931–15946. doi: 10.1523/JNEUROSCI.1818-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Polimeni JR, Granquist-Fraser D, Wood RJ, Schwartz EL. Physical limits to spatial resolution of optical recording: clarifying the spatial structure of cortical hypercolumns. Proc Natl Acad Sci. 2005;102:4158–4163. doi: 10.1073/pnas.0500291102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Orbach HS, Cohen LB. Optical monitoring of activity from many areas of the in vitro and in vivo salamander olfactory bulb: a new method for studying functional organization in the vertebrate central nervous system. J Neurosci. 1983;3:2251–2262. doi: 10.1523/JNEUROSCI.03-11-02251.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Das A, Gilbert CD. Long-range horizontal connections and their role in cortical reorganization revealed by optical recording of cat primary visual cortex. Nature. 1995;375:780–784. doi: 10.1038/375780a0. [DOI] [PubMed] [Google Scholar]
- 27.Kodandaramaiah SB, et al. Assembly and operation of the autopatcher for automated intracellular neural recording in vivo. Nat Protoc. 2016;11:634–654. doi: 10.1038/nprot.2016.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goldey GJ, et al. Removable cranial windows for long-term imaging in awake mice. Nat Protoc. 2014;9:2515–2538. doi: 10.1038/nprot.2014.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holtmaat A, et al. Long-term, high-resolution imaging in the mouse neocortex through a chronic cranial window. Nat Protoc. 2009;4:1128–1144. doi: 10.1038/nprot.2009.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang G, Pan F, Parkhurst CN, Grutzendler J, Gan WB. Thinned-skull cranial window technique for long-term imaging of the cortex in live mice. Nat Protoc. 2010;5:201–208. doi: 10.1038/nprot.2009.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goltstein PM, Montijn JS, Pennartz CMA. Effects of isoflurane anesthesia on ensemble patterns of Ca2+ activity in mouse V1: reduced direction selectivity independent of increased correlations in cellular activity. PLoS One. 2015;10:e0118277. doi: 10.1371/journal.pone.0118277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Villeneuve MY, Casanova C. On the use of isoflurane versus halothane in the study of visual response properties of single cells in the primary visual cortex. J Neurosci Methods. 2003;129:19–31. doi: 10.1016/s0165-0270(03)00198-5. [DOI] [PubMed] [Google Scholar]
- 33.Shtoyerman E, Arieli A, Slovin H, Vanzetta I, Grinvald A. Long-term optical imaging and spectroscopy reveal mechanisms underlying the intrinsic signal and stability of cortical maps in V1 of behaving monkeys. J Neurosci. 2000;20:8111–8121. doi: 10.1523/JNEUROSCI.20-21-08111.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kalatsky VA, Stryker MP. New paradigm for optical imaging: temporally encoded maps of intrinsic signal. Neuron. 2003;38:529–545. doi: 10.1016/s0896-6273(03)00286-1. [DOI] [PubMed] [Google Scholar]
- 35.Malonek D, Grinvald A. Interactions between electrical activity and cortical microcirculation revealed by imaging spectroscopy: implications for functional brain mapping. Science. 1996;272:551–554. doi: 10.1126/science.272.5261.551. [DOI] [PubMed] [Google Scholar]
- 36.Nauhaus I, Ringach DL. Precise alignment of micromachined electrode arrays with V1 functional maps. J Neurophysiol. 2007;97:3781–3789. doi: 10.1152/jn.00120.2007. [DOI] [PubMed] [Google Scholar]
- 37.Ratzlaff EH, Grinvald A. A tandem-lens epifluourescence macroscope: hundred-fold brightness advantage for wide field imaging. J Neurosci Methods. 1991;36:127–137. doi: 10.1016/0165-0270(91)90038-2. [DOI] [PubMed] [Google Scholar]
- 38.Silasi G, Xiao D, Vanni MP, Chen ACN, Murphy TH. Intact skull chronic windows for mesoscopic wide-field imaging in awake mice. J Neurosci Methods. 2016;267:141–149. doi: 10.1016/j.jneumeth.2016.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schuett S, Bonhoeffer T, Hübener M. Mapping retinotopic structure in mouse visual cortex with optical imaging. J Neurosci. 2002;22:6549–6559. doi: 10.1523/JNEUROSCI.22-15-06549.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bandettini PA, Jesmanowicz A, Wong EC, Hyde JS. Processing strategies for time-course data sets in functional mri of the human brain. Magn Reson Med. 1993;30:161–173. doi: 10.1002/mrm.1910300204. [DOI] [PubMed] [Google Scholar]
- 41.Engel S, Glover GH, Wandell BA. Retinotopic organization in human visual cortex and the spatial precision of functional MRI. Cereb Cortex. 1997;7:181–192. doi: 10.1093/cercor/7.2.181. [DOI] [PubMed] [Google Scholar]
- 42.Oommen BS, Stahl JS. Eye orientation during static tilts and its relationship to spontaneous head pitch in the laboratory mouse. Brain Res. 2008;1193:57–66. doi: 10.1016/j.brainres.2007.11.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.