Abstract
Background
Increasing evidence suggests that the intestinal microbiota plays an important role in liver diseases. However, the dynamics of the intestinal microbiota during liver transplantation (LT) and its potential role in clinical course remain unknown.
Methods
We prospectively analyzed the intestinal microbiota of 38 patients who underwent LT in Kyoto University Hospital. We characterized the microbial compositions of fecal specimens from LT patients using a metagenomics approach by an Illumina MiSeq platform. We analyzed the diversity of microbiota sequentially from pretransplantation until 2 months after LT and also compared the microbiota during an episode of acute cellular rejection (ACR) and bloodstream infections (BSI) to the microbial composition of time-matched fecal specimens obtained from patients who did not experience ACR or BSI, respectively.
Results
Three hundred twenty fecal specimens were analyzed. Dynamic changes were observed in the microbial composition of LT recipients during the perioperative period. Over the course of LT, the mean diversity index decreased during the first 3 weeks after LT and gradually increased during our observation period. The loss of intestinal microbiota diversity was associated with high Child-Pugh scores, high model for end-stage liver disease scores, ACR, and BSI. At the family level, Bacteroides, Enterobacteriaceae, Streptococcaceae, and Bifidobacteriaceae were increased whereas Enterococcaceae, Lactobacillaceae, Clostridiaceae, Ruminococcaceae, and Peptostreptococcaceae were decreased in ACR patients.
Conclusions
The microbiota of LT patients was associated with the severity of liver diseases and the presence of ACR and BSI. These results lay the groundwork for more comprehensive investigations of microbiota characteristics to identify diagnostic markers for transplant health and to guide intervention strategies to improve transplant outcomes.
The assessment and characterization of intestinal microbiota has become a major research area in human disease. Recent reports suggest that human disease is attributable not only to single pathogens but also to global changes in our microbiome.1-3 Recently, the roles of the microbiota in alcoholic liver disease, nonalcoholic fatty liver disease, cholesteric liver diseases, viral hepatitis, hepatocellular carcinoma, and cirrhosis have been extensively investigated.1,4-7 In the field of liver transplantation (LT), a previous study used an animal model to demonstrate that a shift in the intestinal microbial after LT predicted early-phase acute cellular rejection (ACR) before severe aggravation of graft function. According to the study results, Ren et al8 concluded that a rat’s intestinal microbial profile was a bio-marker that predicted liver injury or rejection after LT. However, information on the intestinal microbiota and how alterations in it can affect LT outcomes, including early infections and ACR as well as long-term complications, is scarce because of the limited number of human trials and their small sample sizes.9
The aims of this study were to characterize the intestinal microbiome of LT recipients, investigate the composition of the intestinal microbiota during perioperative periods and longitudinally evaluate changes in the intestinal microbiota in patients who experience ACR and infections.
MATERIALS AND METHODS
Study Population
This study was a prospective analysis of patients who underwent LT at Kyoto University Hospital, Japan, from June 2013 to September 2014. Liver transplant recipients who were 20 years or older and who consented to our study were included in the analysis, regardless of whether they received a graft from a deceased or living donor. We excluded fulminant hepatitis patients and patients who underwent retransplantation because the characteristics of the intestinal microbiota in the pretransplantation period were considered to be different from those of patients with cirrhosis. The selection criteria for the recipients and the surgical techniques used in both the donor and recipient have been described in detail elsewhere.10-13 All medical records were reviewed to obtain data on demographics, underlying diseases, model for end-stage liver disease (MELD) score, Child-Pugh score (CPS), transplant type, clinical characteristics, microbiological culture results, types and sites of infections, and medical and surgical complications during the hospital stay.
Bloodstream infections were defined using the criteria proposed by the Centers for Disease Control and Prevention.14 ACR cases were identified by performing liver biopsies on patients based on clinical suspicion. A high MELD score was defined as a MELD score over 20, and a high CPS was defined as a score over 10. The experimental protocol was approved by the Ethics Review Committee of Kyoto University Hospital (E1734) in accordance with the ethical standards established in the 1964 Declaration of Helsinki. Informed consent was obtained from all patients before specimen collection.
Transplantation Practices
Perioperative prophylaxis consisted of treatment with ampicillin and cefotaxime for 48 hours. The administered immunosuppressants included tacrolimus, mycophenolate mofetil and low-dose steroids, and these regimens were started within 24 hours after LT in all patients. In ABO-incompatible cases, the recipients were administered rituximab (300 mg) more than two weeks before LT and received mycophenolate mofetil preoperatively. Plasma exchange was also performed if the titer of anti-A or anti-B antibodies did not decrease after treatment with rituximab.13,15 The perioperative nutritional therapy program that was used has been described in detail in previous reports. Briefly, prebiotics, including rich branched-chain amino acids (BCAA), zinc, dietary fiber, and oligosaccharides (GFO, Otsuka Pharmaceutical Co., Ltd., Tokushima, Japan); probiotics, such as Clostridium butyricum MIYAIRI (MIYA-BM; Miyarisan Pharmaceutical Co., Ltd., Tokyo, Japan); and fermented milk containing Lactobacillus casei strain Shirota (Yakult, Tokyo, Japan) were administered before, during and after LT. One pack of GFO (15 g) has 36 kcal and contains 3 g of glutamine, 5 g of dietary fiber, 1.5 g of oligosaccharide, and 1.2 mg of sodium was given 3 times daily. MIYA-BM is commercially available and contains 20 mg of Clostridium butyricum MIYAIRI in each tablet. Three tablets of MIYA-BM per day were administered in this study. Each 65 ml bottle of fermented milk contained a minimum of 6.5 × 109 live cells of Lactobacillus casei strain Shirota. Enteral tube feeding was administered immediately after LT if there were no complications involving the digestive organs. Nutrition therapy was started within 24 hours after LT via enteral tube and continued until the patient could perform oral intake.
Fecal Sample Collection
Fecal specimens were collected from each patient within 2 weeks before LT and every 7 days after LT during the first and second months. We also collected fecal specimens when adverse events, such as fever, diarrhea, reoperation, ACR and infection, occurred. We compared the microbial composition of the fecal specimens collected from patients during an episode of ACR or blood stream infection (BSI) to the microbial composition of time-matched fecal specimens obtained from the patients who did not experience ACR or BSI.
DNA Isolation and Analysis of the Sequencing Data
DNA was isolated from liquid cultures by applying the bead beating technique to stool samples. The V3-V4 region of the 16S rRNA gene was amplified via PCR using barcoded primers, and PCR products were sequenced using the paired-end technique (Illumina MiSeq platform), as previously described.16 Sequencing data were then processed, and diversity trends were analyzed using QIIME v1.8.0.16,17 Phylogenetic classification was used to describe the intestinal composition of each subject over the time course before and after the transplant. Hierarchical clustering of the specimens was performed based on genus-level phylogenies. Microbial diversity was estimated by calculating the Shannon diversity index (SDI).
Statistical Analysis
Crude comparisons were performed using the χ2 or Fisher exact tests for percentages, as appropriate, and the Mann-Whitney U test for continuous variables. Continuous variables were expressed as the mean ± standard deviation or median (interquartile range). Relative operational taxonomic units (OTU) abundances were calculated using QIIME (www.qiime.org).18,19 Community richness and diversity were examined in each sample using biodiversity indices, including SDI, which was calculated from OTUs. The relative microbial abundance between patients was compared at the phylum, class, order, family, genus, and OTU levels. Unpaired and paired t tests were used to identify significant differences in biodiversity and the abundance of a microbial lineage among all patients and also between each patient before and after transplantation. Both weighted and unweighted UniFrac distances were calculated to analyze beta diversity, using Student t test. The results were transformed using principal coordinate analysis and visualized with Emperor. A P value less than 0.05 was considered significant. Statistical data were generated using PASW for Windows, version 18.0 (SPSS Inc., Chicago, IL).
RESULTS
Characteristics of Liver Transplant Recipients
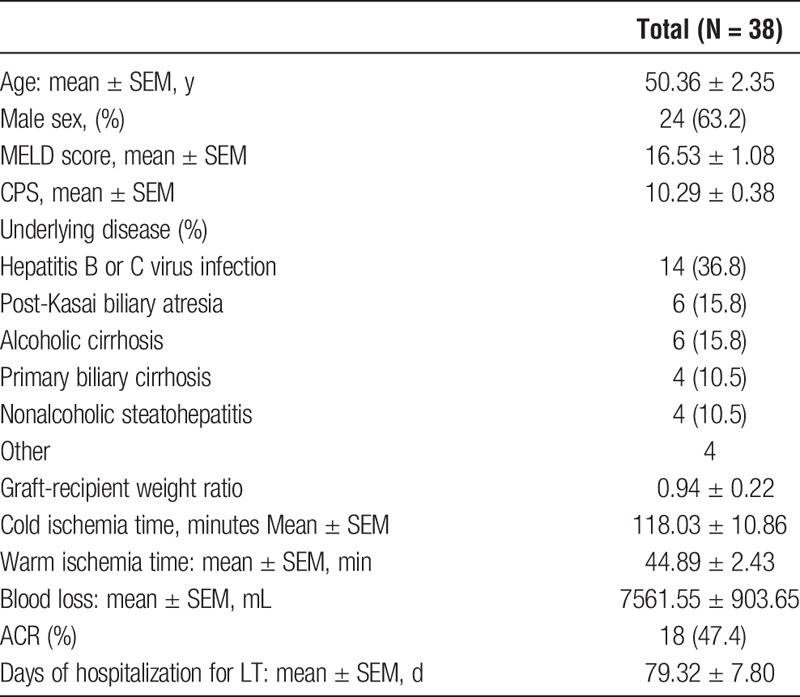
Thirty-eight consecutive adult patients who underwent LT at our institute between July 2013 and September 2014 were analyzed in this study. Table 1 summarizes the baseline characteristics of the patients. The indications for LT were viral hepatitis in 14 (36.8%) patients followed by alcoholic liver cirrhosis and post-Kasai biliary atresia in 6 patients (15.8%) each. The median MELD score was 16 (range, 6-33), and the median CPS was 11 (range, 5-20). Thirty of 38 patients received broad-spectrum antibiotics within 4 weeks postoperatively. The median number of hospitalization days after LT was 54 days (range, 27-272). Eighteen patients (47.4%) suffered ACR and 8 patients (21.0%) had bloodstream infection during their hospital stay.
TABLE 1.
Characteristics of patients who underwent LT
16s rRNA Deep Sequencing
A total of 320 fecal specimens were collected, including 3 to 22 specimens per patient.
From these specimens, a total of 21 165 900 high-quality 16S rRNA-encoding sequences were identified. The mean number of sequences obtained per specimen was 60 647.3 (range, 4057-218 757).
Pretransplant Intestinal Microbiota
Figure 1 shows the pre-LT intestinal microbiota of the 38 patients. The microbiota of these LT recipients was dominated by the following 3 bacterial phyla: Firmicutes (56.6%), Bacteroidetes (21.2%), and Actinobacteria (14.4%). A minor detected phyla was Proteobacteria (6.5%). We found the following bacterial orders predominated in pretransplant samples: Clostridiales (mean, 34.3%; median, 33.6%) Bacteroidales (mean, 21.2%; median, 18.3%), Lactobacillales (mean, 21.1%; median, 9.5%), Bifidobacteriales (mean, 12.5%; median, 10.0%), and Enterobacteriales (mean, 5.0%; median, 1.4%).
FIGURE 1.
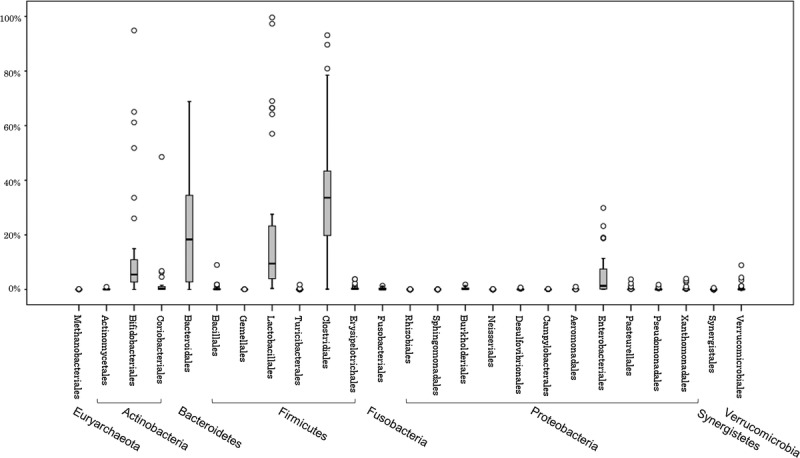
The intestinal microbiota of 38 patients before LT. The phylotypes of the liver transplant recipients are shown. The phylotypes with median relative abundances greater than 0.1% of the total abundance in the patients are included. The boxes represent the IQR from the first and third quartiles, and the inside line represents the median. The whiskers indicate the lowest and highest values within 1.5 IQR of the first and third quartiles. The circles represent outliers beyond the whiskers. IQR, interquartile range.
Temporal Changes in the SDI During LT
There was considerable variation of the SDI values (Figure 2). There was no difference in diversity between samples obtained during postoperative days 0 to 7 and pretransplantation samples. However, over the course of LT, the mean diversity index decreased and gradually increased during our observation period (Figures 2A and B). Similar decreases were observed in other measures of microbial diversity (Figure S1, SDC, http://links.lww.com/TXD/A35).
FIGURE 2.
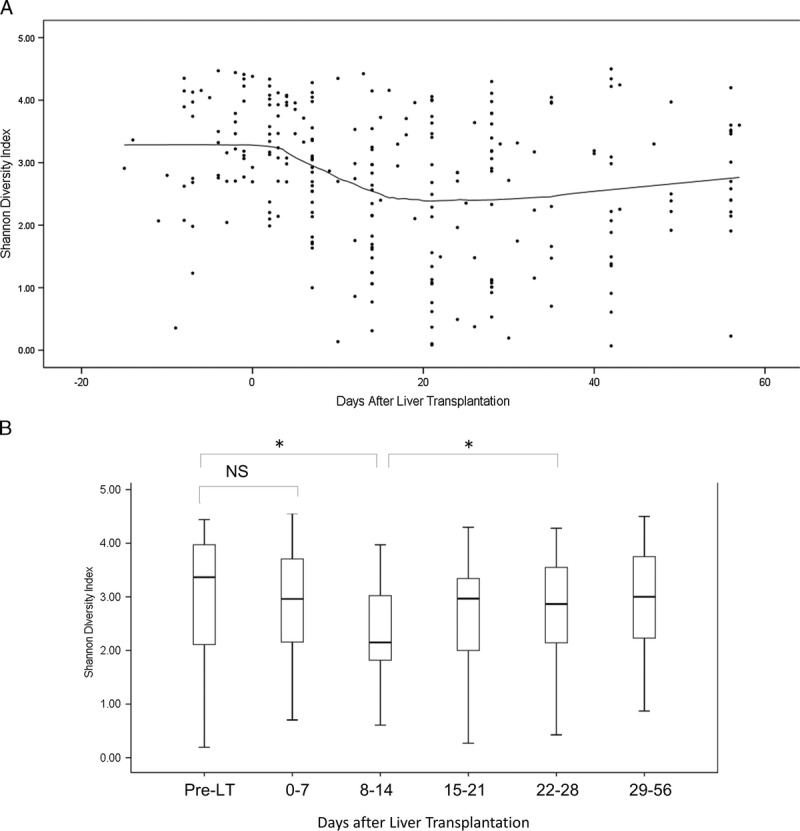
Changes in the microbial diversity of fecal specimens during LT. A, The microbial diversity, as quantified by the SDI, was calculated for each fecal specimen from each patient and plotted as a function of day of transplant (circles). A diversity trend (solid black line) was constructed using Locally Weighted Scatterplot Smoother analysis. B, Microbial diversity was quantified using the SDI and calculated for each fecal specimen from each patient. The boxes represent the IQRs from the first and third quartiles, and the inside line represents the median. The X-axis indicates the day of specimen collection from the transplantation event as the reference day. The Y-axis indicates the SDI. * P < 0.05. NS, not statistically significant.
Relationship Between the Diversity of Intestinal Microbiota and the Severity of Cirrhosis
The overall bacterial diversity, determined using the SDI, showed that significantly less diversity was observed in patients with high MELD scores and high CPS scores in the pretransplant period. The mean SDI of the microbiota in patients with high MELD scores was 2.37, whereas the mean SDI in those with low MELD scores was 2.71 (P = 0.01). This trend was also observed by statistical analysis using observed species, whole tree analysis, and Chao-1. At the phylum level, there was no significant difference between the 2 groups. At the order level, Lactobacillales and Enterobacteriales were increased and Bacteroidales and Clostridiales were decreased in patients with high MELD scores. At the family level, Enterococcaceae and Enterobacteriaceae were increased and Bacteroidaceae, Lachnospiraceae, and Ruminococcaceae were decreased in patients with high MELD score. At the genus level, Enterococcus was increased whereas Bacteroides, Parabacteroides, Ruminococcus, Streptococcus, Blautia, and Faecalibacterium were decreased. (Figure S2, Table S1, SDC, http://links.lww.com/TXD/A35).
In contrast to results for MELD scores, patients with a high CPS did not display significantly less diversity but did display a tendency toward a lower SDI (mean, 2.56 for high CPS, and mean, 2.80 for low CPS) (P = 0.09). However, this trend was statistically significant in other statistical analyses using observed species, whole tree analysis, and Chao-1. At the phylum level, Firmicutes and Proteobacteria increased, and Bacteroidetes was decreased in patients with high CPS. At the order level, Lactobacillales and Enterobacteriales were increased and Bacteroidales and Clostridiales were decreased in patients with high MELD scores. At the family level, Enterococcaceae and Lactobacillaceae were increased, and Bacteroidaceae, Lachnospiraceae, and Ruminococcaceae were decreased in patients with high CPS. At the genus level, Enterococcus and Lactobacillus were increased whereas Bacteroides, Parabacteroides, Lachnospiraceae, Streptococcus, Blautia, Ruminococcus, and Faecalibacterium were decreased (Figure S3, Table S2, SDC, http://links.lww.com/TXD/A35).
There was no difference between the patients with high MELD scores and low MELD scores regarding history of antibiotic use in the pretransplant period. This observation was also seen between the patients with high CPS and low CPS.
Posttransplant Intestinal Microbiota and ACR
There were no significant clinical differences between ACR patients and non-ACR (NR) patients (Table S3, SDC, http://links.lww.com/TXD/A35). Five of the 8 ACR patients received antibiotics within 7 days before ACR. In patients with ACR, SDI was significantly lower during the post-transplantation period than during the pretransplantation period (P < 0.01, paired t test). SDI was also lower in the posttransplant samples obtained from ACR patients than in the time-matched fecal specimens obtained from NR patients (P < 0.01, unpaired t test) (Figure 3). In ACR patients, the mean SDI decreased before periods of ACR, remained low during the period of rejection and then recovered after rejection (Figure 3). At the phylum level, Proteobacteria and Actinobacteria increased in ACR patients whereas Firmicutes was decreased. At the family level, the following 9 OTUs showed statistically significant differences in abundance between NR patients and ACR patients: Bacteroides, Enterobacteriaceae, Streptococcaceae and Bifidobacteriaceae were increased in ACR patients, whereas Enterococcaceae, Lactobacillaceae, Clostridiaceae, Ruminococcaceae, and Peptostreptococcaceae were increased in NR patients. (Figure 4, Table S4, SDC, http://links.lww.com/TXD/A35) There was no clustering of those who suffered ACR and those who did not suffer ACR on principal coordinate analysis (Figure S2, SDC, http://links.lww.com/TXD/A35).
FIGURE 3.
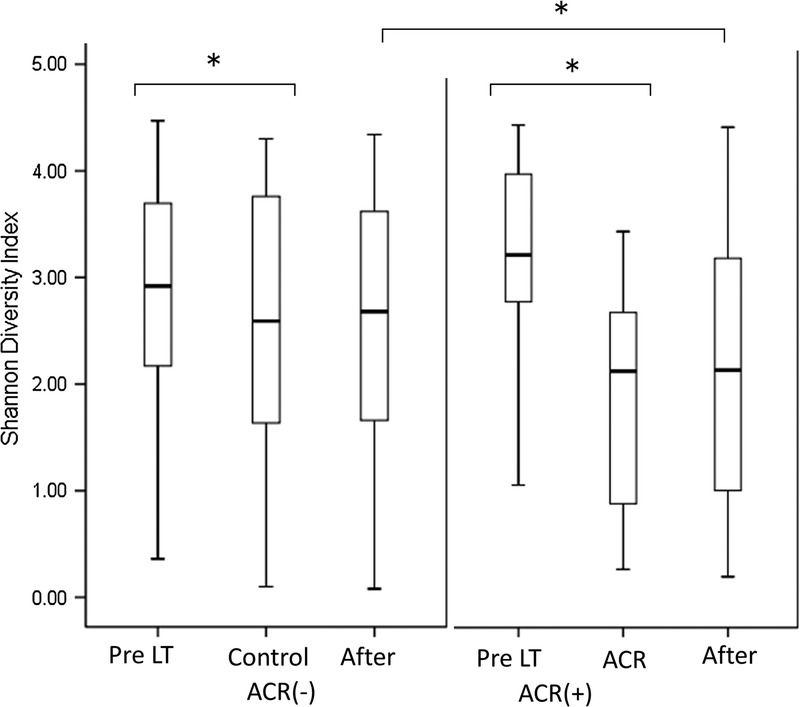
Comparison of microbial diversity between fecal specimens obtained from patients with or without ACR both before/after LT. We compared the SDI of fecal samples obtained from patients during an episode of ACR to time-matched samples obtained from patients who did not suffer ACR. The boxes represent the IQRs from the first and third quartiles, and the inside line represents the median. A P value less than 0.05 was considered to indicate a significant difference between patients with and without ACR. Pre, pretransplantation; control/ACR, at the onset of ACR or time-matched control; after, after the event. (*P < 0.05).
FIGURE 4.
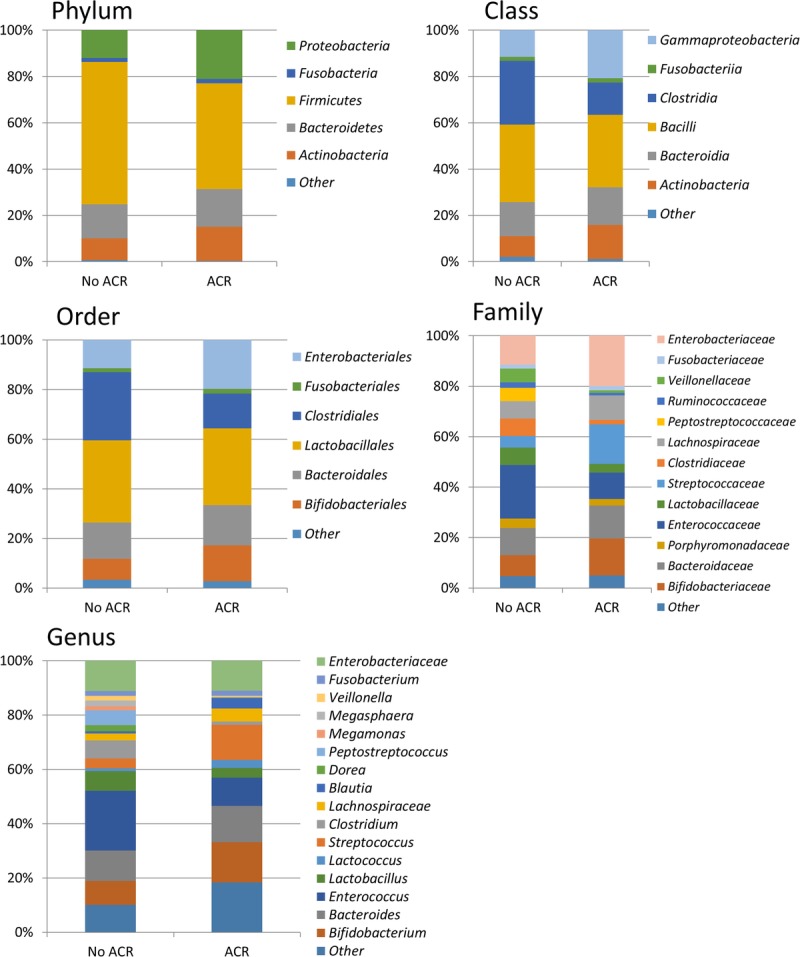
Collective intestinal microbial composition in patients with and without ACR at the phylum, order, family, class, and genus level. Only taxonomic groups 1% or greater are shown.
Posttransplant Intestinal Microbiota and Bloodstream Infection
Of the 38 included recipients, 8 suffered from BSI after LT. One patient experienced 2 episodes of BSI during the study period. The causative pathogens and foci included the following: 3 episodes of Staphylococcus aureus surgical site infections, 3 episodes of Enterococcus faecium cholangitis, 2 episodes of intra-abdominal infections caused by Escherichia coli, an intra-abdominal abscess caused by E. faecium, a ventilator-associated pneumonia caused by E. coli, a urinary tract infection caused by Pseudomonas aeruginosa. One patient had a polymicrobial BSI that was caused by an intra-abdominal infection with E. faecium, E. gallinarum. There were no significant differences in clinical characteristics between patients with and without BSI (Table 5 SDC, http://links.lww.com/TXD/A35). In BSI patients, SDI was significantly lower at the onset of BSI than in the pretransplantation period (P = 0.026, paired t test). In the posttransplant samples, SDI was also lower in BSI patients than in time-matched fecal specimens obtained from non-BSI patients (P = 0.040, unpaired t test) (Figure 5).
FIGURE 5.
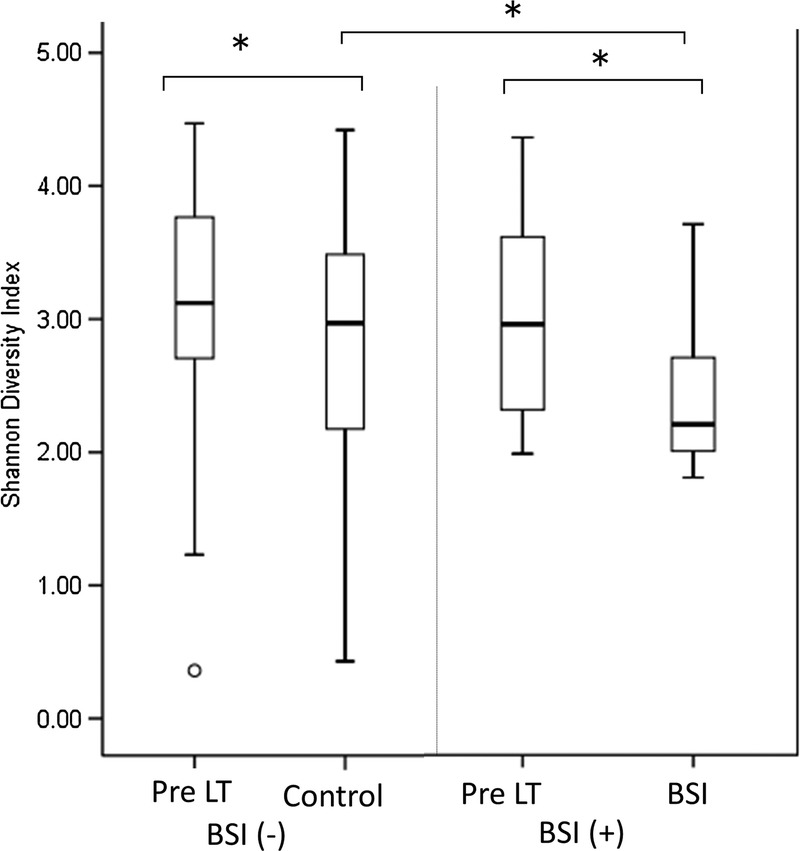
Comparison of the microbial diversity in fecal specimens with or without BSI before/after LT. We compared the SDI of fecal samples obtained from patients during an episode of BSI (BSI) to those of samples obtained from time-matched patients who did not experience BSI. The boxes represent the IQRs from the first and third quartiles, and the inside line represents the median. A P value less than 0.05 was considered to indicate a significant difference between patients with and without BSI. Pre, pretransplantation samples; control/BSI, at the onset of BSI or time-matched control.
DISCUSSION
The intestinal microbiome is now recognized as the most important micro-ecosystem in and a major metabolic organ that maintains a symbiotic relationship with the body. To the best of our knowledge, this is the first study to use a metagenomics approach to longitudinally analyze the microbiota of patients who underwent LT.
Pretransplant Intestinal Microbiota
Several studies recently reported that qualitative changes (dysbiosis) in the intestinal microbiome were associated with chronic liver disease in humans. In previous studies that analyzed the composition of the intestinal microbiota and its association with cirrhosis, the most abundant species that were observed in liver cirrhosis patients belonged primarily to the genera Bacteroides, Streptococcus, and Veillonella.18 In addition, previous studies revealed that cirrhosis is associated with a potentially deleterious shift in the structure of the intestinal microbial community, including a decrease in the abundance of multiple beneficial bacterial groups, such as members of the autochthonous families (eg, Lachnospiraceae and Ruminococcaceae), lactic acid bacteria, and Bifidobacterium species, and an increase in the abundance of potentially pathogenic organisms, especially Enterobacteriaceae and Enterococcus species.20,21 Our results revealed that Clostridiales was the most dominant group but that species with a buccal origin were not abundant. Enterobacteriaceae and Enterococcus were not major microbes in our patients, whereas Lactobacillales and Bifidobacteriales were relatively abundant in the LT population. The differences between our and previous results probably arose because we used of perioperative nutrition therapy that contained synbiotics, and possibly because there were differences in ethnicity or other characteristics between end-stage liver patients in our and previous studies.
In this study, a high MELD score and a high CPS were associated with less microbial diversity. The intestinal microbiota of patients suffering from severe liver disease often decreases in diversity, resulting in domination by a smaller range of microbes, such as Enterobacteriaceae and Enterococcus spp., which can enter the bloodstream and cause septicemia, particularly in the presence of mucosal barrier injury.9 Another study showed that Bifidobacterium and Enterococcus were independent predictors of MELD scores.22 We could not determine the cause of this tendency in our patients, because previous studies were performed using culture-based methods or quantitative PCR. However, we found that the abundance of Blautia, Coprococcus, and Ruminococcus was lower in patients with high MELD scores, in line with the results of previous studies. It is reasonable to suggest that decreases in the abundances of these potentially beneficial bacteria reflect changes in the intestinal microbiota in end-liver stage patients.9,22,23
Diversity of Intestinal Microbiota Posttransplantation
During LT, the surgical procedure damages the intestinal epithelium and mucosal barrier, while the simultaneous administration of broad-spectrum antibiotics and immunosuppressants dramatically alters the microbiota within a short time frame. We found that microbial diversity was not significantly altered during postoperative days 0 to 7 but that it began to reflect these influences during postoperative days 8 to 14. Then, diversity was restored during post-operative days 28 to 56, perhaps as a result a decrease in antibiotic pressure, ACR and other adverse events. In this study, we found that ACR was associated with a loss of intestinal microbial diversity regardless of the presence of other known predictors, such as pretransplant comorbidity and disease status. The composition of the intestinal microbiota and its contribution to allograft survival are a logical area of investigation, and several studies have suggested that there is a relationship between the intestinal microbiota and graft rejection.24,25 Intestinal domination, such as by Enterococcus in hematopoietic stem cell transplantation, an increase in the Proteobacteria/Firmicutes ratio in small bowel transplantation, and other microbial shifts that lead to a loss of diversity have an impact on clinical outcomes.9,24,26-28 We do not know whether these changes occur as a result of ACR or whether these changes are induced by ACR. However, microbial changes are presumed to be correlated with higher endotoxin levels and increased bacterial translocation. Previous reports revealed that the intestinal microbiota promotes liver tumorigenesis or inflammatory reactions through the activity of proinflammatory microorganism-associated molecular patterns and bacterial metabolites, both of which reach the liver via the portal vein.29,30 Further analysis of the roles and functions of the dominant bacteria that were observed during ACR might be a next step toward increasing our understanding of the relationship between ACR and the microbiota in human models. Characterizing the shifts that occur in the microbial community during ACR could improve our understanding of this disease's etiology and help us identify predictive bacterial signatures that are associated with ACR, which could yield potential diagnostic markers.
The diversity of the intestinal microbiota was also lower in the posttransplant samples of BSI patients than in time-matched fecal specimens obtained from non-BSI patients. In BSI patients, the mean SDI decreased before the onset of BSI and continued to decrease during and after BSI. The decrease in SDI that was observed during/after BSI was probably caused by the administration of broad-spectrum antimicrobial agents. However, the observed tendency toward a lower SDI before BSI onset suggests that a decrease in microbiota diversity may predispose patients to developing BSI. Previous studies revealed that disruptions to the diversity and stability of the intestinal flora were associated with subsequent bacteremia.31 Host-derived immune and inflammatory responses are an important driving force that shape the composition of the microbial community and, when altered, this may contribute to dysbiosis. In addition to microbial regulation by innate immunity, inflammation (with its complex set of mediators) may also contribute to a milieu that favors the outgrowth of specific bacteria. Accordingly, a bloom in Enterobacteriaceae has been observed in a variety of inflammatory disease models and patients with chronic inflammation.30,32,33 In our study population, there were 8 cases of BSI that originated in intra-abdominal infections, and the relative abundance of specific bacteria was not significantly increased in patients with BSI perhaps due to the heterogeneity of the causative pathogens. However, previous studies reported that the fecal abundance of Enterococcus was associated with urinary tract infections caused by Enterococcus in kidney transplant recipients and enterococcal bacteremia in bone marrow transplant patients.26,31,34,35 Further analyses are needed to identify the association between microbiota and intra-abdominal infections. Because the changes observed in this study in intestinal microbiota were related to the consequences of postoperative infections, interventions aimed at maintaining intestinal diversity may lead to improved outcomes in LT patients. Incorporating prebiotics, probiotics, or microbiota transplants into proper diets may restore the normal flora in such patients, thereby reducing microbial-induced inflammation.
One of the limitations of this study is that the results observed in the adverse event group may have been confounded by differences in antibiotic regimens. In this study, patients were administered antibiotics and synbiotics for a variety of reasons, including surgical prophylaxis and treatments that could be either targeted or empirical. Interpretations of these results may be not straightforward because the influence of diet and synbiotics as well as broad-spectrum antibiotics may confound results depending on the situation under which they were administered. In addition, due to the heterogeneity of the included patients’ underlying conditions (ABO-incompatible patients receiving Rituximab pretransplant, deceased vs living donors, and underlying diseases), the results might be confounded by each other and difficult to interpret. Furthermore, the present data demonstrated only the relative abundance of the microbiota. Quantitative analyses and investigations of the functions of the flora that were found to predominate in our study are rational next steps that will increase our understanding of how intestinal microbiota influence ACR and infections.
CONCLUSIONS
In this study, we showed that a variety of factors, including MELD scores, CPS, ACR and BSI, affected or were affected by the composition of the intestinal microbiota in LT recipients. These factors serve as both potential causes of complications and targets for optimal management strategies during the perioperative period in patients who undergo LT.
Supplementary Material
Footnotes
Published online 10 March, 2017.
The authors declare no funding.
The authors of this manuscript have conflicts of interest to disclose as described by the Journal Transplantation Direct. K.M., K.O. and M.T. are employees of Miyarisan Pharmaceutical. Co., Ltd. All other authors have no financial relationships to disclose.
K.K., M.N., and T.K. contributed to the study design, draft, and article writing. M.Y. and Y.M. contributed to the collection of data. K.M., K.O., M.T., S.U. and S.I. participated in the analysis and interpretation of data.
Supplemental digital content (SDC) is available for this article. Direct URL citations appear in the printed text, and links to the digital files are provided in the HTML text of this article on the journal’s Web site (www.transplantjournal.com).
REFERENCES
- 1.Chassaing B, Etienne-Mesmin L, Gewirtz AT. Microbiota-liver axis in hepatic disease. Hepatology. 2014;59:328–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eckburg PB, Bik EM, Bernstein CN, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–848. [DOI] [PubMed] [Google Scholar]
- 4.Ahluwalia V, Betrapally NS, Hylemon PB, et al. Impaired gut-liver-brain axis in patients with cirrhosis. Sci Rep. 2016;6:26800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bajaj JS, Betrapally NS, Gillevet PM. Decompensated cirrhosis and microbiome interpretation. Nature. 2015;525:E1–E2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brenner DA, Paik YH, Schnabl B. Role of gut microbiota in liver disease. J Clin Gastroenterol. 2015;49(Suppl 1):S25–S27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Macnaughtan J, Jalan R. Clinical and pathophysiological consequences of alterations in the microbiome in cirrhosis. Am J Gastroenterol. 2015;110:1399–1410. [DOI] [PubMed] [Google Scholar]
- 8.Ren Z, Jiang J, Lu H, et al. Intestinal microbial variation may predict early acute rejection after liver transplantation in rats. Transplantation. 2014;98:844–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doycheva I, Leise MD, Watt KD. The intestinal microbiome and the liver transplant recipient: what we know and what we need to know. Transplantation. 2016;100:61–68. [DOI] [PubMed] [Google Scholar]
- 10.Kaido T, Ogawa K, Fujimoto Y, et al. Impact of sarcopenia on survival in patients undergoing living donor liver transplantation. Am J Transplant. 2013;13:1549–1556. [DOI] [PubMed] [Google Scholar]
- 11.Nagao M, Fujimoto Y, Yamamoto M, et al. Epidemiology of invasive fungal infections after liver transplantation and the risk factors of late-onset invasive aspergillosis. J Infect Chemother. 2016;22:84–89. [DOI] [PubMed] [Google Scholar]
- 12.Yamamoto M, Takakura S, Iinuma Y, et al. Changes in surgical site infections after living donor liver transplantation. PLoS One. 2015;10:e0136559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takada Y, Ueda M, Ito T, et al. Living donor liver transplantation as a second-line therapeutic strategy for patients with hepatocellular carcinoma. Liver Transpl. 2006;12:912–919. [DOI] [PubMed] [Google Scholar]
- 14.Garner JS, Jarvis WR, Emori TG, et al. CDC definitions for nosocomial infections, 1988. Am J Infect Control. 1988;16:128–140. [DOI] [PubMed] [Google Scholar]
- 15.Raut V, Uemoto S. Management of ABO-incompatible living-donor liver transplantation: past and present trends. Surg Today. 2011;41:317–322. [DOI] [PubMed] [Google Scholar]
- 16.Hammad A, Kaido T, Ogawa K, et al. Perioperative changes in nutritional parameters and impact of graft size in patients undergoing adult living donor liver transplantation. Liver Transpl. 2014;20:1486–1496. [DOI] [PubMed] [Google Scholar]
- 17.Hammad A, Kaido T, Uemoto S. Perioperative nutritional therapy in liver transplantation. Surg Today. 2015;45:271–283. [DOI] [PubMed] [Google Scholar]
- 18.Klindworth A, Pruesse E, Schweer T, et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013;41:e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuczynski J, Stombaugh J, Walters WA, et al. Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Curr Protoc Microbiol. 2012;Chapter 1:Unit 1E.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qin N, Yang F, Li A, et al. Alterations of the human gut microbiome in liver cirrhosis. Nature. 2014;513:59–64. [DOI] [PubMed] [Google Scholar]
- 21.Kakiyama G, Pandak WM, Gillevet PM, et al. Modulation of the fecal bile acid profile by gut microbiota in cirrhosis. J Hepatol. 2013;58:949–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu J, Wu D, Ahmed A, et al. Comparison of the gut microbe profiles and numbers between patients with liver cirrhosis and healthy individuals. Curr Microbiol. 2012;65:7–13. [DOI] [PubMed] [Google Scholar]
- 23.Grąt M, Hołówko W, Gałecka M, et al. Gut microbiota in cirrhotic liver transplant candidates. Hepatogastroenterology. 2014;61:1661–1667. [PubMed] [Google Scholar]
- 24.Chen Y, Zhao Y, Cheng Q, et al. The role of intestinal microbiota in acute graft-versus-host disease. J Immunol Res. 2015;2015:145859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bromberg JS, Fricke WF, Brinkman CC, et al. Microbiota—implications for immunity and transplantation. Nat Rev Nephrol. 2015;11:342–353. [DOI] [PubMed] [Google Scholar]
- 26.Fricke WF, Maddox C, Song Y, et al. Human microbiota characterization in the course of renal transplantation. Am J Transplant. 2014;14:416–427. [DOI] [PubMed] [Google Scholar]
- 27.Vindigni SM, Surawicz CM. The gut microbiome: a clinically significant player in transplantation? Expert Rev Clin Immunol. 2015;11:781–783. [DOI] [PubMed] [Google Scholar]
- 28.Wang W, Xu S, Ren Z, et al. Gut microbiota and allogeneic transplantation. J Transl Med. 2015;13:275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dapito DH, Mencin A, Gwak GY, et al. Promotion of hepatocellular carcinoma by the intestinal microbiota and TLR4. Cancer Cell. 2012;21:504–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu LX, Yan HX, Liu Q, et al. Endotoxin accumulation prevents carcinogen-induced apoptosis and promotes liver tumorigenesis in rodents. Hepatology. 2010;52:1322–1333. [DOI] [PubMed] [Google Scholar]
- 31.Taur Y, Xavier JB, Lipuma L, et al. Intestinal domination and the risk of bacteremia in patients undergoing allogeneic hematopoietic stem cell transplantation. Clin Infect Dis. 2012;55:905–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwabe RF, Jobin C. The microbiome and cancer. Nat Rev Cancer. 2013;13:800–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh B, Qin N, Reid G. Microbiome regulation of autoimmune, gut and liver associated diseases. Inflamm Allergy Drug Targets. 2015;14:84–93. [DOI] [PubMed] [Google Scholar]
- 34.Lee JR, Muthukumar T, Dadhania D, et al. Gut microbial community structure and complications after kidney transplantation: a pilot study. Transplantation. 2014;98:697–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee JR, Muthukumar T, Dadhania D, et al. Gut microbiota and tacrolimus dosing in kidney transplantation. PLoS One. 2015;10:e0122399. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


