Abstract
Blumeria graminis (Ascomycota) includes fungal pathogens that infect numerous grasses and cereals. Despite its economic impact on agriculture and its scientific importance in plant–pathogen interaction studies, the evolution of different lineages with different host ranges is poorly understood. Moreover, the taxonomy of grass powdery mildew is rather exceptional: there is only one described species (B. graminis) subdivided in different formae speciales (ff.spp.), which are defined by their host range. In this study we applied phylogenomic and population genomic methods to whole genome sequence data of 31 isolates of B. graminis belonging to different ff.spp. and reconstructed the evolutionary relationships between different lineages. The results of the phylogenomic analysis support a pattern of co-evolution between some of the ff.spp. and their host plant. In addition, we identified exceptions to this pattern, namely host jump events and the recent radiation of a clade less than 280,000 years ago. Furthermore, we found a high level of gene tree incongruence localized in the youngest clade. To distinguish between incomplete lineage sorting and lateral gene flow, we applied a coalescent-based method of demographic inference and found evidence of horizontal gene flow between recently diverged lineages. Overall we found that different processes shaped the diversification of B. graminis, co-evolution with the host species, host jump and fast radiation. Our study is an example of how genomic data can resolve complex evolutionary histories of cryptic lineages at different time scales, dealing with incomplete lineage sorting and lateral gene flow.
Keywords: phylogenomics, demographic inference, co-evolution, host jump, Blumeria graminis, grass powdery mildew
Introduction
Blumeria graminis (grass powdery mildew) is a fungal pathogen that attacks grass species belonging to the family of Poaceae. It is considered one of the most important fungal pathogens because of its economic impact on cereal crops (especially wheat and barley) and represents a model system to study biotrophic pathogens (Dean et al. 2012). Like other powdery mildews B. graminis is an obligate biotroph, depending on living host to complete its life cycle. Grass powdery mildew is considered a single species, the sub-specific taxonomical category forma specialis (f.sp.) is used to distinguish between forms which show only minimal (or no) morphological differences but are distinct because they occur on different host species (table 1). Following this definition different formae speciales (ff.spp) can normally not mate because of the strict host specialization, but they can occasionally mate on alternate hosts (Schulze-Lefert and Panstruga 2011). However, experimental crosses have been successful only for some of the ff.spp (e.g. B.g. tritici and B.g. triticale and B.g. tritici and B.g. secalis) (Hiura 1965, 1978; Troch et al. 2014; Menardo et al. 2016).
Table 1.
Isolates of B. graminis used in this study a
| Taxon | Number of isolates | Host of origin b |
|---|---|---|
| B.g. avenae | 1 | Avena sativa |
| B.g. dactylidis | 1 | Dactylis glomerata |
| B.g. hordei | 3 | Hordeum vulgare |
| B.g. on Lolium c | 1 | Lolium perenne |
| B.g. poae | 1 | Poa pratensis |
| B.g. secalis | 5 | Secale cereale |
| B.g. tritici1 d | 13 | Triticum aestivum and Triticum dicoccoides |
| B.g. tritici2 d | 6 | Triticum dicoccoides |
See Material and Methods for the accession numbers.
Plants on which the isolates were collected.
B. graminis growing on Lolium sp. was never formally designated as a f.sp.
These two groups were found to be genomically divergent despite a common host range (Ben-David et al. 2016; Menardo et al. 2016).
Based on a review of studies that assessed host ranges of different forms of B. graminis Troch and colleagues (2014) partially contested the validity of the concept of ff.spp., proposing to retain it only for forms infecting cereals which show a stronger host specialization compared to forms infecting wild grasses. Despite the importance of B. graminis as a pathogen and model for research on biotroph life style the evolutionary relationship between ff.spp. of B. graminis and even the validity of the category of forma specialis in evolutionary analysis are topics of intense debate (Panstruga and Spanu 2014). Due to the very simple morphology of B. graminis and to the absence of polymorphic traits between different lineages, evolutionary analyses of ff.spp are limited to phylogenetic studies performed on molecular data. Most of these studies are based on one or few sequenced genes, and led to discordant results and contrasting interpretations: the results of Inuma and colleagues (2007) based on 4 nuclear genes suggested a pattern of co-evolution between Blumeria graminis and its host with some exceptions (i.e. host jumps). However Troch et al. (2014) found in a phylogeny of the β-tubulin that all ff. spp. that grow on cereals (including f.sp avenae) cluster together despite their hosts being distantly related (oat belongs to the tribe Avenae while rye, wheat and barley belong to the Triticeae). These data suggest a recent origin of ff.spp. that infect cereals and they contradict the hypothesis of plant-pathogen coevolution. Similar results supporting evolutionary mechanisms that do not involve co-evolution have been observed in other pathogens (Vienne et al. 2013; Choi and Thines 2015). It is known that the analysis of few gene topologies between closely related species or in this case supposedly sub-specific taxa can lead to contrasting or uninformative results. In particular, gene trees can be different from the species tree because of incomplete lineage sorting (ILS) and lateral gene flow between lineages. A high level of ILS is normally expected between recently diverged lineages while gene flow could be substantial between sub-specific taxa (Maddison 1997; Rosenberg and Nordborg 2002; Sousa and Hey 2013; Posada 2016).
Recently the genomes of the ff.spp. hordei and tritici were sequenced and their divergence time was estimated to be approximately 6 Ma based on a molecular clock (Spanu et al. 2010; Wicker et al. 2013), two Myr younger than the estimated divergence between wheat and barley (Middleton et al. 2014) supporting the hypothesis of host tracking, a form of co-evolution in which the speciation of the pathogen is delayed compared to the speciation of the host. However the accuracy of these results depends on a correct assumption of the molecular clock rate. The largest study on B. graminis that made use of genome-wide data was conducted by Menardo et al. (2016). In that study we discovered that B.g. triticale, a f.sp. that can infect the artificial hybrid crop triticale, originated through a hybridization of isolates of the f. sp. tritici and of the f. sp. secalis. This finding underlined the importance of reticulate evolution and lateral gene flow in B. graminis. Moreover this study identified two distinct lineages of B. graminis infecting wheat. In addition, infection tests revealed that one lineage was specific to tetraploid wheat and defined it as the new f.sp. dicocci. However, Ben-David and colleagues (2016) found that the isolates of the f.sp. dicocci can grow on some hexaploid wheat genotypes that were not tested by Menardo et al. (2016), indicating that the specialization of the two lineages is not complete and that they belong to the same f.sp. In this study we will refer to B.g. tritici1 to identify the lineage named tritici in Menardo et al. (2016), to B.g. tritici2 to identify the lineage named dicocci in Menardo et al. (2016) and to B.g. tritici when we refer to both lineages.
B. graminis can infect at least 4 different tribes of Pooideae (Bromeae, Triticeae, Aveneae and Poaea), however until now all studies based on genome-wide data analyzed mildew growing on a phylogenetically limited set of hosts, barley, rye, wheat and triticale, which are all domesticated plants belonging to the Triticeae tribe. Furthermore all studies based on a large set of mildew isolates from phylogenetically distant host species made use of a small number of molecular markers which resulted in contradictory results, probably due to the poor performance of phylogenetic inference methods in presence of ILS and lateral gene flow between lineages.
Here we used genome sequences of 31 B. graminis isolates collected on 8 different plant species belonging to three of the tribes attacked by grass powdery mildew. We aimed to reconstruct the evolutionary relationships between different lineages of B. graminis and used phylogenomic methods to infer the species tree, the divergence time between lineages and to identify conflicts between gene trees. Moreover when we found discordance between gene trees we applied a coalescent based approach to identify the most likely species tree and tested for the presence of lateral gene flow between lineages. We reconstructed the evolutionary history of grass powdery mildew finding evidence for co-evolution between host and pathogen, host jumps or host range expansions and fast radiation. Our results highlight the importance of using a diverse set of methods that can deal with different levels of isolation and divergence between lineages.
Materials and Methods
Sampling
We included in the dataset for the phylogenomic analysis all the currently available powdery mildew genomes (3 B.g. hordei isolates: GCA_000151065.1, AOLT00000000 and AOIY01000000; 19 B.g. tritici isolates: PRJNA183607 and SRP062198; 5 B.g. secalis isolates: SRP062198; one isolate of Golovinomyces orontii (Arabidopsis powdery mildew): PRJEA50317; one isolate of Erysiphe pisi (pea powdery mildew): PRJEA50315; and the reference genome of Neurospora crassa as outgroup: PRJNA13841) (Galagan et al. 2003; Spanu et al. 2010; Hacquard et al. 2013; Wicker et al. 2013; Menardo et al. 2016). Additionally we sampled and sequenced powdery mildew on Poa pratensis (f.sp. poae), on Avena sativa (f.sp. avenae) on Lolium perenne (not formally described as a f.sp.) and on Dactylis glomerata (f.sp. dactylidis). Infected plants of Poa pratensis and Lolium perenne were obtained from the Agroscope Research station in Reckenholz (Zurich, Switzerland), infected plants of Dactylis glomerata were collected in Albisrieden, (Zurich, Switzerland). Single infected plants have been collected and kept in an isolated climate chamber at 20 °C and in 16 h light/8 h dark conditions. Spores were collected every 5 days. Oat leaves infected with powdery mildew were kindly provided by Dr Okon and Prof. Kowalczy (University of Lubklin, Poland). Oat powdery mildew was maintained on detached leaf segments with fresh spores and the infected leaf segments were kept on benzimidazole agar plates at 20 °C, 70% humidity and in 16 h light/8 h dark conditions.
DNA Extraction and Sequencing
DNA was extracted from spores as described by Bourras et al. (2015). 125 bp paired-end libraries were created and sequenced with Illumina Hi-Seq at the Functional Genomics Center of Zürich obtaining about 123, 77, 109, and 83 millions of paired reads for B.g. poae, avenae, dactiliydis and B. graminis infecting Lolium, respectively, all sequences are deposited at the sequence read archive with accession number SRP062198. Bad quality reads were filtered out with sickle 1.33 (Joshi and Fass 2011) with standard parameters. All de novo assemblies were performed with CLC Genomic Workbench 8 with standard parameters and minimum contig size of 200 bp.
Identification of Homologous Genes, Alignments and Gene Tree Discordance Analysis
In a previous study we identified a set of 206 single copy genes that are suitable for phylogenetic analysis in powdery mildew (Menardo et al. 2016). We could retrieve 93 of these genes in all newly sequenced genomes with gmap (version 2013-07-20) (Wu and Nacu 2010), using the genes of the B.g. tritici reference isolate 96224 as template. The concatenated alignment resulted to be 239,655 bp long and contained a minimum of 91,299 bases for G. orontii. All alignments were performed with muscle 3.8.31 (Edgar 2004). In Menardo et al. (2016) we also identified 4,556 homologous single copy genes in the ff. spp. tritici, secalis, hordei, and triticale. These genes could not be used in the phylogenomic analysis because they are too divergent or absent in the outgroups N. crassa, G. orontii and E. pisi. However we retrieved those 4556 single copy genes in the genome of one isolate for each of the lineages of Blumeria graminis (96224 for B.g. tritici1, 220 for B.g. tritici2, the reference isolate DH14 for B.g. hordei and S-1201 for B.g. secalis, for all other lineages there was only one isolate available). We could find 4,057 of these genes as a single copy in all analyzed genomes and aligned them with muscle 3.8.31 (Edgar 2004). Gene trees were inferred with RAXML 8.0.22 (Stamatakis 2014) using a GTR + GAMMA model (Yang 1993, 1994). We used Newick Utilities (Junier and Zdobnov 2010) to identify and summarize topology patterns.
Bayesian Phylogeny and Divergence Time Estimation
With the concatenated alignment (93 genes and 34 taxa), we performed a phylogenetic analysis with MrBayes 3.2.2 (Ronquist et al. 2012) using the independent gamma rate clock model (Lepage et al. 2007). Variation of substitution rates across sites was modeled with a discretized (4 categories) gamma (Γ) distribution (Yang 1993, 1994). The chains have been let free to sample all models of the GTR model family using reversible jump Monte Carlo Markov Chain (Huelsenbeck et al. 2004). We ran 10 independent analyses of 5 million generations each, sampling every 10,000 generations and discarding the first 1,250,000 generation as burn-in. The analysis was repeated several times with different number of generation (5 and 25 million with 25% burn-in) giving very similar results in all the analyses. To calibrate the tree, we set three calibration points using uniform priors: the first is the divergence between Leotiomycetes (to whom belong the powdery mildew) and Sordariomycetes (to whom belongs N. crassa) and was defined as the narrowest range including all the different estimations of Prieto and Wedin (2013) and Beimforde et al. (2014) (160–320 Ma) . For the other calibration points we used the estimated divergence between the hosts of different mildews and set it as the oldest possible age for the divergence of the mildews. This is equivalent to a flat prior for the divergence time spanning from the oldest possible divergence of the host to the present. We set the divergence between monocot and dicot as oldest possible divergence between B. graminis and the dicot powdery mildew G. orontii and E. pisi (200 Ma, Chaw et al. 2004). The origin of Pooideae was used as oldest possible radiation of the ff. spp. of B. graminis (in particular the split between B.g. poae and the other ff.spp.) (57 Ma, Bouchenak-Khelladi et al. 2010). All calibration points are secondary calibration points, meaning that they are estimations obtained from trees that were calibrated with fossil age estimations.
Mapping, SNP Calling and Principal-Component Analysis (PCA)
We observed that all isolates of the ff. spp. tritici, dactylidis and secalis cluster together in the phylogenomic analysis and are much more closely related between them than the other ff.spp. We therefore mapped the raw Illumina reads for all isolates of these ff. spp. on the B.g. tritici reference genome (Wicker et al. 2013) using Bowtie2 (Langmead and Salzberg 2012) with option—score-min L, −0.6, −0.25 (this option allows for approximately four mismatches every 100 bp). We used the following command in SAMtools 0.1.19 (Li et al. 2009) to convert formats and collect information about single genomic positions: view, sort, mpileup -q 15 (only reads with mapping quality greater than 15 were considered). Finally, we used bcftools to generate a VCF file that was parsed with in-house Perl scripts (available upon request). We considered as high-confidence SNPs only positions with a minimum mapping score of 20, a minimum coverage of 8× and a minimum frequency of the alternative call of 0.9. The principal component analysis was performed with the R package GAPIT (Lipka et al. 2012).
Fastsimcoal2
To make a more efficient use of the genome-wide data for the clade that include B.g. tritici, B.g. secalis, and B.g. dactylidis and to test for gene flow between these different lineages we used a method that fits demographic models to the observed multi-dimensional site frequency spectra implemented in fastsimcoal 2.5.2.8 (Excoffier et al. 2012). We selected unlinked SNPs (at least 2,500 bp apart, average r 2 between adjacent SNPs in B.g. tritici = 0.12, computed with VCFtools 0.1.14 Danecek et al. 2011). Additionally, to obtain presumably neutral SNPs we selected only position at least 2,500 bp far from the closest gene. Finally we selected only SNPs without missing data. After filtering we obtained 19,270 SNPs. The folded site frequency spectrum (FSFS) was calculated with a home-made Perl script (available upon request). Since fastsimcoal2 cannot perform model search we limited our analysis to the three most commonly observed tree topologies. To test for introgression between the different lineages we modified the three basic models adding one bidirectional introgression between all possible pairs of lineages (6 additional models for each of the basic trees). To distinguish between introgression and migration we modified the basic tree models allowing migration between a pair of lineages for all possible pairs combinations (6 additional models for each of the basic trees). Finally we tested the hypotheses that B.g. tritici1 originated through hybridization of B.g. tritici2 and an unsampled lineage (as proposed in Menardo et al. 2016) implementing this scenario for each of the basic trees (3 additional models). In total, we tested 42 different models. We optimized the likelihood of each model in 100 independent runs, each using 100,000 simulations for every cycle of the conditional maximization algorithm (ECM). We set a minimum of 10 ECM cycles and a maximum of 40, entries in the FSFS with less than 10 observations were not considered in the likelihood computation. Control files of the fastsimcoal2 analysis are provided in Supplementary Material, model comparison was performed with the Akaike information criterium (AIC).
Results
Phylogenomic Analysis
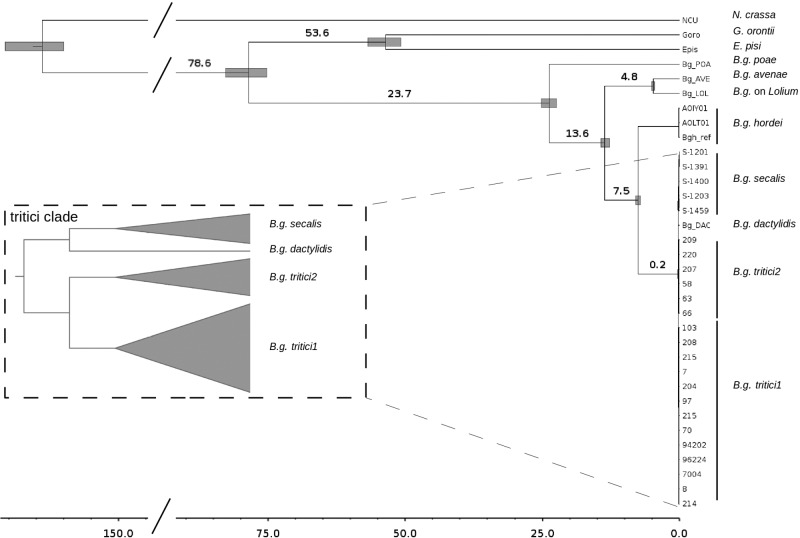
To reconstruct the phylogenetic relationships between different lineages of B. graminis we used 93 single copy genes (total alignment of 239,655 bp) in a bayesian partitioned analysis. We found that the ff. spp. of B. graminis cluster as a monophyletic group that diverged from the mildews infecting dicots between 75 and 83 Ma (95% c.i.) (fig. 1). The diversification of the ff. spp. started between 22.4 and 25.1 Ma (95% c.i.) with the divergence of B.g. poae. Poa was the first lineage to diverge among the grasses infected by lineages of B. graminis analyzed in this study, 26-39 Ma (95% c.i.) (Bouchenak-Khelladi et al. 2008, 2010), the topological patterns and the divergence time estimations of B.g. poae and its host coincide and support the hypothesis of coevolution between B.g. poa and Poa. Between 12.8 and 14.3 Ma (95% c.i.) the lineages of B.g. avenae and B. graminis growing on Lolium diverged. In this case the topology of the pathogen is only partially concordant with the topology of the host, the tribe Aveneae is the sister taxon of the Poeae tribe and not of the Triticeae to whom Lolium belongs. This could be the result of a host jump or host range expansion of B. graminis from Triticeae to the Avenae. Between 7.1 and 8.0 Ma (95% c.i.) B.g. hordei diverged from the tritici clade. The divergence between barley and wheat was estimated to be about 8 Ma by Middleton et al. (2014), also here the topological patterns and the divergence time estimations of B.g hordei and Hordeum coincide and support the hypothesis of coevolution. Finally we found a recent radiation between 170,000 and 280,000 years ago (95% c.i.) that led to the origin of the ff. spp. infecting Secale, Triticum and Dactylis (named tritici clade from now on). Dactylis belongs to the Poeae while Secale and Triticum belong to the Triticeae, suggesting another very recent host jump or host range expansion from Triticeae tribe to Poeae tribe. Overall the phylogenomic analysis supported the hypothesis of co-evolution of some lineages of B. graminis and their hosts, but also revealed topological patterns which suggest possible host jumps or host range expansions (B.g. avenae and B.g. dactylidis) and a recent radiation of the ff. spp. secalis, tritici and dactylidis.
Fig. 1.—
Bayesian consensus tree of powdery mildew strains. Labels on branches represent the median age estimate for the divergence time, gray bars represent the 95% confidence interval of the divergence time, the scale is in million years. In the dashed panel a zoom of the tritici clade is shown (not in scale). All visible ramifications have maximum posterior probabilities.
Genome-Wide Gene Topologies Analysis
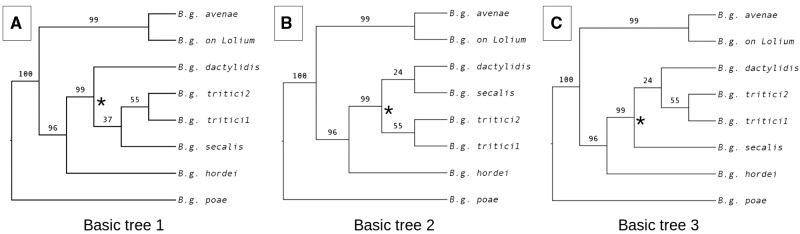
To investigate the occurrence of incomplete lineage sorting (ILS), reticulate evolution or lateral gene flow in B. graminis that could be overlooked by the concatenated phylogenomic analysis we identified 4,057 homologous single copy genes in B. graminis and inferred the maximum likelihood tree for each of them singularly using one isolate for each lineage of B. graminis. We found 75 different topologies among the 4,057 gene trees with the 14 most frequent topologies observed in more than 95% of the trees. We computed the proportion of gene trees that support the clades observed in the three most frequently inferred topologies (found for 53.7% of genes) and found that the incongruence between topologies are localized in the tritici clade (fig. 2). Conflicts between gene topologies could be due to ILS, to gene flow or lack of nucleotide variation.
Fig. 2.—
The first (a) second (b) and third (c) most frequently observed gene tree topologies, representing 19.6%, 18.3% and 15.8%, respectively of the 4,057 single copy gene trees. The percentage of single gene tree topologies that support a clade is reported on the relative branch, most of the discordances between tree topologies is localized in the tritici clade (marked with *), in particular regarding the relative positions of B.g. secalis and B.g. dactylidis.
PCA and Demographic Inference with Fastsimcoal2
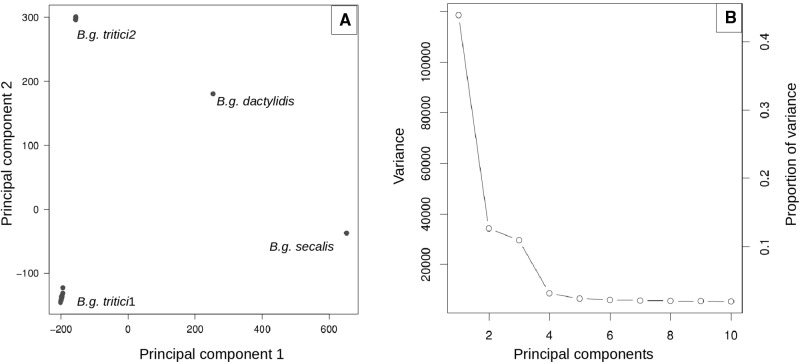
Population genomics methods based on SNP data can be more informative on closely related lineages than phylogenomic analysis. We profited from the similarity between the isolates belonging to the tritici clade and mapped the raw Illumina reads on the B.g. tritici reference isolate 96224 with comparable overall alignment rate for all isolates (∼70%). After filtering we obtained 684,338 SNPs, that were used in a PCA and found that isolates of B.g. secalis, tritici1 and tritici2 cluster in three different groups as described in Menardo et al. (2016). The B.g. dactylidis isolate does not cluster with any of these groups indicating that it is part of a different lineage (fig. 3). In this study we included all available B. graminis genomes except B.g. triticale sequences which were object of a previous study. To exclude the possibility that B.g. dactylidis and B.g. triticale are genomically similar and belong to the same lineage we repeated the PCA analysis after addition of two B.g. triticale isolates. We found that they cluster in a separate group that does not include B.g. dactylidis (supplementary Appendix S1, Supplementary Material online).
Fig. 3.—
a) Principal component analysis of B. graminis isolates based on 684,338 SNPs. There are four distinct groups that correspond to the four lineages of the tritici clade identified with the phylogenomic analysis. b) Proportion of the variance explained by different principal components, the first and the second together explain 56.6% of the variance, indicating a high level of genetic structure in the dataset.
SNP data can be used to test models for different evolutionary history of closely related lineages. We identified 19,270 neutral (at least 2,500 bp far from the closest gene) unlinked SNPs to compute the folded multidimensional site frequency spectrum (FSFS) of the four lineages (B.g. tritici1, tritici2, secalis and dactylidis) and used fastsimcoal2 to fit different demographic models to the observed FSFS. Due to the high dimensionality of the models evaluated by fastsimcoal2 and to the computational requirement it is not possible to exhaustively search the model space. Therefore we decided to test the three most frequent gene tree topologies (shown in fig. 2). We found that the model representing the topology in figure 2a (basic tree 1) was the most likely followed by the model representing the topology in figure 2c (basic tree 3), and the model representing the topology in figure 2b (basic tree 2) being the least likely (table 2). We then tested for gene flow between lineages. We modified each of the three basic trees adding introgression or migration between all possible pairs of lineages (12 additional models for each basic tree). We found that the majority or all the models with gene flow performed better than basic tree 1, 2 and 3 (9, 12 and 8 models, respectively), and that introgression models generally performed better than migration models (i.e. the best 7 models were introgression models) (table 2). Additionally, we tested the hypothesis that B.g. tritici1 originated from a hybridization between B.g. tritici2 and an unsampled lineage based on Menardo et al. (2016). We implemented this scenario for each of the three basic trees and found that the hybridization models are always more likely than the basic tree models but less likely than other models with introgression and migration (table 2). We found that the most likely among the tested model was basic tree 3 with introgression between B.g. tritici2 and B.g. secalis, (basic_tree3_intro12 in table 2 and fig. 4). Overall these results suggest the occurrence of gene flow between lineages in the tritici clade, especially between B.g. tritici1, B.g. tritici2, and B.g. secalis).
Table 2.
Results of the fastsimcoal2 analysis
| basic_tree1 |
basic_tree2 |
basic_tree3 |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Model name a | logL b | N. of parameters c | AIC d | Model name a | logL b | N. of parameters c | AIC d | Model name a | logL b | N. of parameters c | AIC d |
| basic_tree1_intro01 | −38198 | 13 | 76421 | basic_tree2_intro12 | −38314 | 14 | 76656 | basic_tree3_intro12 | −37181 | 13 | 74387 |
| basic_tree1_intro12 | −38227 | 13 | 76480 | basic_tree2_intro02 | −38602 | 14 | 77232 | basic_tree3_intro02 | −38364 | 13 | 76753 |
| basic_tree1_intro02 | −38367 | 13 | 76760 | basic_tree2_intro01 | −39396 | 14 | 78820 | basic_tree3_mig12 | −39104 | 12 | 78233 |
| basic_tree1_mig01 | −38909 | 12 | 77841 | basic_tree2_mig02 | −39429 | 12 | 78882 | basic_tree3_intro01 | −39124 | 13 | 78275 |
| basic_tree1_mig12 | −39120 | 12 | 78265 | basic_tree2_mig01 | −39590 | 12 | 79204 | basic_tree3_mig02 | −39255 | 12 | 78533 |
| basic_tree1_H | −39132 | 13 | 78290 | basic_tree2_mig12 | −39635 | 12 | 79294 | basic_tree3_mig01 | −39785 | 12 | 79595 |
| basic_tree1_mig13 | −39150 | 12 | 78324 | basic_tree2_intro13 | -40017 | 13 | 80060 | basic_tree3_H | −40006 | 13 | 80037 |
| basic_tree1_intro13 | −39303 | 13 | 78632 | basic_tree2_H | −40425 | 13 | 80876 | basic_tree3_intro23 | −40146 | 13 | 80319 |
| basic_tree1_intro03 | −39316 | 13 | 78658 | basic_tree2_intro23 | −40453 | 13 | 80931 | basic_tree3_mig23 | −40284 | 12 | 80591 |
| basic_tree1_mig02 | −39342 | 12 | 78707 | basic_tree2_intro03 | −40493 | 14 | 81014 | basic_tree3 | −40306 | 10 | 80632 |
| basic_tree1 | −39439 | 10 | 78898 | basic_tree2_mig23 | −40658 | 12 | 81340 | basic_tree3_intro13 | −40355 | 13 | 80736 |
| basic_tree1_mig03 | −39440 | 12 | 78904 | basic_tree2_mig03 | −40786 | 12 | 81596 | basic_tree3_mig03 | −40368 | 12 | 80760 |
| basic_tree1_mig23 | −39462 | 12 | 78947 | basic_tree2_mig13 | −40935 | 12 | 81895 | basic_tree3_intro03 | −40390 | 13 | 80807 |
| basic_tree1_intro23 | −39483 | 13 | 78992 | basic_tree2 | −41382 | 10 | 82784 | basic_tree3_mig13 | −40434 | 12 | 80891 |
Models tested with fastsimcoal2 are listed in three columns divided by topology and ranked from the most to the least likely. Basic_tree1 (column 1), basic_tree2 (column 2) and basic_tree3 (column 3) indicate the topologies shown in figure 2a–c, respectively. The suffixes intro and mig indicate that introgression or migration was modeled between the two lineages reported at the end of the model name (0 = B.g. tritici1, 1 = B.g. tritici2, 2 = B.g. secalis, 3 = B.g. dactylidis), the suffix H indicate a hybridization model in which B.g. tritici1 originated from a hybridization between B.g. tritici2 and an unsampled lineage. The best model is highlighted in dark grey.
Log-likelihood of the model.
Number of parameters of the model.
Akaike information criterium.
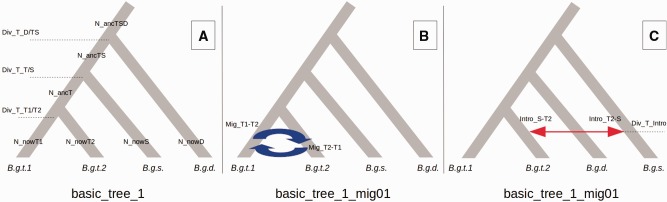
Fig. 4.—
Examples of the demographic models and their parameters used to test different evolutionary hypothesis for the tritici clade. a) The model basic_tree1 resulted to be the most likely among the models without lateral gene flow. The parameters of this class of models (models without gene flow, basic_tree1, basic_tree2 and basic_tree3) are also included in the more complex models with migration or introgression. N_now parameters represent the population sizes of the contemporary lineages B.g. tritici1 (B.g.t.1), B.g. tritici2 (B.g.t.2), B.g. secalis (B.g.s.) and B.g. dactylidis (B.g.d.). N_anc parameters represent the population sizes of the ancestral lineages. Div_T parameters represent the divergence time between lineages. b) The model basic_tree1_mig01 resulted to be the most likely among the models with migration between two lineages. This class of models (models with migration between two lineages) has two additional parameters: Mig parameters represent the migration rate between two lineages. c) The model basic_tree3_intro12 resulted to be the most likely among models with introgression between lineages and the most likely among all tested models. This class of models (models with introgression between two lineages) has three additional parameters: Intro parameters represent the proportion of genealogies that move between two lineages at the introgression time, Div_T_intro represent the time of the admixture event.
Discussion
Co-evolution between Some Lineages of B. Graminis and Its Host
The phylogenomic analysis presented here shows an identity of phylogenetic topologies and divergence times between some lineages of B. graminis (B.g. poae and hordei) and their host species, suggesting that they have co-evolved (fig. 5). The inference of absolute time divergence in the absence of phylogenetically closely related fossils for calibration or accurate estimate of the mutation rate is very challenging (Benton and Donoghue 2007). Unfortunately this is the case for both B. graminis and its host. Moreover the estimation of divergence time of B. graminis lineages, the divergence of Poa and Lolium from the Triticeae and the divergence between barley and wheat are based on three datasets that differ drastically in size (93 nuclear genes, three plastidial genes and whole chloroplast genome respectively) (Bouchenak-Khelladi et al. 2010; Middleton et al. 2014). Therefore, the comparison of divergence time between fungal and plant lineages should be taken with caution. Nevertheless, we found that our estimation of the divergence between B.g. hordei and the tritici clade is very similar to the estimation of divergence between their hosts, wheat and barley (7.1–8.0 and 6.0–10.3 Ma, respectively) and to the estimation obtained by Wicker et al. (2013) assuming a molecular clock (5.2–7.4 Ma) . The divergence time between B.g. poae and the other lineages of B. graminis is slightly younger than the divergence time between Poa and the Triticeae (22–25 Ma and 26–39 Ma) while the divergence between the lineage of B. graminis infecting Lolium and the tritici clade is 8 Myr younger than the divergence between Lolium and the Triticeae (13–14 Ma and 22–33 Ma). We noticed that the only divergence time estimated by both Middleton et al. (2014) and Bouchenak-Khelladi et al. (2010), between barley and wheat, was overestimated by Bouchenak-Khelladi et al. (2010) compared to Middleton et al. (2014) (8–9 Ma and 10–20 Ma). This could explain the discrepancies in divergence times between lineages of B. graminis growing on Poa and Lolium and their hosts. The accuracy of divergence time obtained in this study depends on the correctness of the calibration points, in particular the divergence between Leotiomycetes and Sordariomycetes, nevertheless our results are comparable with the estimation of Wicker et al. (2013) based on a molecular clock.
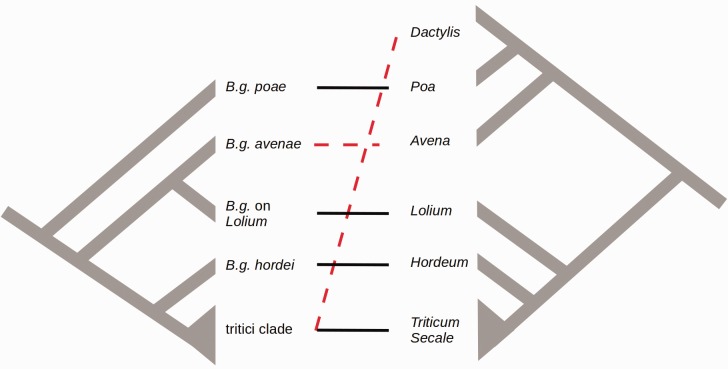
Fig. 5.—
Model of the co-evolution between B. graminis and the Pooideae. The phylogenetic tree of B. graminis is shown on the left, a simplified version of the phylogenetic tree of the Pooideae is on the right (modified from Bouchenak-Kelladi et al. 2008). Pathogen and host co-evolved (black solid line) with the exception of B.g dactylidis in the tritici clade and B.g. avenae (red dashed line)
Two Major Host Jumps or Host Range Expansions of B. Graminis
Not all lineages of B. graminis were found to follow a pattern of co-evolution. Specifically the phylogenetic positions of the ff.spp. avenae and dactylidis do not correspond to the positions of their hosts (Avena and Dactylis) on the phylogenetic tree of the Pooideae, suggesting two host jumps or host range expansion (fig. 5). Moreover the radiation of the tritici clade is very recent (170,000–280,000 years ago) and differs from the divergence time of the Triticeae (wheat and rye diverged about 4 Ma). Previous phylogenetic studies, based on four nuclear genes, identified a clade of closely related isolates, which included mildew collected on Triticum spp., Secale cereale, Agropyron spp., Elymus libanoticus, Aneurolepidium chinense and Aegilops tauschi (Inuma et al. 2007; Troch et al. 2014). This clade corresponds to the tritici clade observed in this study. Additionally, we found an isolate of the f.sp. dactylidis that belong to this clade. This is in contradiction with the study of Inuma et al. (2007) that found that isolates infecting Dactylis glomerata clustered outside the tritici clade. Since this was observed for all four genes analyzed by Inuma et al. (2007) it seems possible that there are phylogenetically different lineages that can infect Dactylis, and therefore the f.sp. dactylidis is probably not a monophyletic group.
Gene Flow between Recently Diverged Lineages of the Tritici Clade
Both the phylogenomic analysis and the PCA revealed a monophyletic group of four lineages that differentiated very recently (less than 280,000 years ago). Using coalescent simulation to fit different demographic models to the observed folded site frequency spectrum we found evidence for lateral gene flow between lineages of the tritici clade. We also found that the tree topology recovered by the phylogenomic analysis is the least likely of all tested models. These findings confirm the importance of considering ILS and lateral gene flow in the reconstruction of evolutionary history of lineages, in particular between closely related ones (Nater et al. 2015). The occurrence of later gene flow is supported by observations on lack of reproductive barriers between ff. spp. Several lineages of the tritici clade can mate and produce fertile progeny (ff. spp. secalis - tritici, tritici – agropyri, Hiura 1965). Moreover Menardo et al. (2016) found that triticale powdery mildew originated from a hybridization of B.g. secalis and B.g. tritici. On the contrary all attempts to cross lineages of the tritici clade with other older lineages of B. graminis failed (Hiura 1978; Troch et al. 2014). We want to point out that the method that we used to infer the probability of different demographic models does not allow for extensive search of the model space. It is possible that more complex not tested models have a higher likelihood. Moreover, the results of Inuma et al. (2007) suggest that there are additional lineages in the tritici clade attacking different wild grasses which we did not sample in this study. More sequencing efforts, which will have to include multiple isolates for each of the plant host species, are needed to draw a complete picture of the composition and evolution of the tritici clade. It is possible that the analysis of genomics data obtained from a larger sample’s set will result in a different interpretation of the evolutionary history of B. graminis in general and of the tritici clade in particular. However based on the available data we speculate that the tritici clade is composed of several radiating lineages with different host ranges, these lineages can exchange genetic material between them through introgression and hybridization. This results in a high potential for the emergence of new pathogens with new virulence spectra.
Formae Speciales in B. G raminis and Evolutionary Analysis
The classification of B. graminis in different formae speciales was introduced for the first time by Marchal (1902) and it is used to define “forms” that belong to the same species, are morphologically not distinguishable but infect different plant species (Schulze-Lefert and Panstruga 2011). According to this definition a f.sp. does not necessarily represent a distinct evolutionary unit (lineage). However the specialization on different hosts implies, at least in theory, barriers to gene flow between different ff. spp. and therefore defines ff. spp. as separately evolving lineages which is the only necessary property of a species according to the unified species concept (de Queiroz 2007).
Nevertheless here we focus on the systematic status of ff. spp. in B. graminis in the light of the genomics data we presented in this paper. We consider first the ff. spp. that do not belong to the tritici clade (hordei, poae, avenae and the not formally defined form growing on Lolium). These ff.spp. represent different lineages that we estimated to be separated by at least 4 million years of independent evolution, even if we cannot exclude the presence of some degree of gene flow between them. This observation together with the evidence for reproductive barriers between these ff.spp. (all crosses attempted failed to produce fertile chasmotecia, Troch et al. 2014), suggests that they lack the characteristics of sub specific categories and that it would be more indicated to refer to them as species. Since for these ff.spp. we sequenced only one or few individuals and there is evidence that B. graminis growing on Lolium and on Avena are not monophyletic groups (Inuma et al. 2007; Troch et al. 2014) we refrain from modifying the current taxonomy in order to avoid confusion and leave this task to future studies which will use whole genome data from several individuals of each f .sp.
Lineages belonging to the tritici clade (B.g. tritici1, B.g. tritici2, B.g. secalis and B.g. dactylidis) are more closely related than the others and exchange genetic material between them. Thus, for them a sub-specific classification is justified. However these ff.spp. cannot always be considered as evolutionary lineages because we found two genetically different lineages belonging to the same f. sp. (tritici1 and tritici2). We therefore recommend caution when using the concept of f.sp. in B. graminis. We suggest that this should be strictly limited to define all isolates growing on the same host and no evolutionary implications should refer to this concept.
Conclusions
The advent of next generation sequences provided researchers that study species evolution and attempt to reconstruct the tree of life with a great amount of data. One consequence of this has been the full recognition of the difference between gene trees and species trees and of the processes that cause it (ILS and lateral gene flow) (Posada 2016). These processes have different relevance in different systematic groups and at different timescales in the same group. Our work shows how one can reconstruct evolutionary histories with genomics data using a diverse set of methods that are suited for lineages with a different level of divergence and isolation. The application of these methods to the grass powdery mildew B. graminis allowed us to disentangle a complex evolutionary trajectory that includes co-evolution between pathogen and host, host jumps and fast radiations.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Dr Franz Schubiger (Reckenholz Agroscope research station, Zurich, Switzerland), Dr Okon and Prof. Kowalczy (University of Lubklin, Poland) for providing plant and fungal material used in this study. We thank the Functional Genomics Center Zurich for performing the sequencing, Linda Lüthi, Helen Zbinden and Geri Herren for technical support. We are grateful to Dr Bouchenak-Kelladi for providing access on data regarding the phylogeny and divergence time of grasses. This work was supported by the University Priority Program “Evolution in action” of the University of Zurich and the grant 310030-163260 from the Swiss National Science Foundation.
Literature Cited
- Beimforde C, et al. 2014. Estimating the Phanerozoic history of the Ascomycota lineages: Combining fossil and molecular data. Mol. Phylogenet. Evol. 77:307–319. [DOI] [PubMed] [Google Scholar]
- Ben-David R, et al. 2016. Differentiation among Blumeria graminis f. sp. tritici isolates originating from wild versus domesticated Triticum species in Israel. Phytopathology. doi:10.1094/PHYTO-07-15-0177-R. [DOI] [PubMed] [Google Scholar]
- Benton M. J., Donoghue P. C. 2007. Paleontological evidence to date the tree of life. Molecular biology and evolution, 24:(1)26–53. [DOI] [PubMed] [Google Scholar]
- Bouchenak-Khelladi Y, et al. 2008. Large multi-gene phylogenetic trees of the grasses (Poaceae): progress towards complete tribal and generic level sampling. Mol. Phylogenet. Evol. 47:488–505. [DOI] [PubMed] [Google Scholar]
- Bouchenak-Khelladi Y, Verboom GA, Savolainen V, Hodkinson TR. 2010. Biogeography of the grasses (Poaceae): A phylogenetic approach to reveal evolutionary history in geographical space and geological time. Bot. J. Linn. Soc. 162:543–557. [Google Scholar]
- Bourras S, et al. 2015. Multiple Avirulence Loci and Allele-Specific Effector Recognition Control the Pm3 Race-Specific Resistance of Wheat to Powdery Mildew. Plant Cell 27:2991–3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaw SM, Chang CC, Chen HL, Li WH. 2004. Dating the monocot-dicot divergence and the origin of core eudicots using whole chloroplast genomes. J. Mol. Evol. 58:424–441. [DOI] [PubMed] [Google Scholar]
- Choi YJ, Thines M. 2015. Host jumps and radiation, not co‐divergence drives diversification of obligate pathogens. A case study in downy mildews and Asteraceae. PloS One 10:e0133655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, et al. 2011. The variant call format and VCFtools. Bioinformatics 27:2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean R, et al. 2012. The top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13:414–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Dupanloup I, Huerta-Sanchez E, Sousa VC, Foll M. 2012. Robust demographic inference from genomic and SNP Data. PLoS Genet. 9:e1003905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galagan JE. et al. 2003. The genome sequence of the filamentous fungus Neurospora crassa . Nature 422:859–868. [DOI] [PubMed] [Google Scholar]
- Hacquard S. et al. 2013. Mosaic genome structure of the barley powdery mildew pathogen and conservation of transcriptional programs in divergent hosts. Proc. Natl. Acad. Sci. U S A. 110:E2219–E2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiura U. 1978. Genetic basis of formae speciales. In: The Powdery Mildew. Spencer, D.M. p. 101–128.
- Hiura U. 1965. Sexual compatibility between form species of Erysiphe graminis DC, and variability of the ascospore derived from the inter-form-specific hybridization. Nogaku Kenkyu 51:67–73. [Google Scholar]
- Huelsenbeck JP, Larget B, Alfaro ME. 2004. Bayesian phylogenetic model selection using reversible jump Markov chain Monte Carlo. Mol. Biol. Evol. 21:1123–1133. [DOI] [PubMed] [Google Scholar]
- Inuma T, Khodaparast SA, Takamatsu S. 2007. Multilocus phylogenetic analyses within Blumeria graminis, a powdery mildew fungus of cereals. Mol. Phylogenet. Evol. 44:741–751. [DOI] [PubMed] [Google Scholar]
- Joshi NA, Fass JN. 2011. Sickle: A sliding-window, adaptive, quality-based trimming tool for FastQ files (Version 1.33) [Software]. Available at https://github.com/najoshi/sickle.
- Junier T, Zdobnov EM. 2010. The Newick utilities: high-throughput phylogenetic tree processing in the UNIX shell. Bioinformatics 26:1669–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepage T, Bryant D, Philippe H, Lartillot N. 2007. A general comparison of relaxed molecular clock models. Mol. Biol. Evol. 24:2669–2680. [DOI] [PubMed] [Google Scholar]
- Li H, et al. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipka AE, et al. 2012. GAPIT: genome association and prediction integrated tool. Bioinformatics 28:2397–2399. [DOI] [PubMed] [Google Scholar]
- Maddison WP. 1997. Gene trees in species trees. Syst Biol. 46:523–536. [Google Scholar]
- Marchal E. 1902. De la specialisation du paristisme chez l’Erysiphe graminis. Acad. Sci. Paris 135:210–212. [Google Scholar]
- Menardo F, et al. 2016. Hybridization of powdery mildew strains gives raise to pathogens on novel agricultural crop species. Nat. Genet. 48:201–205. [DOI] [PubMed] [Google Scholar]
- Middleton CP, et al. 2014. Sequencing of chloroplast genomes from wheat, barley, rye and their relatives provides a detailed insight into the evolution of the Triticeae tribe. PLoS One 9:e85761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nater A, Burri R, Kawakami T, Smeds L, Ellegren H. 2015. Resolving evolutionary relationships in closely related species with whole-genome sequencing data. Syst. Biol. 64:1000–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panstruga R, Spanu PD. 2014. Powdery mildew genomes reloaded. New Phytol. 202:13–14. [DOI] [PubMed] [Google Scholar]
- Posada D. 2016. Phylogenomics for systematic biology. Syst. Biol. 65:353–356. [DOI] [PubMed] [Google Scholar]
- Prieto M, Wedin M. 2013. Dating the diversification of the major lineages of Ascomycota (Fungi). PLoS One 8:e65576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queiroz KD. 2007. Species concepts and species delimitation. Syst. Bot. 56:879–886. [DOI] [PubMed] [Google Scholar]
- Ronquist F, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61:539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg NA, Nordborg M. 2002. Genealogical trees, coalescent theory and the analysis of genetic polymorphisms. Nat. Rev. Genet. 3:380–390. [DOI] [PubMed] [Google Scholar]
- Schulze-Lefert P, Panstruga R. 2011. A molecular evolutionary concept connecting nonhost resistance, pathogen host range, and pathogen speciation. Trends Plant Sci. 16:117–125. [DOI] [PubMed] [Google Scholar]
- Sousa V, Hey J. 2013. Understanding the origin of species with genome-scale data: modelling gene flow. Nat. Rev. Genet. 14:404–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanu PD, et al. 2010. Genome expansion and gene loss in powdery mildew fungi reveal tradeoffs in extreme parasitism. Science 330:1543–1546. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troch V, et al. 2014. Formae speciales of cereal powdery mildew: close or distant relatives?. Mol. Plant Pathol. 15:304–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vienne DM, et al. 2013. Cospeciation vs host‐shift speciation: methods for testing, evidence from natural associations and relation to coevolution. New Phytol. 198:347–385. [DOI] [PubMed] [Google Scholar]
- Wicker T, et al. 2013. The wheat powdery mildew genome shows the unique evolution of an obligate biotroph. Nat. Genet. 45:1092–1096. [DOI] [PubMed] [Google Scholar]
- Wu TD, Nacu S. 2010. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 26:873–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 1994. Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: Approximate methods. J. Mol. Evol. 39:306–314. [DOI] [PubMed] [Google Scholar]
- Yang Z. 1993. Maximum-Likelihood Estimation of Phylogeny from DNA Sequences When Substitution Rates Differ over Sites. Mol. Biol. Evol. 1:1396–1401. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.