Abstract
With recent approvals for multiple therapeutic antibodies that block cytotoxic T lymphocyte associated antigen 4 (CTLA4) and programmed cell death protein 1 (PD1) in melanoma, non-small-cell lung cancer and kidney cancer, and additional immune checkpoints being targeted clinically, many questions still remain regarding the optimal use of drugs that block these checkpoint pathways. Defining biomarkers that predict therapeutic effects and adverse events is a crucial mandate, highlighted by recent approvals for two PDL1 diagnostic tests. Here, we discuss biomarkers for anti-PD1 therapy based on immunological, genetic and virological criteria. The unique biology of the CTLA4 immune checkpoint, compared with PD1, requires a different approach to biomarker development. Mechanism-based insights from such studies may guide the design of synergistic treatment combinations based on immune checkpoint blockade.
Interactions between the immune system and cancer are governed by a complex network of biological pathways. Despite expectations that the immune system should automatically reject cancer cells as ‘foreign’, based on their unique and often extensive mutational profiles, the overriding natural balance between the immune system and cancer is tolerance, in which cancer cells are seen as ‘self’. Tolerance is maintained by multiple mechanisms, including regulatory immune cells, immunosuppressive cytokines and chemokines, and so-called ‘immune check-point’ pathways that down-modulate immune functions. The programmed cell death protein 1 (PD1; also known as PDCD1)–PD1 ligand 1 (PDL1) receptor–ligand pair is a dominant immune checkpoint pathway operative in the tumour microenvironment (TME); its normal function in controlling immune homeostasis is induced in cancer cells to evade immune attack1,2. Monoclonal antibodies (mAbs) that block this pathway have emerged as powerful weapons in the oncological armamentarium. Durable objective (partial or complete) responses following anti-PD1 therapy in patients with advanced melanoma (31–44% of patients)3–7, non-small-cell lung cancer (NSCLC; 19–20%)8–10 and renal cell carcinoma (RCC; 22–25%)11,12, accompanied by extended overall survival compared with conventional therapies, supported recent regulatory approvals by the US Food and Drug Administration (FDA) for the use of two different anti-PD1 drugs (nivolumab and pembrolizumab) in these indications. Further approvals are anticipated as experience accumulates in treating other cancer types such as bladder cancer, Hodgkin lymphoma and head and neck cancer13. However, anti-PD1 drugs are not effective against all cancer types, nor in every patient within a ‘responsive’ cancer type14. Unusual response patterns, including delayed or mixed tumour regression, pose further clinical challenges. Therefore, biomarkers are needed to guide patient selection for both monotherapy and combination therapy, and to provide early on-treatment indicators of response, based on our evolving scientific understanding of the biological mechanisms underlying blockade of the PD1–PDL1 pathway (BOX 1).
Box 1. General considerations for biomarker development.
The recent proliferation of new drugs and treatment combinations in oncology, with increasing costs to the consumer, has generated intense interest in identifying biomarkers to guide patient selection based on predicted efficacy and/or toxicity (US Food and Drug Administration guidance on biomarker use in management of cancer patients).
Tumour biology may predict that only ‘marker-positive’ patients will respond to therapy, such as patients receiving mutant oncogene-targeted drugs. For example, the common BRAF-V600E mutation in melanoma identifies a subset of patients who are likely to respond to highly selective BRAF inhibitors. This example typifies targeted kinase inhibitors as a drug class, but may not apply to immunotherapies that target molecules broadly expressed across a dynamic and context-dependent range. Thus, biomarker paradigms for oncogene-targeted inhibitors cannot simply be transferred to immunotherapies.
Biomarkers may be useful adjuncts for drugs with an unfavourable risk–benefit balance that is, drugs for which the rate of potentially serious side effects approximates to or outweighs potential benefit in the unselected patient population. For example, ipilimumab (anti-cytotoxic T lymphocyte associated antigen 4 (CTLA4)) confers long-term survival benefit in approximately 20% of patients with advanced melanoma in the unselected population, but its rate of serious side-effects is also approximately 20%; therefore, the impact of ipilimumab could be improved by biomarkers for efficacy or toxicity.
Biomarkers gleaned from ‘exceptional responders’ may identify a small subset of patients who are likely to respond to therapy, within a much larger unresponsive population. For instance, advanced colorectal cancers are generally unresponsive to anti-programmed cell death protein 1 (PD1) therapy, but a subset with genomic instability and high tumour mutational burden is very responsive93.
Biomarkers that are not absolutely predictive of response may nevertheless identify patients with a greater likelihood of response, thereby guiding clinical decision-making for treatment sequencing (that is, first-line or later-line therapy). This appears to be the case for PD1 ligand 1 (PDL1) immunohistochemistry testing in the context of anti-PD1 PDL1 therapies against several cancer types13. However, with the expanding activity profile of PD1 blockers across numerous cancers, it seems that the next level of biomarker research should address the characteristic biologies of different tumour types.
In addition to the PD1 pathway, there are dozens of other immune-modulating receptor–ligand interactions at the interface between cancer cells and host immune cells that can be targeted clinically with monotherapies and treatment combinations1,2,15. A mAb, ipilimumab, that blocks the prototypical immune checkpoint cytotoxic T lymphocyte associated antigen 4 (CTLA4) was approved by the FDA as monotherapy for advanced melanoma in 2011, based on a response rate of 11% and prolonged overall survival in 22% of patients16,17. Furthermore, a treatment regimen for melanoma combining anti-CTLA4 (ipilimumab) with anti-PD1 (nivolumab) was approved by the FDA in October 2015, highlighting the need for biomarkers for combination regimens7. Unlike PD1–PDL1, the CTLA4 immune checkpoint predominantly functions early in the life cycle of the immune response, during T cell priming and activation, and it enhances the immunosuppressive activity of regulatory T cells (Treg cells). As such, it has a global impact on the immune system; therefore, biomarkers of response and resistance to anti-CTLA4 therapy may differ from other immune checkpoint inhibitors that have different mechanisms of action (MOAs)15. Although this Review focuses on biomarker development for anti-PD1–PDL1 therapies, the distinct phenotypic and functional properties of the PD1 and CTLA4 immune checkpoints enable informative comparisons of biomarker development for therapeutic agents that target these molecules and potentially other immune checkpoints in the future (TABLE 1).
Table 1.
Phenotypic and functional properties of the PD1 and CTLA4 immune checkpoints: family members with distinct characteristics
| Characteristic | PD1 | CTLA4 |
|---|---|---|
| Cluster of differentiation (CD), gene name | CD279, PDCD1 | CD152, CTLA4 |
| Cellular expression | T, B and NK cells | T cells |
| Ligands | PDL1 (also known as B7H1 or CD274) and PDL2 (also known as B7DC or CD273) | B7-1 (also known as CD80) and B7-2 (also known as CD86) |
| Ligand expression | Activated APCs, haematopoietic and parenchymal cells in an inflammatory microenvironment, placenta and cancer cells (diverse solid and haematological tumours) | APCs, activated T and B lymphocytes and T and B cell malignancies |
| Mouse genetic knockout phenotypes | Organ-specific and strain-specific inflammation in adult mice122,123 | Death within 3 weeks of birth due to uncontrolled lymphoproliferation124,125 |
| Blocking antibodies approved for human use | Nivolumab and pembrolizumab | Ipilimumab |
| Currently approved therapeutic indications* | Unresectable metastatic melanoma and treatment-refractory metastatic non-small-cell lung cancer and renal cell carcinoma | Unresectable metastatic melanoma |
APC, antigen presenting cell; CTLA4, cytotoxic T lymphocyte associated antigen 4; NK, natural killer; PD1, programmed cell death protein 1; PDL, PD1 ligand.
Response rates of various cancer types to these drugs are described in the text.
MOA-based understanding of biomarkers
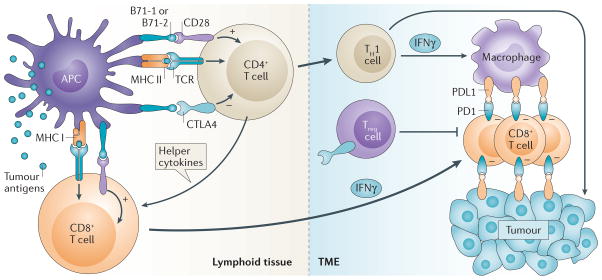
Biomarker exploration should be rationally conducted based on a knowledge of the MOA of the pathway targeted by the administered drug. In cancer, the main site of action for the PD1 pathway seems to be within the tumour itself, where cancer-specific immune effector cells meet their cognate target, and the major effector cells seem to be CD8+ cytolytic (killer) T cells18,19. However, before events occur at the tumour–immune interface, immune cells specific for a tumour antigen must first be activated at sites of antigen priming (lymph nodes) and then they must traffic to sites of antigen display (tumours). Each of these major events — priming, trafficking and target engagement — presents an opportunity for anti-PD1 therapeutic biomarker development (FIG. 1). For antigen priming, it is important to consider the nature of the recognized tumour antigens (mutant or non-mutated) and their diversity (is a crucial threshold of antigenic complexity needed to generate sufficient antitumour immunity?). While the issue of whether antigen-primed T cells actually reach their specific tumour target has been difficult to address experimentally in humans, new or increased CD8+ T cell infiltrates following anti-PD1 therapy in melanoma have been associated with tumour regression4,19,20. Finally, activation of tumour-specific lymphocytes (T and B cells) and natural killer (NK) cells is accompanied by PD1 co-inhibitory receptor display, as part of a normal mechanism to down-modulate immunity once the immune response has done its job, thereby avoiding normal tissue inflammation that might otherwise occur as a by-product of an uncontrolled response. This has focused attention on expression of the PD1 ligands, PDL1 and PDL2, in the tumour milieu as potential biomarkers of therapeutic response. It stands to reason that anti-PD1 therapies might be irrelevant for tumours devoid of PD1 ligand expression14. However, even in the presence of PD1 and its ligands, many parallel checkpoint pathways could potentially support resistance to anti-PD1 therapies1. Therefore, a broad immune profiling of the TME is warranted for the discovery of biomarkers relevant to these drugs. Notably, studies to identify biomarkers from circulating peripheral blood lymphocytes (PBLs) in patients treated with anti-PD1 therapy have not been particularly revealing, as might have been expected based on the MOA of the PD1 pathway predominantly within tumours20,21.
Figure 1. Opportunities for biomarker development based on mechanistic nodes in immune checkpoint pathways.
The nature of recognized tumour antigens, immune cell migration into tumours and the expression of immune checkpoint receptors and ligands all provide opportunities for biomarker development. CD8+ T effector (Teff) cells are thought to be the major type of immune cell affected by the programmed cell death protein 1 (PD1) immunosuppressive checkpoint pathway. In contrast, cytotoxic T lymphocyte associated antigen 4 (CTLA4) predominantly regulates the activity of CD4+ T cells, both effector and regulatory (Treg) subtypes. Priming of T cells requires their recognition of processed tumour antigens presented by antigen presenting cells (APCs) such as monocytes and dendritic cells, through a unique T cell receptor (TCR) that binds to a major histocompatibility complex (MHC) molecule and tumour-derived peptide antigens. Such antigens may be generated from mutant or non-mutated tumour-associated proteins. Priming of T cells generally occurs in lymphoid tissue and CD4+ T cells provide help for CD8+ T cell priming in the form of cytokines. Both CD4+ and CD8+ T cells are then activated through co-stimulatory pathways such as CD28 B7-1 (also known as CD80) and CD28 B7-2 (also known as CD86), causing them to proliferate, secrete inflammatory cytokines, acquire cytolytic properties and migrate to sites of antigen display, that is, tumour deposits. CTLA4 has a major role in regulating the amplitude of CD4+ T cell priming and CD4+ T cell help for CD8+ T cell priming in lymphoid tissue. Within hours to days, activated T cells also begin to express the co-inhibitory receptor PD1. CD4+ T helper 1 (TH1) cells and CD8+ T cells in the tumour microenvironment (TME) produce interferon-γ (IFNγ), which, on the one hand, activates tumour killing by macrophages and antigen display by tumour cells, but, on the other hand, induces PDL1 expression by these same macrophages and tumour cells. Tumour-specific PD1+ CD8+ T cells encountering PDL1+ cells within the TME (tumour or stromal cells) will be functionally disabled. Additionally, CTLA4 expressed by Treg cells in the TME enhances their ability to suppress CD8+ T cell-dependent cytokine production and direct tumour cell killing. Drugs blocking the immune checkpoints CTLA4, PD1 and PDL1 interrupt these immunosuppressive interactions and restore the ability of T cells to eliminate antigen-expressing cancer cells.
Unlike PD1, which modulates antigen-experienced effector cells, CTLA4 is a global immune checkpoint engaged in priming immune responses via down-modulation of CD4+ T effector (Teff) cells and enhancement of Treg cell activity1,2,15 (TABLE 1). Therefore, many biomarker studies of anti-CTLA4 therapies (ipilimumab and tremelimumab) have focused on the diversity, phenotype and function of PBLs before and after therapy, instead of on tumour biopsies. Increased diversity and expression of activation markers on PBLs have been reported following anti-CTLA4 therapy21,22. At least two independent studies noted that a rise in the absolute lymphocyte count in peripheral blood correlated with a higher rate of response to ipilimumab23,24. CD8+ T cells may be the most relevant subset in this analysis, as CTLA4 blockade can enhance CD8+ T cell-mediated immune responses indirectly by enhancing the activity of CD4+ T helper cells25. Furthermore, patients with melanoma who developed CD4+ and CD8+ PBLs with specificity against the NY-ESO-1 cancer testis antigen also demonstrated significant tumour shrinkage or stabilization26. In contrast, other factors in peripheral blood, such as high levels of soluble CD25 (also known as IL2Rα), have been correlated with resistance to anti-CTLA4 therapy27. Local factors in the pretreatment TME, such as PDL1 expression, are generally not associated with clinical response to anti-CTLA4 therapy7, although one study raised the possibility that patients with an inflamed TME before treatment were more likely to respond to anti-CTLA4 therapy28. Increased expression of the co-stimulatory molecule inducible T cell co-stimulator (ICOS) on PBLs and tumour-infiltrating lymphocytes (TILs) has also been observed following CTLA4 blockade in patients with various tumour types29,30. Furthermore, an increased Teff cell:Treg cell ratio in tumour tissues has been observed31,32. Despite these correlations, no predictive biomarker for selection of patients to receive ipilimumab, nor any on-treatment pharmacodynamic marker, has yet proved sufficiently robust to be used clinically.
Immunological biomarkers
The first candidate biomarkers to be explored for immune checkpoint-blocking therapies were immunological, as a logical extension of our knowledge about the biology of the targeted pathways. Immunological biomarkers offer the potential advantage of applicability across multiple tumour types that are amenable to these therapies.
Intratumoural lymphoid infiltrates
Virchow commented on the interaction between leukocytes and tumour cells in his monograph of 1863 (REFS 33,34). Today, we are still unravelling the complexities of the interaction between cancer and the host immune system. Different components of the immune system can either promote or combat tumour growth. The potential for the adaptive immune system, in particular CD8+ cytotoxic T lymphocytes, to control or eradicate tumours has been shown in laboratory models. In a study of human colorectal carcinoma (CRC) specimens detailing the relationship between T cell densities at the invasive tumour margin and those in the centre of the tumour, high densities of CD3+CD8+CD45RO+granzyme+ T cells (that is, antigen-experienced cytolytic Teff cells) were associated with a lower likelihood of tumour relapse and improved overall survival35. Moreover, coordinated pathological analysis of CD3+ T cell densities at both the central aspect and the invasive edge of the tumour outperformed internationally accepted clinical staging criteria (Union for International Cancer Control (UICC)-TNM) in predicting disease-free survival and overall survival in multivariate analysis35. In a survey of the literature on the prognostic importance of T cell infiltrates in different tumour types, the association of CD8+CD45RO+ T cells with improved prognosis was seen in 97% (58/60) of the reports analysed36. In some instances, the host response may be highly organized, with the development of a tertiary lymphoid structure, which is thought to facilitate T cell recruitment and coordinate the local adaptive antitumour immune response, further contributing to improved patient outcomes37–39. Specifically, in the context of anti-PD1 therapy for melanoma, CD8+ T cell density at the invasive tumour edge has been correlated with response to anti-PD1 treatment19.
Intratumoural PDL1 expression
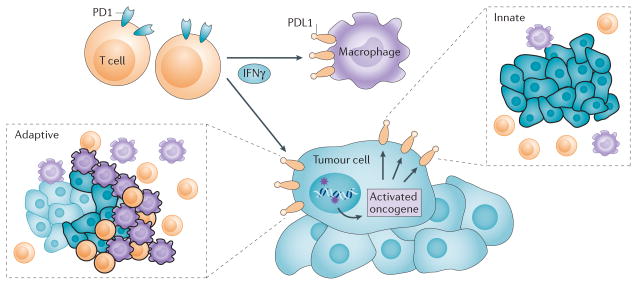
Beyond enumerating, localizing and phenotyping CD8+ Teff cells and other tumour-infiltrating immune cell types, the next layer of biomarker investigation involves examining specific defence mechanisms that tumours can use to guard against immune attack. A dominant mechanism that is relevant to anti-PD1 drug response is PDL1 expression. PDL1 is normally expressed by a subset of macrophages and can be induced on activated lymphocytes (T, B and NK cells), endothelial cells and other non-malignant cell types in an inflammatory microenvironment, as part of a physiological process to down-modulate ongoing host immune responses in peripheral tissues40–42. However, tumour cells and associated stromal cells can also express this checkpoint, thereby turning off Teff cells41. In some cancers, PDL1 expression is driven constitutively by aberrant signalling pathways or chromosomal alterations. For example, PTEN mutations causing PI3K–AKT pathway activation in a subset of glioblastoma cases43 and 9p24.1 gene translocations or amplifications in certain lymphomas44,45 can result in broad expression of PDL1 and PDL2 on the surface of the majority of the tumour cells (FIG. 2). PDL1 may also be expressed in the TME by a mechanism termed adaptive immune resistance1. The first scientific support for the concept of adaptive immune resistance was provided by Taube and colleagues in a study focused on human melanoma samples46. In this study, four distinct archetypes of tumour–host interaction were demonstrated: broad, constitutive tumour cell PDL1 expression in the absence of a substantial host immune response, consistent with the aforementioned genetically driven constitutive PDL1 upregulation; PDL1 expression focally and geographically associated with the host anti-tumour immune response; tumours that had immune cell infiltrates but lacked PDL1 expression; and tumours that lacked both an immune response and PDL1 expression. When immune infiltrates in PDL1+ versus PDL1− melanomas were compared by mRNA expression profiling, a CD8+ T cell–T helper 1 (TH1) cytokine signature characterized by interferon-γ (IFNγ) expression was identified in PDL1+ melanomas46,47. PDL1+ melanomas were associated with overexpression of PD1. Notably, intratumoural PDL1 expression was generally associated with improved overall survival in patients with melanoma46, contrary to expectations based on its known immunosuppressive function. This paradox is explained by a mechanism in which tumour-infiltrating cytotoxic T lymphocytes upregulate PD1 expression and secrete IFNγ when they encounter tumour antigens, followed by an adaptive response to IFNγ, whereby tumour cells and infiltrating immune cells in the vicinity upregulate PDL1 expression. This PDL1 induction in the TME creates a ‘shield’ against attack from activated PD1+ Teff cells (FIG. 2). Thus, PDL1 expression in this context can be considered as a marker of an active host antitumour immune response. Since the initial description of adaptive immune resistance in melanoma, an adaptive pattern of PDL1 expression has been described in other tumour types, including Merkel cell carcinoma (MCC), NSCLC and breast cancer48–51. In each instance, PDL1 expression associated with tumour-infiltrating immune cells was found to be a positive prognostic feature.
Figure 2. Mechanisms for intratumoural PDL1 expression.
Constitutive broad (innate) expression of membranous programmed cell death 1 ligand 1 (PDL1) by tumour cells is thought to be driven by dysregulated signalling pathways such as PI3K AKT, or chromosomal alterations and amplifications such as are found in Hodgkin lymphoma. In contrast, adaptive focal expression of PDL1 by tumour cells and macrophages occurs at the interface of tumour cell nests with immune infiltrates secreting pro-inflammatory factors such as interferon-γ (IFNγ; the ‘immune front’). The ligation of PDL1 with programmed cell death protein 1 (PD1) molecules will down-modulate T cell function, essentially creating a negative feedback loop that dampens antitumour immunity. The innate and adaptive mechanisms for PDL1 induction are not mutually exclusive: constitutive oncogene-driven PDL1 expression may be further upregulated by inflammatory cytokines. In boxed insets, tumour cells are shown as blue, macrophages are purple and T cells are orange; black outlining of cells indicates PDL1 protein expression, such as would be demonstrated with immunohistochemistry.
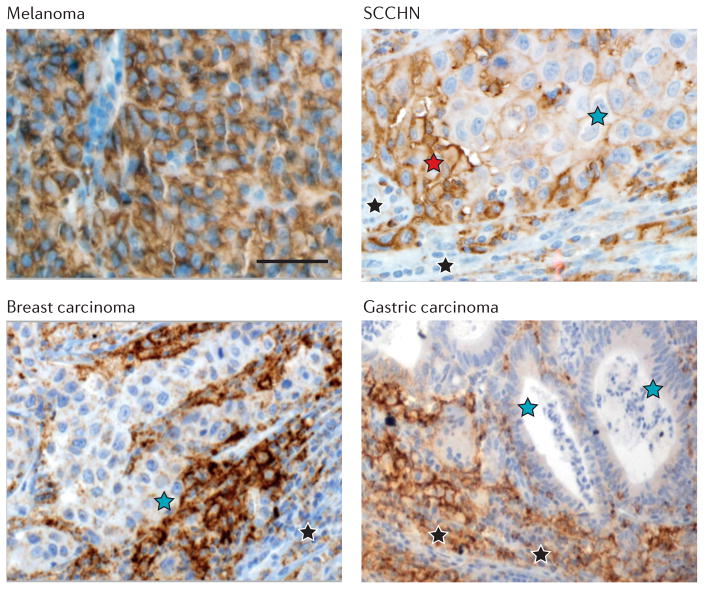
More recent studies in several cancer types have revealed variations on this theme. It has become clear that constitutive and adaptive mechanisms of intratumoural PDL1 display are not mutually exclusive. For example, in squamous cell carcinomas of the head and neck (SCCHNs), PDL1 expression may be driven by both oncogenic and adaptive immune resistance mechanisms in the same tumour52 (FIG. 3). Along the same lines, potential associations between oncogenic driver mutations and PDL1 expression have been examined. A subset of KRAS mutant lung adenocarcinomas may demonstrate heightened PDL1 expression and denser inflammation compared with wild-type tumours53. However, in melanoma, the presence of the common oncogenic BRAF-V600E mutation does not seem to correlate with PDL1 expression54. As discussed below, the total burden of mutations in a given tumour might have a greater role than individual driver mutations in shaping the tumour immune microenvironment; this is particularly relevant for carcinogen-induced tumours such as melanoma and smoking-associated lung cancers, and for tumours with defects in DNA damage responses55,56. Further complexity has arisen with regard to the cell type (or types) expressing PDL1 (FIG. 3). Certain cancers, such as SCCHN, melanoma, breast cancer and RCC, frequently express PDL1 on the surface of tumour cells as well as on infiltrating immune cells46,51,52,57,58. In contrast, in other tumours such as CRC and gastric carcinoma, PDL1 is observed almost exclusively on tumour-infiltrating immune cells and is only rarely expressed on tumour cells themselves56,59,60. Furthermore, tumours such as MCC show intermediate phenotypes on this spectrum48. Such differential expression may reflect the variable susceptibility of tumour cells and infiltrating immune cells to cytokines and other stromal factors in the tumour milieu47.
Figure 3. PDL1 expression patterns in different types of cancer.
In all panels, brown staining indicates programmed cell death 1 ligand 1 (PDL1) protein expression, detected with immunohistochemistry. Upper left panel: a melanoma specimen shows broad PDL1 expression on all malignant cells, independent of an immune infiltrate. This pattern, seen in approximately 1% of melanocytic lesions, suggests oncogene-driven constitutive PDL1 expression. Upper right panel: a squamous cell carcinoma of the head and neck (SCCHN) shows broad areas of low-level PDL1 expression on tumour cells (blue star), with heightened expression at the boundary of tumour nests (red star) with immune infiltrates (black stars); these features suggest a combination of oncogene-driven PDL1 expression and adaptive immune resistance. Lower left panel: a breast carcinoma shows PDL1 expression on malignant cells (blue star) confined to areas immediately adjacent to immune cells (T cells and associated macrophages, black star), many of which also express PDL1. This pattern is seen in approximately 20% of breast cancers. Lower right panel: a gastric carcinoma shows PDL1 expression on infiltrating immune cells (black stars) immediately adjacent to tumour cells, but not on the surface of tumour cells themselves (blue stars). A similar pattern of PDL1 expression has also been described in colorectal cancers. Distinct patterns of PDL1 expression may be observed not only in different tumour types, but also in individual cases within the same tumour type. Scale bar, approximately 50 μm.
Although PDL2 expression has been observed in tumours, its role in mediating immune resistance and thus its predictive importance for response to PD1 pathway blockade is unknown. A subset of Hodgkin lymphomas, mantle cell lymphomas and gastric cancers have a chromosomal amplification of 9q24.1, the location of the genes that encode PDL1 and PDL2 (CD274 and PDCD1LG2, respectively). These tumours express substantial levels of PDL2 (REFS 44,45). Most PDL2 in solid tumours is expressed on myeloid cells in the TME47. Analysis of a limited number of solid tumours demonstrated expression of PDL2 in approximately 20% of cases, but its expression did not enhance the predictive power of PDL1 expression for responsiveness to anti-PD1 therapy58. For cases in which PDL2 might have a role in PD1-dependent immune resistance, anti-PD1 therapy would be predicted to be superior to anti-PDL1 therapy, which would not block the PD1–PDL2 interaction.
Developing PDL1 IHC and other immunological biomarkers for clinical use
The dynamic nature of antitumour immune reactivity predicts that developing immune-based biomarkers for outcomes following immune checkpoint blockade will be challenging. The initial hint that assessment of intratumoural PDL1 protein expression with immunohistochemistry (IHC) could predict anti-PD1 therapy response was provided in the first-in-human study of nivolumab in 39 patients with several solid tumour types20. This finding was recapitulated in a larger nivolumab study showing that patients with PDL1+ tumours, defined as having at least 5% of tumour cells with cell surface PDL1 protein expression, were twice as likely to respond to treatment compared with the overall study population14. Interestingly, variable expression of PDL1 was observed in multiple tumour biopsies collected over time and/or from different anatomical sites in individual patients14, illuminating a potential pitfall of developing PDL1 IHC as an absolute biomarker based on a single tumour specimen per patient (BOX 2). Since these initial reports, expanded investigations in several solid tumour types, including NSCLC, melanoma, SCCHN, RCC and bladder cancer, using several different PDL1 IHC assays and cut-off criteria for positivity, have validated the general conclusion that PDL1 expression in pretreatment tumour specimens portends a greater likelihood of response to anti-PD1 and anti-PDL1 drugs13,61. Of note, however, subsequent studies have also revealed a lower but finite response rate in patients with PDL1− tumours, calling into question the use of this marker as an absolute selection criterion for therapy13. In a recent analysis of multiple reports, the objective response rate to PD1 pathway blockade averaged 29% across 15 studies in various solid tumour types; when analysed according to PDL1 expression, the response rate in patients with PDL1+ tumours was 48%, compared with 15% in patients with PDL1− tumours61. Although the relationship between PDL1 expression and long-term outcomes from anti-PD1 therapy such as progression-free survival and overall survival has yet to be firmly established, some reports suggest that survival is prolonged in patients whose tumours demonstrate PDL1 expression8,11,62. In patients with multiple treatment options, PDL1 expression might be used to prioritize treatment sequencing; that is, patients with PDL1+ tumours might be advised to receive anti-PD1–PDL1 as first-line therapy, whereas patients with PDL1− tumours could receive it as second- or later-line therapy.
Box 2. Pitfalls of using PDL1 immunohistochemistry as a biomarker test for anti-PD1–PDL1 therapy.
Focal programmed cell death 1 ligand 1 (PDL1) expression in some tumours may be missed in small biopsy specimens, such as needle biopsies
PDL1 expression among multiple tumour lesions from individual patients can vary over time and by anatomical site
PDL1 expression in tumour biopsies collected months or years earlier might not accurately reflect PDL1 status at the time of treatment initiation; therapies given after biopsy but before administration of programmed cell death protein 1 (PD1) pathway blockade (radiation therapy, chemotherapy or kinase inhibitors) may alter PDL1 expression
PDL1 epitopes detected by some antibodies are potentially unstable with prolonged specimen fixation or inadequate tissue handling before fixation (see NCI guidelines for tissue handling)
Antibodies used for PDL1 detection have different affinities and specificities
PDL1 protein expression can be membranous and/or cytoplasmic; however, only membranous PDL1 is functionally relevant, by contacting PD1+ T cells
PDL1 can be expressed by multiple cell types within the tumour microenvironment, which poses challenges for scoring and interpretation
In October 2015, a PDL1 IHC test was approved by the FDA as a companion diagnostic for pembrolizumab in treating advanced NSCLC (PDL1 IHC 22C3 pharmDx). This approval was based on results from a large Phase II trial that included patients with squamous and non-squamous NSCLC subtypes, showing that patients whose tumours were ≥50% PDL1+ (approximately 20% of NSCLC cases) had a higher response rate to anti-PD1 therapy and prolonged progression-free survival and overall survival, compared with patients with lower PDL1 expression levels8. Accordingly, the PDL1 diagnostic status of positive for this assay uses a threshold of >50%. However, two Phase III trials of a different anti-PD1 drug, nivolumab, in patients with squamous or non-squamous NSCLC using another PDL1 IHC test with a unique mAb (PDL1 IHC 28-8 pharmDx) showed that PDL1 expression on tumour cells in pretreatment tumour specimens correlated with improved overall survival in patients with non-squamous NSCLC, but not in those with squamous NSCLC9,10. Interestingly, the PDL1 IHC 28-8 pharmDx assay was approved by the FDA in October 2015, as a complementary but not required diagnostic test for nivolumab in lung cancer. It was subsequently approved in January 2016 as a complementary test for nivolumab in melanoma7. This assay uses a threshold of >1% for a ‘positive’ PDL1 result for both melanoma and non-squamous NSCLC. Differences in the current recommendations for using the IHC 22C3 and IHC 28-8 PDL1 tests reflect the available supportive clinical data. The use of PDL1 IHC as a diagnostic test will continue to evolve as more information regarding clinical correlations from randomized trials becomes available. For example, there are now contradictory data from clinical trials of PD1 pathway blockers in RCC, with some trials showing a positive correlation of intratumoural PDL1 expression with response rates and progression-free survival and overall survival11,63, whereas others do not12. A cross-industry initiative, termed the Blueprint Working Group, has been established to provide an analytical comparison of several PDL1 IHC tests currently in use or in development that use different antibodies and different scoring criteria for PDL1 positivity.
Accurate measurement and scoring of PDL1 protein expression are plagued by various technical and biological pitfalls (BOX 2). Proprietary automated IHC tests currently in use or in development use different mAbs for PDL1 detection, and have been developed in isolation without cross-comparisons. They have been applied to tumour specimens collected days, months or years before initiation of anti-PD1 treatment; specimens taken a long time before treatment might not reflect PDL1 status at the time of therapy. Specimens can be collected by various procedures including surgical resection or needle biopsy; focal PDL1 expression as described above for the adaptive immune resistance phenomenon could be missed in small tumour specimens, resulting in a false-negative PDL1 evaluation64. Furthermore, not all anti-PDL1 mAbs produce similar staining results46. In addition to mAbs that target different epitopes in the PDL1 protein, IHC techniques have many variables, including antigen retrieval conditions and temperatures, mAb concentrations and incubation times, and detection systems. The type and duration of tissue fixation are also well established variables that affect IHC determinations65, and the pathologist’s interpretation of tissue staining is yet another important variable. PDL1 can be expressed in the cytoplasm and/or on the cell surface by diverse cell types including tumour cells, activated lymphocytes, tumour-associated macrophages and rare dendritic cells within the tumour; the relative contributions of these factors to local tumour immunosuppression are still incompletely understood40,41,56. In some situations, accurately identifying individual cell types by morphology alone is challenging, as both malignant cells and tumour-associated macrophages can show considerable cytological pleiomorphism. Additionally, whereas studies often report IHC results as simply plus or minus, PDL1 expression levels are continuous and the most useful thresholds for determining positive or negative expression as they relate to patient survival or therapeutic response have yet to be determined. Lastly, patients eligible for clinical trials have often received multiple previous cancer treatments, which could alter the immune microenvironment of the tumour66,67.
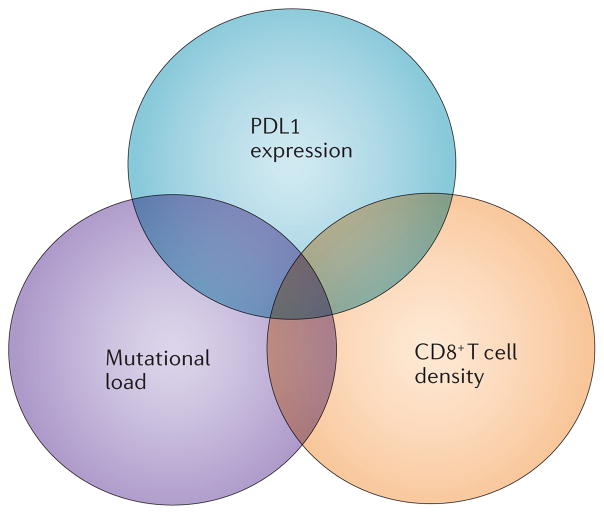
Beyond PDL1 expression, additional intratumoural factors have been nominated as predictive biomarkers for outcomes of anti-PD1 therapy. Factors such as tumour mutational burden and CD8+ T cell density are functionally related to PDL1 expression and to each other, but are potentially easier to detect and quantify on pathological specimens than PDL1 expressed on cell membranes (FIG. 4). Furthermore, multifactorial markers may have enhanced predictive power compared with individual tests. For instance, Tumeh et al.19 demonstrated the increased predictive value of complex factors such as close proximity of PD1+ to PDL1+ cells, CD8+ T cell activation (CD8 and Ki-67 co-staining) and markers of signalling for the IFNγ pathway. Interestingly, expression of the PD1 receptor on lymphocytes, which is the direct target for anti-PD1, does not seem to be more predictive of clinical response than PDL1 expression58. This is consistent with the notion that adaptive PDL1 expression functionally reflects a TH1 cytokine milieu that is advantageous for tumour rejection.
Figure 4. Multifactorial biomarkers of clinical response to PD1 pathway blockade.
Tumour mutational load, the intensity of intratumoural CD8+ T cell infiltrates and tumour programmed cell death ligand 1 (PDL1) expression have each been proposed as distinct biomarkers of response to anti-programmed cell death protein 1 (PD1) PDL1 therapies. However, these factors are functionally interrelated and are often found coordinately in individual tumour specimens. Multifactorial biomarker panels incorporating these and other variables may provide stronger predictive value than individual markers for therapeutic outcomes.
Genetic biomarkers
Oncogenic mutations as biomarkers for anti-PD1 therapies
The revolution in cancer genetics and genomics (that is, the analysis of genetic changes in cancer on a genomic scale) has led to the definition of several activating mutations in driver oncogenes that have been successfully targeted by specific drugs68. In the case of BRAF, epidermal growth factor receptor (EGFR) and anaplastic lymphoma kinase (ALK), drugs that target these oncoproteins are active only when specific mutations or rearrangements are present. This has created a classic paradigm for genetic biomarker-driven therapy, whereby drug choice is absolutely dependent on the presence of a specific mutation that is detected by a DNA-based test.
Given that activation of specific oncogenic pathways can have broad effects on gene expression, it is reasonable to imagine that the genetic make-up of cancer cells could have major effects on the immune TME, by driving specific immune resistance pathways. This could be through induction of immune checkpoints, secretion of specific inhibitory cytokines or production of chemokines and other factors that recruit suppressive cell types. For example, melanomas with constitutive BRAF signalling, such as those driven by the BRAF-V600E mutation (found in ~50% of melanomas), were shown to secrete soluble factors that inhibit dendritic cell activation; silencing of mutant BRAF eliminated the secretion of these factors69.
For some tumours, such as glioblastoma, it has been demonstrated that PDL1 expression is driven by constitutive oncogenic signalling in the tumour cell. PDL1 expression on glioblastomas is enhanced upon deletion or silencing of PTEN, implicating the PI3K–AKT pathway in the upregulation of PDL1 expression43. Similarly, constitutive ALK signalling, observed in certain lymphomas and occasionally in lung cancer, has been reported to drive PDL1 expression via signal transducer and activator of transcription 3 (STAT3) activation70. More recently, a genetically engineered mouse model of mutant EGFR-driven lung cancer71, as well as lung cancer driven by dual loss of liver kinase B1 (LKB1; also known as STK11) and PTEN72, demonstrated induction of PDL1. However, a recent clinical study of anti-PD1 therapy (pembrolizumab) in NSCLC found no significant difference in PDL1 expression (by IHC) between tumours expressing mutant EGFR and those expressing wild-type EGFR8. In the same clinical study, there was a suggestion that KRAS mutant NSCLC expressed higher levels of PDL1, compared with KRAS wild-type tumours, but this requires further investigation.
To date, no specific oncogenic driver or tumour suppressor gene has been associated with response to anti-PD1 therapy as an independent variable. In melanoma, the response to anti-PD1 therapy seems to be similar in BRAF-V600E and BRAF wild-type tumours73,74. In lung cancer, the anti-PD1 response is lower in EGFR-mutant adenocarcinomas (meeting abstract) 75. However, this might reflect the association of mutant EGFR with never-smoker status, which correlates with a considerably lower response rate to anti-PD1 than smoking-associated lung cancer, independent of driver oncogene mutations. This could be because of increased tumour immunogenicity from a higher mutational load in smoking-associated lung cancer (see below). Importantly, there is still a paucity of information on correlations between clinical response to immune checkpoint blockade and specific oncogene, tumour suppressor gene or overall oncogenic pathway-associated mutations. Development of large data sets integrating response to immunotherapy with cancer and host genetics will potentially identify associations of mechanistic relevance and with biomarker implications.
Mutational load as a potential biomarker for response to checkpoint blockade
Beyond their effects on the biological behaviour of cancer cells, mutations, genetic rearrangements, insertions and deletions have the capacity to encode neoantigens that are specific to the tumour relative to normal somatic cells. Because T cells recognize processed peptides presented by host major histocompatibility complex (MHC) molecules (these are termed human leukocyte antigen (HLA) in humans), a cancer mutation can produce a T cell neoantigenic peptide via multiple mechanisms76: it can confer de novo peptide binding to a MHC molecule expressed by the host, by encoding an anchor residue favoured by that MHC molecule; it can encode a new T cell receptor (TCR) contact residue in a peptide that can already bind to a host MHC molecule; it can introduce a preferred proteasomal cleavage site that enables more efficient processing by intracellular machinery than would occur with the wild-type sequence; and it can destroy a proteasomal cleavage site in the middle of an amino acid sequence, thereby preserving a peptide for presentation by host MHC molecules. Because peptide processing steps for MHC I presentation and MHC II presentation are completely different, mutations will differentially affect neoantigen generation for CD8+ T cells and CD4+ T cells, respectively77,78. DNA rearrangements, deletions and insertions can also encode neoantigenic peptides via two mechanisms — they create new sequences at the rearrangement, deletion or insertion junctions and, if out of frame, will create new amino acid sequences until a stop codon is reached. Because humans express three to six HLA I and three to six HLA II molecules, a single DNA alteration can generate multiple epitopes that are potentially recognized by T cells.
Importantly, the antigenicity of a neopeptide is not related to its function; a passenger mutation with no functional role can be a perfectly good tumour antigen. This suggests that tumours with a greater mutational load could possess more neoantigens, and thus the patient may possess a larger repertoire of extant tumour-specific T cells. Given that immune checkpoint blockers essentially ‘unleash’ endogenous antitumour T cell responses, one could imagine that tumours with a higher mutational burden, irrespective of driver mutations, will be more susceptible to immune checkpoint blockade.
The first hint that high mutational load might predict responsiveness to immune checkpoint blockade came from observing the response rates of various cancer types to monotherapy with anti-PD1–PDL1 drugs. On the basis of large databases of mutational load in different cancer types55, cancers with the highest median mutational loads (melanoma, NSCLC, SCCHN, bladder cancer and gastric cancer) demonstrated response rates to anti-PD1 or anti-PDL1 of greater than 15%14,62,79. Melanoma, a carcinogen-induced cancer with one of the highest mutational loads among human tumours, has a particularly high response rate to anti-PD1 therapy (~30–40%). It is also the single cancer in which anti-CTLA4 (ipilimumab) monotherapy produces a significant response rate, and correlations between mutational load and anti-CTLA4 response in melanoma have recently been reported80,81. In contrast, cancer types with relatively low median mutational loads, such as pancreatic and prostate cancer, have shown little response to PD1 pathway-blocking antibodies. However, within distinct cancer types, there is a wide range (typically 2 log-fold) of mutational density among individual cases55,82.
Thus, from a biomarker standpoint, the question is whether mutational load itself, aside from specific oncogene or tumour suppressor gene mutations, will predict response in a given tumour type. A suggestion that this might be the case comes from the findings with PD1 pathway blockade in NSCLC, in which smokers have a higher response rate8. Mutational load in smoking-associated lung cancer is known to be much higher than in non-smoking-associated lung cancer. Analysis of a small group of patients with lung cancer who were receiving anti-PD1 therapy (pembrolizumab) demonstrated that patients deriving clinical benefit had higher mutational densities than those who did not benefit82. However, no clear numerical cut-off in mutational load could be defined that would justify excluding patients from therapy. Indeed there were outliers on both sides, including non-responding patients with very high mutational loads and responders whose tumours had mutational burdens at the low end of the range. Similarly, in two reports of ipilimumab in melanoma, tumours from patients deriving clinical benefit had a statistically significant higher median mutational load than tumours from those who did not, but there was considerable overlap between the two groups, precluding the definition of a cut-off point below which patients would not derive benefit80,81. An initial report that certain motifs of four contiguous amino acids surrounding or encompassing the altered amino acid were highly predictive of clinical benefit80 was not observed in a subsequent larger study81.
Taken together, these early studies correlating mutational load with response of NSCLC to anti-PD1 therapy and of melanoma to anti-CTLA4 therapy are compatible with, but do not directly prove, the notion that mutations can enhance tumour antigenicity, providing the basis for a higher probability of response to immune checkpoint blockade. As a corollary, it has been suggested that mutation-inducing cancer therapies, such as certain chemotherapies or radiation therapy, may predispose to subsequent successful therapy with immune checkpoint blockade83. However, the correlation between mutational burden and clinical response is highly imperfect, suggesting that neoantigenicity is only one factor determining responsiveness to checkpoint blockade and, in these settings, should not be used as a biomarker for patient exclusion.
If mutations generate neoepitopes that confer antigenicity on tumours, why is the correlation between mutational load and response to immune checkpoint blockade so imperfect? There are several factors. The randomness of non-driver mutations in cancer translates to the random generation of peptide epitopes that can be presented by host MHC molecules. Current computational algorithms are highly imperfect in predicting which mutations will generate epitopes recognized by T cells in the host. Despite improvements in recent years, application of current algorithms to cancers with high mutational burden, such as is found in most melanomas and smoking-associated lung cancers, predicts hundreds of possible neoantigenic peptides whereas T cell responses are typically found against very few84,85. For example, recent studies by the Schumacher and Rosenberg groups86,87 have typically identified only 1–3 mutation-associated neoantigens (validated by T cell responses) among the top 30–50 ranked candidates from algorithm predictions. Mass spectrometry of peptides eluted from MHC I molecules on tumours shows that only a small proportion of predicted peptide epitopes are actually found bound in the MHC I groove, indicating that there are additional variables to peptide processing and presentation that are not yet understood. Furthermore, algorithms for predicting MHC II epitope binding are quite poor and thus, the role of CD4+ cells (which bind only MHC II molecules) in responding to tumour neoantigens is under-studied. The demonstration of tumour regression after transfer of CD4+ T cells generated against an HLA II-restricted tumour neoantigen indeed demonstrates the importance of helper T cell reactivity in antitumour immune responses88. It is also possible that by chance, a single ‘perfect’ antigenic peptide could be generated from a relatively small number of mutations, whereas a large number of mutations might not generate any high-quality antigen peptides. Another variable is the T cell repertoire in the host. If a mutation-associated neoantigenic peptide mimics a self-antigen that has already tolerized T cells (by either deletion or anergy induction), there may be no remaining T cells with functional activity to be unleashed even when immune checkpoints such as PD1–PDL1 are blocked. Furthermore, in a given patient, the proportion of the antitumour immune response conferred by T cells specific for ‘private’ mutation-associated neoantigens versus shared non-mutated tumour-associated antigens — such as self-antigens highly upregulated in tumour cells relative to normal cells — is unknown. In fact, early studies of tumour-reactive T cells from melanoma clearly demonstrated that both types of antitumour reactivity exist in many patients89. Thus, epigenetic mechanisms leading to elevated expression of non-mutated self-antigens could confer high tumour antigenicity despite a low number of mutations; as has been hypothesized for RCC, which is generally responsive to immune checkpoint blockade but typically contains only a modest number of mutations11,12,55. Finally, characteristics of the local TME that influence immune effector function are as important as the antigenicity of the tumour and the repertoire of T cells with tumour reactivity. It is well documented that the TME is highly variable between different cancers even within the same histology; in particular, there are many local immune inhibitory mechanisms ranging from cytokines (for example, interleukin-10 (IL-10) and transforming growth factor-β (TGFβ)) to immune checkpoint ligands to metabolic enzymes (for example, indoleamine 2,3-dioxygenase 1 (IDO1) and arginase)2,15. Thus, if the TME is highly immunosuppressive, even large numbers of tumour neoantigen-specific T cells will not function upon blockade of key immune checkpoints such as PD1 and CTLA4.
DNA MMR-deficient cancers: cancer genetics meets immunotherapy
A specific genetic subset associated with multiple different cancer types that results in log-fold increases in tumour mutational burden has recently been explored for responsiveness to anti-PD1 therapy. This subset is defined by defects in one or more of six genes that encode components of a major DNA mismatch repair complex (MMR complex). This group of genes was originally discovered to contain heterozygous mutations in a familial cancer syndrome termed Lynch syndrome (also known as hereditary non-polyposis CRC (HNPCC))90,91. These genes are also found to be mutated, deleted or epigenetically silenced in a subset of sporadic cancers of many different types besides CRC, including gastric, endometrial, ampullary, duodenal and even prostate cancer. In total, approximately 4% of adult solid malignancies in the United States have an MMR-deficient genotype. Identification of this genetic subset can be made either by detection of microsatellite instability (MSI), a hallmark of MMR-deficient cancers, or by the absence of one of the MMR proteins by IHC. Importantly, MMR-deficient cancers possess between 10- and 100-fold the mutational load of their MMR-proficient counterparts. It was thus hypothesized that because MMR deficiency confers such a vast difference in mutational load, this characteristic would be a dominant factor in antitumour immunity despite the many confounding variables described above. Evidence for this notion came from an analysis of the TME of MSI versus microsatellite stable (MSS) colon cancers. Indeed, MSI colon cancers are highly infiltrated with T cells relative to MSS colon cancers; this difference is seen particularly in respect of CD8+ T cells92. CD4+ TILs in MSI colon tumours are skewed towards a TH1 phenotype with high IFNγ production, and CD8+ T cells display an active cytotoxic lymphocyte phenotype. In keeping with the adaptive resistance concept, MSI tumours express high levels of multiple immune checkpoint molecules, including PD1, PDL1, CTLA4 and lymphocyte activation gene 3 (LAG3). In addition, they express high levels of the IFNγ-inducible immune inhibitory metabolic enzyme IDO1 (REF 56). This type of immune microenvironment is certainly suggestive of a tumour that would be responsive to PD1 pathway blockade.
The first hint that MSI status could indeed predict clinical responsiveness to anti-PD1 therapy came from the finding that, despite a generally low response rate in CRC, one durable complete response was observed in a patient with CRC treated in an early trial of anti-PD1 therapy (nivolumab); analysis of this patient’s tumour revealed an MSIhi phenotype20,59. The capacity of the MSI phenotype to predict clinical response to anti-PD1 was formally tested in a three-arm clinical trial of pembrolizumab93. Two arms comprised patients with CRC with either MSI or MSS tumours. The third arm comprised patients with any MSI cancer other than CRC. This trial demonstrated an objective response rate of approximately 60% in MSI CRC, whereas patients with MSS tumours did not respond. The responsiveness of MSI CRC to PD1 blockade was recapitulated in patients with non-CRC MSI tumours, who also demonstrated a response rate of approximately 60%93. This group included patients with chemotherapy-refractory endometrial, duodenal and ampullary cancers. These striking correlations of DNA MMR genotype with clinical response to anti-PD1 therapy, independent of tumour histology, are likely to lead to early adoption of MSI as a biomarker to identify genetically defined subsets of patients for treatment with PD1 blockers across a broad range of cancer types. This strategy is intended for application even in patients with tumour types not generally considered to be responsive to single-agent therapy with PD1 pathway inhibitors, such as prostate cancer and pancreatic cancer.
Virus-associated cancers
Beyond somatic mutations in cancer genomes, integrated oncogenic viruses represent another genetic alteration in cancers that may confer neoantigenicity and thus serve as a molecular biomarker predictive of response to checkpoint blockade. Several human cancers in both immunodeficient and immunocompetent individuals are driven by integrated viruses expressing oncogenes. These include Epstein–Barr virus (EBV), human papillomavirus (HPV), Merkel cell polyomavirus (MCPyV), human T-lymphotropic virus 1 (HTLV-1) and Kaposi sarcoma-associated herpesvirus (KSHV). Additionally, most hepatocellular carcinomas are driven by chronic infection of hepatocytes with either hepatitis B virus (HBV) or HCV, although it is not clear that these viruses encode bona fide oncogenes94. Importantly, in contrast to point mutations or rearrangements of the tumour genome, which are capable of generating a single or limited number of antigenic peptides for T cell recognition, the entire protein product of expressed genes from the virus represents non-self, thereby encoding many potential T cell epitopes. Finally, although the vast majority of somatic mutations in tumours are passengers with no functional role in tumour growth or metastasis, products of viral oncogenes such as HPV-E6 and HPV-E7 drive tumorigenesis and are thus unlikely to be silenced or deleted as a mechanism of immune evasion.
In some cancer types, virtually every patient’s tumour is virus associated; such is the case for nasopharyngeal carcinoma (EBV)95, cervical and anal cancers (HPV)96,97, adult T cell leukaemia (HTLV-1)98 and Kaposi sarcoma (KSHV)99. In other cancers, such as gastric cancer (EBV)100, Hodgkin lymphoma (EBV)101,102, MCC (MCPyV)103 and SCCHN (HPV)104, only a subset of cases are virus associated. For the latter group, the presence or absence of the causative virus could represent a predictive biomarker for response to immune checkpoint blockade. Alternatively, measures of endogenous immune responses to these viruses in patients bearing virus-positive tumours could serve as a predictive biomarker. There are already hints that virus-associated cancers respond at high rates to PD1 pathway blockade. Preliminary data show that advanced MCCs, of which approximately 80% are virus associated, have a response rate of higher than 50% to anti-PD1 therapy (pembrolizumab) (meeting abstract)105. Substantial response rates among both HBV- and HCV-associated hepatocellular carcinomas to anti-PD1 therapy have also been reported (meeting abstract)106. Clinical trials to specifically test the efficacy of PD1 pathway blockade in virus-associated cancers are under way.
Combination therapies
There is considerable interest in developing combination treatment regimens based on drugs blocking immune checkpoints, incorporating additional immune checkpoint blockers or other anticancer therapies. On the basis of preclinical evidence, effective treatment partners for anti-PD1–PDL1 and/or anti-CTLA4 therapy might include radiation therapy, certain chemotherapies, kinase inhibitors or epigenetic drugs, which in themselves have been shown to have immunogenic properties4,107–109. There is also the possibility of combining drugs that target two or more immune checkpoints, administered either concurrently or sequentially, given the array of potential targets and their different MOA110. In melanoma, enhanced response rates to combination anti-CTLA4 and anti-PD1 therapy have been reported, compared with either monotherapy, albeit with a significant rate of high-grade side effects7,111,112. In patients with PDL1− tumours, progression-free survival was greater for patients receiving ipilimumab and nivolumab combination therapy compared with those receiving nivolumab monotherapy; however, in patients with PDL1+ tumours, these treatment options seemed to have equivalent efficacy7. Although statistical analyses have not yet been performed, nor overall survival comparisons been reported, these findings suggest that the PDL1 IHC test might be useful for selecting patients for nivolumab monotherapy, thus avoiding the considerably higher potential for serious toxicity from the nivolumab plus ipilimumab combination. In October 2015, treatment with ipilimumab plus nivolumab was approved by the FDA for patients with advanced BRAF wild-type melanoma, establishing a landmark for the first regulatory approval of an immunotherapy drug combination.
Within the next 5–10 years, the hundreds of ongoing immunotherapy combination trials will undoubtedly produce novel combinations with enhanced clinical activity, thus necessitating further biomarker exploration. Expression of the targeted molecules or tumour antigens (in the case of cancer vaccines) by immune and/or tumour cells provides the most obvious biomarker candidates. Ultimately, validation of any candidate biomarker will rest on correlation with clinical outcome (for example, tumour regression, progression-free survival or overall survival). Thus, all combination trials — even early exploratory ones — should be biomarker rich in their assessment in order to derive the most information as efficiently as possible.
Conclusions and future biomarker considerations
Recent clinical advances with drugs that block immune checkpoints have brought immunotherapy out of the realm of highly specialized therapy and into the mainstream of oncology. To date, anti-CTLA4 therapy has shown reproducible activity only in patients with advanced melanoma. In contrast, anti-PD1–PDL1 drugs blocking a related but distinct immune checkpoint seem to have a broad range of activity extending beyond melanoma to an expanding list of cancers, including those of the lung, kidney, bladder, head and neck, breast, liver and gastrointestinal tract, and certain lymphoma subtypes. However, other cancer types such as prostate cancer and CRC have proved much more resistant to anti-PD1 therapies, underscoring the need for biomarker development. The evolution of our basic mechanistic understanding of immune checkpoint pathways has facilitated the search for pretreatment and on-treatment biomarkers that predict clinical response in different tumour types and the individual patients associated with them. The functional attributes of CTLA4, PD1 and other immune checkpoints will guide future biomarker discovery strategies uniquely suited to these non-redundant regulatory pathways. The predominant impact of the PD1 pathway in peripheral tissues suggests that the tumour site itself contains the most important clues for the development of anti-PD1 therapy biomarkers; in fact, this has been a rich source of promising biomarker leads pertaining to the expression of immunoregulatory molecules, oncogenic driver mutations, mutational burden and cancer-associated viruses. In contrast, the global inhibitory effects of CTLA4 in the immune system are consistent with systemic pharmacodynamic alterations found in circulating lymphocytes.
Whereas CTLA4 and PD1 blockade have reached the stage of regulatory approval, the list of additional checkpoint receptors and ligands being targeted clinically is growing. Examples include LAG3, T-cell immunoglobulin mucin receptor 3 (TIM3; also known as HAVCR2), B7H3 (also known as CD276), CD39, CD73 and the adenosine A2a receptor113–117. Inhibitory metabolic enzymes, such as IDO1, are also being targeted by small molecules118. Most of these immune checkpoints are being targeted in conjunction with PD1 pathway blocking antibodies. As mentioned above, some of these checkpoints are co-expressed with PDL1, providing a rationale for this dual blockade therapy. However, because clinical trials are in their early stages, there are as yet no validated biomarkers to predict which patients will benefit most from dual blockade of these molecules. Although several trials involve analysis of expression of the target in tumour biopsies, it is not yet clear which of these operate predominantly within the TME compared with outside the TME, that is, during T cell activation within lymph nodes. Nonetheless, intensive pharmacodynamic and correlative immune studies will be needed as part of all early-stage clinical trials — this is the only way that biomarker candidates can be identified as combinatorial checkpoint blockade approaches proliferate.
As more is learned about the TME and about regulation of systemic immunity to cancer, it is likely that additional biomarkers will emerge. For example, recent studies on metabolism demonstrate that tumours and T cells compete for glucose and that this competition can affect the glycolysis-dependent function of TILs119, raising the notion that metabolic biomarkers may be crucial factors in antitumour responsiveness. Another potential biomarker is the gut microbiome. Two recent studies speak to this possibility. One study in mice suggested that microbiomes high in Bifidobacter species enhanced tumour response to anti-PDL1 therapy120. A study in humans suggested that Bacteroides species (Bacteroides fragilis and Bacteroides thetaiotaomicron) in the microbiome might enhance the response of patients with melanoma to anti-CTLA4 therapy121. Even though the mechanisms by which these species may enhance systemic antitumour immunity are unknown at this time, these data suggest that microbiome-derived biomarkers may be developed to guide immunotherapy.
Unlike static biomarkers defined by driver mutations directly linked to the activity of targeted kinase inhibitors, the ever-changing immune system poses unique challenges to biomarker development. However, it is the adaptability of the immune system that also underlies its vast potential to keep pace with cancer evolution and provide durable treatment responses that are not achievable with most other forms of cancer therapy.
Acknowledgments
The authors are grateful to M. Hellmann (Memorial Sloan-Kettering Cancer Center, New York, USA), E. Garon (University of California Los Angeles, USA) and J. Brahmer (Johns Hopkins University, Baltimore, Maryland, USA) for helpful discussions. This work was supported by research funding from Bristol-Myers Squibb (S.L.T., J.M.T., R.A.A. and D.M.P.), the Melanoma Research Alliance (S.L.T., J.M.T. and D.M.P.), the US National Cancer Institute NIH (R01 CA142779; S.L.T., J.M.T. and D.M.P.), the Barney Family Foundation (S.L.T. and J.M.T.), the Dermatology Foundation (J.M.T.), the Laverna Hahn Charitable Trust (S.L.T.), the Commonwealth Foundation (D.M.P.), the W.W. Smith Charitable Trust (J.M.T.) and Moving for Melanoma Delaware (S.L.T., J.M.T. and D.M.P.). All authors were also supported by a Stand Up To Cancer—Cancer Research Institute Cancer Immunology Translational Research Grant (SU2C-AACR-DT1012). Stand Up To Cancer is a programme of the Entertainment Industry Foundation administered by the American Association for Cancer Research.
Glossary
- Tolerance
An immunological phenomenon in which antigen-specific T and/or B cells are absent or unresponsive to antigen-bearing cells, as opposed to rejection, in which antigen-specific immune cells eliminate their targets.
- Mixed tumour regression
A therapeutic response pattern in which different metastatic lesions in an individual patient show different responses to therapy, some regressing whereas others progress.
- Regulatory T cells
(Treg cells). A subset of CD4+ T cells characterized by expression of the forkhead box transcription factor FOXP3, which interacts with other immune cells to inhibit immune responses.
- Adaptive immune system
Comprises T and B lymphocytes with unique antigen receptors generated by somatic DNA recombination events, as compared with the innate immune system, which comprises cells with invariant antigen receptors (for example, natural killer cells and macrophages).
- Tertiary lymphoid structure
Ectopic lymphoid tissue that recapitulates some of the structural organization of a lymph node (including T cells, B cells, antigen presenting cells and high endothelial venules) and can support the generation of an adaptive immune response.
- Adaptive immune resistance
A phenomenon in which tumour and stromal cells adapt to attack from infiltrating T cells by expressing the immune inhibitory ligand programmed cell death 1 ligand 1 (PDL1). In this scenario, PDL1 expression is driven by inflammatory cytokines such as interferon-γ secreted by tumour antigen-specific T cells.
- Objective response rate
The rate of significant tumour regressions in patients undergoing cancer therapy, including complete and partial regressions as defined by standard oncological criteria (for example, Response Evaluation Criteria In Solid Tumours (RECIST), or World Health Organization (WHO) criteria).
- Neoantigens
Newly expressed tumour antigens that arise from genetic alterations in tumour cells and are therefore not present in normal cells, such as antigens generated by somatic (non-heritable) mutations or oncoviruses.
- Self-antigen
In the context of tumour antigens, a non-mutated component of tumour cells that is also expressed by some normal cells and can be recognized by the immune system.
- Anergy
A functional state of T cells in which they are hyporesponsive to T cell receptor engagement by cognate antigen, relative to naive or memory T cells.
- DNA mismatch repair complex
(MMR complex). A complex of enzymes that recognizes DNA base mismatches introduced during DNA replication, excises them and replaces them with correctly matched bases.
- Microsatellite instability
(MSI). A hallmark of defects in DNA mismatch repair, characterized by alterations in the frequency of repeated DNA sequences (microsatellites).
- TH1 phenotype
Differentiation state of CD4+ T cells characterized by production of interferon-γ (in addition to other cytokines) upon encountering cognate antigen.
Footnotes
Competing interests statement
The authors declare competing interests: see Web version for details.
FURTHER INFORMATION
Blueprint Working Group: http://www.aacr.org/AdvocacyPolicy/GovernmentAffairs/Pages/industry-working-group-blueprint-proposal.aspx
FDA guidance on biomarker use in management of cancer patients: http://www.fda.gov/downloads/MedicalDevices/DeviceRegulationandGuidance/GuidanceDocuments/UCM262327.pdf
NCI guidelines for tissue handling: http://biospecimens.cancer.gov/bestpractices
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
References
- 1.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12:252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pardoll D. Cancer and the immune system: basic concepts and targets for intervention. Semin Oncol. 2015;42:523–538. doi: 10.1053/j.seminoncol.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Topalian SL, et al. Survival, durable tumor remission, and long-term safety in patients with advanced melanoma receiving nivolumab. J Clin Oncol. 2014;32:1020–1030. doi: 10.1200/JCO.2013.53.0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hamid O, et al. Safety and tumor responses with lambrolizumab (anti-PD-1) in melanoma. N Engl J Med. 2013;369:134–144. doi: 10.1056/NEJMoa1305133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robert C, et al. Pembrolizumab versus ipilimumab in advanced melanoma. N Engl J Med. 2015;372:2521–2532. doi: 10.1056/NEJMoa1503093. [DOI] [PubMed] [Google Scholar]
- 6.Robert C, et al. Nivolumab in previously untreated melanoma without BRAF mutation. N Engl J Med. 2015;372:320–330. doi: 10.1056/NEJMoa1412082. [DOI] [PubMed] [Google Scholar]
- 7.Larkin J, et al. Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N Engl J Med. 2015;373:23–34. doi: 10.1056/NEJMoa1504030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garon E, et al. Pembrolizumab for the treatment of non–small-cell lung cancer. N Engl J Med. 2015;372:2018–2028. doi: 10.1056/NEJMoa1501824. [DOI] [PubMed] [Google Scholar]
- 9.Brahmer JR, et al. Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. N Engl J Med. 2015;373:123–135. doi: 10.1056/NEJMoa1504627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borghaei H, et al. Nivolumab versus docetaxel in advanced nonsquamous non-small-cell lung cancer. N Engl J Med. 2015;373:1627–1639. doi: 10.1056/NEJMoa1507643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Motzer RJ, et al. Nivolumab for metastatic renal cell carcinoma: results of a randomized phase II trial. J Clin Oncol. 2015;33:1430–1437. doi: 10.1200/JCO.2014.59.0703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Motzer RJ, et al. Nivolumab versus everolimus in advanced renal-cell carcinoma. N Engl J Med. 2015;373:1803–1813. doi: 10.1056/NEJMoa1510665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lipson EJ, et al. Antagonists of PD-1 and PD-L1 in cancer treatment. Semin Oncol. 2015;42:587–600. doi: 10.1053/j.seminoncol.2015.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Topalian SL, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366:2443–2454. doi: 10.1056/NEJMoa1200690. The report of this large early-phase clinical study demonstrates a role for anti-PD1 therapy in NSCLC, melanoma and kidney cancer, but not in prostate cancer or CRC; it also provides the first demonstration of PDL1 IHC as a potential biomarker for anti-PD1 therapy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Topalian SL, Drake CG, Pardoll DM. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell. 2015;27:450–461. doi: 10.1016/j.ccell.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hodi FS, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363:711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schadendorf D, et al. Pooled analysis of long-term survival data from phase II and phase III trials of ipilimumab in unresectable or metastatic melanoma. J Clin Oncol. 2015;33:1889–1894. doi: 10.1200/JCO.2014.56.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barber DL, et al. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature. 2006;439:682–687. doi: 10.1038/nature04444. [DOI] [PubMed] [Google Scholar]
- 19.Tumeh PC, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515:568–571. doi: 10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brahmer JR, et al. Phase I study of single-agent anti–programmed death-1 (MDX-1106) in refractory solid tumors: safety, clinical activity, pharmacodynamics, and immunologic correlates. J Clin Oncol. 2010;28:3167–3175. doi: 10.1200/JCO.2009.26.7609. This first-in-human study of MDX-1106 (nivolumab) reports early evidence for clinical activity in multiple cancer types, pharmacodynamics and PDL1 IHC as a predictor of clinical outcomes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robert L, et al. Distinct immunological mechanisms of CTLA-4 and PD-1 blockade revealed by analyzing TCR usage in blood lymphocytes. Oncoimmunology. 2014;3:e29244. doi: 10.4161/onci.29244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maker AV, Attia P, Rosenberg SA. Analysis of the cellular mechanism of antitumor responses and autoimmunity in patients treated with CTLA-4 blockade. J Immunol. 2005;175:7746–7754. doi: 10.4049/jimmunol.175.11.7746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ku GY, et al. Single-institution experience with ipilimumab in advanced melanoma patients in the compassionate use setting: lymphocyte count after 2 doses correlates with survival. Cancer. 2010;116:1767–1775. doi: 10.1002/cncr.24951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang A, et al. CTLA-4 blockade with ipilimumab increases peripheral CD8+ T cells: correlation with clinical outcomes. J Clin Oncol. 2010;28(15 suppl):2555. [Google Scholar]
- 25.Yuan J, et al. CTLA-4 blockade enhances polyfunctional NY-ESO-1 specific T cell responses in metastatic melanoma patients with clinical benefit. Proc Natl Acad Sci USA. 2008;105:20410–20415. doi: 10.1073/pnas.0810114105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yuan J, et al. Integrated NY-ESO-1 antibody and CD8+ T-cell responses correlate with clinical benefit in advanced melanoma patients treated with ipilimumab. Proc Natl Acad Sci USA. 2011;108:16723–16728. doi: 10.1073/pnas.1110814108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hannani D, et al. Anticancer immunotherapy by CTLA-4 blockade: obligatory contribution of IL-2 receptors and negative prognostic impact of soluble CD25. Cell Res. 2015;25:208–224. doi: 10.1038/cr.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ji RR, et al. An immune-active tumor microenvironment favors clinical response to ipilimumab. Cancer Immunol Immunother. 2012;61:1019–1031. doi: 10.1007/s00262-011-1172-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen H, et al. Anti-CTLA-4 therapy results in higher CD4+ ICOShi T cell frequency and IFN-gamma levels in both nonmalignant and malignant prostate tissues. Proc Natl Acad Sci USA. 2009;106:2729–2734. doi: 10.1073/pnas.0813175106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vonderheide RH, et al. Tremelimumab in combination with exemestane in patients with advanced breast cancer and treatment associated modulation of inducible costimulator expression on patient T cells. Clin Cancer Res. 2010;16:3485–3494. doi: 10.1158/1078-0432.CCR-10-0505. [DOI] [PubMed] [Google Scholar]
- 31.Hodi FS, et al. Immunologic and clinical effects of antibody blockade of cytotoxic T lymphocyte-associated antigen 4 in previously vaccinated cancer patients. Proc Natl Acad Sci USA. 2008;105:3005–3010. doi: 10.1073/pnas.0712237105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liakou CI, et al. CTLA-4 blockade increases IFNγ-producing CD4+ICOShi cells to shift the ratio of effector to regulatory T cells in cancer patients. Proc Natl Acad Sci USA. 2008;105:14987–14992. doi: 10.1073/pnas.0806075105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Virchow R. Die Krankhaften Geschwülste. Berlin: 1863. [Google Scholar]
- 34.Balkwill F, Mantovani A. Inflammation and cancer: back to Virchow? Lancet. 2001;357:539–545. doi: 10.1016/S0140-6736(00)04046-0. [DOI] [PubMed] [Google Scholar]
- 35.Galon J, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–1964. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 36.Fridman WH, Pagès F, Sautès-Fridman C, Galon J. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer. 2012;12:298–306. doi: 10.1038/nrc3245. [DOI] [PubMed] [Google Scholar]
- 37.Dieu-Nosjean MC, et al. Long term survival for patients with non-small-cell lung cancer with intratumoral lymphoid structures. J Clin Oncol. 2008;26:4410–4417. doi: 10.1200/JCO.2007.15.0284. [DOI] [PubMed] [Google Scholar]
- 38.Di Caro G, et al. Occurrence of tertiary lymphoid tissue is associated to T-cell infiltration and predicts better prognosis in early-stage colorectal cancers. Clin Cancer Res. 2014;20:2147–2158. doi: 10.1158/1078-0432.CCR-13-2590. [DOI] [PubMed] [Google Scholar]
- 39.Taube JM. Emerging immunologic biomarkers: setting the (TNM-immune) stage. Clin Cancer Res. 2014;20:2023–2025. doi: 10.1158/1078-0432.CCR-14-0328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dong H, Zhu G, Tamada K, Chen L. B7-H1, a third member of the B7 family, co-stimulates T-cell proliferation and interleukin-10 secretion. Nat Med. 1999;5:1365–1369. doi: 10.1038/70932. [DOI] [PubMed] [Google Scholar]
- 41.Dong H, et al. Tumor-associated B7-H1 promotes T-cell apoptosis: a potential mechanism of immune evasion. Nat Med. 2002;8:793–800. doi: 10.1038/nm730. [DOI] [PubMed] [Google Scholar]
- 42.Mazanet MM, Hughes CC. B7-H1 is expressed by human endothelial cells and suppresses T cell cytokine synthesis. J Immunol. 2002;169:3581–3588. doi: 10.4049/jimmunol.169.7.3581. [DOI] [PubMed] [Google Scholar]
- 43.Parsa AT, et al. Loss of tumor suppressor PTEN function increases B7-H1 expression and immunoresistance in glioma. Nat Med. 2007;13:84–88. doi: 10.1038/nm1517. [DOI] [PubMed] [Google Scholar]
- 44.Green MR, et al. Integrative analysis reveals selective 9p24.1 amplification, increased PD-1 ligand expression, and further induction via JAK2 in nodular sclerosing Hodgkin lymphoma and primary mediastinal large B-cell lymphoma. Blood. 2010;116:3268–3277. doi: 10.1182/blood-2010-05-282780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ansell SM, et al. PD-1 blockade with nivolumab in relapsed or refractory Hodgkin’s lymphoma. N Engl J Med. 2015;372:311–319. doi: 10.1056/NEJMoa1411087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taube JM, et al. Colocalization of inflammatory response with B7-H1 expression in human melanocytic lesions supports an adaptive resistance mechanism of immune escape. Sci Transl Med. 2012;4:127ra37. doi: 10.1126/scitranslmed.3003689. This report provides the first evidence for, and articulation of, the adaptive immune resistance hypothesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taube JM, et al. Differential expression of immune-regulatory genes associated with PD-L1 display in melanoma: implications for PD-1 pathway blockade. Clin Cancer Res. 2015;21:3969–3976. doi: 10.1158/1078-0432.CCR-15-0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lipson EJ, et al. PD-L1 expression in the Merkel cell carcinoma microenvironment: association with inflammation, Merkel cell polyomavirus and overall survival. Cancer Immunol Res. 2013;1:54–63. doi: 10.1158/2326-6066.CIR-13-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Velcheti V, et al. Programmed death ligand-1 expression in non-small cell lung cancer. Lab Invest. 2014;94:107–116. doi: 10.1038/labinvest.2013.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schalper KA, et al. In situ tumor PD-L1 mRNA expression is associated with increased TILs and better outcome in breast carcinomas. Clin Cancer Res. 2014;20:2773–2782. doi: 10.1158/1078-0432.CCR-13-2702. [DOI] [PubMed] [Google Scholar]
- 51.Cimino-Mathews A, et al. PD-L1 (B7-H1) expression and the immune tumor microenvironment in primary and metastatic breast carcinomas. Hum Pathol. 2016;47:52–63. doi: 10.1016/j.humpath.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lyford-Pike S, et al. Evidence for a role of the PD-1:PD-L1 pathway in immune resistance of HPV-associated head and neck squamous cell carcinoma. Cancer Res. 2013;73:1733–1741. doi: 10.1158/0008-5472.CAN-12-2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Skoulidis F, et al. Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities. Cancer Discov. 2015;5:860–877. doi: 10.1158/2159-8290.CD-14-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rodic N, et al. PD-L1 expression in melanocytic lesions does not correlate with the BRAF V600E mutation. Cancer Immunol Res. 2015;3:110–115. doi: 10.1158/2326-6066.CIR-14-0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lawrence MS, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Llosa NJ, et al. The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov. 2015;5:43–51. doi: 10.1158/2159-8290.CD-14-0863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thompson RH, et al. Tumor B7-H1 is associated with poor prognosis in renal cell carcinoma patients with long-term follow-up. Cancer Res. 2006;66:3381–3385. doi: 10.1158/0008-5472.CAN-05-4303. [DOI] [PubMed] [Google Scholar]
- 58.Taube JM, et al. Association of PD-1, PD-1 ligands, and other features of the tumor immune microenvironment with response to anti-PD-1 therapy. Clin Cancer Res. 2014;20:5064–5074. doi: 10.1158/1078-0432.CCR-13-3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lipson EJ, et al. Durable cancer regression off-treatment and effective reinduction therapy with an anti-PD-1 antibody. Clin Cancer Res. 2013;19:462–468. doi: 10.1158/1078-0432.CCR-12-2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thompson ED, et al. Patterns of PD-L1 expression and CD8 T cell infiltration in gastric adenocarcinomas and associated immune stroma. Gut. 2016 doi: 10.1136/gutjnl-2015-310839. http://dx.doi.org/10.1136/gutjnl-2015-310839. [DOI] [PMC free article] [PubMed]
- 61.Sunshine J, Taube JM. PD-1/PD-L1 inhibitors. Curr Opin Pharmacol. 2015;23:32–38. doi: 10.1016/j.coph.2015.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Herbst RS, et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515:563–567. doi: 10.1038/nature14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McDermott DF, et al. Atezolizumab, and anti-programmed death-ligand 1 antibody, in metastatic renal cell carcinoma: long-term safety, clinical activity and immune correlates from a phase Ia study. J Clin Oncol. 2016;34:833–842. doi: 10.1200/JCO.2015.63.7421. [DOI] [PubMed] [Google Scholar]
- 64.Kitazono S, et al. Reliability of small biopsy samples compared with resected specimens for the determination of programmed death-ligand 1 expression in non-small-cell lung cancer. Clin Lung Cancer. 2015;16:385–390. doi: 10.1016/j.cllc.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 65.Dabbs DJ. In: Diagnostic immunohistochemistry. Dabbs DJ, editor. Churchill Livingstone; 2002. pp. 29–39. [Google Scholar]
- 66.Von Mehren M, et al. The influence of granulocyte macrophage colony-stimulating factor and prior chemotherapy on the immunological response to a vaccine (ALVAC-CEA B7.1) in patients with metastatic carcinoma. Clin Cancer Res. 2001;7:1181–1189. [PubMed] [Google Scholar]
- 67.Emens LA, Middleton G. The interplay of immunotherapy and chemotherapy: harnessing potential synergies. Cancer Immunol Res. 2015;3:436–443. doi: 10.1158/2326-6066.CIR-15-0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Garraway LA. Genomics-driven oncology: framework for an emerging paradigm. J Clin Oncol. 2013;31:1806–1814. doi: 10.1200/JCO.2012.46.8934. [DOI] [PubMed] [Google Scholar]
- 69.Sumimoto H, Imabayashi F, Iwata T, Kawakami Y. The BRAF-MAPK signaling pathway is essential for cancer-immune evasion in human melanoma cells. J Exp Med. 2006;203:1651–1656. doi: 10.1084/jem.20051848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Marzec M, et al. Oncogenic kinase NPM/ALK induces through STAT3 expression of immunosuppressive protein CD274 (PD-L1, B7-H1) Proc Natl Acad Sci USA. 2008;105:20852–20857. doi: 10.1073/pnas.0810958105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Akbay EA, et al. Activation of the PD-1 pathway contributes to immune escape in EGFR-driven lung tumors. Cancer Discov. 2013;3:1355–1363. doi: 10.1158/2159-8290.CD-13-0310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xu C, et al. Loss of Lkb1 and Pten leads to lung squamous cell carcinoma with elevated PD-L1 expression. Cancer Cell. 2014;25:590–604. doi: 10.1016/j.ccr.2014.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Larkin J, et al. Efficacy and safety of nivolumab in patients with BRAF V600 mutant and BRAF wild-type advanced melanoma: a pooled analysis of 4 clinical trials. JAMA Oncol. 2015;1:433–440. doi: 10.1001/jamaoncol.2015.1184. [DOI] [PubMed] [Google Scholar]
- 74.Weber JS, et al. Nivolumab versus chemotherapy in patients with advanced melanoma who progressed after anti-CTLA-4 treatment (CheckMate 037): a randomized, controlled, open-label phase 3 trial. Lancet Oncol. 2015;16:375–384. doi: 10.1016/S1470-2045(15)70076-8. [DOI] [PubMed] [Google Scholar]
- 75.Hellmann MD, et al. Efficacy of pembrolizumab in key subgroups of patients with advanced NSCLC. J Thor Oncol. 2015;10(suppl 2) abstract MINI03.05. [Google Scholar]
- 76.Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science. 2015;348:69–74. doi: 10.1126/science.aaa4971. [DOI] [PubMed] [Google Scholar]
- 77.Shastri N, Cardinaud S, Schwab ST, Serwold T, Kunisawa J. All the peptides that fit: the beginning, the middle, and the end of the MHC class I antigen-processing pathway. Immunol Rev. 2005;207:31–41. doi: 10.1111/j.0105-2896.2005.00321.x. [DOI] [PubMed] [Google Scholar]
- 78.Roche PA, Furata K. The ins and outs of MHC class II-mediated antigen processing and presentation. Nat Rev Immunol. 2015;15:203–216. doi: 10.1038/nri3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Brahmer JR, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med. 2012;366:2455–2465. doi: 10.1056/NEJMoa1200694. This article reports the first demonstration of clinical activity of a drug to block PDL1; a companion article in the same journal issue (ref. 14) describes a different drug that blocks PD1; together these papers highlight the PD1–PDL1 pathway as a vital target in cancer immunotherapy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Snyder A, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371:2189–2199. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Van Allen EM, et al. Genomic correlates of response to CTLA4 blockade in metastatic melanoma. Science. 2015;350:207–211. doi: 10.1126/science.aad0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rizvi NA, et al. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Twyman-Saint Victor C, et al. Radiation and dual checkpoint blockade activate non-redundant immune mechanisms in cancer. Nature. 2015;520:373–377. doi: 10.1038/nature14292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gubin MM, et al. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature. 2014;515:577–581. doi: 10.1038/nature13988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yadav M, et al. Predicting immunogenic tumour mutations by combining mass spectrometry and exome sequencing. Nature. 2014;515:572–576. doi: 10.1038/nature14001. [DOI] [PubMed] [Google Scholar]
- 86.Tran E, et al. Immunogenicity of somatic mutations in human gastrointestinal cancers. Science. 2015;350:1387–1390. doi: 10.1126/science.aad1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kvistborg P, et al. Anti-CTLA-4 therapy broadens the melanoma-reactive CD8+ T cell response. Sci Transl Med. 2014;6:254ra128. doi: 10.1126/scitranslmed.3008918. [DOI] [PubMed] [Google Scholar]
- 88.Tran E, et al. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science. 2014;344:641–645. doi: 10.1126/science.1251102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Coulie PG, Van den Eynde BJ, van der Bruggen P, Boon T. Tumour antigens recognized by T lymphocytes: at the core of cancer immunotherapy. Nat Rev Cancer. 2014;14:135–146. doi: 10.1038/nrc3670. [DOI] [PubMed] [Google Scholar]
- 90.Liu B, et al. Analysis of mismatch repair genes in hereditary non-polyposis colorectal cancer patients. Nat Med. 1996;2:169–174. doi: 10.1038/nm0296-169. [DOI] [PubMed] [Google Scholar]
- 91.Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 92.Drescher KM, et al. Lymphocyte recruitment into the tumor site is altered in patients with MSI-H colon cancer. Fam Cancer. 2009;8:231–239. doi: 10.1007/s10689-009-9233-0. [DOI] [PubMed] [Google Scholar]
- 93.Le DT, et al. PD-1 Blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372:2509–2520. doi: 10.1056/NEJMoa1500596. This is the first report linking a genetic marker in cancer (MSI) with clinical outcomes following anti-PD1 therapy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sukowati CH, et al. Significance of hepatitis virus infection in the oncogenic initiation of hepatocellular carcinoma. World J Gastroenterol. 2016;22:1497–1512. doi: 10.3748/wjg.v22.i4.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pearson GR. Epstein-Barr virus and nasopharyngeal carcinoma. J Cell Biochem Suppl. 1993;17:150–154. doi: 10.1002/jcb.240531021. [DOI] [PubMed] [Google Scholar]
- 96.Stanley MA. Human papillomavirus vaccines. Rev Med Virol. 2006;16:139–149. doi: 10.1002/rmv.498. [DOI] [PubMed] [Google Scholar]
- 97.Hoots BE, Palefsky JM, Pimenta JM, Smith JS. Human papillomavirus type distribution in anal cancer and anal intraepithelial lesions. Int J Cancer. 2009;124:2375–2383. doi: 10.1002/ijc.24215. [DOI] [PubMed] [Google Scholar]
- 98.Matsuoka M. Human T-cell leukemia virus type I (HTLV-I) infection and the onset of adult T-cell leukemia (ATL) Retrovirology. 2005;2:27. doi: 10.1186/1742-4690-2-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wen KW, Damania B. Kaposi sarcoma-associated herpesvirus (KSHV): molecular biology and oncogenesis. Cancer Lett. 2010;289:140–150. doi: 10.1016/j.canlet.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chen XZ, Chen H, Castro FA, Hu JK, Brenner H. Epstein–Barr virus infection and gastric cancer: a systematic review. Medicine. 2015;94:e792. doi: 10.1097/MD.0000000000000792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.List AF, Greco FA, Vogler LB. Lymphoproliferative diseases in immunocompromised hosts: the role of Epstein–Barr virus. J Clin Oncol. 1987;5:1673–1689. doi: 10.1200/JCO.1987.5.10.1673. [DOI] [PubMed] [Google Scholar]
- 102.Kapatai G, Murray P. Contribution of the Epstein–Barr virus to the molecular pathogenesis of Hodgkin lymphoma. J Clin Pathol. 2007;60:1342–1349. doi: 10.1136/jcp.2007.050146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Samimi M, et al. Merkel cell polyomavirus in Merkel cell carcinoma: clinical and therapeutic perspectives. Semin Oncol. 2015;42:347–358. doi: 10.1053/j.seminoncol.2014.12.021. [DOI] [PubMed] [Google Scholar]
- 104.Gillison ML, et al. Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J Natl Cancer Inst. 2000;92:709–720. doi: 10.1093/jnci/92.9.709. [DOI] [PubMed] [Google Scholar]
- 105.Nghiem P, et al. Activity of PD-1 blockade with pembrolizumab as first systemic therapy in patients with advanced Merkel cell carcinoma. Ann Oncol. 2015;27(suppl) abstract 22LBA, in the press. [Google Scholar]
- 106.El-Khoueiry AB, et al. Phase I/II safety and antitumor activity of nivolumab in patients with advanced hepatocellular carcinoma (HCC): CA209-040. J Clin Oncol. 2015;33(15 suppl) abstract LBA101. [Google Scholar]
- 107.Galluzzi L, Buque A, Kepp O, Zitvogel L, Kroemer G. Immunological effects of conventional chemotherapy and targeted anticancer agents. Cancer Cell. 2015;28:690–714. doi: 10.1016/j.ccell.2015.10.012. [DOI] [PubMed] [Google Scholar]
- 108.Frederick DT, et al. BRAF inhibition is associated with enhanced melanoma antigen expression and a more favorable tumor microenvironment in patients with metastatic melanoma. Clin Cancer Res. 2013;19:1225–1231. doi: 10.1158/1078-0432.CCR-12-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chiappinelli KB, et al. Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell. 2015;162:974–986. doi: 10.1016/j.cell.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Melero I, et al. Evolving synergistic combinations of targeted immunotherapies to combat cancer. Nat Rev Cancer. 2015;15:457–472. doi: 10.1038/nrc3973. [DOI] [PubMed] [Google Scholar]
- 111.Wolchok JD, et al. Nivolumab plus ipilimumab in advanced melanoma. N Engl J Med. 2013;369:122–133. doi: 10.1056/NEJMoa1302369. This initial report demonstrates the activity and side effect profile of anti-PD1 plus anti-CTLA4 therapy, the first immunotherapy combination to eventually receive FDA approval. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Postow MA, et al. Nivolumab and ipilimumab versus ipilimumab in untreated melanoma. N Engl J Med. 2015;372:2006–2017. doi: 10.1056/NEJMoa1414428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Goldberg MV, Drake CG. LAG-3 in cancer immunotherapy. Curr Top Microbiol Immunol. 2011;344:269–278. doi: 10.1007/82_2010_114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ngiow SF, et al. Anti-TIM3 antibody promotes T cell IFN-gamma-mediated antitumor immunity and suppresses established tumors. Cancer Res. 2011;71:3540–3551. doi: 10.1158/0008-5472.CAN-11-0096. [DOI] [PubMed] [Google Scholar]
- 115.Yi KH, Chen L. Fine tuning the immune response through B7-H3 and B7-H4. Immunol Rev. 2009;229:145–151. doi: 10.1111/j.1600-065X.2009.00768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Deaglio S, et al. Adenosine generation catalyzed by CD39 and CD73 expressed on regulatory T cells mediates immune suppression. J Exp Med. 2007;204:1257–1265. doi: 10.1084/jem.20062512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zarek PE, et al. A2A receptor signaling promotes peripheral tolerance by inducing T-cell anergy and the generation of adaptive regulatory T cells. Blood. 2008;111:251–259. doi: 10.1182/blood-2007-03-081646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Uyttenhove C, et al. Evidence for a tumoral immune resistance mechanism based on tryptophan degradation by indoleamine 2,3-dioxygenase. Nat Med. 2003;9:1269–1274. doi: 10.1038/nm934. [DOI] [PubMed] [Google Scholar]
- 119.Ho PC, et al. Phosphoenolpyruvate is a metabolic checkpoint of anti-tumor T cell responses. Cell. 2015;162:1217–1228. doi: 10.1016/j.cell.2015.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sivan A, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350:1084–1089. doi: 10.1126/science.aac4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Vetizou M, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015;350:1079–1084. doi: 10.1126/science.aad1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Nishimura H, et al. Development of lupus-like autoimmune diseases by disruption of the PD-1 gene encoding an ITIM motif-carrying immunoreceptor. Immunity. 1999;11:141–151. doi: 10.1016/s1074-7613(00)80089-8. [DOI] [PubMed] [Google Scholar]
- 123.Nishimura H, et al. Autoimmune dilated cardiomyopathy in PD-1 receptor-deficient mice. Science. 2001;291:319–322. doi: 10.1126/science.291.5502.319. [DOI] [PubMed] [Google Scholar]
- 124.Tivol EA, et al. Loss of CTLA-4 leads to massive lymphoproliferation and fatal multiorgan tissue destruction, revealing a critical negative regulatory role of CTLA-4. Immunity. 1995;3:541–547. doi: 10.1016/1074-7613(95)90125-6. [DOI] [PubMed] [Google Scholar]
- 125.Waterhouse P, et al. Lymphoproliferative disorders with early lethality in mice deficient in Ctla-4. Science. 1995;270:985–988. doi: 10.1126/science.270.5238.985. [DOI] [PubMed] [Google Scholar]