Abstract
Whole genome duplication has played an important role in plant evolution and diversification. Sugarcane is an important crop with a complex hybrid polyploid genome, for which the process of adaptation to polyploidy is still poorly understood. In order to improve our knowledge about sugarcane genome evolution and the homo/homeologous gene expression balance, we sequenced and analyzed 27 BACs (Bacterial Artificial Chromosome) of sugarcane R570 cultivar, containing the putative single-copy genes LFY (seven haplotypes), PHYC (four haplotypes), and TOR (seven haplotypes). Comparative genomic approaches showed that these sugarcane loci presented a high degree of conservation of gene content and collinearity (synteny) with sorghum and rice orthologous regions, but were invaded by transposable elements (TE). All the homo/homeologous haplotypes of LFY, PHYC, and TOR are likely to be functional, because they are all under purifying selection (dN/dS ≪ 1). However, they were found to participate in a nonequivalently manner to the overall expression of the corresponding gene. SNPs, indels, and amino acid substitutions allowed inferring the S. officinarum or S. spontaneum origin of the TOR haplotypes, which further led to the estimation that these two sugarcane ancestral species diverged between 2.5 and 3.5 Ma. In addition, analysis of shared TE insertions in TOR haplotypes suggested that two autopolyploidization may have occurred in the lineage that gave rise to S. officinarum, after its divergence from S. spontaneum.
Keywords: sugarcane, R570, genome evolution, autopolyploidization, homo/homeologues expression
Introduction
Whole genome duplication (WGD, polyploidization) is an important driving force for the emergence of evolutionary novelties, mainly through pseudo or neo-functionalization processes of duplicated genes and consequent rewiring of regulatory networks (Ohno 1970; Lynch and Conery 2000).
Angiosperms underwent several WGD events during their evolution (paleopolyploidization; Renny-Byfield and Wendel 2014) followed by genomic reorganization (i.e., translocation, chromosome fusion or loss, insertion of transposable elements, gene loss, and transposition), which is part of a frequent process known as diploidization, that drives polyploids back to a diploid state, where chromosomes tend to present diploid behavior with disomic pairing during meiosis (Chen and Ni 2006).
Allopolyploidization (derived from interspecific cross) or autopolyploidization (same species cross) events may have been involved in plants WGDs (Comai 2005). The meiotic behavior of chromosomes is a fundamental difference between auto and allopolyploids (Tate et al. 2005). The increased similarity between autopolyploid homologous chromosomes leads to frequent multivalent and random pairings, while in allopolyploids bivalent and preferential pairing are favored depending on the divergence of the parental genomes (Chen 2007; Renny-Byfield and Wendel 2014).
Synthetic or recent allopolyploids show structural remodelation of the combined genomes and changes in gene expression (Chen and Ni 2006). Differentiation of homeologous chromosomes can overcome meiotic instabilities that generate aneuploid gametes and unviable zygotes leading to restablished fertility. Changes in gene expression can recover the homeostasis lost with the increased gene dosage and relaxation of imprinted genes (Levy and Feldman 2002; Chen 2007; Renny-Byfield and Wendel 2014). However, evidences suggest that autopolyploids experience less genome rearrangements and changes in gene expression than allopolyploids in the process of adaptation to WGD (Albertin et al. 2005; Church and Spaulding 2009; Parisod et al. 2010). Besides, many crops (potato, banana, cotton, wheat, and canola) are stable recent polyploids (neopolyploid) that have not diploidized.
Modern cultivars of sugarcane harbor two sub-genomes with different basic chromosome numbers. They are interspecific hybrids of Saccharum officinarum (2n = 8x = 80) and Saccharum spontaneum (2n = 5x − 16x = 40–128) (Grivet et al. 2004), both species formed by two or more events of autopolyploidization (D’Hont et al. 1996). Thus, sugarcane cultivars are highly polyploid presenting frequent aneuploidy.
Recently, Kim et al. (2014) suggested that (1) Saccharum and Miscanthus share an ancestral allopolyploidization event that probably led to the divergence of Saccharinae and Sorghinae sub-tribes and (2) Saccharum passed through an autopolyploidization after diverging from Miscanthus. This sequence of allo and autopolyploidization events is consistent with the occurrence of chromosomal random pairing, with some degree of preferential pairing, observed in sugarcane cultivars (Grivet et al. 1996; Hoarau et al. 2001; Jannoo et al. 2004).
Although it seems clear that sugarcane genome did not undergone a major reshaping as a consequence of polyploidization (Jannoo et al. 2007; Le Cunff et al. 2008; Wang et al. 2010; Aitken et al. 2014; de Setta et al. 2014), the polyploidization processes that shaped the Saccharum genome still need to be better defined.
Another important issue in sugarcane is whether the expression of genes is dosage-dependent or -independent, which is critical to maintain protein balance in regulatory complex (Veitia et al. 2008). Homo/homeologuous gene-specific expression was investigated for the Sugarcane Loading Stem Gene (ScLSG) and revealed that multiple homo/homeologues are expressed and exhibit different patterns of expression across tissues (Moyle and Birch 2013), which was interpreted as reflecting tissue subfunctionalization of homo/homeologues. Subfunctionalization consists in the subdivision of ancestral function and is therefore reported as an important process for copie retention, particularly of dosage-dependent genes (Chaudhary et al. 2009). Nevertheless, how gene expression is balanced in sugarcane, as well as the meiotic behavior of chromosomes are still poorly answered.
An approach to get new insights into the questions regarding genome structure and evolution in polyploids is to investigate into detail the range of polymorphism (haplotypes) at specific nuclear loci within and between species. Among the members of Andropogoneae tribe with complete genome available, sorghum (diploid) exhibits the closest synteny with sugarcane and, therefore, is the best model for sugarcane comparative genomics (Jannoo et al. 2007; Le Cunff et al. 2008; de Setta et al. 2014).
Here we sequenced homo/homeologous BACs carrying the putative single-copy genes LFY (Leafy), PHYC (Phytochrome C), and TOR Kinase (Target of Rapamycin) from the sugarcane R570 cultivar. We analyzed the microsynteny between the homo/homeologous BACs and orthologous regions of sorghum and rice to understand patterns of divergence in fine detail. Expression analyses allowed us to quantify the relative expression of homo/homeologous alleles. Since LFY, PHYC, and TOR are key genes in plant development and members of complex regulatory networks, these data shed new light on sugarcane genome evolution and function and how gene expression homeostasis is established in complex polyploids.
Materials and Methods
Identification of LFY, PHYC, and TOR Sequences in Sugarcane
In order to evaluate whether LFY, PHYC, and TOR are putative single-copy genes in sugarcane, we used Arabidopsis thaliana sequences as queries against a database developed in our laboratory containing several predict proteomes of green plants (Viridiplantae 1.0; Papini-Terzi et al. 2009) and the sugarcane ESTs database SUCEST. Redundant sequences, including splice variants (sequences differ by the presence or absence of small domains), were eliminated and the remaining sequences aligned (MAFFT; Katoh et al. 2002) to generate phylogenetic trees by maximum likelihood method (PhylML 3.0; Guindon and Gascuel 2003).
BAC Library Screening
We used specific pairs of primers (supplementary table S1, Supplementary Material online) for screenning the R570 BAC library using PCR reactions and a 3D pool as template (Adam-Blondon et al. 2005). BACs positive for PCR amplification were purified using QIAGEN® Large-Construct Kit and sequenced by 454 sequencing technology (Roche). BAC assembly was conducted as described in de Setta et al. (2014).
Haplotype Determination
We compared the sequence of all selected homo/homeologous BACs to identify possible redundant sequences derived from the same sugarcane haplotype. MAFFT online alignment tool (http://mafft.cbrc.jp/alignment/software/; last accessed December 28, 2016) and GEvo (Genome Evolution Analysis—https://genomevolution.org/CoGe/GEvo.pl.; last accessed December 28, 2016), from online CoGe platform (Comparative Genomics Platform), were used for alignment.
When the entire sequences of two BACs had total overlap with nucleotide identity above 99.8%, they were considered to be the same haplotype and just one BAC was considered in subsequent analyses. When BACs did not overlap completely but had more than 99.8% of identity over a minimum overlap window of 20 kb in their boarders, they were considered as parts of the same haplotype. In this case, the unaligned sequences were incorporated to one of the BACs giving rise to a new larger single sequence haplotype named “BAC 1 + 2” (e.g., BAC 202G24 + 013O24). In both cases, the potential SNPs (Single Nucleotide Polymorphisms) (<0.2%) were preserved and may represent recent alleles or technical artifacts.
BAC Sequence Annotation and Comparisons
Automatic BAC annotation was carried out using the GNPAnnot Community Annotation System (Guignon et al. 2012) available on the South Green Bioinformatics platform (http://www.southgreen.fr; last accessed December 28, 2016). Artemis: Genome Browser and Annotation Tool (Rutherford et al. 2000; Berriman and Rutherford 2003) and ACT: Artemis Comparison Tool (Carver et al. 2005) were used to manually correct the annotation of genes and transposable elements (TEs) as described by Garsmeur et al. (2011). Sorghum, rice, and maize genome annotations (Phytozome v5.0; Goodstein et al. 2012), as well as CENSOR online software tool (Kohany et al. 2006) and NCBI (National Center for Biotechnology Information) databases were used to manually annotate genes and TEs, respectively. BACs sequences and annotatios were deposited in NCBI under GenBank accession numbers KF184957, KF184754, KF184931, and KX608891–KX608914 (supplementary table S2, Supplementary Material online). BACs graphic representations were made using GENOGRAPH (not published) developed by Olivier Garsmeur (CIRAD, Structure et Evolution des Génomes, Montpellier, France).
Haplotype Origin
To identify the origin of haplotypes, based on SNPs, around 25 million 100 bp paired-end reads of S. officinarum and 23 million 100 bp paired-end reads of S. spontaneum (Grativol et al. 2014) were mapped against LFY, PHYC, and TOR coding sequences using Bowtie 2 software with default parameters in sensitive mode (Langmead and Salzberg 2012). The resulting ∼8200 and ∼7000 mapped reads of S. officinarum and S. spontaneum, respectively, were analyzed by QualitySNPng (Nijveen et al. 2013) aiming to identify specific SNPs of S. officinarum and S. spontaneum that are present in R570 haplotypes of LFY, PHYC, and TOR genes, including exons and introns.
Indels (insertions/deletions) and amino acids polymorphisms shared by 043C15 and 156D23 TOR haplotypes were identified by alignments using MAFFT online tool and MEGA 5.05 software. We sequenced TOR 27th and 43th exons, which harbor these polymorphisms, from one accession of S. officinarum and one of S. spontaneum. The exons were amplified using specific primers, cloned in pGEM®-T Easy vector (Promega) and sequenced by Sanger methodology. Six clones of each accession were sequenced, obtaining at least four good quality sequences of each exon for both species.
Evolutionary Analysis of Genes
Phylogenetic trees were built by neighbor-joining method (Saitou and Nei 1987) with nucleotide distances calculated with Jukes–Cantor model (Jukes and Cantor 1969), in MEGA 5.05 software (Tamura et al. 2011). Different models of distance estimation, like p-distance, Kimura 2-parameter (Kimura 1980) and Tajima–Nei distance (Tajima and Nei 1984), were tested to build the trees and the resulting topologies were highly consistent.
The number of nonsynonymous substitutions per nonsynonymous site (dN) and the number of synonymous substitutions per synonymous site (dS) were calculated for LFY, PHYC, and TOR genes using Nei–Gojobori method with Jukes–Cantor correction, implemented in MEGA 5.05 software. Divergence times were estimated by T = d/2r, were “T” corresponds to divergence time, “d” corresponds to the number of substitutions between two sequences and “r” is the substitution rate. When estimating divergence times of alleles (gene haplotypes), “d” was replaced by dS and “r” by the widely used monocot substitution rate 6.5 × 10− 9 (Gaut et al. 1996) substitutions per site per year or the specific synonymous substitution rate of each gene.
Synonymous substitution rates specific for LFY, PHYC, and TOR genes were estimated using the average divergence time between sorghum and sugarcane (7 Ma) (Al-Janabi et al. 1994; Jannoo et al. 2007; Kim et al. 2014) to calibrate the molecular clock.
Haplotype-Specific Expression
Multiple sequence alignment was performed using LFY, PHYC, and TOR haplotypes to estimate the allele dosage for SNP loci using CLUSTALW version 2.0 (Larkin et al. 2007). To estimate the relative expression of alleles, about 52 million of RNA-Seq short reads (72 bp paired-end reads) from sugarcane cultivar R570 were mapped against LFY (011C13), PHYC (038J02), and TOR (007C22) haplotypes using Bowtie2 software (Langmead and Salzberg 2012). SNP detection was performed using Freebayes software 0.9.5 (Garrison and Marth 2012), considering 10 reads per SNP position, minimum of two reads supporting the variant SNP, minimum variant haplotype frequency of 0.02 and quality 30 at the central base. The genomic dosage and the relative expression level of each SNP were compared by exact binomial test, while Pearson’s correlation test was performed to evaluate the global expression level of SNPs and the haplotypes dosages, estimating R and P-value. The RNA-Seq data from this study have been deposited in the NCBI Sequence Read Archive (SRA) under accession number SRX1822635.
Evolutionary Analysis of TEs
Divergence times of insertions (TEs or other undefined insertions) shared by different BACs were estimated by T = d/2r. The entire insertions sequences were used to calculate the pairwise distances (d) and “r” was replaced by the mutation rate 1.3 × 10− 8 mutations per site per year as proposed by Ma and Bennetzen (2004). The distance “d” was determined using Kimura two-parameters model, available in MEGA 5.05 software (Tamura et al. 2011). Estimation of insertion ages of LTR retrotransposons was based on the accumulated number of substitutions between the two LTRs (d) (SanMiguel et al. 1998), using the mutation rate of 1.3 × 10− 8 mutations per site per year.
Results
BAC Clones Selection and Annotation
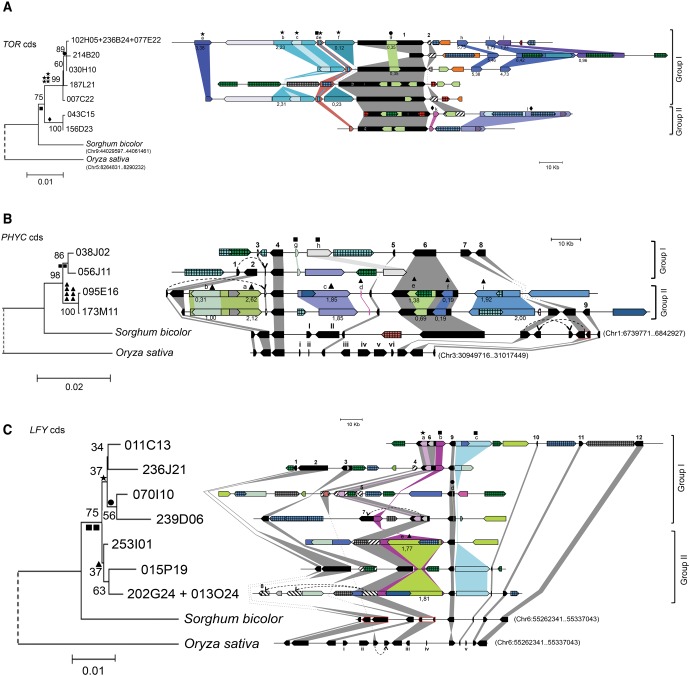
Genomic regions of Leafy (LFY), Phytochrome C (PHYC), and Target of rapamycin kinase (TOR) genes were chosen to analysis because they play an important role on plant development and, therefore, their expression must be well regulated to maintain the stoichiometry of the protein regulatory complexes in which they are involved. Most importantly, LFY, PHYC, and TOR are single-copy genes in several grasses and dicots (Mathews et al. 1995; Mathews and Sharrock 1996; Howe et al. 1998; Chujo et al. 2003; Wullschleger et al. 2006; Hamès et al. 2008; Moyroud et al. 2009; Agredano-Moreno et al. 2007; Robaglia et al. 2012). That is especially true in the sorghum lineage, which has not experienced any WGD because the paleoduplication common to all grasses around 70 Ma (Paterson et al. 2009). Blast searches in sugarcane ESTs database (SUCEST) did not detect redundancy, indicating that these genes are also likely to be single-copy in sugarcane. This precaution ensures that BACs harboring these genes represent indeed homo/homeologous regions and not duplicated regions containing paralogous genes in sugarcane genome.
The strategy of screening by 3D pool PCR of the R570 BAC library, using specific pairs of primers for each gene, led to the isolation of 10 positive BACs for LFY, seven for PHYC, and 10 for TOR. All of them were sequenced and assembled. Up to 12 different homo/homeologous BACs would be expected for the three genes based on described R570 genome structure (D’Hont et al. 1998; Ha et al. 1999).
Redundancy was found among the 27 sequenced BACs and those with more than 99.8% of nucleotide identity over their alignment were considered to represent the same (if totally overlapped) or parts of the same haplotype (if partially overlapped) (details see Materials and Methods). After merging the partially overlapped BACs, we obtained seven different haplotypes, i.e., homo/homeologous BACs, for LFY genomic region, four for PHYC and seven for TOR. Considering that sugarcane varieties (including R570) are likely to carry between six to 14 homo/homeologous loci with eight to ten being the most expected (D’Hont et al. 1996; Garcia et al. 2013), the uncovered seven haplotypes for TOR and LFY loci are therefore within the range of expected number of homo/homeologous loci. Moreover, based on SNPs calling from R570 RNAseq data, for the 2 kb sequence common to the seven TOR haplotypes (see hereafter), 26 SNPs (∼90%) matched one of the seven haplotypes and only three (∼10%) did not (data not shown). Thus, based on the proportion of SNPs matching BACs haplotypes (∼3.7 SNPs per BAC), it may be concluded that we most likely missed one TOR haplotype.
Two of the TOR haplotypes do not carry the complete gene sequence, BACs 043C15 and 214B20 have 69.7% and 27.2% of TOR coding sequence, respectively. This is not unexpected, because the gene has 58 exons, reaching more than 30 000 nucleotides (nt) in closely related species such as Brachypodium distachyon and Sorghum bicolor (fig. 1A and supplementary fig. S1 C, Supplementary Material online).
Fig. 1.—
Phylogenetic analysis and schematic synteny of TOR (A), PHYC (B), and LFY (C) genomic regions for sugarcane haplotypes (homo/homeologous), sorghum and rice. TOR BACs have just one gene and a noncollinear pseudogene. Phylogenies were generated by neighbor-joining analysis of LFY, PHYC, and TOR nucleotide coding sequences (cds) alignments. The scale bar represents the relative genetic distance (number of substitutions per site). Numbers close to the branches are bootstrap values. Genes are represented by black arrows and pseudogenes by textured arrows. Collinear genes are linked by gray strips. Transposable elements (TEs) are represented by colored arrows: blue arrows are gypsy-like TEs; green arrows are copia-like TEs; red and orange arrows are nonLTR TEs; pink arrows are DNA transposons; gray and white arrows are undefined insertions. Shades of the same color represent similarity between TEs or undefined insertions. Textured and colored arrows are TEs and undefined insertions that have no similarity with any other in the genomic region. Shared TEs are linked by strips of the TE color and identified from “a” to “l” according to supplementary table S4, Supplementary Material online. Predicted insertion times of shared LTR-retrotransposons are indicated below the respective arrows. Predicted insertion events of most shared TEs are reported in the phylogenetic tree using symbols (black circles, stars, squares, diamonds and triangles). Numbers 1–12 indicate sugarcane annotated genes (table 1); I and II indicate sorghum noncollinear genes; i–vi indicate rice noncollinear genes. LFY is gene number 9, PHYC is number 4 and TOR is number 1. Dashed curved arrows connect duplicated genes. Red lines outline sorghum genes edited into one gene, based on the structure of their orthologous counterparts in other grasses.
Annotation of LFY, PHYC, and TOR BACs consisted in identifying and annotate the genes by comparison to other grasses predicted proteomes and characterize the TEs based on structure patterns (presence of LTRs and IRs) and similarity with repetitive elements databases (CENSOR and NCBI) (fig. 1).
PHYC and TOR genes did not show differences in exon number and length among grasses. However, TOR haplotype from BAC 043C15 has a frameshift mutation in exon 35, probably a recent mutation or an artifact generated during BAC replications in Escherichia coli, because the mutation was confirmed to be present in the BAC clone by ressequencing using Sanger method. LFY from S. bicolor has one additional exon compared with other grasses (supplementary fig. S1, Supplementary Material online) and Arabidopsis thaliana. LFY is the only one of these genes showing exon length variation between haplotypes (genes versions) caused by indels of 6–18 nt (supplementary fig. S2, Supplementary Material online).
Overall, the homo/homeologous BACs showed great heterogeneity regarding the number of TEs and, to a lower extent, the number of protein-coding genes (fig. 1 and supplementary table S3, Supplementary Material online). LTR retroelements (RTEs) of Gypsy-like family were the most abundant TE observed in PHYC and TOR surrounding regions (supplementary table S3, Supplementary Material online), comprising about 21% and 30% respectively, of the combined BAC length. Copia-like LTR RTEs, which comprises 10% and 14% of PHYC and TOR total BAC length, was the most abundant in LFY region (21%).
Comparative Genomics between Sugarcane, Sorghum, and Rice
The arrangement of genes in the sugarcane BACs and their corresponding orthologous regions in sorghum and rice were found, as expected (Jannoo et al. 2007; Wang et al. 2010; Garsmeur et al. 2011), to be highly collinear (table 1). TOR was the only gene in its BACs together with a noncollinear pseudogene, thus synteny analysis with sorghum (Chr9:44029597.44061461) and rice (Chr5:8264831.8290232) was impossible.
Table 1.
Annotated Genes in Sugarcane BACs and Their Orthologous Relationships with Sorghum and Rice Genes
| Genomic Region | Gene No. | Functional Annotation | BLASTX E-Value | Orthologous Locus in Sorghum | Orthologous Locus in Rice |
|---|---|---|---|---|---|
| LFY | 1 | Brachypodium distachyon COBW domain-containing protein 1-like | 0.0E+00 | Sb06g027386 | Os04g51100 |
| 2 | Oryza sativa Indica Group hypothetical protein OsI_17248 | 0.0E+00 | Sb06g027382+ Sb06g027384c | Os04g51090 | |
| 3 | Zea mays scramblase family protein | 0.0E+00 | Sb06g027380 | Os04g51080 | |
| 4 | Zea mays WAK53a–OsWAK receptor-like protein kinase precursor–Pseudogene | 0.0E+00 | Sb01g013060 | Os04g51040 and Os04g51050d | |
| 5a | Zea mays wall-associated receptor kinase 1-like isoform X2–Pseudogene | – | – | – | |
| 6 | Brachypodium distachyon wall-associated receptor kinase-like 17-like | 0.0E+00 | Sb06g027350+ Sb06g027360c | Os04g51030 | |
| 7b | Brachypodium distachyon wall-associated receptor kinase-like 17-like | – | – | – | |
| 8b | Brachypodium distachyon wall-associated receptor kinase-like 17-like - Pseudogene | – | – | – | |
| 9 | Zea mays floricaula/leafy-like 2 | 5.0E−171 | Sb06g027340 | Os04g51000 | |
| 10 | Zea mays 60S ribosomal protein L12 | 1.0E−108 | Sb06g027330 | Os04g50990 | |
| 11 | Zea mays seed specific protein Bn15D1B | 6.0E54− | Sb06g027320 | Os04g50970 | |
| 12 | Sorghum bicolor hypothetical protein | 0.0E+00 | Sb06g027315 | Os04g50960 | |
| PHYC | 1 | Hordeum vulgare voltage-dependent outwardly rectifying plasma membrane K+ channel KCO1/TPK1 | 0.0E+00 | Sb01g007830 | Os03g54100 |
| 2 | Zea mays meiotic recombination protein SPO11 | 0.0E+00 | Sb01g007840 | Os03g54091 | |
| 3b | Hordeum vulgare voltage-dependent outwardly rectifying plasma membrane K+ channel KCO1/TPK1–Pseudogene | – | – | – | |
| 4 | Zea mays phytochrome C1 apoprotein | 0.0E+00 | Sb01g007850 | Os03g54084 | |
| 5 | Sorghum bicolor hypothetical protein | 3.0E−27 | Sb01g007870 | Os03g54050 | |
| 6 | Eulaliopsis binata embryonic flower 2 protein | 0.0E+00 | Sb01g007878 | Os09g13630 | |
| 7 | Zea mays glutathione transporter | 0.0E+00 | Sb01g007880 and Sb01g007900d | Os03g54000 | |
| 8 | Oryza sativa Japonica Group hypothetical protein | 0.0E+00 | Sb01g007890 and Sb01g007910+ Sb01g007920c, d | Os03g53990 | |
| 9 | Sorghum bicolor hypothetical protein | 8.0E−69 | Sb01g007930 | Os03g53980 | |
| TOR | 1 | Setaria italica predicted DNA primase small subunit-like–Pseudogene | – | – | – |
| 2 | Setaria italica predicted serine/threonine-protein kinase TOR-like | 0.00E+00 | Sb09g017790 | Os05g14550 |
The pseudogene does not have orthologous counterpart in sorghum and rice genomic region.
Duplicated gene.
Sorghum genes edited into one gene based on the structure of their orthologous counterparts in rice (Oryza sativa), maize (Zea mays), and Brachypodium (Brachypodium distachyon). These loci were named Sb06g027350 + Sb06g027360, Sb06g027382 + Sb06g027384, and Sb01g007910 + Sb01g007920.
The predicted gene in sugarcane has orthology with two distinct genes in the related species (fig. 1).
PHYC genomic region is highly conserved between sugarcane, sorghum (Chr1:6739771.6842927), and rice (Chr3:30949716.31017449). However, the orthologous regions of sorghum and rice have a set of exclusive genes (fig. 1B , genes I–II and i–vi for sorghum and rice, respectively). For the LFY genomic region, all the eight annotated sorghum genes (Chr6:55262341.55337043) have collinear orthologous in sugarcane, while rice (Chr4:30167235.30267397) has five (i–v—fig. 1C ) noncollinear genes with sorghum and sugarcane. Besides the relative orientation of LFY in rice is inverted (gene number 9 in fig. 1C ).
Evolutionary Analysis
Phylogenetic trees generated from aligned nucleotide coding sequences (cds) of TOR haplotypes (homo/homeologous TOR genes of sugarcane) allowed organizing TOR-containing BACs in two distinct groups: Group I with five haplotypes (102H05 + 236B24 + 077E22, 214B20, 030H10, 187L21, and 007C22) and Group II with two (043C15 and156D23) (fig. 1A ).
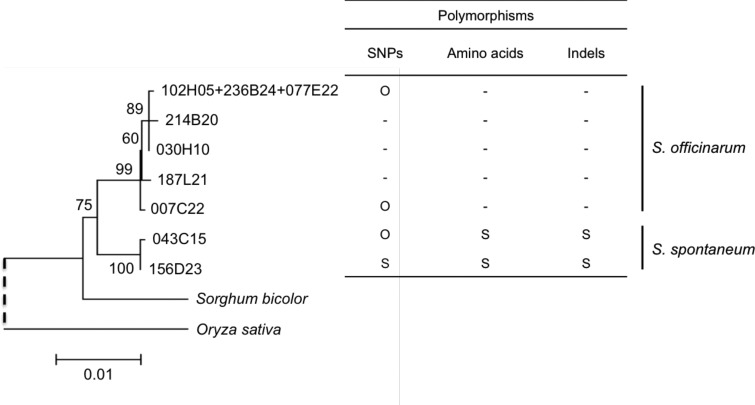
Most of R570 genome derives from S. officinarum (80-90%) with a small portion from S. spontaneum (10–20%) (Cuadrado et al. 2004; D’Hont 2005; Piperidis et al. 2010). Based on this imbalance, we infer that Group I most likely contains haplotypes originated from S. officinarum and Group II from S. spontaneum (fig. 1).
To obtain additional evidence about haplotypes origin, we used gDNA Illumina reads from S. officinarum and S. spontaneum (Grativol et al. 2014) to identify TOR species-specific single nucleotide polymorphisms (SNPs) present in R570 haplotypes. This approach linked the haplotypes 007C22, 214B20, 102H05 + 236B24 + 077E22 (Group I), and 043C15 (Group II) to S. officinarum and the haplotype 156D23 (Group II) to S. spontaneum (supplementary table S5, Supplementary Material online). Additional polymorphisms between haplotypes of Group I and II (fig. 1A ), consisting of one indel and three amino acid changes in exons 27 and 43, were also explored. We amplified and sequenced these exons from one S. officinarum and one S. spontaneum accession/genotype and discriminate the 043C15 and 156D23 haplotypes as derived from S. spontaneum (supplementary fig. S3, Supplementary Material online). Despite the ambiguity about 043C15 origin in terms of these polymorphisms (i.e., one SNP from S. officinarum vs. one indel and two amino acids from S. spontaneum) the phylogenetic analysis clearly suggests that 043C15 and 156D23 originated from the same ancestor, because they are very close related with minimal differences in TOR cds (fig. 1). Thus, the evidences presented here indicate that haplotypes 043C15 and 156D23 likely come from S. spontaneum (fig. 2) and enhance the idea that the two phylogenetic groups represent the ancestor species. LFY and PHYC phylogenetic trees also show two clear groups (fig. 1B and C ). Based on species-specific SNPs in LFY and PHYC genes, the LFY haplotype 239D06 from Group I and PHYC haplotype 056J11 from Group I possibly originated from S. officinarum (fig. 1B and C and supplementary table S5, Supplementary Material online). Considering the phylogeny topology, we suggest Group I from LFY and PHYC may represent haplotypes derived from S. officinarum (fig. 1B and C ).
Fig. 2.—
Origin of TOR haplotypes based on shared insertions and sequence polymorphisms consistent with tree topology. “O” indicates the evidence for S. officinarum origin, “S”, S. spontaneum origin and “-” undetermined origin (for details see text, supplementary fig. S3 and table S5, Supplementary Material online).
To further characterize how LFY, PHYC, and TOR haplotypes are evolving in sugarcane, synonymous substitutions per synonymous site (dS) were calculated by pairwise comparison between sugarcane haplotypes and between sugarcane haplotypes and their sorghum and rice orthologues. The haplotypes of each gene, LFY, PHYC, and TOR, have similar dS values when comparing to the respective orthologues of sorghum or rice, indicating that all haplotypes are evolving homogeneously (table 2). However, dS values vary between genes, which indicate these genes are evolving at different rates. These rates are not proportionally maintained when comparing dS values between sugarcane and sorghum and sugarcane and rice, indicating gene-specific variation in rates of evolution among these grass lineages (table 2).
Table 2.
Estimated dS, dN, and dN/dS for LFY, PHYC, and TOR Genes
| Sugarcane × Sorghum |
Sugarcane × Rice |
||||||
|---|---|---|---|---|---|---|---|
| Gene | BACs | dS | dN | dN/dSa | dS | dN | dN/dSa |
| TOR | 007C22 | 0.064 | 0.007 | 0.108 | 0.545 | 0.033 | 0.060 |
| 030H10 | 0.062 | 0.008 | 0.124 | 0.551 | 0.033 | 0.060 | |
| 102H05 | 0.061 | 0.008 | 0.126 | 0.556 | 0.033 | 0.060 | |
| 187L21 | 0.062 | 0.007 | 0.116 | 0.554 | 0.033 | 0.060 | |
| 043C15 | 0.070 | 0.005 | 0.075 | 0.568 | 0.031 | 0.054 | |
| 156D23 | 0.069 | 0.005 | 0.071 | 0.569 | 0.030 | 0.053 | |
| Average | 0.065 | 0.007 | 0.103 | 0.557 | 0.032 | 0.058 | |
| σ | 0.004 | 0.001 | 0.022 | 0.009 | 0.001 | 0.003 | |
| PHYC | 038J02 | 0.066 | 0.014 | 0.206 | 0.414 | 0.056 | 0.136 |
| 056J11 | 0.070 | 0.013 | 0.183 | 0.418 | 0.055 | 0.132 | |
| 095E16 | 0.077 | 0.014 | 0.187 | 0.414 | 0.056 | 0.136 | |
| 173M11 | 0.075 | 0.014 | 0.190 | 0.414 | 0.056 | 0.136 | |
| Average | 0.072 | 0.014 | 0.192 | 0.415 | 0.056 | 0.135 | |
| σ | 0.004 | 0.000 | 0.009 | 0.002 | 0.000 | 0.002 | |
| LFY | 011C13 | 0.087 | 0.008 | 0.097 | 0.290 | 0.072 | 0.249 |
| 015P19 | 0.096 | 0.012 | 0.123 | 0.302 | 0.073 | 0.240 | |
| 070I10 | 0.093 | 0.009 | 0.101 | 0.302 | 0.075 | 0.249 | |
| 202G24 | 0.089 | 0.012 | 0.132 | 0.296 | 0.073 | 0.245 | |
| 239D06 | 0.095 | 0.013 | 0.135 | 0.308 | 0.073 | 0.236 | |
| 236J21 | 0.085 | 0.013 | 0.158 | 0.285 | 0.076 | 0.267 | |
| 253I01 | 0.076 | 0.010 | 0.127 | 0.287 | 0.068 | 0.235 | |
| Average | 0.089 | 0.011 | 0.125 | 0.296 | 0.073 | 0.246 | |
| σ | 0.006 | 0.002 | 0.019 | 0.008 | 0.002 | 0.010 | |
All dN/dS values are statistically significant for purifying selection (dN/dS ≪ 1; Z-test P-value = 0).
Based on dS values and using the substitution rates calculated for Adh (Gaut et al. 1996) and for TOR, the divergence time estimated between TOR haplotypes probably derived from S. spontaneum and S. officinarum is between 2.5 and 3.5 Ma (supplementary table S6 a and b, Supplementary Material online).
Furthermore, all haplotypes of the three genes are under purifying selection (dN/dS ≪ 1; Z-test P-value = 0.0) (table 2), suggesting that all of them are functional and possibly expressed. This latter hypothese was further investigated.
Expression Analysis
In order to evaluate if all haplotypes of LFY, PHYC and TOR genes are expressed, we quantified the expression of the homo/homeologous copies in RNA-seq data from R570 leaves (unpublished data) based on mapped read counts of discriminatory SNPs previously identified.
Analysis of 27 SNPs present among TOR haplotypes revealed a global high correlation of SNP genomic dosage and their relative expression level (R 2 = 0.8857; P-value < 0.01), suggesting that no gross preferential expression pattern is prevailing. Closer examination of TOR double dose SNPs revealed that all of them correspond to the two S. spontaneum haplotypes, which indicates that haplotypes of S. officinarum and S. spontaneum origin are expressed (table 3 and supplementary table S7, Supplementary Material online). Yet, the single dose SNP from 007C22 TOR haplotype of S. officinarum origin was not found expressed (SNP 7356; table 3 and supplementary table S7, Supplementary Material online) as well as the single dose SNP from 043C15 haplotype of S. spontaneum (SNP 6315; table 3 and supplementary table S7, Supplementary Material online) that is barely expressed. Thus, we can conclude that not all haplotypes of TOR are equally expressed in R570 leaves. Similar analysis was performed with 33 SNPs of PHYC and no correlation was observed between SNP genomic dosage and expression (R 2 = 0.0023; P-value = 0.7007). In fact, 173M11 and 056J11 haplotypes are barely expressed in R570 leaves (SNPs 2619, 2961, 3207, and 3353; table 3 and supplementary table S7, Supplementary Material online). For LFY there were not enough mapped reads to perform the expression analysis, probably because its expression is mainly restricted to floral tissues during development.
Table 3.
Correlation between SNP Genomic Dosage and Relative Expression Level of TOR and PHYC Haplotypes
| Haplotype dosage (Genomic data) |
Read counts RNA-seq |
RNA-seq depth (# mapped reads) | Haplotype relative dosage (gDNA) |
Haplotype relative expression (mRNA) |
Exact binomial test p-value | Haplotypes with alternative SNP |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | SNP position | Main haplotype | Alternative haplotype | Main haplotype | Alternative haplotype | Main haplotype | Alternative haplotype | Main haplotype | Alternative haplotype | Main haplotype | Alternative haplotype | 030H10 | 214B20 | 187L21 | 102H05 | 007C22 | 043C15 | 156D23 | ||
| TOR | 5463 | C | T | 6 | 1 | 34 | 6 | 40 | 0.86 | 0.14 | 0.85 | 0.15 | 0.823 | X | ||||||
| 5887 | G | A | 5 | 2 | 48 | 18 | 66 | 0.71 | 0.29 | 0.73 | 0.27 | 0.892 | X | X | ||||||
| 6315 | G | A | 6 | 1 | 30 | 1 | 31 | 0.86 | 0.14 | 0.97 | 0.03 | 0.117 | X | |||||||
| 6604 | A | T | 6 | 1 | 65 | 9 | 74 | 0.86 | 0.14 | 0.88 | 0.12 | 0.740 | X | |||||||
| 6646 | G | T | 5 | 2 | 44 | 15 | 59 | 0.71 | 0.29 | 0.75 | 0.25 | 0.667 | X | X | ||||||
| 6924 | C | T | 6 | 1 | 96 | 11 | 107 | 0.86 | 0.14 | 0.90 | 0.10 | 0.271 | X | |||||||
| 7295 | T | C | 4 | 3 | 102 | 67 | 169 | 0.57 | 0.43 | 0.60 | 0.40 | 0.437 | X | X | X | |||||
| 7356 | C | T | 6 | 1 | 216 | 0 | 216 | 0.86 | 0.14 | 1.00 | 0.00 | 7.84E-15 | X | |||||||
| 7380 | T | C | 4 | 3 | 117 | 58 | 175 | 0.57 | 0.43 | 0.67 | 0.33 | 0.009 | X | X | X | |||||
| Haplotype dosage (Genomic data) |
Read counts RNA-seq |
RNA-seq depth (# mapped reads) | Haplotype relative dosage (gDNA) |
Haplotype relative expression (mRNA) |
Exact binomial test p-value | Haplotypes with alternative SNP |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | SNP position | Main haplotype | Alternative haplotype | Main haplotype | Alternative haplotype | Main haplotype | Alternative haplotype | Main haplotype | Alternative haplotype | Main haplotype | Alternative haplotype | 095E16 | 173M11 | 038J02 | 056J11 | ||
| PHYC | 2484 | G | T | 2 | 2 | 30 | 24 | 54 | 0.50 | 0.50 | 0.56 | 0.44 | 0.497 | X | X | ||
| 2531 | C | T | 3 | 1 | 52 | 15 | 67 | 0.75 | 0.25 | 0.78 | 0.22 | 0.675 | X | ||||
| 2619 | T | C | 3 | 1 | 99 | 0 | 99 | 0.75 | 0.25 | 1.00 | 0.00 | 7.48E-13 | X | ||||
| 2871 | G | A | 2 | 2 | 62 | 64 | 126 | 0.50 | 0.50 | 0.49 | 0.51 | 0.929 | X | X | |||
| 2961 | A | G | 3 | 1 | 139 | 3 | 142 | 0.75 | 0.25 | 0.98 | 0.02 | 6.18E-14 | X | ||||
| 3207 | G | A | 3 | 1 | 86 | 1 | 87 | 0.75 | 0.25 | 0.99 | 0.01 | 9.03E-10 | X | ||||
| 3353 | T | C | 3 | 1 | 110 | 2 | 112 | 0.75 | 0.25 | 0.98 | 0.02 | 1.48E-11 | X | ||||
Note.—Gray filled cells indicate statistically significant correlation between the genomic dosage of SNP loci and the SNP RNA-seq frequency (P-value < 0.05, Exact Binomial Test). “X” indicates the haplotypes containing the alternative SNP.
Together these data indicate that in leaves the expression of TOR and PHYC genes involve differential expression of the haplotypes. Although the mechanism accounting for these differences in expression is unknown, we can speculate that sequence variation in promoter regions and/or epigenetic silencing might be responsible for the low or nonexpression of some haplotypes.
Evaluation of PHYC and TOR putative cis-regulatory sequences (1 kb upstream start codon) showed that sequence conservation among all haplotypes is limited to ∼600 bp and ∼460 bp, respectively, and then is disrupted by TE and other insertions (supplementary fig. S4, Supplementary Material online). This pattern of conservation would be compatible with some similarity of expression among haplotypes, while TE insertions could promote diversification of expression.
Analysis of Shared Insertions between BACs
For the three genomic regions analyzed BACs that share collinear insertions were found (i.e., TEs or other undefined sequences inserted in the exact same position in different BACs). All collinear insertions were found to be highly conserved, presenting more than 93% of identity. These shared insertions may represent footprints of WGD (Whole Genome Duplication—defined in this work as synonym of autopolyploidization) of S. officinarum or S. spontaneum genome evolution and might help reconstruct the evolutionary history of the haplotypes that coexist in modern sugarcane genome.
The divergence time (T = d/2r) of these shared insertions were estimate by comparing their entire sequences and applying a mutation rate of 1.3 × 10− 8 substitutions per site per year (Ma and Bennetzen 2004), which should, at least for some insertions, define the approximate moment when genome duplication occurred. The insertions are shared between 2–5 homo/homeologous genomic regions and all of them, except two (TE “d”, in TOR genomic region, and TE “b”, in LFY genomic region; fig. 1A and C ), diverged less than 2.6 Ma (supplementary table S4, Supplementary Material online) suggesting these haplotypes may have diverged after the separation of S. officinarum and S. spontaneum (2.5–3.5 Ma). Besides, the insertion shared by the most likely S. officinarum and S. spontaneum haplotypes of TOR region (TE “d”, fig. 1A and supplementary table S4, Supplementary Material online) diverged around 3.2 Ma, which is close to the estimated time of S. officinarum and S. spontaneum separation.
For a RTE inserted into the 25th intron of TOR gene in BACs 030H10 and 102H05 + 236B24 + 077E22 (TE “g”, fig. 1A and supplementary table S4, Supplementary Material online) and an uncharacterized small insertion (∼500 bp) in the second intron of LFY gene, in BACs 070I10 and 239D06 (TE “d”, fig. 1C and supplementary table S4 and fig. S5, Supplementary Material online), we designed specific primers (supplementary table S8 and fig. S6, Supplementary Material online) to check for their presence in species of the “Saccharum complex” and therefore evaluate their origin.
The amplifications confirmed that the “d” insertion shared by LFY BACs 070I10 and 239D06 and the insertion “g” shared by TOR BACs 030H10 and 102H05 + 236B24 + 077E22 are present only in S. officinarum and S. robustum (the wild ancestor of S. officinarum) accessions (supplementary fig. S7, Supplementary Material online), supporting the possibility that these insertions took place after the separation of S. officinarum and S. spontaneum. These results indicate that at least one WGD of S. officinarum lineage happened after divergence of S. officinarum and S. spontaneum.
Discussion
The evolutionary changes in polyploid genomes that allow adaptation and structural stability enabling the success of these organisms are broad and still unclear. A consensus is that both structural and regulatory changes in polyploid are highly complex and species-specific (Chen and Ni 2006; Chen 2007; Otto 2007; Doyle et al. 2008; Renny-Byfield and Wendel 2014). Studies of synteny between sugarcane and sorghum at the scale of BACs (∼12.3 to 259.2 kb) showed that this highly polyploid genome does not seem to have undergone any major genomic reshaping (Jannoo et al. 2007; Le Cunff et al. 2008; Wang et al. 2010; de Setta et al. 2014). Comparative mapping with sorghum and maize (Aitken et al. 2014), and the data presented here, regarding the sugarcane single copy genes LFY, PHYC, and TOR, also confirm these conclusions. Genomic regions encompassing these three genes presented high conservation of gene structure and collinearity between the homo/homeologous haplotypes and with sorghum and rice. However, in contrast to rice and sorghum, a number of TEs have inserted in both intergenic and intronic sequences of these sugarcane genomic regions (fig. 1), which partly supports the idea presented that sugarcane monoploid genome expanded as compared with sorghum (de Setta et al. 2014).
The polyploid and aneuploid genome of sugarcane cultivars is composed by 80–90% of chromosomes derived from S. officinarum (2n = 8x = 80) and 10–20% derived from S. spontaneum (2n = 5x to 16x = 40–128) (Cuadrado et al. 2004; D’Hont 2005; Piperidis et al. 2010). This hybrid genomic architecture was exemplified in the TOR genomic region analysis, for which, based on specific SNPs and indels (fig. 2), two and five haplotypes were found to have a probable S. spontaneum and S. officinarum ancestry, respectively (Group II and I in fig. 1). The divergence time estimated for TOR haplotypes derived from S. officinarum and S. spontaneum (Group I and II in fig. 1A ) indicates that these two species diverged between 2.5 and 3.5 Ma.
Tracing back the origin of the haplotypes is fundamental to understand the interactions between S. officinarum and S. spontaneum sub-genomes in the hybrid genome of sugarcane cultivars. A crucial aspect is how homo/homeologous genes expression pattern in sugarcane is established to maintain the functionality of regulatory networks.
LFY, PHYC, and TOR are important genes in plant development and integrate regulatory networks in which proteins balance must be precisely regulated to guarantee the homeostasis of interactions. The fact is that the maintenance of these three genes as single-copy gene in most angiosperms possibly represents a mean to control efficiently their expression levels. Therefore, these genes are good candidates to evaluate the extent of expression patterns reshaping after polyploidization.
Our results indicate that all haplotypes of the three genes are under purifying selection and are, therefore, likely to be functional and potentially expressed. TOR haplotypes exhibit a significative correlation between their genomic dosage and their relative expression level in leaves. Both S. spontaneum- and S. officinarum-derived TOR haplotypes are expressed, but not all of them are expressed in the same proportion (table 3 and supplementary table S7, Supplementary Material online). However, PHYC haplotypes display a clear nonadditive pattern of expression with no correlation between haplotype dosage and relative expression level. These data indicate a nonequivalent mode of expression of the different haplotypes, which could partly be related to the insertion of TE and the resulting epigenetic changes in the promoter sequences of these genes (cis polymorphism; Rebollo et al. 2012; Lisch 2013; Song and Chen, 2015). These data also imply that compensatory processes must operate to maintain the right balance of expression of key genes. The expression variations observed between the haplotypes may also reflect a process of subfunctionalization as has been proposed for the sugarcane ScLSG gene (Moyle and Birch 2013).
Another important issue in sugarcane genome evolution is the polyploidization events that shaped S. officinarum and S. spontaneum genomes. Based on dS values between paralogous exons, Kim et al. (2014) suggested that Saccharum and Mischanthus share an allopoliploydization event and that Saccharum has experienced a WGD event after separation from Miscanthus. Shared and possibly derived TE insertion events should be informative in tracing back duplication events, thus helping in defining the chronology of these duplications. Several of such putative insertions appear in pairs or in three, four, or even five of the BACs covering the genomic regions of LFY, PHYC, and TOR. Most of these insertions occurred after the estimated separation of S. officinarum and S. spontaneum (2.5–3.5 Ma) (supplementary table S4, Supplementary Material online). Moreover, according to TOR phylogeny, some of these insertions are restricted to S. officinarum haplotypes (fig. 1A , supplementary fig. S6 and table S4, Supplementary Material online), further supporting the notion that they took place after the separation of S. officinarum and S. spontaneum. These shared insertions most likely are witnesses of at least one autopolyploidization event of S. officinarum. Additionally, we found four TOR haplotypes most likely originated from S. officinarum that harbor a set of shared insertions (TEs “a”, “b”, “c”, “e”, “f”, “h”, “i”, and “j”; haplotypes 102H05 + 236B24 + 077E22, 214B20, 030H10, and 007C22; fig. 1A and supplementary table S4, Supplementary Material online), as well as four LFY haplotypes (011C13, 236J21, 070I10, and 239D06) that share one TE (TE “a”; fig. 1C and supplementary table S4, Supplementary Material online), reveal the potential occurrence of two events of WGD independent from S. spontaneum. No evidence for an older allopolyploidization event shared with Mischanthus was detected using either dS values (table 2) or ancestral shared insertions (supplementary table S4, Supplementary Material online), in discordance with previous literature (Kim et al. 2014). Thus, our results point to the possibility that S. officinarum and its wild ancestor, S. robustum, went through two autopolyploidizations to generate the octaploid stage after separation from S. spontaneum. Such scenario implies that S. spontaneum went through independent and variable rounds of polyploidizations, a hypothesis that is supported by the difference in the basic chromosome number between S. officinarum (x = 8) and S. spontaneum (x = 10) (D’Hont et al. 1998).
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
The authors acknowledge the Brazilian institutions FAPESP (Fundação de Amparo à Pesquisa do Estado de São Paulo—BIOEN Program, grant number 2010/02610-2 and 2008/5207-0) and CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico - INCT Bioetanol Program, grant number 141074/2010-8) for financial support and fellowships. We also acknowledge Angélique D’Hont and members of her group in CIRAD (Carine Charron, Catherine Hervouet and Olivier Garsmeur) for genetic material, PCR of transposable elements insertions and expertise in analyzing BAC sequences.
Literature Cited
- Adam-Blondon A-F, Bernole A, Faes G, Lamoureux D, Pateyron S. 2005. Construction and characterization of BAC libraries from major grapevine cultivars. Theor Appl Genet. 110:1363–1371. [DOI] [PubMed] [Google Scholar]
- Agredano-Moreno LT, de la Cruz HR, Martínez-Castilla LP, Jiménez ES. 2007. Distinctive expression and functional regulation of the maize (Zea mays L.) TOR kinase ortholog. Mol Biosyst. 3(11):794–802. [DOI] [PubMed] [Google Scholar]
- Aitken KS, et al. 2014. Comparative mapping in the Poaceae family reveals translocations in the complex polyploid genome of sugarcane. BMC Plant Biol. 14:190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertin W, et al. 2005. Autopolyploidy in cabbage (Brassica oleracea L.) does not alter significantly the proteomes of green tissues. Proteomics 5:2131–2139. [DOI] [PubMed] [Google Scholar]
- Al-Janabi SM, McClelland M, Petersen C, Sobral BW. 1994. Phylogenetic analysis of organellar DNA sequences in the Andropogoneae: Saccharinae. Theor Appl Genet. 88(8):933–944. [DOI] [PubMed] [Google Scholar]
- Berriman M, Rutherford K. 2003. Viewing and annotating sequence data with Artemis. Brief Bioinform. 4:124–132. [DOI] [PubMed] [Google Scholar]
- Carver TJ, et al. 2005. ACT: the Artemis comparison tool. Bioinformatics 21:3422–3423. [DOI] [PubMed] [Google Scholar]
- Chaudhary B, et al. 2009. Reciprocal silencing, transcriptional bias and functional divergence of homeologs in polyploid cotton (gossypium). Genetics 182(2):503–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZJ. 2007. Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annu Rev Plant Biol. 58:377–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZJ, Ni Z. 2006. Mechanisms of genomic rearrangements and gene expression changes in plant polyploids. Bioessays 28(3):240–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chujo A, Zhang Z, Kishino H, Shimamoto K, Kyozuka J. 2003. Partial conservation of LFY function between rice and Arabidopsis. Plant Cell Physiol. 44(12):1311–1319. [DOI] [PubMed] [Google Scholar]
- Church SA, Spaulding EJ. 2009. Gene expression in a wild autopolyploid sunflower series. J Hered. 100:491–495. [DOI] [PubMed] [Google Scholar]
- Comai L. 2005. The advantages and disadvantages of being polyploid. Nat Rev Genet. 6(11):836–846. [DOI] [PubMed] [Google Scholar]
- Cuadrado A, Acevedo R, Moreno Diaz de la Espina S, Jouve N, de la Torre C. 2004. Genome remodelling in three modern S. officinarum x S. spontaneum sugarcane cultivars. J Exp Bot. 55(398):847–854. [DOI] [PubMed] [Google Scholar]
- D’Hont A. 2005. Unravelling the genome structure of polyploids using FISH and GISH: examples of sugarcane and banana. Cytogenet Genome Res. 109:27–33. [DOI] [PubMed] [Google Scholar]
- D’Hont A, et al. 1996. Characterization of the double genome structure of modern sugarcane cultivars (Saccharum spp.) by molecular cytogenetics. Mol Gen Genet. 250:405–413. [DOI] [PubMed] [Google Scholar]
- D’Hont A, Ison D, Alix K, Roux C, Glaszmann JC. 1998. Determination of basic chromosome numbers in the genus Saccharum by physical mapping of ribosomal RNA genes. Genome 41:221–225. [Google Scholar]
- De Setta N, et al. 2014. Building the sugarcane genome for biotechnology and identifying evolutionary trends. BMC Genomics 15:540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, et al. 2008. Evolutionary genetics of genome merger and doubling in plants. Annu Rev Genet. 42:443–461. [DOI] [PubMed] [Google Scholar]
- Garcia AA, et al. 2013. SNP genotyping allows an in-depth characterisation of the genome of sugarcane and other complex autopolyploids. Sci Rep. 3:3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison E, Marth G. 2012. Haplotype-based variant detection from short-read sequencing. arXiv 1207.3907. [Google Scholar]
- Garsmeur O, et al. 2011. High homologous gene conservation despite extreme autopolyploid redundancy in sugarcane. New Phytol. 189:629–642. [DOI] [PubMed] [Google Scholar]
- Gaut BS, Morton BR, McCaig BC, Clegg MT. 1996. Substitution rate comparisons between grasses and palms: synonymous rate differences at the nuclear gene Adh parallel rate differences at the plastid gene rbcL. Proc Natl Acad Sci U S A. 93(19):10274–10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstein DM, et al. 2012. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40:D1178–D1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grativol C, et al. 2014. Sugarcane genome sequencing by methylation filtration provides tools for genomic research in the genus Saccharum . Plant J. 79(1):162–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivet L, et al. 1996. RFLP mapping in cultivated sugarcane (Saccharum spp): genome organization in a highly polyploid and aneuploid interspecific hybrid. Genetics 142:987–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivet L, Daniels C, Glaszmann JC, D’Hont A. 2004. A review of recent molecular genetics evidence for sugarcane evolution and domestication. Ethnobot Res Appl. 2:9–17. [Google Scholar]
- Guignon V, et al. 2012. Chado controller: advanced annotation management with a community annotation system. Bioinformatics 28(7):1054–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 52(5):696–704. [DOI] [PubMed] [Google Scholar]
- Ha S, et al. 1999. Quantitative chromosome map of the polyploid Saccharum spontaneum by multicolor fluorescent in situ hybridization and imaging methods. Plant Mol Biol. 39:1165–1173. [DOI] [PubMed] [Google Scholar]
- Hamès C, et al. 2008. Structural basis for LEAFY floral switch function and similarity with helix-turn-helix proteins. EMBO J. 27(19):2628–2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoarau JY, et al. 2001. Genetic dissection of a modern cultivar (Saccharum spp). I. Genome mapping with AFLP. Theor Appl Genet. 103:84–97. [Google Scholar]
- Howe GT, et al. 1998. Evidence that the phytochrome gene family in black cottonwood has one PHYA locus and two PHYB loci, but lacks members of the PHYC/F and PHYE subfamilies. Mol Biol Evol. 15:160–175. [DOI] [PubMed] [Google Scholar]
- Jannoo N, et al. 2007. Orthologous comparison in a gene-rich region among grasses reveals stability in the sugarcane polyploid genome. Plant J. 50(4):574–585. [DOI] [PubMed] [Google Scholar]
- Jannoo N, Grivet L, David J, D’Hont A, Glaszmann JC. 2004. Differential chromosome pairing affinities at meiosis in polyploidy sugarcane revealed by molecular markers. Heredity 93:460–467. [DOI] [PubMed] [Google Scholar]
- Jukes TH, Cantor CR. 1969. Evolution of protein molecules. New York: Academic Press; p. 21–132. [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30(14):3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C, et al. 2014. Comparative analysis of Miscanthus and Saccharum reveals a shared whole-genome duplication but different evolutionary fates. Plant Cell 26(6):2420–2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 16(2):111–120. [DOI] [PubMed] [Google Scholar]
- Kohany O, Gentles AJ, Hankus L, Jurka J. 2006. Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. BMC Bioinformatic 7:474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg S. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23(21):2947–2948. [DOI] [PubMed] [Google Scholar]
- Le Cunff L, et al. 2008. Diploid/polyploid syntenic shuttle mapping and haplotype-specific chromosome walking toward a rust resistance gene (bru1) in highly polyploid sugarcane (2n ∼ 12x ∼ 115). Genetics 180(1):649–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy AA, Feldman M. 2002. The impact of polyploidy on grass genome evolution. Plant Physiol. 130:1587–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisch D. 2013. How important are transposons for plant evolution?. Nat Rev Genet. 14:49–61. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS. 2000. The evolutionary fate and consequences of duplicate genes. Science 290:1151–1155. [DOI] [PubMed] [Google Scholar]
- Ma J, Bennetzen JL. 2004. Rapid recent growth and divergence of rice nuclear genomes. Proc Natl Acad Sci U S A. 101:12404–12410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews S, Lavin M, Sharrock RA. 1995. Evolution of the phytochrome gene family and its utility for phylogenetic analyses of angiosperms. Ann Missouri Bot Gard. 82:296–321. [Google Scholar]
- Mathews S, Sharrock RA. 1996. The phytochrome gene family in grasses (Poaceae): A phylogeny and evidence that grasses have a subset of the loci found in dicot angiosperms. Mol Biol Evol. 13:1141–1150. [DOI] [PubMed] [Google Scholar]
- Moyle RL, Birch RG. 2013. Diversity of sequences and expression patterns among alleles of a sugarcane loading stem gene. Theor Appl Genet. 126:1775–1782. [DOI] [PubMed] [Google Scholar]
- Moyroud E, Tichtinsky G, Parcy F. 2009. The LEAFY floral regulators in angiosperms: conserved proteins with diverse roles. J Plant Biol. 52:177–185. [Google Scholar]
- Nijveen H, van Kaauwen M, Esselink DG, Hoegen B, Vosman B. 2013. QualitySNPng: a user-friendly SNP detection and visualization tool. Nucleic Acids Res. 41:W587–W590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. 1970. Evolution by gene duplication. Heidelberg, Germany: Springer-Verlag. [Google Scholar]
- Otto SP. 2007. The evolutionary consequences of polyploidy. Cell 131(3):452–462. [DOI] [PubMed] [Google Scholar]
- Papini-Terzi FS, et al. 2009. Sugarcane genes associated with sucrose content. BMC Genomics 10:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisod C, Holderegger R, Brochmann C. 2010. Evolutionary consequences of autopolyploidy. New Phytol. 186:5–17. [DOI] [PubMed] [Google Scholar]
- Paterson AH, et al. 2009. The Sorghum bicolor genome and the diversification of grasses. Nature 457:551–556. [DOI] [PubMed] [Google Scholar]
- Piperidis G, Piperidis N, D'Hont A. 2010. Molecular cytogenetic investigation of chromosome composition and transmission in sugarcane. Mol Genet Genomics 284:65–73. [DOI] [PubMed] [Google Scholar]
- Rebollo R, Romanish MT, Mager DL. 2012. Transposable elements: an abundant and natural source of regulatory sequences for host genes. Annu Rev Genet. 46:21–42. [DOI] [PubMed] [Google Scholar]
- Renny-Byfield S, Wendel JF. 2014. Doubling down on genomes: Polyploidy and crop plants. Am J Bot. 101(10):1711–1725. [DOI] [PubMed] [Google Scholar]
- Robaglia C, Thomas M, Meyer C. 2012. Sensing nutrient and energy status by SnRK1 and TOR kinases. Curr Opin Plant Biol. 15:301–307. [DOI] [PubMed] [Google Scholar]
- Rutherford K, et al. 2000. Artemis: sequence visualisation and annotation. Bioinformatics 16:944–945. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 4(4):406–425. [DOI] [PubMed] [Google Scholar]
- SanMiguel P, Gaut BS, Tikhonov A, Nakajima Y, Bennetzen JL. 1998. The paleontology of intergene retrotransposons of maize. Nat Genet. 20(1):43–45. [DOI] [PubMed] [Google Scholar]
- Song Q, Chen ZJ. 2015. Epigenetic and developmental regulation in plant polyploids. Curr Opin Plant Biol. 24:101–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F, Nei M. 1984. Estimation of evolutionary distance between nucleotide sequences. Mol Biol Evol. 1(3):269–285. [DOI] [PubMed] [Google Scholar]
- Tamura K, et al. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28(10):2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate JA, Soltis PS, Soltis DE. 2005. Polyploidy in plants In: Gregory TR, ed. The evolution of the genome. San Diego: Elsevier; p. 371–426. [Google Scholar]
- Veitia RA, Bottani S, Birchler JA. 2008. Cellular reactions to gene dosage imbalance: genomic, transcriptomic and proteomic effects. Trends Genet. 24:390–397. [DOI] [PubMed] [Google Scholar]
- Wang J, et al. 2010. Microcollinearity between autopolyploid sugarcane and diploid sorghum genomes. BMC Genomics 11:261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN. 2006. TOR signaling in growth and metabolism. Cell 124:471–484. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.