Abstract
The “hallmarks” of pancreatic ductal adenocarcinoma (PDAC) include proliferative, invasive and metastatic tumor cells and an associated dense desmoplasia comprised of fibroblasts, pancreatic stellate cells, extracellular matrix and immune cells. The oncogenically-activated pancreatic epithelium and its associated stroma are obligatorily interdependent, with the resulting inflammatory and immune-suppressive microenvironment contributing greatly to the evolution and maintenance of PDAC. The peculiar pancreas-specific tumor phenotype is a consequence of oncogenes hacking the resident pancreas regenerative program, a tissue specific repair mechanism regulated by discrete super enhancer networks. Defined as genomic regions containing clusters of multiple enhancers, super enhancers play pivotal roles in cell/tissue specification, identity and maintenance. Hence, interfering with such super enhancer driven repair networks should exert a disproportionately disruptive effect on tumor versus normal pancreatic tissue. Novel drugs that directly or indirectly inhibit processes regulating epigenetic status and integrity, including those driven by histone deacetylases, histone methyltransferase and hydroxylases, DNA methyltransferases, various metabolic enzymes, and bromodomain and extra-terminal motif proteins (BETs) have shown the feasibility of disrupting super enhancer-dependent transcription in treating multiple tumor types, including PDAC. The idea that pancreatic adenocarcinomas rely on embedded super enhancer transcriptional mechanism suggests a vulnerability that can be potentially targeted as novel therapies for this intractable disease.
Introduction
Pancreatic cancer is a devastating neoplastic disease that is becoming increasingly common and is projected to surpass breast cancer as a cause of death in the next few years. In the USA alone, 53,670 people are diagnosed with pancreas cancer and 43,090 die each year (118 people/day) (1). Worldwide, the disease claims >300,000 lives per year and has the worst 5-year survival of any cancer (7%) (1). As devastating as these statistics are, they do not encompass the considerable pain and suffering associated with pancreatic ductal adenocarcinoma (PDAC), the commonest form of pancreatic cancer. Despite intense efforts, progress in improving survival rates for patients with metastatic pancreatic cancer has been modest at best (2).
Like most adult solid tumors, PDACs are driven by mutations that disrupt the intra- and extracellular networks that normally restrain untoward growth, proliferation, survival and invasion. As might be expected of any pathology arising through random mutation, DNA sequence analysis of PDACs reveals them to comprise many, highly genetically diverse clonal clades (3–5). Such diversity, together with the well-known proclivities of pancreatic cancers to adapt, evolve and relapse in response to treatment, has occasioned a huge effort to catalog and categorize them – with the holy grail being effective personalized therapy designed around each tumor’s unique qualities. However, appearances can be misleading: this recent focus on the endless differences between individual pancreatic adenocarcinomas has overshadowed their even more remarkable uniformity – almost all PDACs share the same signature genetic lesions (activation of the KRas oncoprotein and inactivation of the p16INK4/CDKN2A, p53 and SMAD4/DPC4 tumor suppressors) and exhibit the same signature phenotypic features (metastasis, resistance to radio and chemotherapy, highly avascular, inflammatory and desmoplastic). Indeed, the phenotypic signatures of PDACs are so universal that arduous genomic maneuvers are needed just to discriminate between them. Of equal note, while some of the shared signature PDAC features are patently tumor cell-autonomous, many relate to the histology and behavior of the accompanying (and genetically normal) stroma. This implies deep evolutionary canalization of the disease at the level of the entire neoplastic tissue - both tumor cells themselves and their associated stroma. Of course, this remarkable phenotypic idiosyncrasy is not restricted to pancreatic adenocarcinomas: indeed, the differential diagnosis of almost all individual types of solid cancer is based upon the molecular and phenotypic attributes that each type shares with other tumors of the same type, and which can differ profoundly in cancers arising in other tissues or cells of origin. It is quite the paradox: if tumor evolution is a mutational free-for-all, why do cancers of a particular type end up looking so alike and so different from other cancer types?
The commonalities across pancreatic adenocarcinomas prompt some intriguing questions. The recurring nature of PDAC signature genetic lesions (3) implies the existence of common mechanistic bottlenecks peculiar to the evolution of all PDACs. If so, this could mean that all PDACs, despite their apparent diversity, are at heart driven by, maintained and built upon a limited platform of common oncogenic mechanisms. Could targeting this common platform form a therapeutic strategy for effectively treating all PDAC patients, without the need for expensive personalization? And what mechanisms could possibly be responsible for so tightly constraining the evolutionary trajectories and phenotypes (both tumor cells and stroma) of PDACs? Where does the peculiarly metastatic, avascular, drug resistant and desmoplastic phenotype of PDAC come from and why is it so distinct from other epithelial cancers driven by many of the same oncogenic mutations? And more generally, why do all epithelial cancers exhibit such distinctive, tissue-specific phenotypes?
Super enhancers define tissue and tumor identity
The integrity of multicellular organisms relies on the consistent and robust specification of cell type and function during ontogeny and during regeneration after injury – somatic cells need to know “who” they are, “where” they are and “how” to behave. Critical to this is cell type-specific regulation of gene expression, which is strictly determined by the interplay of regulatory events at gene promoters, orientation-specific cis-acting elements lying immediately upstream of transcriptional start sites, and at gene-distal, orientation and position-independent regulatory elements called enhancers. Enhancers play critical roles in regulating precise spatial and temporal control of transcriptional programs in tissue- and cell-type specific manners (6, 7). Recently, studies have identified genomic regions, dubbed “super enhancers,” where higher order spatial re-organization of chromatin clusters multiple enhancer elements close to key genes that determine cell identity (Figure 1) (8, 9). The first super enhancers were identified bioinformatically based on data from chromatin immunoprecipitation and high throughput sequencing (“ChIP-Seq”) studies in mouse embryonic stem cells. Such enhancer clusters were found to map close to genes encoding “master” stem cell transcription factors such as Oct4, Sox2, and Nanog and were associated with high concentrations of the Mediator 1 (MED1) transcriptional co-activator complex-binding (9). Since then, other super enhancers have been identified in multiple distinct cell types and tissues based on their signature histone modification marks, such as histone H3 mono- and di-methyl lysine 4 (H3K4me1, H3K4me2) and histone H3 acetyl lysine 27 (H3K27Ac), their corresponding open chromatin architecture, RNA Pol II loading that generates diagnostic bidirectional transcripts called eRNAs (10–13), and their associations with cell-type and lineage-specific transcription factors and co-activators. It is now clear that super enhancers play particularly pivotal roles in cell/tissue specification, identity and maintenance, while their peculiar dependency on distinctive transcriptional co-factors (e.g. BET bromodomain-containing proteins such as BRD4) (8, 9, 14, 15) may make them selectively vulnerable to pharmacological disruption. This is especially intriguing, given that a recent study of association between single nucleotide polymorphisms (SNPs) and a diverse range of phenotypic traits and disease, in multiple cell- and tissue-types, revealed that many trait-associated SNPs map to super enhancers. Consequently, super-enhancer disruption might have therapeutic uses in those diseases where aberrations in cell specification and identity are pivotal – notably immune dysfunction, developmental and metabolic disorders, ageing and degeneration, and cancer (8).
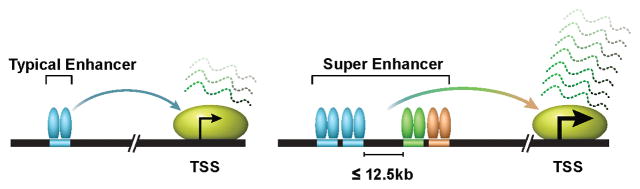
Figure 1. A simplified view of typical enhancers versus super enhancers.
Enhancers are orientation and position-independent cis-acting regulatory elements distally located from transcription start sites (TSS). Enhancers are typically bound by multiple transcription factors to regulate gene expression outcomes. Regions of chromatin incorporating multiple enhancers, defined by ChIP-Seq (e.g. H3K27Ac, Med1, BRD4) within 12.5 kb, are referred to as super enhancers. Super enhancers are typically an order of magnitude larger than typical enhancers in size, have higher transcription factor density and greater ability for transcriptional activation.
As in ontogeny, cell type specification and identity are crucially important also in the regeneration and repair of tissues damaged by injury or infection. Different tissues have very distinct architectures, functions and regenerative capacities and differing vulnerabilities to physical or chemical assaults, infection, inflammation and neoplasia. Hence, while the same core, cell-autonomous processes of cell proliferation, migration and apoptosis are probably shared by all cell types, the tightly choreographed dance between damaged epithelium and stroma needed to rebuild each must be tightly tailored to its unique structure and function. It is only logical to presume that such tissue-specific regenerative programs are also controlled by discrete meta-transcriptional super enhancer networks. Indeed, since tissue regeneration requires coordination of multiple epithelial and stromal cell types, each of the contributing lineages - epithelial, mesenchymal, endothelial, inflammatory and immune –presumably has its own cell-intrinsic network of super enhancers, all of which are presumably coordinated by a web of reciprocal signaling between the epithelium and stroma.
Distinct tissue-specific regenerative programs, each with their own bespoke interaction between epithelial-stromal interactions tailored to the rebuilding of that tissue look suspiciously similar to tissue-specific tumor phenotypes, each with their own signature epithelial-stromal interactions. Moreover, the same tissue-specific signature features are frequently shared by pathologies involving chronic tissue injury and regeneration and tumors, pancreatitis and PDAC being an excellent example where the similarity is so marked as to frequently complicate unambiguous diagnosis (16). Normal repair after injury is driven by spatially and temporally regulated mitogenic signals and attenuates when those signals subside. Cancers, by contrast, are driven by relentless activating mutations in those same mitogenic processes and truly are wounds that never heal (17).
Our data are most consistent with the notion that driver oncogenic mutations, rather than themselves specifying the phenotypes of each distinct cancer, instead all hack into the resident, pre-configured, super enhancer regenerative program of the target tissue. This idea fits well with a number of puzzling observations. First, if tumor phenotypes are principally a property of the tissue being hacked rather than of the diverse drivers that do the hacking, it makes sense that distinct cancer types, although arising by random mutation, share the same signature features. The obligatory intertwining of the mechanisms underpinning tumorigenesis with those mediating inflammation and immunity also explains why there is such remarkable overlap, in both form and function, between wounds and tumors in any tissue type. The dependence of tumor phenotypes on tissue of origin rather driver oncogenes also explains how the same, restricted set of recurring oncogenic hubs – such as Ras, Myc, p53, E2F/Rb/CDKN2, TGFβ, NF-κB – can be associated with so many diverse cancer phenotypes. However, the most profound implication of the hacking concept is that the evolutionary trajectories of cancers, including which and when specific oncogenic mutations confer a selective clonal advantage, are deeply constrained by the inherent properties of each target tissue. Understanding these constraints, and how and why they are imposed, could reveal all sorts of novel potential vulnerabilities in cancers, not least that it suggests that cancers might be unusually sensitive to perturbation of the regenerative super enhancer networks that drive and maintain them.
The origins of the peculiar pancreas cancer phenotype
A window into the innate regenerative program of normal pancreas is provided by the histopathology of chronic pancreatitis, a persistent injury/regeneration/re-injury pathology of diverse etiologies that affects 7-10/100,000 patients a year (18). Macroscopically, the affected organ displays characteristic irregular contours with atrophied ductal epithelium, abundant and poorly vascularized fibrosis heavily infiltrated with activated, α-smooth muscle actin (α-SMA) positive stellate cells, macrophages, various other inflammatory cells and nerve vestiges, as well as sporadic areas of cyst and necrosis (16). Critical in the establishment of this signature phenotype are the pancreatic stellate cells which, in direct response to pancreatic injury, rapidly trans-differentiate into highly proliferative and migratory α-SMA-positive myofibroblastic phenotype. These secrete abundant ECM, cytokines and growth factors necessary to rebuild damaged pancreas (19, 20), notably Sonic Hedgehog (21, 22). Activated stellate cells also trigger extensive fibrosis, a component of the prototypical pancreas regenerative program that presumably evolved to isolate the damaged region and its payload of digestive enzymes and protect the remaining pancreatic and other nearby tissues.
Although the discrete regenerative component of chronic pancreatitis is somewhat confused by the superposition of repeated re-injury, it nonetheless clearly shares such provocative similarities with its oncogenically hacked sibling, pancreatic ductal adenocarcinoma, that disentangling which is which is often diagnostically challenging (16). The hallmarks of late stage, aggressive PDAC are its proliferative, invasive and metastatic tumor cells – directly driven by their signature oncogenic mutations – but also its associated (and genetically normal) dense desmoplasia, which forms the bulk of the tumor mass and comprises fibroblasts, pancreatic stellate cells, extracellular matrix and an assortment of other cell types including macrophages and neutrophils, immune suppressive cells such as myeloid-derived suppressor cells and Tregs, and residual nerve fibers. Overall, the tumors are generally hypovascular and hypoxic (20). A collateral consequence of this localized inflammatory signaling storm is the marked elevation in the plasma of PDAC patients of diverse inflammatory markers such as IL-6, IL-8, IL-10 and the IL-1 receptor-antagonist IL-1ra. And just as in pancreatitis, the PDAC epithelium and stroma are obligatorily interdependent: oncogenically activated pancreatic cancer cells signal to stellate cells and recruit macrophages and immune suppressor cells; in turn, activated pancreatic stellate cells secrete a variety of factors that directly promote tumor cell proliferation and migration and suppress their apoptosis, while mounting evidence attests to the critical role played by the complex inflammatory and immune suppressive environment in evolution and maintenance of PDAC (23, 24). The jury may still be undecided as to whether the net effect of the dense PDAC desmoplasia is to promote or retard tumor growth and therapeutic sensitivity (25–30). However, its deeply embedded role in the regenerative programs of both normal and neoplastic pancreas is indisputable.
Ras and Myc oncogenes switch on regenerative super enhancer programs
The deep relevance of the Ras and Myc oncogenes to human cancers is incontestable. Ras proteins are generic, self-attenuating, apical GTPase switches that, in response to ligation of upstream receptors, engage a network of downstream signaling molecules that eventually elaborate out into a wide variety of transcriptional responses. Mutations that directly activate one of the three members of the Ras family (K, N and H-Ras) are causally implicated in some 25% of all human tumors, and as many as 90% of PDACs, while indirect activation of Ras signaling by mutations in upstream Ras effectors or downstream effectors is implicated in most of the rest (31). The three Myc proteins (Myc, NMyc and LMyc) are sequence-specific bHLHZip DNA-binding transcription factors and over 7000 Myc binding sites have been identified in the mammalian genome implicating Myc as a prolific promoter of gene expression. In normal tissues, expression of the short-lived Myc protein is tightly and continuously dependent upon mitogenic or developmental signals. By contrast, Myc protein expression is deregulated, aberrantly persistent and/or over-expressed in almost all human cancers (32) including pancreatic adenocarcinoma (33).
In normal cells, Ras and Myc act physiologically as common downstream effectors for the diverse mitogenic signaling pathways that drive proliferation of cells and regeneration of tissues. Oncogenic activation of Ras and Myc short-circuits the normal requirement for mitogenic signals, leading to the persistent engagement of tissue regenerative programs. Interestingly, deregulated Ras and Myc are each, alone, relatively weak oncogenes. However, when combined they potently synergize to drive tumorigenesis (34), albeit through a cooperative mechanism that is still not understood.
Perhaps most intriguingly, the oncogenic capacity of Ras+Myc is tissue-agnostic: in experimental mouse models Ras+Myc drives tumorigenesis in all tested tissues. However, the tumors they induce typically display the tissue-specific phenotypes of spontaneous cancers arising in those same tissues. Hence, the same basal oncogenic Ras+Myc combination switch can engage widely disparate, tissue-specific oncogenic phenotypes, consistent with the hypothesis that oncogenic mutations hack into resident tissue-specific regenerative programs. To explore the mechanism by which they do this, we made use of two existing tissue-specific cancer models driven by oncogenic KRasG12D – one in lung (35) and one in pancreas (36). Both exhibit slow and inefficient evolution of tumors but, upon co-activation of a switchable Myc allele, they immediately progress to invasive carcinomas that exhibit the signature tumor and stromal characteristics of spontaneous tumors arising in each tissue type. Thus, Ras+Myc lung adenocarcinomas are highly invasive, angiogenic, inflammatory, heavily infiltrated with macrophages and with relatively little desmoplasia. By contrast, Ras+Myc-driven pancreatic adenocarcinomas are highly desmoplastic, invasive, avascular and hypoxic and heavily infiltrated with macrophages and neutrophils. Perhaps one of the most interesting features of these models is that although Myc is activated only in the epithelial compartment, tumors form within a few days that are complex interwoven neoplasms comprising both Myc-expressing epithelium and the coordinated recruitment, migration and proliferation of diverse, but tissue-specific, stromal elements. This concept is illustrated in Figure 2, which depicts the signature phenotype of PDAC as an aberrant consequence of an oncogenically hijacked regenerative response that evolved physiologically to integrate the individual regenerative super enhancer programs of each of the diverse epithelial, mesenchymal, endothelial and hematopoietic cells needed, together, to rebuild the peculiar structure and function of pancreatic tissue.
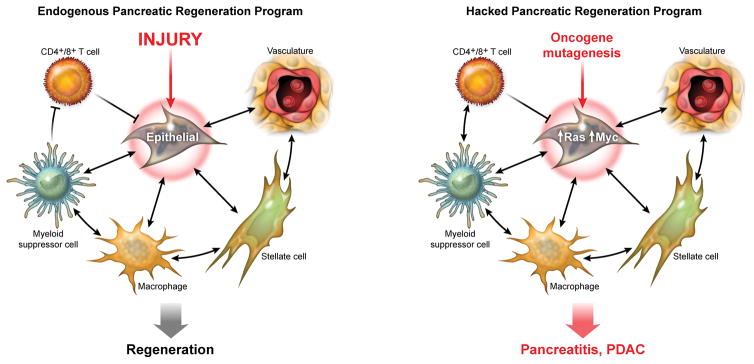
Figure 2. Oncogenes driving pancreatic cancer hack the endogenous pancreas regenerative program.
The regeneration of damaged pancreas requires the coordinated and interdependent activities of multiple cell lineages to rebuild the characteristic structure and function of the damaged tissue. We posit that cell-specific super enhancer networks are integrated into a carefully choreographed whole through continuous exchange of signals (shown much simplified). Both the super enhancer networks and the signals that integrate them offer novel targets for therapeutic intervention.
With respect to the hypothesis that Ras+Myc hack into each tissue’s resident regenerative program, our studies in pancreas are especially compelling since the signature features that Myc induces are not only signature features of invasive PDAC (37) but also of pancreatitis. It therefore seems likely that the distinct characteristics of PDAC are indeed determined by a latent program inherent to pancreatic epithelium – a program we guess is super enhancer-dependent and that in normal pancreas is driven by damage-induced mitogenic signaling and by relentless oncogenic mutations in PDAC.
But how might oncogenes like Ras and Myc hack into this program? As well established previously (36), Ras alone drives only indolent and slowly evolving pancreatic intraepithelial neoplasia (PanINs), progressing rarely to overt PDAC only upon activation of secondary sporadic events. Activation of Myc alone has no discernible impact on pancreatic tissue, underscoring the requirement for Ras and Myc to cooperate in hacking each tissue’s regenerative program. KRas activation appears to set up each tissue to be permissive for Myc to hack into each tissue’s resident regenerative program. One possibility, therefore, is that Ras signaling establishes the epigenetic configuration of latent pancreas regenerative super enhancer program and Myc is the switch that then engages and maintains it. Such a role as a super enhancer switch fits with emerging data that Myc can act as a direct modulator of the epigenome, presumably in addition to its role as a conventional sequence-specific transcription factor. Early studies demonstrated that acute Myc activation triggers rapid global changes in chromatin, markedly increasing histone acetylation (H3 and H4) and methylation (H3K4me3) and enhancing DNA accessibility (38). Myc has since been shown to interact with multiple epigenetic modulators, including TRRAP (an adaptor that then recruits multiple histone acetylases (HATs) such as GCN5, TIP60, p300 and CBP that open chromatin for transcription (39, 40) and the ASHL2 subunit of the KMT2 histone methyl transferase that trimethylates lysine 4 of histone 3 (H3K4me3) and antagonizes the transcriptionally repressive methylation of H3K27me (41). Myc also promotes transcription at accessible genes by suppressing promoter pausing of RNA polymerase (42), in part by increasing accessibility of DNA to binding by the BET co-activators. This, in turn, recruits P-TEFb, whose component CDK9 activates RNA pol II by phosphorylating its C-terminal domain.
Intriguingly, the Myc gene is itself controlled by its own suite of super enhancers, perhaps reflecting the pivotal role that Myc plays as the generic master switch that engages the diverse regenerative sub-programs peculiar to each cell/tissue type. Intriguingly, Myc gene super enhancers come in various guises, depending on cell/tissue type or tumor origin. For example, in HCT116 colorectal cancer cells the Myc-associated super enhancer lies ~500kb upstream of the transcription start site, whereas it is located close to the transcription termination site in pancreatic adenocarcinoma cells, and ~1Mb downstream of the MYC gene in T cell acute lymphoblastic leukemia (8, 43, 44). Hence, there is no single, pre-configured super enhancer format by which Myc is co-opted, which underscores the idea that super enhancers are not universally hardwired but assembled in a bespoke manner, when needed, according to cell and tissue type (Figure 3).
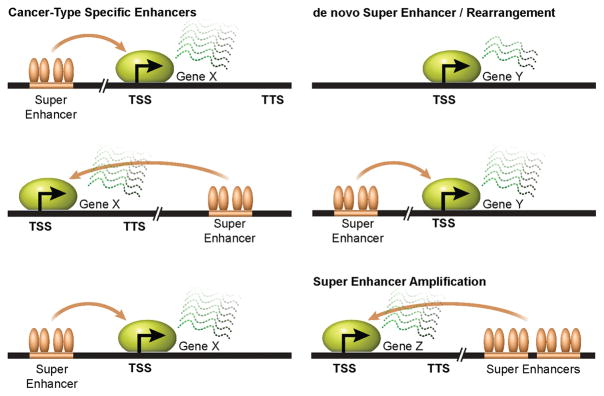
Figure 3. Super enhancers can be altered in a cancer-type specific manner.
The same gene can be regulated by cancer-type specific super enhancers in different cell types to promote malignancy. Transcriptional activation of oncogenic genes can be modulated by the activities of super enhancers, which can be gained as a consequence of extracellular signals, genomic rearrangements, as well as genomic locus and focal amplifications.
Curing pancreatic cancers by crashing super enhancers
Because Myc and Ras serve as convergent downstream effectors for the diverse upstream driver mutations that cause cancer, targeting them offers a therapeutic strategy, potentially synergizing with the current therapeutics and ongoing clinical trials discussed in this CCR Focus section (2, 5, 45). Indeed, our own and others’ studies using switchable variants of oncogenic Myc and Ras in multiple tumor types (46–53) demonstrate that de-activation of either Myc or KRasG12D triggers rapid and profound regression in many diverse types of experimental tumors in mice, including PDAC (54–56). Importantly, in every instance, both epithelial and stromal compartments regress in tandem, underscoring their tight and continuous interdependency. Unfortunately, both Myc and Ras have proven intractable pharmacological targets, so our only recourse at present is to target them, and what they do, indirectly.
However, if pancreatic cancers are indeed a consequence of oncogenes hacking the resident pancreas regenerative super enhancer network, interfering with such super enhancer networks should exert a disproportionately disruptive effect on tumor versus normal pancreatic tissue. To this end, a range of novel drugs have recently emerged that directly (57) or indirectly (58) inhibit processes regulating epigenetic status and integrity, including those driven by histone deacetylases, methylases and hydroxylases, DNA methyltransferases, various metabolic enzymes, and bromodomain and extra-terminal motif proteins (BETs) such as BRD4. BETs, in particular, appear especially important in the operation of super enhancers, whose high occupancy by BRD4 is critical for their recruitment of transcriptional co-factors to appropriately acetylated histones. Several BET inhibitors are currently in development but initial studies using the BRD2/4 inhibitor tool compound JQ1 have already shown the feasibility of disrupting super enhancer-dependent transcription of several oncogenes (59) including Myc (60–63) and in treating multiple tumor types, including PDAC (64–66). And given the intimate relationship between cancer and regeneration, it is no surprise that BET inhibition is also finding uses in the treatment of pathologies driven by aberrant inflammatory/regenerative programs (67–70). While reported side effects of BET inhibitors including neutropenia/thrombocytopenia may limit their clinical use as single agents, their therapeutic indices will likely be increased through combination treatments with traditional chemotherapies. For example, although histone deacetylase inhibitors cause hyponatremia, neutropenia, and anaemia, they are currently in Phase III trails for breast cancer in combination with exemestane, an aromatase inhibitor (71). An alternative, complementary strategy to disrupting super enhancers directly is to target the web of signaling molecules that integrate the regenerative super enhancer-driven sub-programs in each of the epithelial and stromal compartments that participate in the hacked repair program (Figure 4). Being extracellular, such tissue/tumor-specific organizing signals may not only present more tractable pharmacological targets but might also double as sensitive biomarkers for disease.
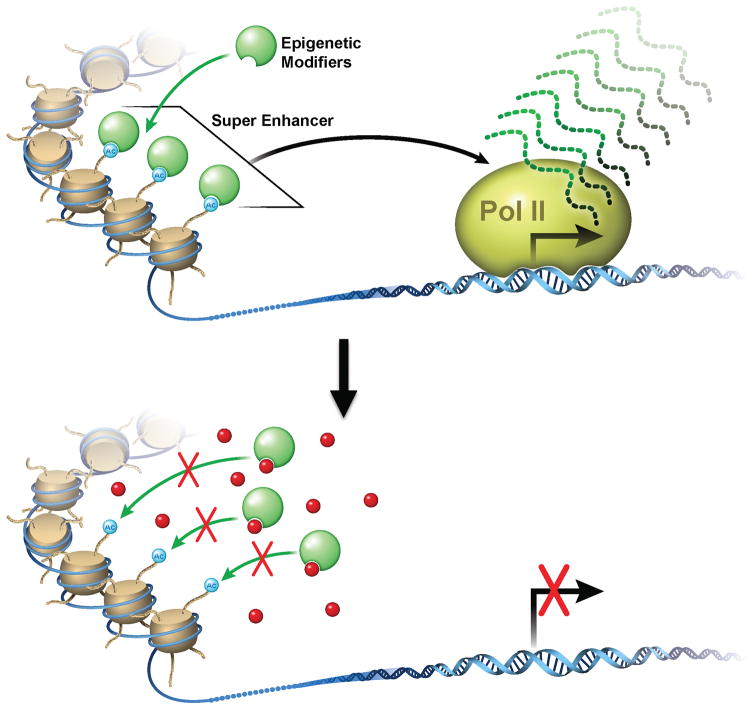
Figure 4. Putative therapeutic interventions directly or indirectly target super enhancers and epigenetic networks in multiple cell lineages in pancreatic regenerative program.
A range of novel drugs (shown in red) directly or indirectly disrupt super enhancers by targeting epigenetic modifiers that integrate the regenerative super enhancer-driven sub-programs in each of the epithelial and stromal compartments that participate in the hacked repair program.
The radical idea that pancreatic adenocarcinomas, notwithstanding their extensive heterogeneity and tumor-stromal and immune complexities, all rely on the same, embedded super enhancer transcriptional mechanism is unorthodox. But it is also consistent with decades of observations and data. It is exciting because shared mechanism may expose shared vulnerability that could be exploited to build effective, durable and generally applicable novel treatments for this awful disease.
Acknowledgments
RME and GIE are supported in part by a Stand Up To Cancer-Cancer Research UK-Lustgarten Foundation Pancreatic Cancer Dream Team Research Grant (Grant Number: SU2C-AACR-DT-20-16). Stand Up To Cancer is a program of the Entertainment Industry Foundation. Research grants are administered by the American Association for Cancer Research, the scientific partner of SU2C. RME is an investigator of the Howard Hughes Medical Institute and March of Dimes Chair in Molecular and Developmental Biology at the Salk Institute and is supported by a grant from the Lustgarten Foundation. GIE is also supported by Cancer Research UK (Program Grant A12077).
Footnotes
Conflicts of Interest
No potential conflicts of interest are disclosed by the authors
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA: a cancer journal for clinicians. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Manji GA, KO, Saenger Y, Oberstein P. Current and Emerging Therapies in Metastatic Pancreatic Cancer. Clinical Cancer Research. 2017:23. doi: 10.1158/1078-0432.CCR-16-2319. [DOI] [PubMed] [Google Scholar]
- 3.Bailey P, Chang DK, Nones K, Johns AL, Patch A-M, Gingras M-C, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531:47–52. doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 4.Chang DK, Grimmond SM, Biankin AV. Pancreatic cancer genomics. Current Opinion in Genetics & Development. 2014;24:74–81. doi: 10.1016/j.gde.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 5.Dreyer SB, Chang DK, Biankin AV. Pancreatic Cancer Genomes: implications for clinical management and therapeutic development. Clinical Cancer Research. 2017:23. doi: 10.1158/1078-0432.CCR-16-2411. [DOI] [PubMed] [Google Scholar]
- 6.Bulger M, Groudine M. Functional and mechanistic diversity of distal transcription enhancers. Cell. 2011;144:327–39. doi: 10.1016/j.cell.2011.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Plank JL, Dean A. Enhancer function: mechanistic and genome-wide insights come together. Molecular cell. 2014;55:5–14. doi: 10.1016/j.molcel.2014.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–47. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–19. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:21931–6. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hah N, Murakami S, Nagari A, Danko CG, Kraus WL. Enhancer transcripts mark active estrogen receptor binding sites. Genome research. 2013;23:1210–23. doi: 10.1101/gr.152306.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Natoli G, Andrau JC. Noncoding transcription at enhancers: general principles and functional models. Annual review of genetics. 2012;46:1–19. doi: 10.1146/annurev-genet-110711-155459. [DOI] [PubMed] [Google Scholar]
- 13.Zentner GE, Tesar PJ, Scacheri PC. Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome research. 2011;21:1273–83. doi: 10.1101/gr.122382.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chapuy B, McKeown MR, Lin CY, Monti S, Roemer MG, Qi J, et al. Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer cell. 2013;24:777–90. doi: 10.1016/j.ccr.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parker SC, Stitzel ML, Taylor DL, Orozco JM, Erdos MR, Akiyama JA, et al. Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:17921–6. doi: 10.1073/pnas.1317023110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klöppel G, Adsay NV. Chronic pancreatitis and the differential diagnosis versus pancreatic cancer. Arch Pathol Lab Med. 2009;133:382–7. doi: 10.5858/133.3.382. [DOI] [PubMed] [Google Scholar]
- 17.Virchow R. Die Cellularpathologie in ihrer Begründung auf physiologische und pathologische Gewebelehre. Berlin: Verlag von August Hirschwald; 1859. [Google Scholar]
- 18.Klöppel G. Chronic pancreatitis, pseudotumors and other tumor-like lesions. Mod Pathol. 2007;20:S113–S31. doi: 10.1038/modpathol.3800690. [DOI] [PubMed] [Google Scholar]
- 19.Apte MV, Wilson JS, Lugea A, Pandol SJ. A Starring Role for Stellate Cells in the Pancreatic Cancer Microenvironment. YGAST. 2013;144:1210–9. doi: 10.1053/j.gastro.2012.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rasheed ZA, Matsui W, Maitra A. Pathology of pancreatic stroma in PDAC. In: Grippo PJ, Munshi HG, editors. Pancreatic Cancer and Tumor Microenvironment. Trivandrum India: Transworld Research Network; 2012. [PubMed] [Google Scholar]
- 21.Fendrich V, Esni F, Garay MVR, Feldmann G, Habbe N, Jensen JN, et al. Hedgehog Signaling Is Required for Effective Regeneration of Exocrine Pancreas. Gastroenterology. 2008;135:621–31. e8. doi: 10.1053/j.gastro.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strobel O, Rosow DE, Rakhlin EY, Lauwers GY, Trainor AG, Alsina J, et al. Pancreatic Duct Glands Are Distinct Ductal Compartments That React to Chronic Injury and Mediate Shh-Induced Metaplasia. YGAST. 2010;138:1166–77. doi: 10.1053/j.gastro.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clark CE, Hingorani SR, Mick R, Combs C, Tuveson DA, Vonderheide RH. Dynamics of the Immune Reaction to Pancreatic Cancer from Inception to Invasion. Cancer Research. 2007;67:9518–27. doi: 10.1158/0008-5472.CAN-07-0175. [DOI] [PubMed] [Google Scholar]
- 24.Wörmann SM, Diakopoulos KN, Lesina M lHAu. The immune network in pancreatic cancer development and progression. 2013;33:2956–67. doi: 10.1038/onc.2013.257. [DOI] [PubMed] [Google Scholar]
- 25.Gore J, Korc M. Pancreatic Cancer Stroma: Friend or Foe? Cancer cell. 2014;25:711–2. doi: 10.1016/j.ccr.2014.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee JJ, Perera RM, Wang H, Wu DC, Liu XS, Han S, et al. Stromal response to Hedgehog signaling restrains pancreatic cancer progression. Proceedings of the National Academy of Sciences. 2014;111:E3091–E100. doi: 10.1073/pnas.1411679111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olive KP, Jacobetz MA, Davidson CJ, Gopinathan A, McIntyre D, Honess D, et al. Inhibition of Hedgehog Signaling Enhances Delivery of Chemotherapy in a Mouse Model of Pancreatic Cancer. Science. 2009;324:1457–61. doi: 10.1126/science.1171362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Özdemir BC, Pentcheva-Hoang T, Carstens JL, Zheng X, Wu C-C, Simpson TR, et al. Depletion of Carcinoma-Associated Fibroblasts and Fibrosis Induces Immunosuppression and Accelerates Pancreas Cancer with Reduced Survival. Cancer cell. 2014;25:719–34. doi: 10.1016/j.ccr.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rhim AD, Oberstein PE, Thomas DH, Mirek ET, Palermo CF, Sastra SA, et al. Stromal Elements Act to Restrain, Rather Than Support, Pancreatic Ductal Adenocarcinoma. Cancer cell. 2014;25:735–47. doi: 10.1016/j.ccr.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watt J, Kocher HM. The desmoplastic stroma of pancreatic cancer is a barrier to immune cell infiltration. OncoImmunology. 2014;2:e26788–3. doi: 10.4161/onci.26788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prior IA, Lewis PD, Mattos C. A Comprehensive Survey of Ras Mutations in Cancer. Cancer Research. 2012;72:2457–67. doi: 10.1158/0008-5472.CAN-11-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tansey WP. Mammalian MYC Proteins and Cancer. New Journal of Science. 2014;2014:1–27. [Google Scholar]
- 33.Schleger C, Verbeke C, Hildenbrand R, Zentgraf H, Bleyl U. c-MYC activation in primary and metastatic ductal adenocarcinoma of the pancreas: incidence, mechanisms, and clinical significance. Mod Pathol. 2002;15:462–9. doi: 10.1038/modpathol.3880547. [DOI] [PubMed] [Google Scholar]
- 34.Land H, Parada LF, Weinberg RA. Tumorigenic conversion of primary embryo fibroblasts requires at least two cooperating oncogenes. Nature. 1983;304:596–602. doi: 10.1038/304596a0. [DOI] [PubMed] [Google Scholar]
- 35.Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15:3243–8. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, Jacobetz MA, et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer cell. 2003;4:437–50. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- 37.Hruban R. Pathology of Genetically Engineered Mouse Models of Pancreatic Exocrine Cancer: Consensus Report and Recommendations. Cancer Research. 2006;66:95–106. doi: 10.1158/0008-5472.CAN-05-2168. [DOI] [PubMed] [Google Scholar]
- 38.Knoepfler PS, Zhang X-y, Cheng PF, Gafken PR, McMahon SB, Eisenman RN. Myc influences global chromatin structure. The EMBO journal. 2006;25:2723–34. doi: 10.1038/sj.emboj.7601152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hessmann E, Schneider G, Ellenrieder V, Siveke JT. MYC in pancreatic cancer: novel mechanistic insights and their translation into therapeutic strategies. 2015;35:1609–18. doi: 10.1038/onc.2015.216. [DOI] [PubMed] [Google Scholar]
- 40.McMahon SB, Wood MA, Cole MD. The essential cofactor TRRAP recruits the histone acetyltransferase hGCN5 to c-Myc. Molecular and cellular biology. 2000;20:556–62. doi: 10.1128/mcb.20.2.556-562.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ullius A, Luscher-Firzlaff J, Costa IG, Walsemann G, Forst AH, Gusmao EG, et al. The interaction of MYC with the trithorax protein ASH2L promotes gene transcription by regulating H3K27 modification. Nucleic Acids Research. 2014;42:6901–20. doi: 10.1093/nar/gku312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, et al. c-Myc Regulates Transcriptional Pause Release. Cell. 2010;141:432–45. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ahmadiyeh N, Pomerantz MM, Grisanzio C, Herman P, Jia L, Almendro V, et al. 8q24 prostate, breast, and colon cancer risk loci show tissue-specific long-range interaction with MYC. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:9742–6. doi: 10.1073/pnas.0910668107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Herranz D, Ambesi-Impiombato A, Palomero T, Schnell SA, Belver L, Wendorff AA, et al. A NOTCH1-driven MYC enhancer promotes T cell development, transformation and acute lymphoblastic leukemia. Nat Med. 2014;20:1130–7. doi: 10.1038/nm.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johnson BA, 3rd, Yarchoan M, Lee V, Laheru D, Jaffee EM. Strategies for Increasing Pancreatic Tumor Immunogenicity. Clinical Cancer Research. 2017:23. doi: 10.1158/1078-0432.CCR-16-2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chin L, Tam A, Pomerantz J, Wong M, Holash J, Bardeesy N, et al. Essential role for oncogenic Ras in tumour maintenance. Nature. 1999;400:468–72. doi: 10.1038/22788. [DOI] [PubMed] [Google Scholar]
- 47.Felsher DW. MYC Inactivation Elicits Oncogene Addiction through Both Tumor Cell-Intrinsic and Host-Dependent Mechanisms. Genes & Cancer. 2010;1:597–604. doi: 10.1177/1947601910377798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pelengaris S, Khan M, Evan GI. Suppression of Myc-induced apoptosis in beta cells exposes multiple oncogenic properties of Myc and triggers carcinogenic progression. Cell. 2002;109:321–34. doi: 10.1016/s0092-8674(02)00738-9. [DOI] [PubMed] [Google Scholar]
- 49.Podsypanina K, Politi K, Beverly LJ, Varmus HE. Oncogene cooperation in tumor maintenance and tumor recurrence in mouse mammary tumors induced by Myc and mutant Kras. Proceedings of the National Academy of Sciences. 2008;105:5242–7. doi: 10.1073/pnas.0801197105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sodir NM, Swigart LB, Karnezis AN, Hanahan D, Evan GI, Soucek L. Endogenous Myc maintains the tumor microenvironment. Genes & Development. 2011;25:907–16. doi: 10.1101/gad.2038411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Soucek L, Whitfield J, Martins CP, Finch AJ, Murphy DJ, Sodir NM, et al. Modelling Myc inhibition as a cancer therapy. Nature. 2008;455:679–83. doi: 10.1038/nature07260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Soucek L, Whitfield JR, Sodir NM, Masso-Valles D, Serrano E, Karnezis AN, et al. Inhibition of Myc family proteins eradicates KRas-driven lung cancer in mice. Genes & Development. 2013;27:504–13. doi: 10.1101/gad.205542.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tran PT, Fan AC, Bendapudi PK, Koh S, Komatsubara K, Chen J, et al. Combined Inactivation of MYC and K-Ras Oncogenes Reverses Tumorigenesis in Lung Adenocarcinomas and Lymphomas. PLoS ONE. 2008;3:e2125. doi: 10.1371/journal.pone.0002125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Collins MA, Bednar F, Zhang Y, Brisset J-C, Galbán S, Galbán CJ, et al. Oncogenic Kras is required for both the initiation and maintenance of pancreatic cancer in mice. J Clin Invest. 2012;122:639–53. doi: 10.1172/JCI59227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Collins MA, Brisset J-C, Zhang Y, Bednar F, Pierre J, Heist KA, et al. Metastatic Pancreatic Cancer Is Dependent on Oncogenic Kras in Mice. PLoS ONE. 2012;7:e49707–13. doi: 10.1371/journal.pone.0049707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McAllister F, Bailey JM, Alsina J, Nirschl CJ, Sharma R, Fan H, et al. Oncogenic Kras Activates a Hematopoietic-to-Epithelial IL-17 Signaling Axis in Preinvasive Pancreatic Neoplasia. Cancer cell. 2014;25:621–37. doi: 10.1016/j.ccr.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jones PA, Issa J-PJ, Baylin S. Targeting the cancer epigenome for therapy. Nat Rev Genet. 2016;17:630–41. doi: 10.1038/nrg.2016.93. [DOI] [PubMed] [Google Scholar]
- 58.Yu, Xin F, Wang F, Yu Z, Xu W. Epigenetics and Cellular Metabolism. GEG. 2016;2016:43–9. doi: 10.4137/GEG.S32160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–34. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bandopadhayay P, Bergthold G, Nguyen B, Schubert S, Gholamin S, Tang Y, et al. BET Bromodomain Inhibition of MYC-Amplified Medulloblastoma. Clinical Cancer Research. 2014;20:912–25. doi: 10.1158/1078-0432.CCR-13-2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, et al. BET Bromodomain Inhibition as a Therapeutic Strategy to Target c-Myc. Cell. 2011;146:904–17. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Grayson AR, Walsh EM, Cameron MJ, Godec J, Ashworth T, Ambrose JM, et al. MYC, a downstream target of BRD-NUT, is necessary and sufficient for the blockade of differentiation in NUT midline carcinoma. 2013;33:1736–42. doi: 10.1038/onc.2013.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fowler T, Ghatak P, Price DH, Conaway R, Conaway J, Chiang C-M, et al. Regulation of MYC Expression and Differential JQ1 Sensitivity in Cancer Cells. PLoS ONE. 2014;9:e87003. doi: 10.1371/journal.pone.0087003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Garcia PL, Miller AL, Kreitzburg KM, Council LN, Gamblin TL, Christein JD, et al. The BET bromodomain inhibitor JQ1 suppresses growth of pancreatic ductal adenocarcinoma in patient-derived xenograft models. Oncogene. 2015:1–13. doi: 10.1038/onc.2015.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mazur PK, Herner A, Mello SS, Wirth M, Hausmann S, Sanchez-Rivera FJ, et al. Combined inhibition of BET family proteins and histone deacetylases as a potential epigenetics-based therapy for pancreatic ductal adenocarcinoma. Nature Medicine. 2015:1–15. doi: 10.1038/nm.3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sahai V, Kumar K, Knab LM, Chow CR, Raza SS, Bentrem DJ, et al. BET Bromodomain Inhibitors Block Growth of Pancreatic Cancer Cells in Three-Dimensional Collagen. Molecular Cancer Therapeutics. 2014;13:1907–17. doi: 10.1158/1535-7163.MCT-13-0925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Belkina AC, Nikolajczyk BS, Denis GV. BET Protein Function Is Required for Inflammation: Brd2 Genetic Disruption and BET Inhibitor JQ1 Impair Mouse Macrophage Inflammatory Responses. The Journal of Immunology. 2013;190:3670–8. doi: 10.4049/jimmunol.1202838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ferri E, Petosa C, McKenna CE. Bromodomains: Structure, function and pharmacology of inhibition. Biochemical Pharmacology. 2016;106:1–18. doi: 10.1016/j.bcp.2015.12.005. [DOI] [PubMed] [Google Scholar]
- 69.Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nature Publishing Group. 2014;14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Khan YM, Kirkham P, Barnes PJ, Adcock IM. Brd4 Is Essential for IL-1β-Induced Inflammation in Human Airway Epithelial Cells. PLoS ONE. 2014;9:e95051. doi: 10.1371/journal.pone.0095051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.NCI Website. https://http://www.cancer.gov/about-cancer/treatment/clinical-trials/search/view?cdrid=760055 - link/TrialDescription_CDR0000760055.