Abstract
The Oka vaccine is a live attenuated vaccine for the prevention of varicella. Although the vaccine differs from the progenitor virus by over 40 mutations, only three of these are fixed, the rest being a mixture of the wild-type and the vaccine allele. To examine the extent of this variability between two of the three commercially available vaccine preparations, we analysed the vaccine/wild-type allele frequencies present at fifteen vaccine loci in five preparations each from two different manufacturers of the vOka vaccine. Our results suggest that differences in manufacturing processes between the two companies have resulted in significant variation in the frequencies of the vaccine/wild-type alleles in their vaccines. Yet despite these differences, the allele frequencies in the vaccines from the two companies are strongly correlated. We discuss the significance of these findings and the role of evolutionary processes that influence the production of this live attenuated vaccine.
Keywords: Varicella-zoster virus, Oka vaccine, Pyrosequencing, Comparative sequence alignment, Batch, Variation, Genetic diversity
1. Introduction
The live attenuated Oka vaccine is licensed for the prevention of varicella in children in the USA, Japan, Korea, Germany, Greece, Australia and Canada and for targeted immunisation of seronegative health care workers in several countries, including the UK. Over 15 years of experience has demonstrated that VZV vaccines are safe and well tolerated [1–4]. Universal vaccination in the USA has massively decreased the incidence of varicella in both vaccinated individuals and, by herd immunity, unvaccinated individuals, with significant reduction of associated hospitalization and mortality (reviewed in [5]). The commonest adverse event is a mild rash occurring following vaccination, which occurs in around 5% of children and a higher proportion of adults. Life threatening vOka (vaccine Oka) infection has only been described in subjects with undiagnosed cell mediated immunodeficiencies.
The original Oka vaccine strain was derived by serial passage of a wild-type Japanese VZV isolate, pOka (parental Oka) which had been obtained from a child with chickenpox [6]. Three main preparations derived from the original vaccine seed stock are licensed for use, the Biken vaccine, Varivax (Merck) and Varilrix (GSK). In addition, combined vOka with live attenuated measles, mumps and rubella, is marketed by both Merck and GSK. Comparative Sanger sequencing of the Biken strain vOka and pOka, the wild-type progenitor, genomes identified 42 loci at which vaccine substations have occurred, 20 of which are non-synonymous [7]. In addition, length polymorphisms in the known variable repeat R1, R3 and R4 repeat regions and the viral origin of replication OriS were observed and five loci identified that were fixed for the vaccine polymorphism, the rest being polymorphic for vaccine and wild-type single nucleotide polymorphisms (SNPs). Sequencing of 10 clones of the Biken vaccine ORF62 confirmed the Biken vaccine to contain a mixture of related but distinct haplotypes [8]. Subsequently, both the Merck and GSK vOka vaccine preparations were also found to be heterogeneous [9–11].
Only three loci in the ORF 62 gene, which codes for the major immediate transactivating protein, are fixed for the vaccine SNP in all three vaccine preparations, the remainder being either wild-type or mixtures of vaccine and wild-type SNPs [7,11–13]. Overall the GSK vaccine has been found to carry more loci which are fixed for vaccine mutations and fewer which are wild-type than either the Merck or Biken vaccines [11]. Merck and Biken vaccines also differ from one another, with the former having at least one SNP that was not previously identified [9]. However, unlike the Merck vaccine [10,13], there has been no analysis of vaccine allele frequencies in the GSK vaccine. Differences between Merck and GSK vaccines could have arisen due to differences in the downstream manufacturing process by each company (Fig. 1). For example, the GSK vaccine was subjected to five rounds of terminal dilution in an attempt to generate a genetically uniform vaccine strain [14]. Somewhat surprisingly this process did not appear to generate new vaccine mutations [11]. Limited analysis of different batches of vOka vaccines suggests little variation, [13], however, this has not been extensively characterised. To further examine variability between companies and among batches/lots, we analysed vaccine/wild-type allele frequencies present at 15 vaccine loci in five preparations each of the Merck and GSK Oka vaccines, which are currently available for vaccine programmes in Europe and the USA.
Fig. 1.
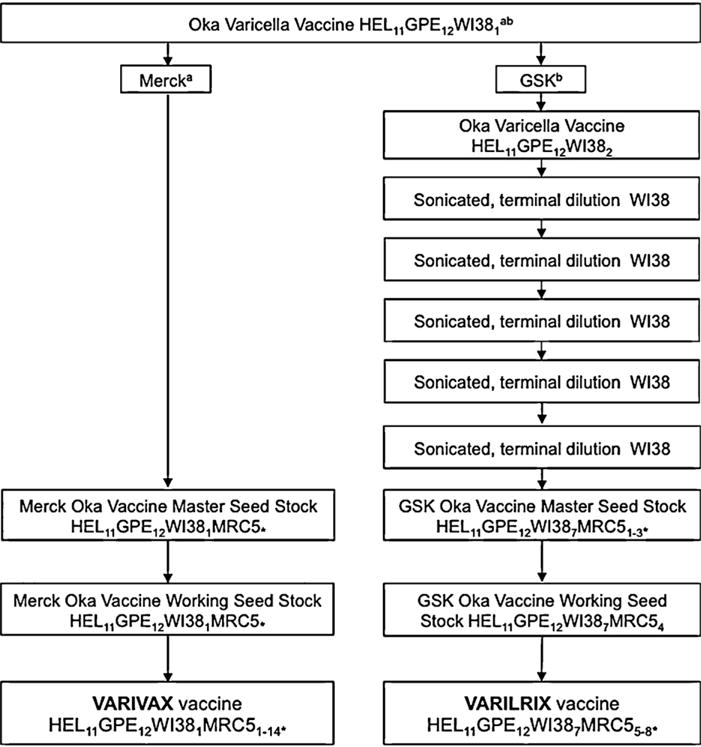
Overview of varicella vaccine production. HEL, human embryonic lung cell cultures; GPE, embryonic guinea pig cell cultures; WI38, human diploid cell cultures; MRC5, human diploid cell cultures. Subscript numbers indicate passage levels. aInformation obtained from Merck Product information IPC-VRV3-I-032008. bInformation obtained from D’Hondt et al. [14]. *Unknown number of passage levels in MRC5 cells. Where passage levels are given next to a “*”, these have been inferred from a and b. World Health Organisation guidelines indicate a limit for Oka-strain varicella vaccine at passage level 38, hence final levels of passage in MRC5 cells can be estimated [14].
2. Methodology
2.1. Vaccine batches and SNPs
Five lots each of GSK and Merck vaccines derived in each case from three batches were available for analysis (Table 1). Fifteen loci at which vaccine substitutions have been identified were selected for analysis. Nine of these were located in ORF62, which contains over 30% of all the vaccine SNPs identified—several of these SNPs (105169, 105705, 105724, 107136 and 107797) have been identified as being of potential biological importance for vaccine attenuation and as immune targets [13,15]. Two of these were previously identified as fixed vaccine SNPs. The remainder were chosen as they have previously been found to be variable and therefore informative [10,13] and to consistently be present in ORFs across the VZV genome.
Table 1.
Vaccines used in this study.
| Company | Vaccinea | Year (country) of purchase | Number of SNPs analysed |
|---|---|---|---|
| Merck | Batch 1: Lot 1 (A) |
2003 (U.K.) | 35 |
| Batch 2: Lot 1 (B) |
2004 (U.K.) | 15 | |
| Batch 2: Lot 2 (C) |
2005 (U.K.) | 15 | |
| Batch 3: Lot 1 (D) |
2005 (U.S.) | 15 | |
| Batch 3: Lot 2 (E) |
2005 (U.S.) | 15 | |
| GSK | Batch 1: Lot 1 (1) |
2003 (U.K.) | 35 |
| Batch 2: Lot 1 (2) |
2008 (U.K.) | 15 | |
| Batch 2: Lot 2 (3) |
2008 (U.K.) | 15 | |
| Batch 2: Lot 3 (4) |
2008 (U.K.) | 15 | |
| Batch 3: Lot 1 (5) |
2010 (U.K) | 15 |
Vaccine lot identifiers were anonymised for the purpose of this study. Numbers and letters in parenthesis are abbreviated identifiers for each vaccine.
2.2. Nucleic acid extractions
Total DNA was extracted using a QIAamp® DNA Mini Kit (Qiagen LTD., UK) according to the manufacturers instructions. The lot numbers, their date of issue and the number of loci analysed in each are shown in Table 1. Prior to extraction, lyophilised vaccine preparations were resuspended in the 500 μl of accompanying buffer. DNA from each preparation was extracted from 2×200 μl and 1×100 μl (mixed with 100 μl of PBS) aliquots. The 3 ×200 μl aliquots of eluted DNA were merged to form a 600 μl DNA pool, which was used for subsequent experiments. A negative control was used during the extraction process by replacing sample material with 200 μl of water.
2.3. PCR reactions
All PCRs were performed in a GeneAmp 2700 thermocycler (Perkin Elmer, UK), initiated by hot start activation at 95 °C for 10 min and ended with a step of 72 °C for 10 min before being incubated at 4 °C. For direct sequencing, reactions included 35 cycles of amplification (95 °C, 1 min; primer annealing temperature (supplementary Table 1), 1 min, 72 °C, 1 min). For pyrosequencing, reactions included 45 cycles of amplification (95 °C, 15 s; primer annealing temperature (supplementary Table 2), 30 s, 72 °C, 15 s). PCR reactions included 5 μl of template DNA, 5 μl of PCR buffer II (Perkin Elmer, UK), 1.5 mM MgCl2 (Perkin Elmer, UK), 200 μM of each deoxynucleotide triphosphate (Advanced Biotechnologies, UK), 12 pmol of each primer and 1 unit of AmpliTaq Gold DNA polymerase (Perkin Elmer, UK). Final reaction volumes were adjusted to 50 μl using DNAse/RNAse free water (Sigma, UK). For PCR reactions with each primer set, two negative controls were employed. The first consisted of PCR reaction mix, in which template DNA was replaced with water. The second consisted of PCR reaction mix, in which template DNA was replaced with the water used as a negative control during the DNA extraction process. PCR with each set of CSA and pyrosequencing primers also included a reaction containing DNA homogenous for the wild-type allele. For pyrosequencing primers a second reaction containing DNA homogenous for the vaccine allele was also included.
2.4. Determining allele frequencies
In the past we have used two methods for determining allele frequencies, comparative sequence analysis (CSA) and pyrosequencing, using primers directed against 35 of 42 vaccine SNPs identified by Gomi et al. [7]. To determine the accuracy of CSA and pyrosequencing, we used primer sets which amplify fragments containing the nucleotide present at position 108838 (supplementary Tables 1 and 2). These fragments were cloned into the EcoRI site of a pUC based pCR2.1 vector using a TA Cloning Kit in accordance with the manufacturer’s instructions (Invitrogen, UK) and transformed into One Shot competent TOP10 Escherichia coli (Invitrogen, UK). Clones containing the vector plus insert were isolated and the DNA was purified using a QIAprep Spin Miniprep Kit (Qiagen, UK) according to the manufacturer’s instructions. The DNA concentration was measured on a Gene Spec I Spectrophotometer (Nalca Instruments Ltd., UK) and Tris–EDTA (10 mM) was used to normalise the concentrations (i.e. DNA from clones containing the wild-type allele and clones containing the vaccine allele were both diluted to a concentration of 200 ng/μl each). DNA from clones containing the wild-type SNP at 108838, and clones containing the vaccine SNP at 108838, were then mixed in ratios of 100:0, 90:10, 70:30, 50:50, 30:70, 10:90 and 5:95. The allele frequencies at position 108838 were analysed in triplicate by CSA and pyrosequencing using published methods [13,16]. For mixtures where the one allele was present at <10%, the experiment was repeated, giving six readings per SNP rather than just three. Relative SNP percentages were calculated automatically for the pyrosequencing, by measuring the area under the curve for loci with dual peaks. The relative percentages for CSA were calculated according to the method described by Mattocks et al. [16].
2.5. Statistical methods
The significance of the genetic differences among the various vaccines was evaluated using generalized linear modelling of the arcsine transformed allele frequencies.
3. Results
3.1. Comparison of CSA and pyrosequencing for allelic quantification
The accuracy of CSA and pyrosequencing for allele mixtures were comparable (Fig. 2), but the pyrosequencing was significantly closer to the true proportions (P = 0.012): the standard deviation around the known frequency being 0.016 (max. 0.03) compared to 0.027 (max. 0.05) for CSA. When the mixtures of allele frequencies were at their most different, the pyrosequencing still detected the lowest frequency allele, whereas CSA did not (Fig. 2). Because of this greater sensitivity and reproducibility, pyrosequencing was used for further analyses. When quantifying alleles in the vaccines, if the wild-type allele was detected at ≤10%, the PCR and relevant assay was repeated.
Fig. 2.
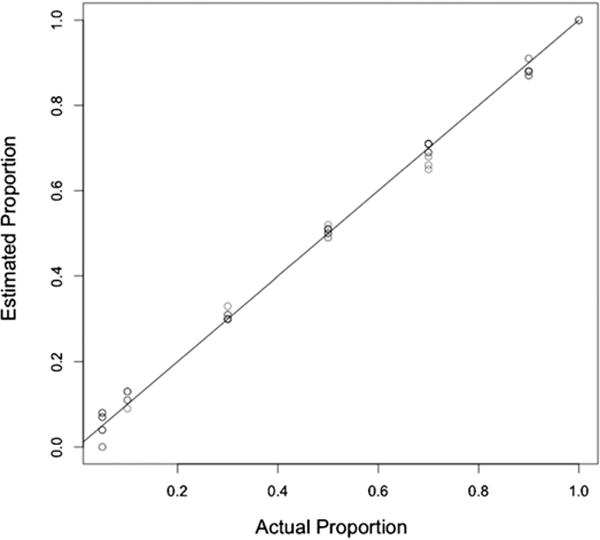
Calibration of the two different methods for determining the frequency of alleles at polymorphic loci. The three estimates obtained by pyrosequencing (black symbols) and three obtained by CSA (grey symbols) are plotted for each mixture, along with a 1:1 line illustrating the known frequency.
3.2. Variation in lots of Oka/GSK and Oka/Merck
The allele frequencies observed at the fifteen loci analysed in all of the GSK and Merck lots are given in Table 2. Three of the loci were fixed for the novel vaccine allele (560, 106262 and 107252). With the exception of positions 89734 which retained the ancestral allele in all Merck batches, and 105724 which retained the ancestral allele in all GSK batches, the remaining loci were a mixture of the of the ancestral (wild-type) allele and the vaccine allele in varying proportions.
Table 2.
Variation in lots of Oka/GSK and Oka/Merck.
| SNPa | ORF | P-Okab | Oka/GSK lotsc,e
|
Oka/Merck lotsd,e
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | 1 | 2 | 3 | 4 | 5 | |||
| 560 | 5′ non-cod | T | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 19431 | 14 | T | 61 | 57 | 57 | 57 | 64 | 79 | 81 | 80 | 85 | 83 |
| 58595 (530) | 31 | A | 71 | 67 | 67 | 68 | 78 | 45 | 46 | 48 | 47 | 49 |
| 87280 | 50 | A | 100 | 84 | 80 | 74 | 100 | 100 | 85 | 85 | 85 | 87 |
| 89734 | 51 | A | 24 | 25 | 25 | 25 | 19 | 100 | 100 | 100 | 100 | 100 |
| 97748 (585) | 55 | G | 45 | 56 | 50 | 58 | 30 | 61 | 56 | 64 | 67 | 60 |
| 105169 | 61/62 | A | 50 | 21 | 21 | 21 | 61 | 56 | 50 | 53 | 53 | 51 |
| 105356 (1260) | 62 | T | 13 | 6 | 0 | 0 | 0 | 34 | 36 | 31 | 33 | 31 |
| 105705 | 62 | T | 11 | 0 | 0 | 0 | 0 | 51 | 0 | 0 | 0 | 0 |
| 105724 (1137) | 62 | A | 100 | 100 | 100 | 100 | 100 | 93 | 81 | 82 | 81 | 100 |
| 106262 (958) | 62 | T | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 107136 | 62 | T | 40 | 0 | 32 | 33 | 0 | 50 | 50 | 48 | 48 | 46 |
| 107252 (628) | 62 | T | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 107797 (446) | 62 | A | 69 | 55 | 56 | 55 | 55 | 70 | 75 | 68 | 68 | 68 |
| 108838 (99) | 62 | A | 100 | 60 | 60 | 63 | 60 | 100 | 76 | 75 | 77 | 75 |
Numbers refer to the nucleotide position in Dumas; numbers in parenthesis refer to the affected amino acid residue within a particular ORF.
P-Oka bases are identical to those in Dumas at every position; P-Oka sequence is from Gomi et al. [7].
Oka/GSK-A was analysed by CSA, Oka/GSK-B, -C & -D were analysed by pyrosequencing.
Oka/Merck-1 was analysed by CSA, Oka/Merck-2, -3, -4 & -5 were analysed by pyrosequencing.
Numbers refer to the percentage wild-type allele.
3.3. Comparison between and within Merck and GSK Oka vaccine preparations
The percentage wild-type alleles present in the vaccine preparations from Merck and GSK for 15 of the 42 loci known to differ between pOka and the Oka Biken vaccine preparation are shown in Fig. 3 (data in Table 2). As can be seen in Fig. 3, there is a strong correlation between the allele frequencies in the vaccines from the two companies (r = 0.79, P = 0.0005). The implications of this result are discussed further (see below). For each vaccine we compared genetic variation between different lots from the same batch, between different batches of the same vaccine and between the two vaccine preparations. There was a significant trend across most loci for the Merck vaccine preparations to have higher frequencies of the wild-type allele (P = 3.422×10−5), as illustrated in Fig. 3. There was significant batch to batch variation between the three Merck batches (P = 7.181×10−7) and between the three GSK batches (P = 9.697×10−10). Although the amount of variation between batches was not significantly different for the two vaccine companies (P = 0.160), there was significantly more variation in allele frequencies among lots from a single batch of GSK vaccine compared to lots from a single batch of Merck vaccine (P = 0.006). The implications of these between and within-vaccine variations are discussed.
Fig. 3.
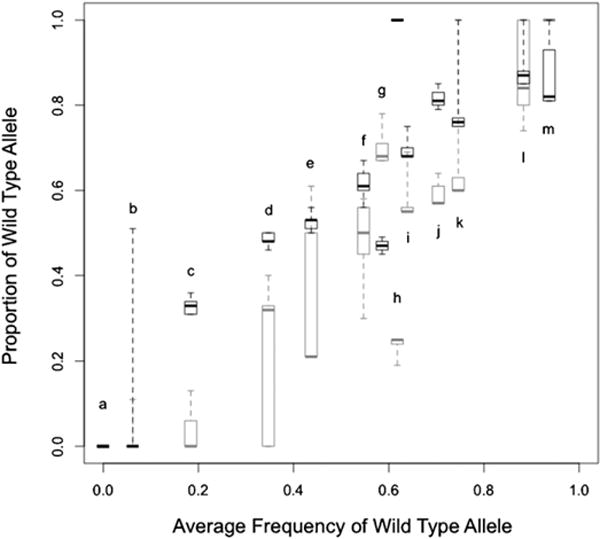
Comparison of range and average allele frequencies at polymorphic sites in Oka/Merck (black) and Oka/GSK (grey) vaccines. The whiskers on the box plots represent the full range of estimates of the wild-type allele, from the three batches obtained from each company; the box illustrates the inter-quartile range. The band inside the box represents the median value. Letters indicate the SNP position as below: a—560, 106262, 107252; b—105705; c—105356; d—107136; e—105169; f—97748; g—58595; h—89734; i—107797; j—19431; k—108838; l—87280; m—105724.
4. Discussion
We and others [10,11,13] have confirmed that the Oka vaccine comprises multiple haplotypes, each of which contains different numbers of vaccine mutations. Our data show three important findings. First, we confirm previous reports of the loss of wild-type alleles in the GSK as compared to Merck at 4 loci (560, 105705, 106262 and 107252), particularly where the wild-type allele was rare (i.e. at low frequency). Additionally, we show that where loci are mixed for vaccine and wild-type SNPs, the frequency of the wild-type allele at these loci is on average lower in the GSK than in the Merck vaccine. These findings are likely to be explained by production differences. Prior to development of the commercial vaccine in the 1980s, Smith Kline Beecham attempted to produce a genetically uniform vaccine—as a part of this the GSK seed lot underwent serial terminal dilutions in WI38 cells before further passages in MRC5s to generate the GSK working seed lot and vaccine stocks (Fig. 1). At the very least, this would be expected to reduce the amount of variability present in the vaccine mixture, if not indeed succeeding in making the vaccine genetically uniform. In contrast, the Merck vaccines are derived directly from the seed stock without any attempted purification before further passages in MRC5 cells ([7], Merck Product information IPC-VRV3-I-032008—summarised in Fig. 1), and are therefore expected to be more of a mixture of multiple haplotypes, as in the original seed stock.
Our second observation is that despite the differences in production processes, the allele frequencies in the Merck and GSK vaccines are strongly correlated (Fig. 3). This is an unexpected observation. If the vaccine alleles were simply neutral in the in vitro environment of vaccine production one would not expect to see a correlation in allele frequencies between the two vaccines given the differences in the manufacturing process (Fig. 1). Taking into consideration the GSK terminal dilution procedures, there are two possible explanations. If we assume that the GSK process succeeded in producing a genetically uniform vaccine, then the vaccine alleles could have arisen by de novo mutation and spread in the GSK production line after purification had occurred. However, two questions then arise: why have the vaccine mutations appeared at the same loci and with approximately the same frequency, as the Merck vaccine, and what would explain the lower frequency of the ancestral alleles in the GSK vaccine? One possibility is that both the Merck and GSK production processes select for the same intermediate allele frequencies, but that the GSK values have had insufficient time to reach this stable “optimum” value through the accumulation of mutations following the loss of the ancestral allele during the process of serial terminal dilutions. An alternative explanation is that the GSK serial dilutions did not result in a genetically uniform population but rather one that remained polymorphic with approximately the same allele frequencies as the original seed mixture. Both explanations would account for the lower frequency of the ancestral allele in GSK batches at those loci where the ancestral allele was initially rare in the seed stock, since it is the rare alleles that would tend to be lost by serial dilution and be prone to ascertainment bias (whereby rare alleles would have been detected if they were slightly more common in the Merck population but not vice versa). The fact that the ancestral allele is rarer in GSK at all but two loci (Fig. 3, 58595 and 87280), suggests a third possibility: that some difference in the Merck production favours selection of the ancestral alleles at the majority of loci.
Finally, we have observed that the Merck vaccine has more similar allele frequencies between lots of the same batch, than does the GSK vaccine. If the variation between lots were due to chance (i.e. random genetic drift), then this observation would imply that the Merck protocols maintain larger sample sizes of virus at each generation than the GSK, as the effects of genetic drift are more pronounced in smaller populations than larger populations. Alternatively, if as suggested earlier, selection is occurring in the Merck production process, this could explain the more similar allele frequencies observed in the Merck vaccines than the GSK.
In summary, our data suggest that evolutionary processes do influence production of live attenuated vaccines and affect their composition. We have focused on variable loci that have previously been identified as being of potential biological importance for vaccine attenuation and as immune targets [13]. However, there is evidence to suggest other loci may also play a key role in attenuation [17]. More extensive sequencing of different batches and lots from each source would give a better idea of the allele frequencies present at all variable loci, allowing us to distinguish between the different hypotheses described here as they make different predictions. If the lower frequency of ancestral alleles in GSK batches were due to ascertainment bias, this trend would disappear once all loci were sequenced. If the trend were due to the loss of rare alleles in the GSK production line, more extensive sequencing would reveal a pattern in which the GSK allele frequencies were more extreme than those in Merck batches, i.e. closer to zero in GSK at loci where they were rare in Merck batches, and closer to one where they were common. Finally, if there were genome-wide selection against ancestral alleles in GSK, that trend should be apparent across most vaccine loci irrespective of their frequency. Our analyses of the differences in allele frequencies between the vaccines provide an insight into the various evolutionary processes that are clearly affecting the composition of live attenuated vaccines. The data suggest that both batch to batch variation and variation between vaccines manufactured by different companies is not insignificant. To date, there is little evidence that the differences between the GSK and Merck vaccines or the batch to batch variation, affect efficacy or safety. However, a recent paper reported more breakthrough infections in children immunized with one dose of the GSK vaccine as compared with the Merck preparation, although the differences were not significant [18]. A previous publication also reported lower antibody titres following vaccination with the GSK preparation as compared to the Merck, albeit in the context of co administration of MMR vaccine [1]. Whether these data reflects greater variation in batch to batch immunogenicity of the GSK vaccine remains to be established. Nonetheless, understanding the evolutionary processes influencing this vaccine is clearly an important consideration for continued quality assurance of production.
Supplementary Material
Acknowledgments
RKK was funded by the Medical Research Council (grant no. G0700814). JB receives funding from the UCL/UCLH Collaborative Biomedical Research Centre. We thank the MRC Centre for Molecular Virology (grant no. G0900950) for infrastructure support.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.vaccine.2011.02.021.
References
- 1.Lau YL, Vessey SJ, Chan IS, Lee TL, Huang LM, Lee CY, et al. A comparison of safety, tolerability and immunogenicity of Oka/Merck varicella vaccine and VARILRIX in healthy children. Vaccine. 2002;20(23–24):2942–9. doi: 10.1016/s0264-410x(02)00245-1. [DOI] [PubMed] [Google Scholar]
- 2.Sharrar RG, LaRussa P, Galea SA, Steinberg SP, Sweet AR, Keatley RM, et al. The postmarketing safety profile of varicella vaccine. Vaccine. 2000;19(7–8):916–23. doi: 10.1016/s0264-410x(00)00297-8. [DOI] [PubMed] [Google Scholar]
- 3.Galea SA, Sweet A, Beninger P, Steinberg SP, Larussa PS, Gershon AA, et al. The safety profile of varicella vaccine: a 10-year review. J Infect Dis. 2008;197(Suppl. 2):S165–9. doi: 10.1086/522125. Review. [DOI] [PubMed] [Google Scholar]
- 4.Goulleret N, Mauvisseau E, Essevaz-Roulet M, Quinlivan M, Breuer J. Safety profile of live varicella virus vaccine (Oka/Merck): five-year results of the European Varicella Zoster Virus Identification Program (EU VZVIP) Vaccine. 2010;28(36):5878–82. doi: 10.1016/j.vaccine.2010.06.056. [DOI] [PubMed] [Google Scholar]
- 5.Quinlivan M, Breuer J. Molecular studies of Varicella zoster virus. Rev Med Virol. 2006 Jul-Aug;16(4):225–50. doi: 10.1002/rmv.502. Review. [DOI] [PubMed] [Google Scholar]
- 6.Takahashi M, Otsuka T, Okuno Y, Asano Y, Yazaki T. Live vaccine used to prevent the spread of varicella in children in hospital. Lancet. 1974;2(7892):1288–90. doi: 10.1016/s0140-6736(74)90144-5. [DOI] [PubMed] [Google Scholar]
- 7.Gomi Y, Sunamachi H, Mori Y, Nagaike K, Takahashi M, Yamanishi K. Comparison of the complete DNA sequences of the Oka varicella vaccine and its parental virus. J Virol. 2002 Nov;76(22):11447–59. doi: 10.1128/JVI.76.22.11447-11459.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gomi Y, Imagawa T, Takahashi M, Yamanishi K. Oka varicella vaccine is distinguishable from its parental virus in DNA sequence of open reading frame 62 and its transactivation activity. J Med Virol. 2000 Aug;61(4):497–503. doi: 10.1002/1096-9071(200008)61:4<497::aid-jmv13>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 9.Quinlivan MA, Gershon AA, Nichols RA, La Russa P, Steinberg SP, Breuer J. Vaccine Oka varicella-zoster virus genotypes are monomorphic in single vesicles and polymorphic in respiratory tract secretions. J Infect Dis. 2006;193(7):927–30. doi: 10.1086/500835. [DOI] [PubMed] [Google Scholar]
- 10.Loparev VN, Rubtcova E, Seward JF, Levin MJ, Schmid DS. DNA sequence variability in isolates recovered from patients with postvaccination rash or herpes zoster caused by Oka varicella vaccine. J Infect Dis. 2007;195(4):502–10. doi: 10.1086/510532. [DOI] [PubMed] [Google Scholar]
- 11.Tillieux SL, Halsey WS, Thomas ES, Voycik JJ, Sathe GM, Vassilev V. Complete DNA sequences of two oka strain varicella-zoster virus genomes. J Virol. 2008 Nov;82(22):11023–44. doi: 10.1128/JVI.00777-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quinlivan ML, Gershon AA, Steinberg SP, Breuer J. Rashes occurring after immunization with a mixture of viruses in the Oka vaccine are derived from single clones of virus. J Infect Dis. 2004;190(4):793–6. doi: 10.1086/423210. [DOI] [PubMed] [Google Scholar]
- 13.Quinlivan ML, Gershon AA, Al Bassam MM, Steinberg SP, LaRussa P, Nichols RA, et al. Natural selection for rash-forming genotypes of the varicella-zoster vaccine virus detected within immunized human hosts. Proc Natl Acad Sci USA. 2007;104(1):208–12. doi: 10.1073/pnas.0605688104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D’Hondt E, Berge E, Colinet G, Duchene M, Peetermans J. Production and quality control of the Oka-strain live varicella vaccine. Postgrad Med J. 1985;61(Suppl. 4):53–6. [PubMed] [Google Scholar]
- 15.Frey CR, Sharp MA, Min AS, Schmid DS, Loparev V, Arvin AM. Identification of CD8+ T cell epitopes in the immediate early 62 protein (IE62) of varicella-zoster virus, and evaluation of frequency of CD8+ T cell response to IE62, by use of IE62 peptides after varicella vaccination. J Infect Dis. 2003;188(1):40–52. doi: 10.1086/375828. [DOI] [PubMed] [Google Scholar]
- 16.Mattocks C, Tarpey P, Bobrow M. Whittaker, comparative sequence analysis (CSA): a new sequence-based method for the identification and characterization of mutations in DNA. J Hum Mutat. 2000 Nov;16(5):437–43. doi: 10.1002/1098-1004(200011)16:5<437::AID-HUMU9>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 17.Zerboni L, Hinchliffe S, Sommer MH, Ito H, Besser J, Stamatis S, et al. Analysis of varicella zoster virus attenuation by evaluation of chimeric parent Oka/vaccine Oka recombinant viruses in skin xenografts in the SCIDhu mouse model. Virology. 2005;332(1):337–46. doi: 10.1016/j.virol.2004.10.047. [DOI] [PubMed] [Google Scholar]
- 18.Spackova M, Wiese-Posselt M, Dehnert M, Matysiak-Klose D, Heininger U, Siedler A. Comparative varicella vaccine effectiveness during outbreaks in day-care centres. Vaccine. 2010;28(3):686–91. doi: 10.1016/j.vaccine.2009.10.086. Erratum in: Vaccine. 2010 May 7;28(21):3754. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


