Abstract
In species with highly heteromorphic sex chromosomes, the degradation of one of the sex chromosomes will result in unequal gene expression between the sexes (e.g. between XX females and XY males) and between the sex chromosomes and the autosomes. Dosage compensation is a process whereby genes on the sex chromosomes achieve equal gene expression. We compared genome-wide levels of transcription between males and females, and between the X chromosome and the autosomes in the green anole, Anolis carolinensis. We present evidence for dosage compensation between the sexes, and between the sex chromosomes and the autosomes. When dividing the X chromosome into regions based on linkage groups, we discovered that genes in the first reported X-linked region, anole linkage group b (LGb), exhibit complete dosage compensation, although the rest of the X-linked genes exhibit incomplete dosage compensation. Our data further suggest that the mechanism of this dosage compensation is upregulation of the X chromosome in males. We report that approximately 10% of coding genes, most of which are on the autosomes, are differentially expressed between males and females. In addition, genes on the X chromosome exhibited higher ratios of nonsynonymous to synonymous substitution than autosomal genes, consistent with the fast-X effect. Our results from the green anole add an additional observation of dosage compensation in a species with XX/XY sex determination.
Keywords: dosage compensation, sex chromosomes, X chromosome, fast-X, RNA-seq, sex-biased expression
Introduction
The green anole, Anolis carolinensis, is a squamate model used to investigate reproductive physiology and behavior (Lovern et al. 2004; Wade 2012), evolutionary developmental biology (Koshiba-Takeuchi et al. 2009; Eckalbar et al. 2013), and tissue regeneration (Hutchins et al. 2014). In the presence of dosage compensation, genes on the sex chromosomes in males and females will be expressed at approximately similar levels, nearing a sex ratio of 1. In the absence of dosage compensation, however, the expression of sex-linked genes is expected to reflect their overall copy number in each sex (i.e. half as much in the hemizygous sex), with an expected sex ratio of 0.5. Studies across species suggest that male-heterogametic sex determination systems (XX/XY) typically undergo chromosome-wide dosage compensation, albeit with unique mechanisms in each convergent system (Wilson and Makova 2009; Graves 2016). In therian mammals, gene dosage is upregulated on the X chromosome in both males and females, and females experience silencing of one X chromosome (Payer and Lee 2008) (although gene-by-gene escape from inactivation can occur, Carrel and Willard 2005); in the platypus, a monotreme mammal with a chromosomal sex determination system that is derived independently of therian mammals, the X chromosome exhibits partial inactivation (Deakin et al. 2008); in Caenorhabditis elegans, the single X chromosome in males is upregulated and the two X chromosomes in hermaphrodites are both partially down-regulated (Kramer et al. 2015; Lau and Csankovszki 2015); and in Drosophila melanogaster gene dosage on the X is upregulated only in males (Conrad and Akhtar 2012). In contrast, ZZ/ZW female-heterogametic systems typically do not exhibit chromosome-wide dosage compensation (Mank 2013), although dosage compensation can occur on a gene-by-gene basis (Dean et al. 2015). This gene-by-gene pattern has been observed in birds (Dean et al. 2015; Itoh and Arnold 2015), snakes (Vicoso et al. 2013), silkworms (Xingfu 2009), and the flatworm Schistosoma mansoni (Vicoso and Bachtrog 2011).
However, it is unknown whether the limited number of taxa that have been studied to date biases perceived trends about dosage compensation. For example, the ZZ/ZW moth Manduca sexta has been found to exhibit complete dosage compensation (Smith et al. 2014), and near-global patterns of dosage compensation have been observed in ZZ/ZW Heliconius butterflies (Walters et al. 2015), suggesting that patterns in dosage compensation may not be linked to male- or female-heterogametic sex determination. In addition, dosage compensation in XX/XY systems may not be as complete as previously thought. About 15–25% of X-linked genes escape inactivation in humans, with the proportion differing greatly among regions of the X chromosome (Carrel and Willard 2005). This pattern is explained in part by the time since gametologous Y-linked alleles were pseudogenized or lost (Wilson Sayres and Makova 2013). Given the variation among the few systems that have been studied, it is necessary to examine dosage compensation in evolutionarily independent chromosomal sex determination systems.
In addition to dosage compensation, natural selection is expected to behave differently on the sex chromosomes vs. the autosomes because of their unique inheritance patterns (Meisel and Connallon 2013). In species with XX/XY sex determination, females have two copies of the X chromosome, which means that, similar to the autosomes, deleterious recessive genes can be shielded from selection by dominant alleles in heterozygous individuals. In contrast, XY males only possess one copy of the X chromosome, so recessive X-linked alleles are directly exposed to natural selection. If most new mutations are recessive, then natural selection will be more efficient on the X chromosome, both removing harmful variants and also increasing the frequency of beneficial X-linked variants. The latter process is expected to generate a higher ratio of non-synonymous to synonymous substitutions on the X chromosome than the autosomes—a phenomenon called the fast-X effect (Vicoso and Charlesworth 2006; Meisel and Connallon 2013). Fast-X (which is equivalently called fast-Z in species with ZZ/ZW sex determination) has been reported across species with male- and female-heterogametic chromosomal sex determination, including fruit flies (Thornton and Long 2002), birds (Mank et al. 2007; Meisel and Connallon 2013), snakes (Vicoso et al. 2013), and mice (Kousathanas et al. 2014). Other factors may result in a similar signature: genetic drift on the Z chromosome relative to the autosomes has also been shown to result in a "fast-X" effect (Mank et al. 2009).
Genomic analyses are crucial for a comprehensive picture of anole sex chromosome differentiation, dosage compensation, and signatures of natural selection. No pseudoautosomal genes have yet been described and pseudoautosomal regions in A. carolinensis are hypothesized to either be very small or completely absent (Rovatsos et al. 2014a), similar to marsupials (Murtagh et al. 2012). The identification of many genomic regions that are haploid in males but diploid in females suggest that the A. carolinensis Y chromosome may be highly degenerated and, if it exists at all, has lost much of the ancestral sex chromosome content during the course of evolution (Rovatsos et al. 2014a). Y-linked degeneration is expected to result in changes in X-linked transcript levels, a form of dosage compensation to achieve similar X-linked expression in males and females (Wilson Sayres and Makova 2013). X chromosome expression, however, has not yet been comprehensively characterized in the green anole. If the anole X and Y chromosomes are as differentiated as preliminary data suggests, and if it follows the trend of other male-heterogaemtic systems, then we expect to observe dosage compensation on the anole X chromosome.
In this study, we conducted analyses to infer additional X-linked loci in the green anole, quantify patterns of sex-biased gene expression across the genome, and characterize the extent of dosage compensation on the anole X chromosome. Based on these analyses, we propose that the green anole X chromosome contains at least 374 genes. In addition, we present evidence for dosage compensation on the anole X chromosome. Finally, we show that X-linked genes have a higher ratio of non-synonymous to synonymous substitution rates than autosomal genes, a pattern consistent with fast-X.
Materials and Methods
Comparative analysis: To identify putative X-linked genes, we conducted bidirectional best hit alignment between the chicken and anole genomes. We acquired a list of all genes from the green anole microchromosome LGb (Alföldi et al. 2011) from Ensembl BioMart (Ensembl v79) (Kinsella et al. 2011) and aligned them to the chicken genome (galGal4, Ensembl v79) (Hillier et al. 2004) using BLAT (Cunningham et al. 2015). Next, we obtained a list of transcript IDs of genes located on the chicken chromosome 15 from Ensembl BioMart (Ensembl v79) (Kinsella et al. 2011) and mapped these transcripts to the anole genome (AnoCar2.0, Ensembl v79). We then aligned genes from each anole scaffold back to the galGal4 genome. For each alignment step, we used a cutoff score of 100 to include genes for further analysis. We included genes in the set of proposed X-linked genes if they were either previously identified (Alföldi et al. 2011), or were located on a scaffold in which at least 60% of the gene content of the scaffold mapped one-to-one between this scaffold and chicken chromosome 15, and contained at least one gene that had been previously identified as X-linked by Q-PCR (Rovatsos et al. 2014a) (supplementary fig. S1, Supplementary Material online). We included genes annotated from both the Ensembl v79 and a previously published annotation using transcriptomes (Eckalbar et al. 2013).
Transcriptome analysis: We analyzed whole transcriptome RNA-seq data from A. carolinensis tail samples for two males (D015 and D026) and two females (D047 and D061), data from which are accessible via BioProject PRJNA253971 (Hutchins et al. 2014). Briefly, regenerating tail samples were collected 25 days post autotomy and sliced into five sections along the proximal–distal axis. This resulted in five biological replicates from each of the four individuals for a total of twenty samples. Total RNA was isolated from each sample separately, all samples were barcoded and multiplexed with paired-end sequencing libraries generated using manufacturer protocols and sequenced on an Illumina HiSeq 2000 (Hutchins et al. 2014). After sequencing the samples were de-multiplexed and analyzed separately. We assessed the quality of the reads with FastQC (Andrews 2010) and further trimmed using Trimmomatic (Bolger et al. 2014) to eliminate a nucleotide bias from the flow cell. We mapped trimmed reads to the A. carolinensis genome using Bowtie2.2.4 (Langmead and Salzberg 2012) and TopHat2.0.14 (Trapnell et al. 2009) with the ASU_Acar_v2.2.1 annotation and the following options: segment-length = 19 – no-coverage-search. We measured RNA-seq statistics using the stats function of bamtools2.4.0 (Trapnell et al. 2012) (supplementary table S1, Supplementary Material online).
Differential expression: We conducted the differential expression analysis using Cuffdiff in Cufflinks2.2.1 (Trapnell et al. 2012) and analyzed the output using R3.2.2 (R Core Team 2014) (readme files, associated scripts, and transcriptome assemblies are available on GitHub: https://github.com/WilsonSayresLab/Anole_expression). By default, Cuffdiff normalizes read count estimates between samples, so models for inter-sample normalization of raw read counts were not applied to read estimates returned by Cuffdiff (Dillies et al. 2013). We called genes as differentially expressed if the q-value—a P value with a Benjamini–Hochberg correction for multiple testing (Trapnell et al. 2012)—is less than 0.05.
Given that the minimum expression threshold can skew results when examining patterns of dosage compensation, we examined the relative expression results with five different FPKM thresholds (0, 1, 2, 3, and 4), similar to Smith et al. (2014) (supplementary table S2, Supplementary Material online). We found that the same trends for autosomal vs. X-linked genes were present at each threshold and here present data using an FPKM threshold of 2 (supplementary fig. S2, Supplementary Material online). We also found that our results were robust when we analyzed expression after accounting to where reads mapped in the chicken genome (galGal4; supplementary table S3, Supplementary Material online).
Diversity analyses: For independent verification of the putative X-linked transcripts, we analyzed patterns of genetic diversity across the genome. To call variants, we first aligned reads from all 20 samples—five replicates each of the four individuals (see Transcriptome Analysis above)—to the AnoCar2 reference genome (Alföldi et al. 2011) using multi-sample 2-pass mapping in STAR (Dobin et al. 2013). Specifically, we first mapped samples individually using default parameters and then did a second pass of mapping with default parameters except for including information about identified splice junctions for all samples using the "–sjdbFileChrStartEnd" command (Li 2011; Li, 1000 Genome Project Data Processing Subgroup et al. 2009). We then sorted bam files using Samtools before adding read group information and removing duplicates with Picard (http://broadinstitute.github.io/picard/; last accessed September 1, 2016). We trimmed intronic tails with the SplitNCigarReads tool in the Genome Analysis Toolkit (GATK) using the parameters "-rf ReassignOneMappingQuality -RMQF 255 -RMQT 60 -U ALLOW_N_CIGAR_READS" (McKenna et al. 2010; DePristo et al. 2011; Van der Auwera et al. 2013). We then called variants individually for each sample with the GATK HaplotypeCaller using the parameters "-dontUseSoftClippedBases -stand_call_conf 20.0 -stand_emit_conf 20.0 –emitRefConfidence GVCF –variant_index_type LINEAR –variant_index_parameter 128000" to output a GVCF file. Lastly, we performed a round of joint genotyping of all 20 samples using GATK’s GenotypeGVCFs with default parameters.
We calculated genetic diversity within each set of biological replicates. For example, population 1 consisted of biological replicate 1 for each of the four individuals, population 2 consisted of biological replicate 2 for each for the four individuals, and so on. This primarily allowed us to control for differences in sequencing coverage and quality among replicates, but also provided five biologically and technically independent estimates of diversity among these individuals. For these analyses, we focused exclusively on biallelic SNPs, to which we applied a series of filters, removing sites at which the sequencing depth was less than 2, Fisher strand bias was greater than 30, the mean mapping quality was less than 30, or the site quality was less than 30. Additionally, we only considered a sample’s genotype call if the sample read depth at that site was greater than or equal to 10. We then only examined sites that were callable in all samples within a given population on scaffolds containing at least 250 callable sites per population.
At each site, we calculated nucleotide diversity, , as the number of pairwise differences among chromosomes sampled divided by the number of comparisons (, where is the number of chromosomes sampled) (Tajima 1989; Charlesworth and Charlesworth 2010). To identify all callable sites in the genome for each sample, including invariant sites, we used GATK’s CallableLoci tool with the parameters "–minMappingQuality 30 –minDepth 10" (McKenna et al. 2010). We then used BEDTools intersect iteratively to identify sites callable in all samples in a given population (Quinlan and Hall 2010). Given this information, we calculated average nucleotide diversity separately for the autosomes and putatively X-linked scaffolds as the arithmetic mean of per site diversity across all callable sites. We estimated confidence intervals for autosomal diversity, X-linked diversity, and the ratio of X to autosomal diversity by sampling 1000 bootstrap replicates with replacement.
Gene ontology: We submitted lists of gene IDs for differentially expressed genes identified by Cuffdiff (q < 0.05) (Trapnell et al. 2012), as well as male-biased and female-biased differentially expressed genes, to g:profiler for enrichment analysis (Reimand et al. 2011). Additionally, we also conducted an analysis of genes with 50 or more reads in males and genes with 50 or more reads in females. Each list was run as a target set with a list of gene IDs for the whole genome as the background set, and we specified that only significant results with a multiple testing corrected P value of 0.05 be returned.
Substitution rate analysis: We downloaded pairwise alignments between the AnoCar2.0 genome and the galGal3 chicken genome from the UCSC genome browser (Meyer et al. 2013). We filtered these alignments using scripts developed in our lab (https://github.com/WilsonSayresLab/AlignmentProcessor; last accessed September 1, 2016). These scripts ensured that the open reading frame was conserved in each species and required alignments to have 50% coverage of nucleotide sequences relative to the anole genome. Gene sequences that did not meet these requirements were discarded from the analysis. Lastly, one script replaced internal and terminal stop codons with gaps. We conducted analysis both including and excluding alignments with internal stop codons to avoid inflating ratios of non-synonymous to synonymous substitution rates by including pseudogenes, and here we present the analysis with genes with internal stop codons excluded. The substitution rates for the filtered alignments were then calculated using KaKs_Calculator2.0 using the default settings (Zhang et al. 2006). We calculated means, medians, and corresponding 95% confidence intervals in R3.2.2 (R Core Team 2014) with the "boot" package (Canty and Brian 2016). We conducted a permutation analysis to calculate P values using 10,000 replicates with a Python script developed in lab (https://github.com/WilsonSayresLab/Anole_expression).
Results and Discussion
Identification of X-Linked Sequences in A. carolinensis
If genes on the green anole X chromosome have evolved dosage compensation between males and females, relative expression values between the sexes will not be sufficient to identify X-linked transcripts. Therefore, to identify putative X-linked genes in the green anole, we used a comparative genomics approach. Chicken (Gallus gallus) and anole genomes share many large syntenic blocks (Alföldi et al. 2011), making the chicken genome an informative point of comparison. Linkage group b (LGb) was previously identified as X-linked in the anole because it is present in two copies in females and only one in males (Alföldi et al. 2011), suggesting that all, or nearly all Y-linked genes from this region are highly differentiated or absent. Twenty-three additional genes were later identified on LGb, bringing the total of known X-linked genes to 87. In a subsequent analysis, quantitative PCR in male and female samples identified an additional 38 genes that are consistent with male-heterogamety (Rovatsos et al. 2014a).
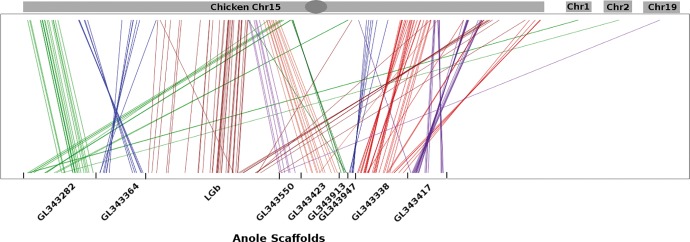
We first investigated the genomic location of previously annotated anole X-linked genes in the chicken genome and found that all mapped to either chicken chromosome 15 or nowhere in the chicken genome (fig. S1; supplementary fig. 1, Supplementary Material online). Given the conservation of gene order between chicken and anole (Alföldi et al. 2011), this suggests that additional undiscovered X-linked genes may also have homologs on chicken (G. gallus) chromosome 15 (GG15). To find additional candidate X-linked genes, we compared GG15 with the anole genome to identify one-to-one orthologs (supplementary fig. S1, Supplementary Material online). We began by using BLAT (Cunningham et al. 2015) to map 399 transcripts on GG15 to assembled chromosomes and unassembled scaffolds from the anole genome, then conducted reciprocal best-mapping from anole to the chicken genome. To retain proposed X-linked scaffolds, we required a minimum of 60% of the genes on the anole scaffold to either map to GG15 or not map anywhere in the chicken genome. Through this reciprocal analysis we determined that 118 genes (corresponding to 127 transcripts) mapped to chicken chromosome 15, and another 165 genes from the unmapped scaffolds in the green anole genome did not have a homolog in the chicken genome (supplementary fig. S1, Supplementary Material online). From these scaffolds, two genes mapped to chicken chromosome 1, one gene mapped to chromosome 2, and one mapped to chromosome 19, for a total of four genes which mapped to other chicken chromosomes.
In total, these 287 genes spanned eight scaffolds (fig. 1). Among these scaffolds, each contains at least one gene previously reported to be X-linked (Rovatsos et al. 2014a), further corroborating that these scaffolds likely belong to the anole X chromosome. Taken together with the 87 genes on LGb, which was previously identified as the X chromosome (Alföldi et al. 2011), and the remaining genes on these scaffolds (some of which were previously described as sex-linked, Rovatsos et al. 2014a), these results suggest there are 374 genes on the anole X chromosome (fig. 1, supplementary table S1, Supplementary Material online).
Fig. 1.—
Location of proposed X-linked sequences in the chicken genome. Chicken genomic regions are shown on the top chromosome, while anole scaffolds that may be X-linked and the previously described X-linked LGb are shown on the bottom. Lines connect orthologous genes between the two genomes. Genes are color-coded by scaffold.
As an additional line of evidence to verify these X-linked scaffolds, we conducted an analysis of genetic diversity on the autosomes and the putative X-linked scaffolds. Diversity is expected to be reduced on X chromosomes relative to autosomes due to a smaller effective population size and more efficient natural selection (Ellegren 2009). We found that the mean diversity () for the autosomal macro-chromosomes 1–6 ranged from 0.000605 to 0.000637 among biological replicates, although the mean diversity for the putative X chromosome ranged from 0.000320 to 0.000397 (supplementary table S4, Supplementary Material online). This corresponds to very low X/A diversity ratios (supplementary table S2, Supplementary Material online) and is consistent with results from other species in which the X chromosome exhibits lower diversity than autosomes, particularly in coding regions, due to a lower effective population size, greater exposure to selection in males, and male-mutation bias (Ellegren 2009; Gottipati et al. 2011; Hvilsom et al. 2012).
Dosage Compensation on the Anole X Chromosome
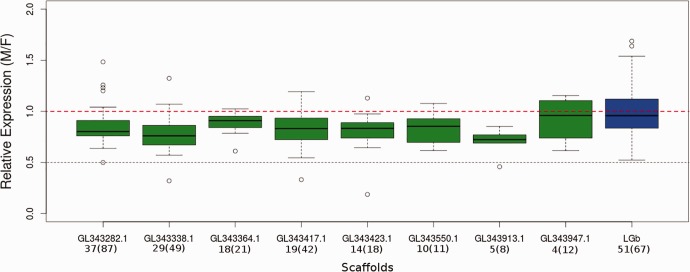
To assess anole sex chromosome dosage compensation, we investigated transcript abundance from the X and autosomes in samples from males and females. In the absence of dosage compensation, the ratio of expression in males vs. females on the X chromosome is expected to be around 0.5. In contrast, if there is dosage compensation, then genes on the X chromosome should exhibit a male:female expression ratio similar to that of the autosomes (median: 1.009; 95% CI: 0.9713–1.022, 1000 bootstrap replicates; table 1). We analyzed anole X-linked genes in two groups: the previously identified LGb, because its clear sex-biased presence suggests that it may be part of an older, more differentiated part of the anole X chromosome, and our newly proposed X-linked genes that may be part of a less-differentiated portion of the anole sex chromosomes (fig. 2).
Table 1.
Male vs. Female Median Gene Expression
| Chromosome | Median Male:Female Expression (95% CI) | Expressed Genes |
|---|---|---|
| Autosomes 1–6 | 1.009 (0.9713, 1.022) | 6595 |
| LGb (X-linked) | 0.9746 (0.9718, 1.058) | 59 |
| X-Linked scaffolds | 0.8226 (0.7772, 0.8296) | 143 |
| Proposed X | 0.8688 (0.8558, 0.8958) | 202 |
Note.—Median male:female expression values for the autosomal macrochromosomes, X-linked linkage group b (LGb), the newly proposed X-linked scaffolds, and the combined X chromosome sequences (LGb and all other proposed X-linked sequences) using a minimum threshold of 2 FPKM (expressed genes). Relative expression could only be computed for genes with detectable expression values for both males and females. 95% confidence intervals were computed with 1000 bootstrap replicates.
Fig. 2.—
Ratio of male:female anole gene expression across proposed X-linked loci. Each box plot represents a different scaffold. Expression levels of genes on the previously determined X-linked linkage group b (LGb) are blue and proposed X-linked genes are plotted in green. The autosomal median expression is shown in a dotted red line. The expected male:female expression ratio in the absence of buffering and dosage compensation (0.5) is plotted in a dotted black line.
Consistent with dosage compensation, the 87 genes previously annotated on LGb as X-linked genes have a median male:female relative expression of 0.9746 (95% CI: 0.9718–1.058, 1000 bootstrap replicates; table 1), which is statistically indistinguishable from the autosomes. Curiously, compared with both the autosomes and the older X-linked LGb, the newly identified X-linked genes have a lower, but still significantly greater than 0.5, median relative expression of 0.8226 (95% CI: 0.7772–0.8296,1000 bootstrap replicates; table 1), consistent with fewer dosage compensated genes, incomplete compensation, or buffering effects. For the entire set of 374 genes we propose for the anole X chromosome, the median ratio of male:female expression is significantly lower than the autosomes: 0.8688 (95% CI: 0.8558–0.8958, 1000 bootstrap replicates; table 1), suggesting dosage compensation across all genes on the X chromosome is not complete, but that many genes are dosage compensated. In cases of single-gene deletions on the autosomes, buffering has been observed (Malone et al. 2012) that results in expression not exactly proportional to gene copy number. However, the ratio of male:female expression on the anole X chromosome is significantly greater than 0.5 and much closer to one than expected if due solely to buffering in one sex (0.7 (Itoh et al 2007; Wright and Mank 2012)). These results indicate that many genes on the anole X chromosome are dosage compensated, but some regions of the anole X chromosome have not yet evolved complete dosage compensation between the sexes and that the process may be still ongoing. Without a fully assembled X chromosome, it is difficult to confidently discern whether there are physical clusters of complete and incomplete dosage compensation.
Gene Expression Suggests Upregulation of the X Chromosome in Males
To infer the mechanism of dosage compensation, we compared absolute expression of X-linked genes with autosomal genes in males and females, allowing us to evaluate two possible alternatives. The first possibility is that expression on one or both of the X chromosomes in females is downregulated to the, presumably, lower level in males. If this is the case, we expect to observe that although the male:female ratio of expression for the X and autosomes is near one, the X:autosomal expression ratio will be significantly lower for both males and females. Alternatively, if the X chromosome is upregulated in males only, then we expect that the average expression for X-linked genes would be similar to the autosomal expression for both males and females. In the absence of dosage compensation, we expect only the male X-linked expression values to be lower than the autosomal values. But, as we have shown, we do observe dosage compensation in the green anole and so expect both male and female expression to exhibit the same pattern.
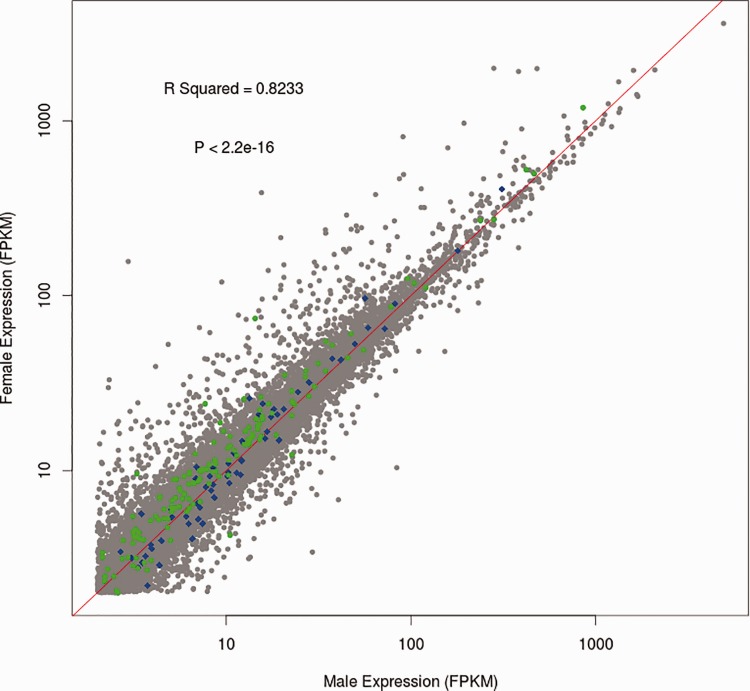
The median expression values for X-linked genes (8.60 FPKM and 9.88 FPKM for males and females, respectively; supplementary table S5, Supplementary Material online) are lower than the median expression on the autosomes (11.8 and 12.0, respectively; supplementary table S5, Supplementary Material online), but are still greater than 50% of the autosomal expression. Mean expression, however, is significantly higher on the X chromosome in males (27.24 FPKM, P = 0.0011; two-sided Wilcoxon Rank Sum test; fig. 3, supplementary table S3, Supplementary Material online), but not females (32.68 FPKM, P = 0.2001; two-sided Wilcoxon Rank Sum test; fig. 3, supplementary table S5, Supplementary Material online), than mean expression on the autosomes (24.12 FPKM in males and 25.36 FPKM in females; supplementary table S5, Supplementary Material online). This is not unexpected as we required a minimum expression of 2 FPKM but had no maximum restriction, which should result in higher means than medians.
Fig. 3.—
Regression plot for male vs. female gene expression. Genes are plotted by FPKM values in male samples and female samples. Genes from LGb are plotted in blue, although all other X-linked genes are shown in green. The regression line calculated between male and female FPKM by gene is shown in red.
Upon further investigation, the two females exhibited different patterns of expression: mean expression was greater on the X chromosome than the autosomes in D61, although expression did not differ in D47 (supplementary table S5, Supplementary Material online). Because D47 exhibited much greater variation in mean expression on the X chromosome than any other sample, we reran analyses excluding the most highly expressed X-linked genes (FPKM > 400 in at least one individual). Here, we found that the overall pattern was reversed, with significantly lower mean and median expression on the X chromosome than autosomes in the two males and D61 (supplementary table S5, Supplementary Material online). However, expression levels remained indistinguishable in D47. Future examination of a larger sample will be required to understand which female exhibited a pattern more typical of female anoles in general.
Taken together, these results are consistent with male upregulation on the X chromosome: both mean and median expression of X-linked genes, with and without the most highly expressed genes, is greater than half that of the autosomes. Further, because many X chromosome transcripts are expressed at levels well below those of autosomal transcripts, it is likely that only a subset of genes are dosage-sensitive. However, our small sample size prevents us from inferring whether compensation is exclusively male based, or if compensatory downregulation occurs in females.
Sex-Biased Gene Expression
In our analysis of expression differences between the sexes, we identified 24 female-biased X-linked genes and four male-biased X-linked genes on the anole X chromosome (supplementary table S4, Supplementary Material online). However, these were only a small fraction of the 1384 genes across the genome that exhibited significant differential expression between males and females (table 2). These differentially expressed genes comprise approximately 10% of all expressed genes and are dispersed throughout the entire genome (table 2). Due to the unique properties of microchromosomes in chicken (Hillier et al. 2004), we further examined the anole micro-chromosomes LGa-LGd and LGf-LGh (there is no LGe). All the microchromosomes exhibited somewhat female-biased expression with the exception of LGd, which showed no observable expression (supplementary table S5, Supplementary Material online).
Table 2.
Location of Genes with Sex-Biased Expression
| General Loci | Specific Locus | Number DE | Percentage DE | Expressed Genes |
|---|---|---|---|---|
| Genome | 1384 | 10.3 | 13,414 | |
| Macrochromosomes | 588 | 8.9 | 6595 | |
| 1 | 124 | 8.3 | 1492 | |
| 2 | 148 | 9.8 | 1515 | |
| 3 | 65 | 6.4 | 1009 | |
| 4 | 99 | 9.8 | 1008 | |
| 5 | 91 | 9.9 | 916 | |
| 6 | 61 | 9.3 | 655 | |
| Microchromosomes | 27 | 13.6 | 198 | |
| LGa | 4 | 26.7 | 15 | |
| LGb | 6 | 10.4 | 59 | |
| LGc | 5 | 10.4 | 48 | |
| LGd | 0 | N/A | 0 | |
| LGf | 12 | 17.4 | 69 | |
| LGg | 0 | 0 | 2 | |
| LGh | 0 | 0 | 5 | |
| Contigs | 69 | 11.1 | 622 | |
| All scaffolds | 700 | 11.7 | 5999 | |
| X-linked scaffolds | 17 | 12.5 | 136 | |
| GL343282.1 | 2 | 2.4 | 84 | |
| GL343338.1 | 7 | 10.9 | 64 | |
| GL343417.1 | 4 | 7.3 | 55 | |
| GL343423.1 | 2 | 8.3 | 24 | |
| GL343550.1 | 0 | 0 | 12 | |
| GL343947.1 | 1 | 9.1 | 11 | |
| GL343913.1 | 1 | 12.5 | 8 | |
| GL343364.1 | 0 | 0% | 26 |
Note.—The number and percentage of genes with differential expression (DE) between male and female anoles are shown across the entire genome and for different subsets of the Assembled and unassembled anole genome. Significance was determined for expressed genes with FPKM values of at least 2. Genes were identified as significant by cuffdiff (Trapnell et al. 2012) if they exhibited a multiple testing corrected q-Value less than or equal to 0.05 (cuffdiff output available at https://github.com/WilsonSayresLab/Anole_expression)
According to gene ontology analyses, genes with male-biased expression are involved in immune and defense response (supplementary table S6, Supplementary Material online), consistent with previous analyses of these samples that found differential expression of immune response transcripts along the proximal–distal axis of regenerating tail tissue (Hutchins et al. 2014). Genes with female-biased expression are enriched for those with functions in tissue development and cell proliferation (supplementary table S6, Supplementary Material online).
The Fast-X Effect
According to the fast-X hypothesis, if most new mutations are recessive, alleles on the X chromosome will experience more efficient positive selection (observable in a higher ratio of non-synonymous to synonymous substitutions) than the autosomes because the X chromosome is hemizygous in males, directly exposing alleles to selection. Alternatively, genetic drift can also result in a signature of fast-X. To test between these hypotheses in the green anole, we conducted substitution rate analysis of the genes on anole chromosomes 1–6, vs. genes on the proposed X chromosome.
We found a significantly higher mean (0.1029 for autosomes, 0.1263 for X-linked genes, P = 0.0002; permutation tests with 10,000 replicates; table 3), but not median (0.0830 for autosomes, 0.0803 for X-linked genes, P =0.8406; permutation tests with 10,000 replicates; table 3), Ka/Ks ratio on the putative X chromosome than the autosomes. When we compared the X-linked genes with the anole’s microchromosomes, we found the same pattern of a significantly higher mean for X-linked genes (0.1263) than the microchromosomes (0.0987, P = 0.0014 permutation tests with 10,000 replicates; supplementary table S7, Supplementary Material online), but not median (X-linked = 0.0803; michrochromosomes = 0.0775; P = 0.1098 permutation tests with 10,000 replicates; supplementary table S7, Supplementary Material online). We did not observe a significant difference in Ka/Ks between female-biased genes, male-biased genes, or unbiased genes on the anole X chromosome (supplementary table S4, Supplementary Material online).
Table 3.
Pairwise Rates of Evolution between Anole and Chicken
| Autosomes (95% CI) 5499 Genes | X Chromosome (95% CI) 192 Genes | P | ||
|---|---|---|---|---|
| Median | Ka | 0.1065 (0.1025, 0.1314) | 0.1170 (0.1115,0.1335) | 0.2403 |
| Ks | 1.257 (1.210, 1.322) | 1.349 (1.279, 1.434) | 0.0197 | |
| Ka/Ks | 0.0830 (0.0805, 0.0998) | 0.0803 (0.0777, 0.0956) | 0.8406 | |
| Mean | Ka | 0.1324 (0.1143, 0.1377) | 0.1522 (0.1270, 0.1548) | 0.0068 |
| Ks | 1.352 (1.294, 1.422) | 1.440 (1.415, 1.543) | 0.0576 | |
| Ka/Ks | 0.1029 (0.0988, 0.1185) | 0.1263 (0.1171 0.1474) | 0.0002 |
Note.—Ka, Ks, and Ka/Ks values were calculated for the six macrochromosomes and the proposed X chromosome. 95% confidence intervals were calculated using 1000 bootstrap replicates. Empirical P values were calculated by permuting genes across the autosomes and X chromosome 10,000 times. All genes with internal stop codons were excluded from this analysis Significant p-values are in italics.
One interpretation of these results, particularly our observation of a significantly higher mean, but not median, Ka/Ks on the X vs. autosomes, is that relatively few genes have experienced strong positive selection on the X and that purifying selection, which is also expected to be more efficient at removing deleterious alleles on the X chromosome, is acting on the majority of X-linked genes. Another possibility is that purifying selection has been relaxed in certain genes, allowing non-synonymous mutations to accumulate at a faster rate. A third possibility is that there is more genetic drift on the X chromosome relative to the autosomes, resulting in the fixation of more nonsynonymous mutations on the X vs. autosomes. Additional factors can affect the presence of, and our ability to detect, fast-X evolution including an effective population size of X that deviates from equilibrium expectations, male mutation bias, and sexually antagonistic genes (Thornton and Long 2002).
Conclusions
We propose that there are 374 genes on the anole X chromosome and provide evidence of dosage compensation in the green anole. Previously identified X-linked LGb sequences exhibited similar expression values in both sexes, consistent with complete dosage compensation for these genes. This is not the case for all genes on the X chromosome; however, as many newly identified X-linked genes do not show evidence of dosage compensation. We further found that genes with sex-biased transcription in the green anole are not clustered on the X chromosome, but are scattered throughout the entire genome, similar to mammals (Lowe et al. 2015). However, despite many genes with sex-biased expression, gene expression is, as expected, broadly similar between the sexes.
We also observed that both previously identified and proposed X-linked genes exhibit higher mean ratios of non-synonymous to synonymous substitution rates than autosomal genes. This pattern is consistent with the fast-X hypothesis, although the Ka/Ks analysis also suggests that purifying selection is strong across both autosomal and X-linked genes.
Further characterization of the green anole's sex chromosomes could be of great importance in understanding the formation of sex chromosomes in general. According to the classic model of sex chromosome evolution, highly diverged sex chromosomes should be heteromorphic; however, most vertebrate species, including many fish and lizards, have homomorphic sex chromosomes (Gamble et al. 2014). Although the green anole has heteromorphic sex chromosomes, many other members of the Anolis genus are homomorphic, and analyzing the green anole's sex chromosomes could provide insights into the formation of sex chromosomes across the genus. Given that previously identified X-linked genes appear to be conserved among anoles (Rovatsos et al. 2014b), it is likely that the newly identified X-linked genes we report here will also be X-linked in other anole species, are conserved as well. If so, the entire proposed X chromosome provides a reference that can be used in comparative studies of X-linked genes in other Anolis species. Additionally, X-linked regions appear to be conserved across Iguania (Rovatsos et al. 2014c), so we would expect to find many of the same X-linked genes on homologous sex chromosomes in various iguanian species. Because sex chromosomes appear to be highly conserved in this group, if other iguanids have highly heteromorphic sex chromosomes, our results predict that they may have also evolved dosage compensation. These results will provide a useful comparison for future studies of sex determination.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We are grateful to the editor and anonymous reviewers for very helpful comments on the manuscript. This work was supported by the School of Life Sciences and the Biodesign Institute at Arizona State University (startup to MAWS); the Mindlin Foundation and the Arizona State University SOLUR program (MAWS and SR); the School of Life Sciences Undergraduate Research Program (SOLUR) through the School of Life Sciences at Arizona State University to SR; the National Center for Research Resources and the Office of Research Infrastructure Programs (NIH grant R21 RR031305 to K.K.); National Institute of Arthritis, Musculoskeletal, and Skin Diseases (NIH grant R21 AR064935 to K.K.); and the Arizona Biomedical Research Commission (Grant 1113 to K.K.).
Literature Cited
- Alföldi J, et al. 2011. The genome of the green anole lizard and a comparative analysis with birds and mammals. Nature 477:587–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. Babraham Bioinformatics. Available from: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ [Google Scholar]
- Bachtrog D, et al. 2014. Sex determination: why so many ways of doing it? PLoS Biol. 12:e1001899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canty A, Ripley B. 2016. boot: bootstrap R (S-Plus) functions. R package version 1.3-18.
- Carrel L, Willard HF. 2005. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature 434:400–404. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Charlesworth D. 2010. Elements of evolutionary genetics. First edition. New York: W. H. Freeman; [Google Scholar]
- Conrad T, Akhtar A. 2012. Dosage compensation in Drosophila melanogaster: epigenetic fine-tuning of chromosome-wide transcription. Nat Rev Genet. 13:123–134. [DOI] [PubMed] [Google Scholar]
- Cunningham F, et al. 2015. Ensembl 2015. Nucleic Acids Res. 43:D662–D669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deakin JE, Hore TA, Koina E, Marshall Graves JA. 2008. The status of dosage compensation in the multiple X chromosomes of the platypus. PLoS Genet. 4: e1000140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean R, Harrison PW, Wright AE, Zimmer F, Mank JE. 2015. Positive selection underlies Faster-Z evolution of gene expression in birds. Mol Biol Evol. 2:2646–2656. msv138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, et al. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 43:491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillies M-A, et al. 2013. A comprehensive evaluation of normalization methods for Illumina high-throughput RNA sequencing data analysis. Brief Bioinform. 14:671–683. [DOI] [PubMed] [Google Scholar]
- Dobin A, et al. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckalbar WL, et al. 2013. Genome reannotation of the lizard Anolis carolinensis based on 14 adult and embryonic deep transcriptomes. BMC Genomics 14:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H. 2009. The different levels of genetic diversity in sex chromosomes and autosomes. Trends Genet. 25:278–284. [DOI] [PubMed] [Google Scholar]
- Gamble T, Geneva A, Glor RE, Zarkower D. 2014. Anolis sex chromosomes are derived from a single ancestral pair. Evolution 68(4):1027–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottipati S, Arbiza L, Siepel A, Clark AG, Keinan A. 2011. Analyses of X-linked and autosomal genetic variation in population-scale whole genome sequencing. Nat Genet. 43:741–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves JAM. 2016. Evolution of vertebrate sex chromosomes and dosage compensation. Nat Rev Genet. 17:33–46. [DOI] [PubMed] [Google Scholar]
- Hillier LW, et al. 2004. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 432:695–716. [DOI] [PubMed] [Google Scholar]
- Hutchins ED, et al. 2014. Transcriptomic analysis of tail regeneration in the lizard Anolis carolinensis reveals activation of conserved vertebrate developmental and repair mechanisms. PLoS One 9:e105004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hvilsom C, et al. 2012. Extensive X-linked adaptive evolution in central chimpanzees. PNAS 109:2054–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh Y, Melamed E, Yang X, Kampf K, Wang S, Yehya N, Van Nas A, Replogle K, Band MR, Clayton DF, Schadt EE, Lusis AJ, Arnold AP. 2007. Dosage compensation is less effective in birds than in mammals. J Biol. 6(1):2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh Y, Arnold AP. 2015. Are females more variable than males in gene expression? Meta-analysis of microarray datasets. Biol Sex Differ. 6:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinsella RJ, et al. 2011. Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database (Oxford) 2011:bar030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshiba-Takeuchi K, et al. 2009. Reptilian heart development and the molecular basis of cardiac chamber evolution. Nature 461:95–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kousathanas A, Halligan DL, Keightley PD. 2014. Faster-X adaptive protein evolution in house mice. Genetics 196:1131–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer M, et al. 2015. Developmental dynamics of X-chromosome dosage compensation by the DCC and H4K20me1 in C. elegans . PLoS Genet. 11:e1005698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau AC, Csankovszki G. 2015. Balancing up and downregulation of the C. elegans X chromosomes. Curr Opin Genet Dev. 31:50–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27:2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, 1000 Genome Project Data Processing Subgroup, et al. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovern MB, Holmes MM, Wade J. 2004. The green anole (Anolis carolinensis): a reptilian model for laboratory studies of reproductive morphology and behavior. ILAR J. 45:54–64. [DOI] [PubMed] [Google Scholar]
- Lowe R, Gemma C, Rakyan VK, Holland ML. 2015. Sexually dimorphic gene expression emerges with embryonic genome activation and is dynamic throughout development. BMC Genomics 16: 295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone JH, et al. 2012. Mediation of Drosophila autosomal dosage effects and compensation by network interactions. Genome Biol. 13:R28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mank JE. 2013. Sex chromosome dosage compensation: definitely not for everyone. Trends Genet. 29:677–683. [DOI] [PubMed] [Google Scholar]
- Mank JE, Axelsson E, Ellegren H. 2007. Fast-X on the Z: rapid evolution of sex-linked genes in birds. Genome Res. 17:618–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mank JE, Nam K, Ellegren H. 2009. Faster-Z evolution is predominantly due to genetic drift. Mol Biol Evol. 27(3):661–670. [DOI] [PubMed] [Google Scholar]
- McKenna A, et al. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel RP, Connallon T. 2013. The faster-X effect: integrating theory and data. Trends Genet. 29:537–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer LR, et al. 2013. The UCSC Genome Browser database: extensions and updates 2013. Nucleic Acids Res. 41:D64–D69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murtagh VJ, et al. 2012. Evolutionary history of novel genes on the tammar wallaby Y chromosome: implications for sex chromosome evolution. Genome Res. 22:498–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payer B, Lee JT. 2008. X chromosome dosage compensation: how mammals keep the balance. Annu Rev Genet. 42:733–772. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26(6):841–842. no. doi:10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2014. R: A Language and Environment for Statistical Computing. Available from: http://www.R-project.org/
- Reimand J, Arak T, Vilo J. 2011. g:Profiler—a web server for functional interpretation of gene lists (2011 update). Nucleic Acids Res. 39:W307–W315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovatsos M, Altmanová M, Pokorná MJ, Kratochvíl L. 2014a. Novel X-linked genes revealed by qPCR in the green anole. Anolis Carolinensis G3:g3.114.014084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovatsos M, Altmanová M, Pokorná M, Kratochvíl L. 2014b. Conserved sex chromosomes across adaptively radiated Anolis lizards. Evolution 68:2079–2085. [DOI] [PubMed] [Google Scholar]
- Rovatsos M, Pokorná M, Altmanová M, Kratochvíl L. 2014c. Cretaceous park of sex determination: sex chromosomes are conserved across iguanas. Biol Lett. 10:20131093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G, Chen Y-R, Blissard GW, Briscoe AD. 2014. Complete dosage compensation and sex-biased gene expression in the moth Manduca sexta . Genome Biol Evol. 6:526–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. 1989. Evolutionary relationship of Dna sequences in finite populations. Genetics 105(2):437–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton K, Long M. 2002. Rapid divergence of gene duplicates on the Drosophila melanogaster X chromosome. Mol Biol Evol. 19:918–925. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25:1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, et al. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protocols 7:562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Auwera GA, et al. 2013. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protocols Bioinform. 11:11.10.1–11.10.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicoso B, Bachtrog D. 2011. Lack of global dosage compensation in Schistosoma mansoni, a female-heterogametic parasite. Genome Biol Evol. 3:230–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicoso B, Charlesworth B. 2006. Evolution on the X chromosome: unusual patterns and processes. Nat Rev Genet. 7:645–653. [DOI] [PubMed] [Google Scholar]
- Vicoso B, Emerson JJ, Zektser Y, Mahajan S, Bachtrog D. 2013. Comparative sex chromosome genomics in snakes: differentiation, evolutionary strata, and lack of global dosage compensation. PLoS Biol. 11:e1001643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade J. 2012. Sculpting reproductive circuits: relationships among hormones, morphology and behavior in anole lizards. Gen Comp Endocrinol. 176:456–460. [DOI] [PubMed] [Google Scholar]
- Walters JR, Hardcastle TJ, Jiggins CD. 2015. Sex chromosome dosage compensation in Heliconius butterflies: global yet still incomplete? Genome Biol Evol. 7:2545–2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MA, Makova KD. 2009. Evolution and survival on eutherian sex chromosomes. PLoS Genetics. 5:e1000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson Sayres MA, Makova KD. 2013. Gene survival and death on the human Y chromosome. Mol Biol Evol. 30:781–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright AE, Mank JE. 2012. Battle of the sexes: conflict over dosage-sensitive genes and the origin of X chromosome inactivation. Proc Natl Acad Sci. 109(14):5144–5145. no. doi:10.1073/pnas.1202905109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xingfu ZQX. 2009. Dosage analysis of Z chromosome genes using microarray in silkworm, Bombyx mori . Insect Biochem Mol Biol. 39:315–321. [DOI] [PubMed] [Google Scholar]
- Zhang Z, et al. 2006. KaKs_Calculator: calculating Ka and Ks through model selection and model averaging. Genomics Proteomics Bioinform. 4:259–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.