Abstract
Regulatory regions harbor multiple transcription factor recognition sites; however, the contribution of individual sites to regulatory function remains challenging to define. We describe a facile approach that exploits the error-prone nature of genome editing-induced double-strand break repair to map functional elements within regulatory DNA at nucleotide resolution. We demonstrate the approach on a human erythroid enhancer, revealing single TF recognition sites that gate the majority of downstream regulatory function.
Transcription regulatory regions harbor the majority of human disease-associated sequence variants,1 rendering them attractive targets for elucidating disease mechanisms via targeted genome engineering2. However, our understanding of the impact of regulatory DNA variation is severely limited by the difficulty of precisely assigning the functional contribution of individual nucleotides to phenotypic outcomes. The error-prone nature of double strand break repair triggered during targeted genome editing typically yields small deletions or, more rarely, insertions, of variable size (1 to >10 nucleotides, typically 2-6 nt). We reasoned that this byproduct of targeted genome editing could be systematically exploited to create a broad spectrum of variant regulatory alleles within a single experimental cycle, and that coupling these alleles to a functional readout (such as protein expression) could pinpoint the contribution of specific nucleotides to regulatory activity (Fig. 1a).
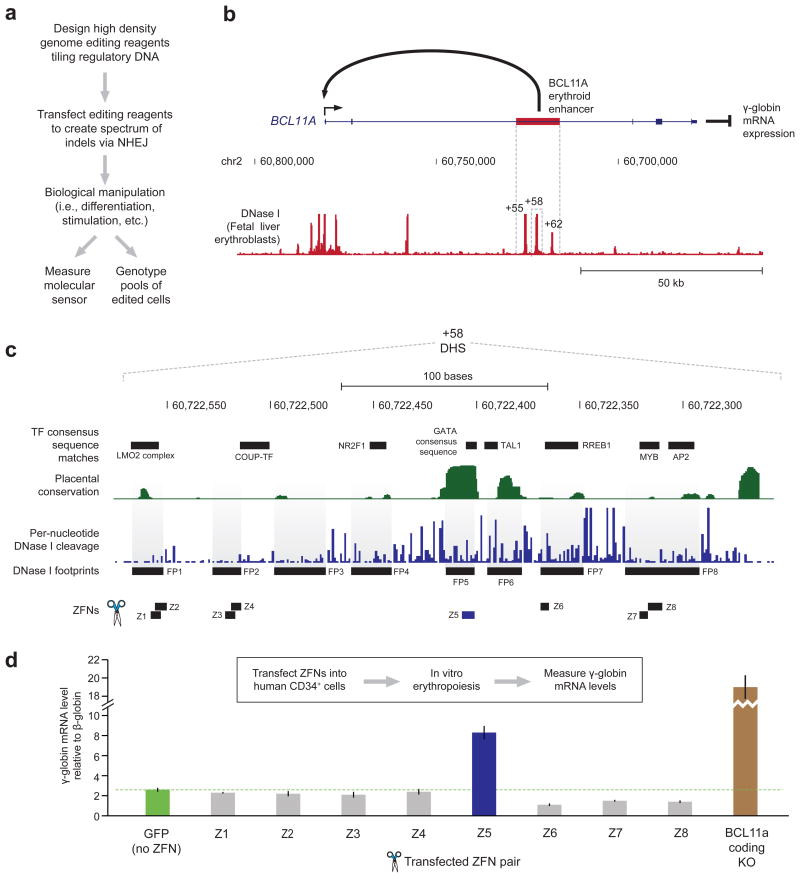
Figure 1. Assessing effects of footprint-targeted genome editing of the BCL11A enhancer on fetal globin mRNA levels in human erythrocytes.
a) Overview of the functional footprinting approach to determine function of regulatory DNA.
b) Schematic of γ-globin gene regulation by BCL11A. An erythroid specific intron enhancer activates BCL11A expression, which represses γ-globin expression.
c) Per-nucleotide conservation, DNase I cleavage and computationally predicted protein-DNA interactions (motifs and footprints) within the +58 DNase I hypersensitive site (DHS) of the BCL11A erythroid enhancer.
d) Effect of engineered zinc-finger nucleases (ZFNs) editing on γ-globin mRNA expression relative to that of β-globin. Both γ-globin and β-globin were measured independently in replicate (each n = 2) and then combinatorially normalized to each other (total n = 4). Bars indicate the mean and error bars indicate the standard deviation of the normalized data.
To test this paradigm (Fig. 1a) we studied the erythroid enhancer region of BCL11A (Fig. 1b), a transcriptional repressor of fetal hemoglobin production in adult erythroid cells3. Naturally-occurring variants within this region are associated with reduced BCL11A expression, with consequent elevation of fetal globin (γ-globin) to levels that are clinically ameliorative for sickle cell disease and beta thalassemia4.
To assess enhancer function in a physiologically relevant context, we first developed an optimized approach for high-efficiency genomic editing in primary human (adult) mobilized CD34+ hematopoietic stem cells, whereby a single exposure to an engineered zinc finger5 or TAL effector6 nuclease could produce 60-80% per-allele editing rates (Supplementary Fig. 1a, Supplementary Tables 1–2, and Online Methods) within the population of edited cells. High-efficiency (70% of alleles) disruption of BCL11A open-reading frame in CD34+ cells followed by erythroid differentiation7 yielded dramatic and highly reproducible elevation of γ-globin mRNA, providing a physiologically relevant readout for loss of BCL11A function (Supplementary Fig. 1a–b). By contrast, control nucleases that drove high-efficiency (>60%) targeted disruption of a neutral locus (AAVS1)8 in the same cell type, coupled with the same differentiation protocol, had no impact on globin expression profiles (Supplementary Fig. 1a–b).
The BCL11A erythroid enhancer region encompasses three DNase I hypersensitive sites (DHSs) located at +55kb, +58kb, and +62kb relative to the transcriptional start site (Fig. 1b). Nucleases targeting each of these DHSs were delivered to human CD34+ cells followed by erythroid differentiation. Importantly, nuclease-treated cells developed morphologically and physiologically indistinguishably from untreated cells, as reviewed by an expert hematologist (T.P.; Supplementary Fig. 2a–b). Deletion of the +55 or +58 DHS reproducibly elevated γ-globin mRNA levels in mature CD34+-derived erythroblasts, while that of the +62 DHS did not (Supplementary Fig. 1c–d). Of note, SNPs in the latter DHS are associated with lower BCL11A expression and elevated γ-globin3; however, it is unclear whether these variants are in fact causal.
We next asked whether it would be possible to identify the specific sequence elements within a DHS that underlie the functional effects of the entire element. For this purpose, we focused on the +58 DHS that showed the most potency (Supplementary Fig. 1d). To map in vivo TF occupancy sites we performed genomic footprinting9 on human erythroblasts (Online Methods), and delineated eight TF footprints (FPs) within the +58 DHS (Fig. 1c). To perform an initial functional scan, we designed zinc-finger nucleases (ZFN) to disrupt each of the most prominent five FPs, and performed editing of CD34+ cells followed by erythroid differentiation and measurement of γ-globin mRNA levels. ZFN-induced lesions within footprint 5 (FP 5) exhibited marked depletion of BCL11A mRNA and corresponding elevation of γ-globin levels irrespective of β-globin expression or normalization strategy (Fig. 1d and Supplementary Fig. 3), indicating that a focused lesion in this footprinted element was sufficient to disrupt BCL11A enhancer function. Of note, FP 5 overlaps a recognition sequence for the master regulator of erythropoiesis GATA110 (Fig. 1d), and ENCODE data show GATA1 occupancy signal over this element in K562 erythroid cells11. Removal of the downstream portion of the +58 DHS using ZFNs provided independent confirmation of the functional role for FP 5 in the regulation of γ-globin expression (Supplementary Fig. 1e–f).
Next, we tiled TAL effector nucleases (TALENs)6 across the +58 DHS (Supplementary Fig. 4a) to address the possibility that our initial ZFN scan missed additional positions in this element critical for enhancer function due either to ZFN nuclease tiling density or to target site-specific variation cleavage activity. Targeted sequencing of edited CD34+ cells reaveled that the dense TALEN scan not only disrupted seven of the eight FPs in the +58 DHS, but that TALEN-triggered NHEJ also comprehensively sampled the intervening sequences between these footprints (Supplementary Fig. 4b and Supplementary Table 3), thus providing complete coverage of the targeted enhancer element. Notably, while we found a weak association between cleavage efficiency and γ-globin mRNA level (Supplementary Fig. 5a–c), specific TALEN pairs were readily identified that induced edits causing increases in γ-globin mRNA expression irrespective of their cleavage rates (Supplementary Fig. 5d and Online Methods). TALEN pair T12 (the cleavage site of which overlaps that of ZFN Z5), corroborated FP 5 as a core functional element within the +58 DHS (Supplementary Fig. 5d). Additionally, edits caused by CD34+ exposure to TALEN T13 and T16 were associated with modest increases in γ-globin mRNA levels (Supplementary Fig. 5d); the lesions triggered by T13 and T16 focused on the predicted binding sites for other well-described erythroid regulators, TAL112 and RREB113 (Supplementary Fig. 4a).
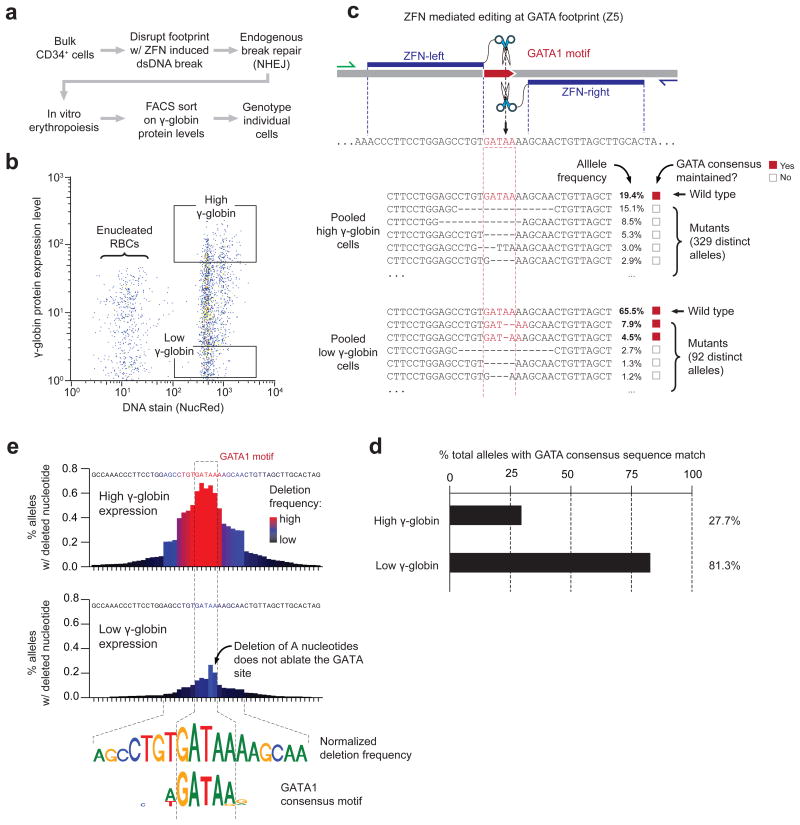
Because the double strand break-repair process is resolved individually (and variably) on each DNA template copy within each cell, we reasoned that subtle variations in the position and extent of nuclease-triggered deletions could be analyzed to quantify the contribution of individual nucleotides to regulatory function. To quantify the spectrum and relative frequency of regulatory alleles, we sorted differentiated cells by their γ-globin protein expression levels, and then performed PCR amplification of DHS +58 within low- and high-γ populations followed by massively parallel sequencing of the reaction products (Fig. 2a–b and Supplementary Table 4). The spectrum of regulatory alleles present in the populations of cells characterized by high vs. low γ-globin expression revealed marked differences in the proportions of GATA1 recognition sequences that were lost as a result of nuclease treatment and subsequent break repair (Fig. 2c–d; 73.3% vs. 18.7%, high vs. low γ-globin). Of note, 46% of the edited targets in the low γ-globin expression population maintained a strong match to the GATA1 consensus due to the presence of a short repeat of adenosine nucleotides trailing the wild-type recognition sequence (Fig. 2c). Shifting the nuclease cleavage site by 1 base pair resulted in a marked (21.8%) decrease GATA1 recognition sequences in edited cells, manifesting in a reduction in BCL11A mRNA levels, and in stronger overall γ-globin induction in bulk cell populations, comparable to that evoked by nucleases that disrupt the BCL11A coding sequence (Supplementary Fig. 6 and Supplementary Table 5). These results indicate that the function of the erythroid-specific BCL11A enhancer is gated on the GATA1 recognition site in the +58 DHS.
Figure 2. Functional footprinting via genome editing and cell phenotyping reveals the precise boundaries of a GATA1 binding site in the BCL11A erythroid enhancer.
a) Experimental outline to link BCL11A enhancer genotypes to molecular phenotypes.
b) Scatterplot of fluorescence-activated cell sorting (FACS) of in vitro generated erythrocytes following targeted disruption of footprint 5 within the +58 DHS using ZFNs. Boxes indicated the FACS gates used to isolate populations of erythroblasts bearing high and low levels of γ-globin protein.
c) Top, diagram of the engineered specificity of ZFN pair Z5. Blue indicates the predicted binding sites for the individual ZFN monomers. Bottom, genotypes derived from the alleles present in the low and high γ-globin protein expressing cells edited with ZFN Z5.
d) Proportion of total alleles genotyped that retain a match to the GATA1 consensus sequence in low and high γ-globin protein expressing cells.
e) Meta-analysis of deep sequencing data on cells containing high vs. low levels of γ-globin reveals the precise boundaries of a GATA1 binding site. The two bar charts plot the frequency for which a given base pair is deleted in high and low γ-globin-containing erythroblasts (top and bottom, respectively). The top panel on the right plots the difference between the data in the left two panels for each base pair. The weblogos (bottom) show the normalized (high minus low γ-globin) nucleotide deletion frequency compared to the idealized GATA1 consensus sequence.
The distinct spectra of allele frequencies observed in high- vs. low-γ-globin expressing cells indicated that γ-globin expression itself could be used as a quantitative molecular sensor for the function of the GATA1 binding site at single base pair resolution. To explore this further, we computed the per-nucleotide editing rate for each position surrounding the GATA1 consensus motif considering the frequencies of all genotypes observed. Edits associated with high γ-globin expression were markedly enriched for positions that covered the core GATA1 consensus motif, and the normalized editing rate observed in the high γ-globin expressing cells strikingly recapitulated an idealized GATA1 consensus sequence (Fig. 2e). We additionally found increased rates of editing in both the upstream and downstream flanking sequences, indicating the presence of additional binding sites (e.g., the GATA1 partner TAL1) that may serve to modulate GATA1 occupancy (Fig. 2e); indeed, the presence of additional factors is supported in the DNase I footprint (FP 5) itself in addition to the clear conservation of its underlying sequence elements (Fig. 1c). As noted above, the vast majority of genome editing events found in low γ-globin-expressing cells created alleles that did not ablate a functional GATA1 binding site.
Our results show that the functional impact of individual TF binding sites within cis-regulatory DNA can be efficiently interrogated by coupling the spectrum of alleles triggered by an engineered nuclease with expression of a downstream transcript or protein target. In the context of the distal BCL11A enhancer, our results pinpoint a single TF recognition site that can be targeted via genome editing to affect a potentially therapeutically meaningful outcome; editing the enhancer has functionally similar consequences on γ-globin mRNA expression to ablating the coding region of BCL11A itself (Supplementary Fig. 6c), both of which do not measurably affect erythroid differentiation (Supplementary Fig. 2).
Functional footprinting thus encompasses a simple and generalizable strategy to dissect the function of individual cis-regulatory elements such that any molecular sensor (i.e., protein, RNA, etc.) can be linked to the function of individual base pairs within non-coding DNA. While methods exist that can assess the function of large libraries of synthetic alleles in both coding and feasibly non-coding DNA in vivo, the low efficiencies allele integration via homology-directed repair (HDR) limit their utility to conventional cell lines due to the requirements for large of amount of starting material and the necessity of selectable markers to enrich for edited cells14. In contrast, functional footprinting does not rely on HDR for the introduction of a library of synthetic alleles, enabling application in primary cells and developmental systems for which material and time are important considerations. Furthermore, this paradigm is agnostic to particular genomic editing and downstream product detection approaches; while the ZFN and TALEN platforms were used herein due to efficiency and downstream considerations such as the potential for therapeutic translation, such efforts could in principle, rely on any genomic editing platform to attain a similar outcome2, subject to inherent limitations in design and efficiency. Similarly, functional footprinting is, in principle, compatible with any molecular readout (i.e., RNA, protein, post-translational modification levels, etc.) that can be efficiently sorted and, critically, the detection of the sensor can occur on fixed (nonviable) cells, permitting use of a wide array of current and emerging technologies such as fluorescence-activated cell sorting on protein or RNA levels15.
Online Methods
Nuclease Design and Validation
Zinc finger nucleases were designed and assembled using an archive of pre-validated two-finger modules5 into expression vectors bearing obligate heterodimer forms of the FokI endonuclease16. TAL effector nucleases were designed as described and cloned into expression vectors bearing truncated forms of the TALE domain6. In brief, the nucleases consist of a triple flag domain, a nuclear localization signal, the engineered DNA binding domain and the FokI nuclease domain containing the obligate heterodimer mutations. The designed sequence recognition domains are provided in Supplementary Tables 1 and 2. Nucleases were first assessed for editing efficiency by transient transfection of expression constructs into K562 cells followed by genotyping of the target locus using the Surveyor/Cel1 endonuclease17. ORFs for maximally active nucleases against each genomic position were re-cloned into an expression vector optimized for mRNA production bearing a 5′ and 3′ UTRs and a synthetic polyA signal. The mRNAs were generated using the mMessage mMachine T7 Ultra kit (Ambion) following the manufacturer's instructions. In vitro synthesis of nuclease mRNAs used either a pVAX-based vector containing a T7 promoter, the nuclease proper and a polyA motif for enzymatic addition of a polyA tail following the in vitro transcription reaction, or a pGEM based vector containing a T7 promoter, a 5′UTR, the nuclease proper, a 3′UTR and a 64 bp polyA stretch.
Purification and Genome Editing Human CD34+ Cells
Human mobilized CD34+ cells (adult) were purchased from AllCells. For small-scale mRNA transfections were performed with a BTX device (Harvard Apparatus), using the CD34+ cell program per manufacturer's instructions. Either 2 μg of mRNA for each ZFN or 4 μg of mRNA for each TALEN was transfected into 200,000 cells in a 100 μl volume. Large-scale transfections were performed using the MaxCyte device according to manufacturer's instructions; using 3 million cells in a total volume of 100 μl and 6 μg of each nuclease mRNA. After transfection, the cells were exposed to transient hypothermia18 for 16 hours and then cultured at 37°C at 5% CO2.
Target Loci Genotyping Following Genome Editing
Forty-eight hours following electroporation, genomic DNA was extracted from 50,000 CD34+ cells using a MasterPure kit (Epicentre). Deletions were genotyped by PCR and non-denaturing PAGE. Targeted locus disruption was measured by Surveyor/Cel117 or deep amplicon sequencing on the Illumina platform. For the latter, the target locus was amplified in a two-step PCR from ∼100 ng genomic DNA. The initial PCR used primers bearing (3′ to 5′) a locus-specific region, a randomized region, and an adapter sequence compatible with the second PCR step; the second PCR uses primers (3′ to 5′) bearing a stretch that anneals to the first-round amplicon, an amplicon-specific barcode, and the Illumina flowcell specific sequences. All generic primer sequences followed manufacturer's instructions for the MiSeq sequencer. Following sequencing, FASTQ sequence reads were filtered via fastq_quality_filter (http://hannonlab.cshl.edu/fastx_toolkit/) for sequences where all bases were Q>=25 (Phred score). Sequences were further filtered for those matching 23 bp on the 5′ and 3′ end of the amplicon to exclude oligonucleotide synthesis-based deletions and primer-dimers before alignment to the intended amplicon. High-quality sequences were then grouped, first according to their indel score (the deviation from wild-type length), and then according to the location of deletions or insertions that account for the difference. Single base pair substitutions are not analyzed since they represent an artifact of the sequencing platform (a bona fide signature of the targeted gene disruption process is a small insertion or deletion).
In Vitro Erythropoiesis
The protocol is based on work from the Douay laboratory7. Following electroporation the cells were treated as follows. Day 0 to day 7: 4 × 104/mL CD34+ cells were cultured in EDM [EDM: IMDM, human holo-transferrin (330 μg/ml); insulin (10 μg/ml); heparin (2 IU/ml), 5% plasma] in the presence of 10−6 M hydrocortisone, 100 ng/mL SCF, 5 ng/mL IL-3, and 3 IU/mL Epo. On day 4, 1 volume of cell culture was diluted in 4 volumes of fresh medium containing SCF, IL-3, Epo, and hydrocortisone. Day 7 to day 11: the cells were resuspended at 4 × 105/mL in EDM supplemented with SCF and Epo. Day 11 to day 15 and out to day 20: the cells are cultured in EDM supplemented with Epo alone. Cell counts are adjusted to between 7.5 × 105 to 1 × 106 on day 11 and harvested on day 13-14 for mRNA analysis and day 20 for immunofluorescent staining of γ-globin and fluorescence-activated cell sorting.
Immunofluorescent staining of γ-globin was modified from a previously described method19. Briefly, cultured erythroid cells were fixed with 4% formaldehyde (Sigma) in PBS and then permeabilized with acetone (Sigma). Cells were washed once with PBS supplemented with 0.5% bovine serum albumin (Sigma), and stained with R-Phycoerythrin conjugated anti-γ-globin antibody (Santa Cruz Biotechnology; cat no. SC-21756PE). Cells were washed and resuspended in NucRed (Life Technologies)-containing PBS-0.5% BSA. Cell sorting was performed using BD Aria III with FACSDiva v6 (BD Biosciences). Erythroblasts were first gated based on positive staining for NucRed. γ-globin high (top ∼10%) and γ-globin low (bottom ∼10%) erythroblasts were sorted and sequenced via amplicon sequencing as described above.
Determining functional effects of nuclease edits
To determine significant effects of nuclease cleavage on γ-globin mRNA expression, we model the relationship between editing efficiency determined by amplicon sequencing and relative γ-globin mRNA levels using a robust linear model using the function lmrob from “robustbase” package in R. We then computed the model residuals for each editing experiment and selected a threshold of 1σ for significance.
Fetal erythroblasts generation
Fetal livers (50–100-day gestation) were obtained from the fetal tissue repository (University of Washington Birth Defects Research Laboratory) with permission of the University of Washington Institutional Review Board. The erythroid culture of dissociated fetal livers and characterization of fetal liver-derived erythroblasts have been described previously20. These cells were subjected to DNase I treatment for profiling for DHSs and transcription factor binding footprints as described21.
Real-time RT-qPCR Measurement of globin mRNA Levels
Whole-cell RNA was isolated from in vitro generated erythrocytes using a High Pure RNA Isolation Kit (Roche). The levels of mRNA for the individual globin genes (α, β, and γ) were then measured by real-time RT-qPCR on an ABI 7300 RT-PCR machine mode using the following manufacturer-provided probesets: α-globin (HBA), Hs00361191_g1; β-globin (HBB), Hs00758889_s1; γ-globin (HBG), Hs00361131_g1; BCL11A , (Hs01093197_m1); 18S rRNA (18S) Hs99999901_s1.
Supplementary Material
Acknowledgments
We thank the Sangamo Production group for assembly of ZFNs and TALENs, and the Cell Process Development group for human CD34+ cell purification. This work was supported by a US National Institute of Health NIDDK grant R01DK101328 to T.P., NHLBI grant P01HL053750 to G.S., and NHGRI grant U54HG007010 to J.A.S.
Footnotes
Author Contributions: K.H.C, S.S.S., Y.Z., C.O., A.S., A.M., N.P. performed the experiments and data collection. J.V., A.R., K.H.C., Y.R., P.L., G.L., M.H., E.R., T.P., G.S. and F.U. analyzed the data. J.V., J.A.S. and F.U. wrote the manuscript with input from A.R., K.H.C., E.R., P.G., T.P. and G.S.
Competing financial interests statement: All Sangamo authors are full-time employees of Sangamo BioSciences, Inc. Patent applications have been filed covering certain aspects of this work.
References
- 1.Maurano MT, et al. Science. 2012;337:1190. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carroll D. Annu Rev Biochem. 2014;83:409. doi: 10.1146/annurev-biochem-060713-035418. [DOI] [PubMed] [Google Scholar]
- 3.Bauer DE, et al. Science. 2013;342:253. doi: 10.1126/science.1242088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bauer DE, Orkin SH. Curr Opin Pediatr. 2011;23:1. doi: 10.1097/MOP.0b013e3283420fd0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Nat Rev Genet. 2010;11:636. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 6.Miller JC, et al. Nat Biotechnol. 2011;29:143. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 7.Giarratana MC, et al. Blood. 2011;118:5071. doi: 10.1182/blood-2011-06-362038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeKelver RC, et al. Genom Res. 2010;20:1133. [Google Scholar]
- 9.Neph S, et al. Nature. 2012;489:83. doi: 10.1038/nature11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ko LJ, Engel JD. Mol Cell Biol. 1993;13:4011. doi: 10.1128/mcb.13.7.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.ENCODE Project Consortium et al. Nature. 2012;489:57. [Google Scholar]
- 12.Shivdasani RA, Mayer EL, Orkin SH. Nature. 1995;373:432. doi: 10.1038/373432a0. [DOI] [PubMed] [Google Scholar]
- 13.Chen RL, Chou YC, Lan YJ, Huang TS, Shen CKJ. J Biol Chem. 2010;285:10189. doi: 10.1074/jbc.M109.049130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Findlay GM, Boyle EA, Hause RJ, Klein JC, Shendure J. Nature. 2014;513:120. doi: 10.1038/nature13695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klemm S, et al. Nat Meth. 2014;11:549. doi: 10.1038/nmeth.2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller JC, et al. Nat Biotechnol. 2007;25:778. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- 17.Guschin DY, et al. Methods Mol Biol. 2010;649:247. doi: 10.1007/978-1-60761-753-2_15. [DOI] [PubMed] [Google Scholar]
- 18.Doyon Y, et al. Nat Meth. 2010;7:459. doi: 10.1038/nmeth.1456. [DOI] [PubMed] [Google Scholar]
- 19.Thorpe SJ, et al. Br J Haematol. 1994;87:125. doi: 10.1111/j.1365-2141.1994.tb04881.x. [DOI] [PubMed] [Google Scholar]
- 20.Chang KH, et al. Stem Cell Rev. 2013;9:397. doi: 10.1007/s12015-012-9355-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.John S, et al. Curr Protoc Mol Biol. 2013;Chapter 27 doi: 10.1002/0471142727.mb2127s103. Unit 21.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.