Abstract
Reading disabilities (RDs) have been associated with chromosome 6p with recent studies pointing to two genes, DCDC2 and KIAA0319. In this study, markers across the 6p region were tested for association with RD. Our strongest findings were for association with markers in KIAA0319, although with the opposite alleles compared with a previous study. We also found association with markers in VMP, but not with DCDC2. Current evidence indicates that differential regulation of KIAA0319 and DCDC2 contributes to RD, thus we used chromatin immunoprecipitation coupled with genomic tiling arrays (ChIP-chip) to map acetylated histones, a molecular marker for regulatory elements, across a 500 kb genomic region covering the RD locus on 6p. This approach identified several regions marked by acetylated histones that mapped near associated markers, including intron 7 of DCDC2 and the 5′ region of KIAA0319. The latter is located within the 70 kb region previously associated with differential expression of KIAA0319. Interestingly, five markers associated with RD in independent studies were also located within the 2.7 kb acetylated region, and six additional associated markers, including the most significant one in this study, were located within a 22 kb haplotype block that encompassed this region. Our data indicates that this putative regulatory region is a likely site of genetic variation contributing to RD in our sample, further narrowing the candidate region.
Keywords: reading disabilities, association, dyslexia, 6p, ChIP-chip
INTRODUCTION
Reading disability (RD) (OMIM 127700; 600202), also known as developmental dyslexia, is a learning disability that affects 3–6% of otherwise normally developing children, and often persists into adulthood. This prevalent disorder is characterized by deficits in word recognition, spelling and decoding abilities despite at least average intelligence and effective classroom instruction [Habib, 2000; Lyon, 2003].
Extensive evidence from twin studies indicates that RD has a substantial genetic component [Pennington, 1990; DeFries and Gillis, 1993; Grigorenko, 2001]. The reading process is composed of a number of partially overlapping cognitive and language processes (phonological awareness, decoding, word identification, orthographic awareness and coding, rapid access and retrieval of names of visual symbols) that show evidence for overlapping heritability. Multivariate analyses indicate that there are genes contributing to multiple reading component processes and some genes contributing to single processes [Olson et al., 1994; Pennington, 1997; Gayan and Olson, 1999, 2001; Olson, 2002]. Twin studies also indicate that the genetic factors that contribute to reading impairment contribute to the normal distribution of reading ability in the population [Harlaar et al., 2005].
Genetic studies have found evidence for linkage, or association, of RD or of reading component skills with chromosomes 1p34-p36 [Rabin et al., 1993; Grigorenko et al., 2001; Tzenova et al., 2004; Bates et al., 2007; Couto et al., 2008], 2p11 [Kaminen et al., 2003], 2p15-16 [Fagerheim et al., 1999; Fisher et al., 2002; Francks et al., 2002; Petryshen et al., 2002], 3p12-q13 [Nopola-Hemmi et al., 2001; Stein et al., 2004; Bates et al., 2007], 6p21.3-22 [Smith and Kimberling, 1991; Cardon et al., 1994; Grigorenko et al., 1997, 2003; Gayan et al., 1999; Kaplan et al., 2002; Turic et al., 2003; Deffenbacher et al., 2004; Francks et al., 2004; Cope et al., 2005], 6q11.2-q12 [Petryshen et al., 2001; Bates et al., 2007], 7q32 [Kaminen et al., 2003; Bates et al., 2007], 11p15.5 [Fisher et al., 2002; Hsiung et al., 2004], 15q [Smith et al., 1983, 1991; Grigorenko et al., 1997; Morris et al., 2000; Bates et al., 2007],18p11.2 [Fisher et al., 2002; Marlow et al., 2003; Bates et al., 2007], and Xq27 [Fisher et al., 2002; de Kovel et al., 2004; Bates et al., 2007]. These studies indicate the possibility of multiple distinct genes contributing to RD.
The unprecedented number of replications for a complex trait supporting linkage in the 6p region prompted association studies, from which the first identified five candidate genes, vesicular membrane protein (VMP) (NM_080723) recently renamed neurensin 1 (NRSN1), doublecortin domain containing 2 (DCDC2) (NM_016356), TRAF and TNF associated protein (TTRAP) (NM_016614), the putative gene KIAA0319 (NM_ 014809) and thioesterase superfamily 2 (THEM2) (NM_018473) [Deffenbacher et al., 2004]. The function and expression pattern of these genes are described in Londin et al. [2003]. Since then, this region on 6p has been the subject of numerous studies in which DCDC2 (three independent samples) and KIAA0319 (four independent samples) have emerged as the two strongest candidates [Francks et al., 2004; Cope et al., 2005; Meng et al., 2005; Harold et al., 2006; Schumacher et al., 2006; Luciano et al., 2007; Paracchini et al., 2008]. Three additional candidate genes, MRSL2 (NM_020662), GPLD1 (NM_001503), ALDH5A1 (NM_001080), located between DCDC2, and KIAA0319 have not shown strong association with RD.
A previous study of KIAA0319 screened for the existence of novel polymorphisms in the coding and part of the predicted promoter region of this gene [Francks et al., 2004]. Twelve DNA changes were identified in the predicted promoter region and first untranslated exon. Three of the 12 variants that were genotyped in their samples (rs2038137, del T (k_pr_del), and rs9467247) were found to be associated with RD. There was also evidence that an associated haplotype constructed from previously reported single nucleotide polymorphism (SNP) markers (rs4504469–rs2038137–rs2143340) [Francks et al., 2004], spanning 70 kb of the candidate region was associated with lowered expression of KIAA0319 in cell lines, suggesting a change in regulation of the gene as a contributor to risk [Paracchini et al., 2006]. Similarly, studies showing association of the gene DCDC2 with RD, have failed to locate a coding region change in the individuals screened that could account for the association, indirectly implicating alterations in regulatory elements [Meng et al., 2005; Schumacher et al., 2006]. Therefore, to find the putative DNA variant(s) that affect expression in individuals with RD it is important to determine where regulatory elements may lie in this large candidate region. However, regulatory elements are difficult to identify because they can be megabases from target promoters, and can even lie in introns or exons of other genes [Kleinjan and van Heyningen, 2005]. Thus, it is critical to focus on regions around and within a gene that are likely to be functionally relevant. Sequence conservation is one approach that can be used, but comparing distantly related species excludes recently evolved elements that might be essential to RD, and comparing sequences from closely related species (such as chimpanzee and human) barely reduces the amount of potentially relevant DNA [Boffelli et al., 2004]. Therefore, to screen for causal variants that confer risk to RD, the location of potential regulatory elements in this entire 6p region is required.
The current study had two aims: The first was to investigate the association of RD to markers in the genes for VMP (NRSN1), DCDC2, KIAA0319, TTRAP, and THEM2, with a specific focus on markers associated in previous studies. The second goal was to map potential regulatory elements in the ~0.5 Mb region encompassing VMP (NRSN1), DCDC2, TTRAP, KIAA0319, and THEM2. For this we exploited the observation that acetylated histones are frequently associated with accessible chromatin at genomic regions containing regulatory elements [Eberharter and Becker, 2002; Kurdistani et al., 2004; Roh et al., 2005; Heintzman et al., 2007; Roh et al., 2007].
The basic unit of chromatin is a nucleosome, consisting of ~147 bp of DNA wound around an octamer of histones (two each of H2A, H2B, H3, and H4). Packaging of DNA into these higher order structures restricts access to regulatory proteins [Kadam and Emerson, 2002]. Covalent histone modifications control access of proteins such as transcription factors [de la Cruz et al., 2005]. For example, acetylation creates accessible chromatin both through effects on charge that alter histone–DNA interactions and through recruitment of regulatory proteins with bromodomains that bind directly to acetylated lysines [Jacobson et al., 2000; Forsberg and Bresnick, 2001]. Genome-wide studies in higher eukaryotes reveal that acetylated histones are enriched in regions of transcriptional competence, and mark regulatory elements such as promoters and enhancers [Eberharter and Becker, 2002; Kurdistani et al., 2004; Roh et al., 2005, 2007; Heintzman et al., 2007]. Therefore, identifying these regions provides functional clues as to the location of genomic sequences involved in gene regulation. This approach identifies putative regulatory regions allowing studies to focus on functionally relevant regions.
Importantly, while the positions of modified histone islands are conserved between humans and mice, the underlying sequences are often not [Bernstein et al., 2005], likely reflecting the fact that regulatory elements consist of combinations of binding sites that are each only a few base pairs long and are degenerate. Further, conservation in function can be maintained even without conservation of sequence due to compensatory changes at other transcription factor binding sites [Ludwig et al., 2000]. Thus, in addition to studying chromatin from a human retinoblastoma cell line (Y79) that has characteristics of neuronal stem cells [Seigel et al., 2007], we tested for positional conservation of histone modifications across a 500 kb region syntenic to human 6p in multiple embryonic mouse tissues. This approach pinpointed ubiquitous and tissue-specific sites of histone acetylation, some of which are of interest because of their proximity to associated markers. At the previously defined 70 kb haplotype at KIAA0319, chromatin analysis cast attention on a 2.7 kb region across the first untranslated exon and this together with genetic analyses here and in previous studies point to its importance in RD.
MATERIALS AND METHODS
Subjects and Assessment
Subjects aged 6–16 that presented with reading problems at local schools were recruited to participate in this study [Wigg et al., 2004; Luca et al., 2007]. The majority of the children had not had a psychological assessment for reading ability prior to study entry. Siblings in the same age range were also recruited, regardless of reading ability. Participants were restricted to families with English as a first language or 5 years in an English language school. The sample is currently made up of 291 nuclear families, of which 165 are trios, 77 are families with 2 children, and 5 families have 3 children. The sample also includes 20 single parent families with 1 child, 23 with 2 children, and 1 with 3. There are, in total, 112 siblings in the sample.
The sample mainly consists of individuals of Northern, Southern, Eastern, and Western European ancestry from the Toronto area. Toronto is the largest city in Canada, and has a high influx of immigrants. The Canadian population of European ancestry reflects diverse waves of immigration from continental Europe and the British Isles. Of the parents in our sample, 68.1% describe their parent’s (child’s grandparents) ethnicity as European or British. This includes individuals who are of mixed European decent (e.g., father from England and mother from Italy). Another 26% describe their parents as “Caucasian Canadians” without specific ethnic backgrounds. Individuals from South America made up 1.8% of the sample, while 2.9% of the sample was of non-European ancestry, and 1.2% was of non-European mixed ethnicity. Of the children in the sample, 38.2% had both parents who came from the same country (e.g., father and mother from Italy), and 61.8% had parents who originated in different countries (e.g., mother from Canada and father from Ireland).
Assessment
The reading measures have been described previously [Wigg et al., 2004; Luca et al., 2007]. Briefly, the structured interview with parents (Children’s Interview for Psychiatric Syndromes) [Weller et al., 2000] and a semi-structured interview with teachers (TTI) [Tannock et al., 2002], supplemented with standardized questionnaires (Conners Parent and Teacher Rating Scales Revised [Conners, 1997] and Ontario Child Health Survey Scales-Revised [Boyle et al., 1993]) were used to obtain information on symptoms of neurological, medical, and psychiatric disorders. Subjects were excluded if they showed evidence of neurological or chronic medical illness, bipolar affective disorder, psychotic symptoms, Tourette syndrome, or chronic multiple tics. Children were also excluded if they scored below 80 on both the Performance and Verbal Scales of the Weschler Intelligence Scale for Children III [Wechsler, 1991].
Single-word reading, phonological decoding skills, and spelling were assessed using the third edition of the Wide Range Achievement Test (WRAT-3), the Woodcock Reading Mastery Tests-Revised (WRMT-R), and Test of Word Reading Efficiency (TOWRE). The WRAT-3 subtests provide assessments of single-word reading and spelling [Wilkinson, 1993]. Two subtests of the WRMT-R were used, one to evaluate phonological decoding (Word Attack), and the other to assess single-word reading (Word ID) [Woodcock, 1987]. The TOWRE is a timed test that examines word identification and phonological decoding [Torgesen et al., 1999]. The scores from the above tests were standardized scores based on age norms. Means and standard deviations for all measures are reported in Supplementary Table 1.
Scores on three standardized reading tests, the WRMT-R Word Attack and Word Identification, and the WRAT-3 reading subtest were used to classify subjects as “affected” for the categorical analysis. These criteria were set to identify a subset of individuals in our sample whose scores on the three core reading measures, on average, fall within the lower 5% tail of normally distributed reading ability in the general population. To be classed as affected, subjects had to score 1.5 standard deviations below the population mean (standard score 78 or lower) on two of these three reading tests, or 1 standard deviation (standard score of 85 or lower) on the average of the three. Out of the 291 probands and 112 siblings, 156 probands and 25 siblings met the criteria and were used in the categorical analysis.
Isolation of DNA and Marker Genotyping
DNA was extracted directly from blood lymphocytes using a high-salt extraction method [Miller et al., 1988]. We initially investigated 37 SNP markers in the genes for VMP (NRSN1), DCDC2, KIAA0319, TTRAP, and THEM2 (Fig. 1 and Table I). Following chromatin immunoprecipitation coupled with microarray (ChIP-chip) analysis, seven additional markers were investigated across the 5′ untranslated region and first intron of KIAA0319 for a total of 44 markers. Assays were either predesigned and tested by Applied Biosystems (ABI, Foster City, CA; Assay-On-Demand by Applied Biosystems®) (Table SIIa), or designed from flanking sequence ascertained from the UCSC database builds 33–35 and sent to Applied Biosystems, who then designed the assays (ABI; Assay-By-Design by Applied Biosystems®) (Table SIIb). Both types of assays were genotyped with the ABI 7900-HT Sequence Detection System® (Applied Biosystems) using the TaqMan 5′ nuclease assay for allelic discrimination. Following the Polymerase Chain Reaction, plates were read on the ABI 7900HT Sequence Detection System (SDS), using the allelic discrimination end-point analysis mode of SDS software package version 2.0 (Applied Biosystems®). The G/T polymorphism, rs2038137, and the A/C polymorphism, rs761100, were genotyped using restriction enzyme analysis. These PCR reactions were performed in a total volume of 20 μl, with 100 ng of each primer for each marker (KIAA0319-8137F: GGTTGGGAAAAGACACTCAA and KIAA0319-8137R: GAC-GACGAGGAGGAACAAGT for rs2038137 and KIAA0319-1100F: AAGCTCTGTGGCTCACCATT and KIAA0319-1100R: CCAGGCAGTAAGGAGTGGAG for rs761100), 0.2 mM dNTP, 1.5 mM magnesium chloride, and 0.5 U Taq polymerase. The PCR reaction started with a denaturing step at 94°C for 3 min, followed by 35 cycles at 94°C for 30 sec, 58°C for 30 sec (60°C for A/C polymorphism), and 72°C for 30 sec. It was then completed by a 10 min extension step at 72°C. PCR product (5–6 μl) for the G/T polymorphism was digested with 5 U of the restriction enzyme BstUI (New England Biolabs, Beverly, MA) at 60°C for 2 h. Alleles were identified on 2% agarose gels. The more frequent allele, G, was cut by the enzyme and was labeled as allele 1. The other allele, T, was not cut and was labeled allele 2. PCR product (2 μl) for the A/C polymorphism was digested with 1 U of the restriction enzyme AjuI (Fermentas International, Inc., Burlington, Canada) at 37°C for 8 h. Alleles were identified on 3% (1.5% Nusieve, 1.5% Agarose) gels. The less frequent allele, C, was cut by the enzyme and was labeled as allele 2. The other allele, A, was not cut and was labeled allele 1.
FIG. 1.
Schematic showing the ~589 kb region on chromosome 6p with the five candidate genes, VMP (NRSN1), DCDC2, KIAA0319, TTRAP, and THEM2 and the position of the markers genotyped in the current study. Untranslated regions (UTR) are drawn as shorter boxes and exons are depicted as longer lines and boxes. The vertical arrows show the approximate location for the initial 37 SNP markers genotyped across the region, starting with rs1419229 (left-most arrow in the 5′ region of VMP/NRSN1). The location of all SNP markers is listed in Table I alongside the rs number. The horizontal arrows show the direction of transcription for each gene.
TABLE I.
TDT Analysis for Markers in the 6p Region and RD
| Gene | Location | Marker | Nuc.1 | Freq. | Trans. | Non-trans. | χ2 (1 df) | P-value |
|---|---|---|---|---|---|---|---|---|
| VMP (NRSN1) | 24,220,516 | rs1419229a | A | 0.46 | 73 | 71 | 0.028 | 0.867 |
| 24,229,973 | C_1809133_10 | G | 0.04 | 7 | 6 | *** | *** | |
| 24,238,542 | rs1936390 | A | 0.97 | 4 | 4 | *** | *** | |
| 24,239,314 | rs9356928 | G | 0.54 | 82 | 57 | 4.496 | 0.034 | |
| 24,240,194 | rs6935378 | C | 0.75 | 58 | 51 | 0.450 | 0.502 | |
| 24,241,681 | rs7455023 | A | 0.57 | 77 | 61 | 1.855 | 0.173 | |
| 24,246,081 | rs4285310 | A | 0.55 | 79 | 56 | 3.919 | 0.048 | |
| 24,253,875 | rs11544636 | A | 0.03 | 6 | 1 | *** | *** | |
| 24,254,651 | rs3829810 | A | 0.26 | 57 | 45 | 1.412 | 0.235 | |
| 24,255,551 | rs3178 | G | 0.49 | 82 | 58 | 4.114 | 0.043 | |
| 24,259,500 | C9373644_10a | C | 0.50 | 79 | 63 | 1.803 | 0.179 | |
| DCDC2 | 24,286,285 | rs1419228a | G | 0.21 | 43 | 40 | 0.108 | 0.724 |
| 24,315,179 | rs793862a,d,e | A | 0.31 | 58 | 51 | 0.450 | 0.502 | |
| 24,369,039 | rs870601a | T | 0.33 | 61 | 59 | 0.033 | 0.856 | |
| 24,381,770 | rs807701e | A | 0.65 | 65 | 62 | 0.071 | 0.790 | |
| 24,399,182 | rs2274305a | T | 0.35 | 64 | 56 | 0.533 | 0.465 | |
| 24,418,623 | rs807685a | A | 0.13 | 39 | 36 | 0.120 | 0.729 | |
| 24,444,623 | rs793704a | C | 0.45 | 74 | 68 | 0.254 | 0.614 | |
| KIAA0319 | 24,477,494 | rs793663a | T | 0.38 | 67 | 64 | 0.069 | 0.793 |
| 24,653,918 | rs807530 | G | 0.58 | 78 | 60 | 2.348 | 0.125 | |
| 24,676,372 | rs12193738 | T | 0.46 | 77 | 73 | 0.107 | 0.744 | |
| 24,692,345 | rs2817200a | G | 0.38 | 68 | 56 | 1.161 | 0.281 | |
| 24,696,863 | rs4504469b,c | A | 0.38 | 69 | 55 | 1.581 | 0.209 | |
| 24,752,301 | rs6935076c | Gf | 0.64 | 74 | 47 | 6.025 | 0.014 | |
| 24,753,399 | rs2038139 | C | 0.34 | 66 | 50 | 2.207 | 0.137 | |
| 24,753,446 | rs2038138 | C | 0.34 | 65 | 52 | 1.444 | 0.230 | |
| 24,753,676 | rs2235676b | C | 0.86 | 35 | 26 | 1.328 | 0.249 | |
| 24,753,922 | rs2038137b,c | T | 0.35 | 67 | 62 | 0.194 | 0.660 | |
| 24,755,198 | rs9467247b | A | 0.21 | 43 | 40 | 0.108 | 0.724 | |
| TTRAP | 24,760,822 | rs2294691 | G | 0.06 | 17 | 11 | 1.286 | 0.257 |
| 24,761,252 | rs2294689 | C | 0.06 | 17 | 14 | 0.290 | 0.590 | |
| 24,763,022 | rs3181238 | A | 0.37 | 74 | 50 | 4.645 | 0.031 | |
| 24,765,461 | rs3212233 | G | 0.34 | 54 | 44 | 1.020 | 0.313 | |
| 24,767,050 | rs2143340b | A | 0.85 | 38 | 27 | 1.862 | 0.172 | |
| 24,773,986 | rs1061925b | A | 0.89 | 32 | 29 | 0.148 | 0.701 | |
| 24,775,778 | rs3181227a | G | 0.21 | 38 | 30 | 0.941 | 0.332 | |
| THEM2 | 24,806,194 | rs7765904 | G | 0.74 | 55 | 47 | 0.627 | 0.429 |
Significant markers from
Nuc1: The nucleotide designation for the over-transmitted allele.
The opposite allele, A, was over-transmitted in the study of Cope et al. [2005].
Markers that were not analyzed because of the small number of informative transmissions.
The genotyping success rate was high (greater than 97%). All data was screened for Mendelian errors using PEDSTATS, and MERLIN to detect any crossovers between markers [Abecasis et al., 2002]. This data set was free of any detectable Mendelian errors and none of the markers used deviated from Hardy–Weinberg equilibrium.
Statistical Analysis
The TDT statistic was calculated using the extended TDT (ETDT) program for the categorical analysis [Sham and Curtis, 1995]. Analysis of the quantitative traits of spelling, single-word recognition, and phonological decoding was carried out using the FBAT program [Laird et al., 2000]. An offset of 100.1 was used to mean center all three traits. Stepwise regression was conducted in STATA (StataCorpLP, College Station, TX) using a package designed by David Clayton (http://www-gene.cimr.cam.ac.uk/clayton/software/) [Cordell and Clayton, 2002]. Haplotype transmission was analyzed using the TRANSMIT program [Clayton and Jones, 1999]. Haplotypes with a frequency of less than 10% were pooled and P values were only reported for those with a frequency greater than 0.10. Genotypes from the Centre d’Etude du Polymorphisme Humaine (CEPH), Utah individuals of Northern and Western European ancestry (CEU), available through the HapMap project were acquired from http://www.hapmap.org (Rel #20/phase II Jan 06) [The International HapMap Consortium, 2005]. These data were used to choose seven SNPs that tagged haplotype variation greater than or equal to 1% within the 22 kb haplotype block. Linkage disequilibrium (LD) blocks were analyzed and visualized using an algorithm by Gabriel et al. [2002] in the Haploview program v3.11 (http://www.broad.mit.edu/mpg/haploview/) [Gabriel et al., 2002; Barrett et al., 2005].
Chromatin Immunoprecipitation (ChIP)
An approach similar to that described by Ni and Bremner [2007] was used for the chromatin immunoprecipitation. Primary tissues from the brain, heart, muscle, lung, and liver of E16 (embryonic day 16) mice were treated with 1% formaldehyde at room temperature for 10 min, after which they were washed twice with cold phosphate buffered saline (PBS), collected in 1 ml PBS and centrifuged at 5,000 rpm for 5 min. Chromatin from the Y79 cell line was prepared similarly. Cells were then resuspended in 1 ml of lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris–HCl (pH 8)) plus proteinase inhibitors (aprotinin, leupeptin, and pepstatin). Subsequently, they were incubated on ice for 10 min and sonicated to an average size of 500 bp (Vibra Cell; Sonics and Materials, Danbury, CT). Chromatin was precleared with 25 μl of Staph A (507862; Calbiochem, San Diego, CA) at 4°C for 15 min. A 100 μl aliquot of sonicated chromatin was then immunoprecipitated (IP) with 2 μg of antibodies for acetylated histone 3 (H3) at lysine residues 9 and 14 (AB 06-599 Upstate Biotechnology, Charlottesville, VA) at 4°C overnight. The IP samples were then centrifuged at 13,200 rpm and the supernatant was incubated with 10 μl of Staph A at room temperature for 15 min. Precipitates were washed sequentially for 3 min in 1×dialysis buffer (2 mM EDTA, 50 mM Tris–HCl (pH 8), and 0.2% sarkosyl) twice, and IP wash buffer (1% Nonidet P-40, 100 mM Tris–HCl (pH 9) 500 mM LiCl 1% and deoxycholic acid) four times. Samples were extracted twice with 150 μl elution buffer (1% SDS and 50 mM NaHCO3) and heated at 65°C overnight to reverse cross-links. DNA fragments were purified with a QIAEX II Extraction kit (catalog no. 20051) and amplified by ligation-mediated PCR (LM-PCR).
Chip Tiling Array Design and Data Analysis
Following amplification, the PCR product was labeled and hybridized to Nimblegen custom built genomic tiling arrays (Nimblegen, Madison WI). Two color hybridizations were performed using input DNA as a reference in each case. The arrays contained 50 mers at minimal intervals of 80–92 bp and covered all non-repetitive sequences across a 500 kb region on human 6p (chr6: 24,141,096–24,884,061) or on the syntenic region on mouse 13qA3.1 (chr13: 24,261,610–24,761,610, March 2005 (mm6)). This region spans the VMP (NRSN1) to C6orf62 genes.
Raw intensities from three independent biological replicates were first assessed for quality visually using Nimblegen SignalMap software (Nimblegen). Data were then quantile normalized [Bolstad et al., 2003] and averaged for the corresponding genomic position of each quadruplicate 50 mer. The Wilcoxon Rank Sum test was used to determine the difference in intensities of the ChIP signal compared to the input DNA signal for probes within a 500 bp sliding window. Genomic positions with statistically higher intensities compared to input DNA (P <10−3) and above a nominal 1.3-fold threshold were merged to form a peak indicating a genomic region enriched for acetylated histones (H3ac). We observed virtually identical results analyzing the data using the same parameters with the program MA2C that normalizes probes for GC content [Song et al., 2007]. The results shown are based on the combined analyses across biological replicates using MA2C. The data for each cell/tissue type were imported into the UCSC genome browser (human assembly (hg18), March 2006, or mouse assembly (mm9), July 2007).
RESULTS
Association was initially investigated using a sample of 291 nuclear families and 37 SNP markers in the genes for VMP (NRSN1), DCDC2, KIAA0319, TTRAP, and THEM2 (Fig. 1). The single marker categorical TDT analysis showed significant evidence for association of rs9356928 (χ2 =4.496, P =0.034), rs4285310 (χ2 =3.919, P =0.048), and rs3148 (χ2 =4.114, P =0.043) in the gene for VMP (NRSN1), rs6935076 (χ2 =6.025, P = 0.014) in the gene for KIAA0319 and rs3181238 (χ2 =4.645, P =0.031) in the gene for TTRAP with RD (Table I). This single marker analysis was conducted using the subset of our sample with a proband that met criteria for RD defined as a categorical trait (156 families). Markers tested in the gene for DCDC2 included rs793862 and rs807701, two polymorphisms most significantly association with RD in the study of Schumacher et al. [2006]. These markers were not associated with RD in our sample. There was also no association with the one marker genotyped in the gene for THEM2 in this sample (Table I).
We examined the LD across this region in our entire set of families to determine if inter-marker LD could account for the observed association with markers in multiple genes. We observed two regions of high LD, one encompassing the genes for VMP (NRSN1) and DCDC2 and the other including KIAA0319 and TTRAP (Fig. 2). In addition, we observed punctate regions of LD between markers in these two regions, notably with markers in the 3′ regions of TTRAP (rs2294691, rs2294689) and VMP (NRSN1) (rs9356928, rs4285310, rs3178) (D′ =0.65–0.85, LOD ≥ 2) (Fig. 2).
FIG. 2.
Graphical view of LD between the 44 markers genotyped across VMP (NRSN1), DCDC2, KIAA0319, TTRAP, and THEM2 on 6p in the RD sample. The locations of VMP (NRSN1), DCDC2, KIAA0319, and TTRAP are marked by the horizontal arrows. The marker rs7765904 is located in THEM2. The rs number for each marker is listed above the LD plot. Red boxes represent inter-marker D′ =1, LOD ≥ 2 and are indicative of high LD. Regions with D′ <1, LOD ≥ 2 are shown in shades of red/pink. Blue squares indicate inter-marker D′ =1, LOD <2. The white boxes show inter-marker D′ <1, LOD <2 and are indicative of low LD. The numbers in the boxes indicate the inter-marker D′ values (multiplied by 100). Markers in the genes for VMP (NRSN1) and TTRAP between which high LD was observed are marked by boxes on the top of the schematic. Arrows indicate regions of LD between markers in VMP (NRSN1) (rs9356928, rs4285310, rs3178) and TTRAP (rs2294691, rs2294689).
The RD locus on 6p has previously been reported to be associated with single-word reading, spelling, phonological decoding, phonological awareness, and orthographic coding, suggesting that it contributes to multiple reading component processes [Deffenbacher et al., 2004; Francks et al., 2004; Harold et al., 2006]. Based on this we investigated the five markers that showed significant evidence for association with RD defined as a categorical trait for association with measures of spelling, phonological decoding and word recognition as quantitative traits. Significant evidence for association was found with the marker rs6935076 in KIAA0319 and spelling (Z = 2.247, P =0.025), with a weak trend for single-word reading (Table II). The marker rs3178 in VMP (NRSN1) showed significant evidence for association with phonological decoding (Z =2.022, P = 0.043), and a trend for association with spelling (Table II). The three remaining makers, rs9356928, rs4285310, and rs3181238 were not associated with the quantitative measures tested (Table II).
TABLE II.
Analysis of Select Quantitative Measures and Markers on 6p
| Gene | Marker | Allele | Freq. | Fam # | S | E (S) | Var (S) | Z | P |
|---|---|---|---|---|---|---|---|---|---|
| WRAT-3 spelling | |||||||||
| VMP (NRSN1) | rs9356928 | G | 0.54 | 183 | −3621.400 | −3420.600 | 24586.403 | −1.281 | 0.200 |
| rs4285310 | A | 0.55 | 176 | −3789.300 | −3642.050 | 24761.676 | −0.936 | 0.349 | |
| rs3178 | G | 0.47 | 179 | −3359.100 | −3092.100 | 23792.988 | −1.731 | 0.083 | |
| KIAA0319 | rs6935076 | G | 0.66 | 157 | −3799.900 | −3453.050 | 23824.343 | −2.247 | 0.025 |
| TTRAP | rs3181238 | A | 0.38 | 165 | −2868.800 | −2679.817 | 23619.677 | −1.230 | 0.219 |
| TOWRE single word reading | |||||||||
| VMP (NRSN1) | rs9356928 | G | 0.54 | 187 | −3849.300 | −3629.050 | 31628.139 | −1.238 | 0.216 |
| rs4285310 | A | 0.55 | 180 | −3821.000 | −3715.400 | 30833.017 | −0.601 | 0.548 | |
| rs3178 | G | 0.47 | 183 | −3521.500 | −3284.500 | 26838.482 | −1.447 | 0.148 | |
| KIAA0319 | rs6935076 | G | 0.66 | 159 | −3746.400 | −3489.100 | 23444.990 | −1.680 | 0.093 |
| TTRAP | rs3181238 | A | 0.38 | 166 | −2892.900 | −2689.367 | 23823.091 | −1.319 | 0.187 |
| TOWRE phonological decoding | |||||||||
| VMP (NRSN1) | rs9356928 | G | 0.54 | 187 | −4293.300 | −4084.050 | 33212.272 | −1.148 | 0.251 |
| rs4285310 | A | 0.55 | 180 | −4317.000 | −4170.900 | 32027.450 | −0.816 | 0.414 | |
| rs3178 | G | 0.47 | 183 | −3995.500 | −3647.000 | 29698.715 | −2.022 | 0.043 | |
| KIAA0319 | rs6935076 | G | 0.66 | 159 | −4183.400 | −3929.600 | 26721.840 | −1.553 | 0.121 |
| TTRAP | rs3181238 | A | 0.38 | 166 | −3257.900 | −3000.533 | 27088.208 | −1.564 | 0.118 |
To exploit the ancestral information provided by the underlying inter-marker LD, we sought to assess more powerful multi-marker associations by analyzing the haplotype structure of the region. Specifically, we analyzed two sets of haplotypes previously reported to be associated with RD [Francks et al., 2004; Cope et al., 2005], and haplotypes constructed from tagging SNPs in VMP (NRSN1). The first was a set of haplotypes from three markers (rs4504469–rs2038137–rs2143340) across KIAA0319 and TTRAP associated with RD in the study in Francks et al. [2004], and replicated by two studies [Luciano et al., 2007; Paracchini et al., 2008]. Haplotypes of these three markers span a 70 kb region and have also been associated with differential expression of KIAA0319 [Paracchini et al., 2006]. We analyzed haplotypes of these three markers for association with RD using TRANSMIT, and found no significant evidence for association (global P =0.327, 3 df) (data not shown).
The second set of haplotypes consisted of alleles from two markers (rs4504469–rs6935076) in KIAA0319 [Cope et al., 2005]. Step-wise regression analysis in that study showed that these two markers were components of a model that best explained the effect of all polymorphisms associated with RD in that sample [Cope et al., 2005]. Two haplotypes of these markers showed evidence for association with RD in that study. The first and most significant finding was for under transmission (trios) and under representation (case–control sample) of one of these haplotypes (2-1 or A-G) [Cope et al., 2005]. The study also reported evidence of association for a second haplotype (1-2 or G-A), though less significant, with over transmission and over representation in these cases. Analysis of these two markers in our sample showed significant evidence for under transmission of the 1-2 (or G-A) haplotype (χ2 =5.566; P =0.018)—the over transmitted haplotype in their study (Table III). No evidence for association was found for the 2-1 haplotype and the global test for association was not significant (χ2 =6.240; P =0.101, 3 df) (Table III).
TABLE III.
Two Marker Haplotype Analysis Based on Cope et al. [2005]
| Marker 1 | Marker 2 | Frequency | Observed | Expected | Var (O-E) | χ2 (1 df) | P-value |
|---|---|---|---|---|---|---|---|
| rs4504469 | rs6935076 | ||||||
| 1(G) | 1(G) | 0.34 | 123.150 | 118.280 | 31.202 | 0.759 | 0.384 |
| 2(A) | 1(G) | 0.32 | 120.850 | 111.510 | 32.763 | 2.666 | 0.103 |
| 1(G) | 2(A) | 0.28 | 79.852 | 93.081 | 31.437 | 5.566 | 0.018 |
| 2(A) | 2(A) | 0.07 | 26.148 | 27.129 | 9.8787 | *** | *** |
Global χ2, 3 df = 6.240; P = 0.101.
The 2-1 haplotype was previously associated with RD in Cope et al. [2005]. This haplotype was underrepresented in the cases and showed biased non-transmission in the trios. The 1-2 haplotype was over transmitted and over represented.
Low frequency haplotype that was not considered in the analyses.
Data from the HapMap project (Based on NCBI build 35) showed that the marker rs3178 in VMP (NRSN1) which was associated to both categorical and quantitative measures of RD exists in a haplotype block, and along with the marker rs3829810, tags common variation greater than 10%. These two markers were chosen to construct haplotypes that were tested for association with RD, using TRANSMIT [Clayton and Jones, 1999]. No significant association was found for any haplotype and RD (global P =0.293, 2 df) (data not shown).
In summary, when the five candidate genes on 6p were analyzed in our sample, evidence for association was found with single markers in VMP (NRSN1), KIAA0319, and TTRAP. In addition, the only significant haplotype association was with markers in KIAA0319.
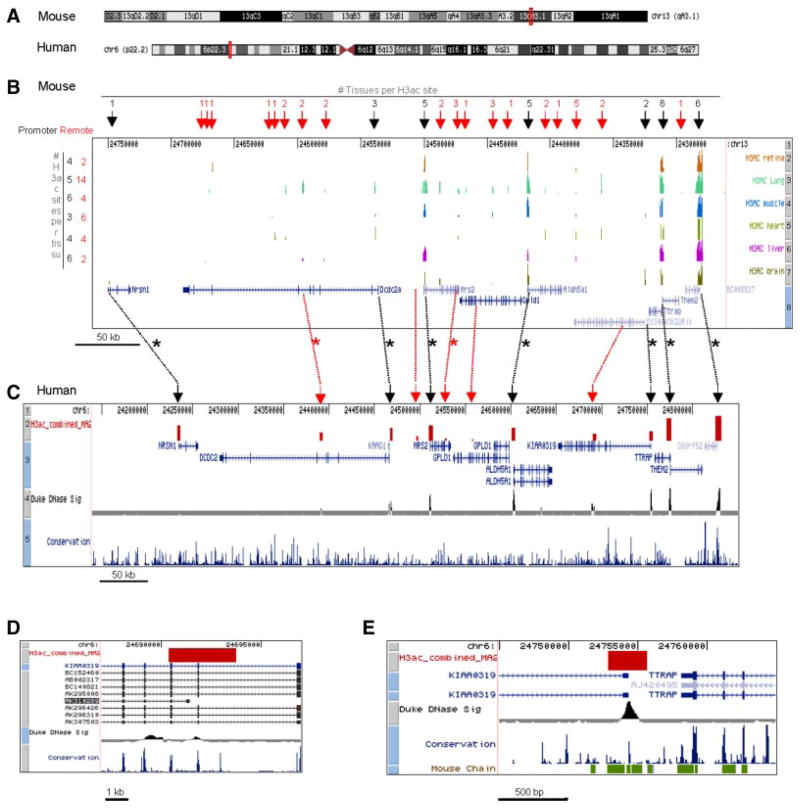
To identify potential regulatory elements around the candidate region on 6p, we mapped the position of acetylated H3 histones (H3ac) at 6p22.2 in the human genome and the corresponding region in the mouse genome, 13qA1.3 (Fig. 3A). The regions analyzed contain 10 genes in humans including VMP (NRSN1), DCDC2, KAAG1 (NM_181337), MRS2, GPLD1, ALDH5A1, KIAA0319 (D130043K22Rik in mice; NM_001081051), TTRAP, THEM2, and C6orf32 (NM_030939; BC005537 in mice; NM-_024473). Of these, all except the small KAAG1 transcript are annotated in mice (Fig. 3B,C). Chromatin immunoprecipitation (ChIP) with anti-H3ac antibodies was performed for six tissues obtained from embryonic day 16 mice (retina, lung, heart, muscle, liver, brain) (Fig. 3B) and the human retinoblastoma cell line, Y79 (Fig. 3C). Y79 shares characteristics with neuronal stem cells [Seigel et al., 2007] and we confirmed expression of KIAA0319 and DCDC2 in this cell line using quantitative PCR (not shown). Enriched and input DNA were differentially labeled and co-hybridized to arrays tiled with oligonucleotides across a ~500 kb region. H3ac peaks that were both highly significant (P <0.001) and also at least 1.3-fold above background were visualized using the UCSC genome browser (Fig. 3B,C). The data are available online at http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr10&hgt.customText=http://vsrp.uhnres.utoronto.ca/jillmousearray_mm9.gff for the mouse data and http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chr13&hgt.customText=http://vsrp.uhnres.utoronto.ca/y79array_chr6.gff for the Y79 data.
FIG. 3.
ChIP-chip analysis. A: Mouse chromosome 13 and human chromosome 6. Red boxes indicate syntenic regions represented on the tiling arrays. B: H3ac ChIP-chip data for six mouse tissues. Arrows indicate the position of promoter (black) or remote (red) H3ac sites, and the number of tissues where that H3ac site was detected is indicated above. The total number of promoter or remote sites per tissue is indicated on the left. Tracks in the browser window indicate 1. Base position on chromosome 13; 2-7 H3ac ChIP-chip peaks for the indicated tissues; 8. Refseq genes. The scale bar is shown on the lower left. C: Human (Y79) ChIP-chip data. Tracks in the browser window indicate 1. Base position on chromosome 6; 2. H3ac ChIP-chip peak; 3. Refseq genes; 4. DNase hypersensitive sites in CD4+T cells; 5. Conservation in 17 vertebrates, including mammalian, amphibian, bird, and fish species. Dotted lines between B and C link the regions in mouse (B) and syntenic H3ac-marked regions in human Y79 cells (C). Red and black dotted lines connect promoter or remote sites, respectively. Asterisks indicate a matching H3ac mark at the corresponding human and mouse positions. D: The alternative KIAA0319 transcript, AK310289, detected in testes cDNA, initiates within a region marked by H3ac in Y79 cells and DNase accessibility in T cells. E: The extent of the KIAA0139 promoter proximal H3ac mark in Y79 human cells. The bottom track shows the syntenic mouse regions; an H3ac mark was also detected at the mouse promoter (see B).
First, we compared the acetylated marks at the promoter regions in the mouse tissues and Y79. TTRAP and THEM2 are located head-to head with overlapping promoter regions, and DCDC2 and KAAG1 are transcribed in the same direction with overlapping promoter regions. Thus of the 10 genes investigated in humans (9 in mice), there are 8 identifiable promoters regions. We observed that seven of the eight proximal promoter regions were marked by H3 acetylation in at least one of the six mouse primary tissues, and these seven regions were also acetylated in the human retinoblastoma cell line, Y79 (black asterisks in Fig. 3B,C), confirming that H3ac flags regulatory elements [Roh et al., 2005; Heintzman et al., 2007]. The GPLD1 promoter was not acetylated in any mouse tissue analyzed or in human Y79 cells. DNase I sensitivity, like H3ac, is a mark of chromatin accessibility, and of the seven promoters where we observed H3ac, six were highly accessible in a recent study that assessed DNase I sensitivity in CD4+ T cells [Boyle et al., 2008]. A prior study found that MRS2, ALDH5A1, TTRAP, and THEM2 are expressed ubiquitously or almost ubiquitously in 20 human adult tissues, whereas VMP (NRSN1), DCDC2, GPLD1, and KIAA0319 are tissue-specific [Londin et al., 2003]. Strikingly, of the four ubiquitously expressed human genes, the orthologous mouse promoter was acetylated in 5/6 or 6/6 embryonic mice tissues and all were also acetylated in human Y79 cells (Fig. 3B,C). Of the four genes with restricted expression patterns in humans, the orthologous mouse promoters were acetylated in 0/6 (Gpld1; NM_008156), 1/6 (Nrsn1; NM_009513), 2/6 (D130043K22Rik), or 3/6 (Dcdc2; NM_177577) of the mouse tissues studied (Fig. 3). Moreover, there was good concordance between the H3ac promoter signals in mouse embryonic tissues and expression in adult human tissues. For example, the D130043K22Rik promoter was acetylated in developing mouse brain and muscle but not liver, heart or kidney (Fig. 3), and its ortholog K1AA0319 is expressed in the human fetal brain, adult muscle, but not liver heart or kidney [Londin et al., 2003]. These data are consistent with H3ac as a marker for active or poised promoters.
Across all mouse tissues examined, we detected a total of 18 distinct remote regions of H3ac (remote defined as >5 kb from a known gene start) and 5 remote H3ac sites in human Y79 cells (Fig. 3B,C). In mouse, most remote sites were detected in lung (14) while fewest were seen in retina (2) and brain (2). Intermediate numbers were observed in muscle (4), heart (6), and liver (4) (Fig. 3). Half of the 18 distinct remote sites detected in mice were found in more than one tissue (including one that was seen in five), suggesting that they mark regulatory features that are utilized in multiple contexts. In a similar vein, two of the five remote sites in human Y79 cells matched sites of DNase I accessibility in CD4+ T cells (Fig. 3C), providing independent evidence that these are accessible chromatin sites and that they may mark regulatory elements used in multiple human cell types; one was located in intron 7 of the large DCDC2 gene, while the second was located at the center of the large KIAA0319 locus (see below for further discussion). The DCDC2 site was also one of two remote Y79 H3ac marks that exhibited acetylation at or very near syntenic mouse regions (red asterisks in Fig. 3). The DCDC2 site was acetylated at the equivalent mouse locus in liver and lung, while another remote H3ac site detected in the second last intron of the human MRS2 gene was 2.5 kb from a region in the last intron which was acetylated in mouse lung, muscle and liver (Fig. 3B,C). The presence of identically positioned remote H3ac sites in multiple mouse tissues, concordance between remote H3ac marks and regions of DNase I hypersensitivity in different human cell types, and conservation of a subset of H3ac regions in human and mouse tissues all serve as indicators that these are important sites of chromatin activity, such as enhancers, silencers, and/or insulators.
“Remote” H3ac marks may, in some cases, flag un-annotated promoters [Heintzman et al., 2007]. The site located between DCDC2 and MRS2 in Y79 (Fig. 3C, chr6: 24,496,400–24,497,725), likely marks the promoter of the recently identified alternative first exon of a DCDC2 transcript (annotated in UCSC as transcript BC050704) that was documented in kidney tissue but not in brain [Schumacher et al., 2006]. Another remote site that may mark an alternative transcript start site is the remote H3ac/ DNase hypersensitive mark at the human KIAA0319 locus (chr6: 24,690,380–24,693,734) overlapping the 5′ end of a transcript detected in testes (AK310289) (Fig. 3D). Open Reading Frame Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) predicted a 483 amino acid protein from this transcript with the start codon in exon 11 of the full transcript. The putative alternative promoter was not acetylated in the syntenic region of any mouse tissue we studied, and currently there is no evidence for an alternative transcript in the mouse genome. Approximately half of all mammalian promoters are associated with CpG islands, but this feature was not at the start of human AK310289, or at the equivalent mouse location. However, using the Evolutionary Conserved Region (ECR) browser (http://ecrbrowser.dcode.org/), we found two non-coding regions conserved between human and mouse: 108 bp at mm9 chr13: 24,954,137–24,954,244 (hg18 chr6: 24,692,222–24,692,329; 70.4-%) and the second, a 131 bp region located 257 bp from the first at position mm9 chr13: 24,953,987–24,954,119 (hg18 chr6: 24,692,350–24,692,480; 71%). Despite this, we did not detect any RT-PCR product in Y79 cells using primers that amplify the region encompassing the predicted first exon of this alternative transcript and the next exon (exon 6 in the full transcript), whereas control primers that amplified across exons 6 and 7 detected the expected product (data not shown). This negative finding does not exclude the possibility that the acetylated peak marks a putative promoter that is active in other cell types, or is poised in Y79 cells.
Of the regions we identified as putative regulatory regions marked by acetylated histones, several stand out as the possible site of genetic variation contributing to RD because of their proximity to association signals in this or other studies. Eight remote H3ac sites were identified in the body of the DCDC2 gene across mouse tissues, although none were in mouse brain. One of the remote sites identified in mouse lung and liver was detected in neuronal Y79 cells as discussed earlier (Fig. 3B,C). This site (chr6: 24,390,434–24,392,587, UCSC May 2004, hg17, NCBI Build 35, and March 2006, hg18, NCBI build 36.1) is located between markers associated previously with RD in several studies. First, it is located between the markers rs807725 (reported as ABI assay C_7454804-_10 in that paper) located at position 24,386,848 in UCSC (March 2004) in intron 7 (previously reported as intron 6 before the identification of the alternative exon 1 [Schumacher et al., 2006-]) and a complex repeat located in intron 3 (previously reported as intron 2) [Meng et al., 2005]. The complex repeat [Meng et al., 2005] was not represented on our microarray because it is a repetitive sequence. Second, the intron 7 H3ac site we detected is 9.5 and 76 kb from intron 8 markers rs807701 and rs793862, respectively and both were associated with RD [Schumacher et al., 2006]. Third, it is also between markers rs870601 (intron 7) and rs2274305 (intron 9) associated to RD in another study [Deffenbacher et al., 2004]. No evidence for association was observed in an independent study for markers in introns 7 and 8 [Harold et al., 2006], and we also found no evidence for association to any marker in DCDC2 for RD in this study. However, we recently identified association with markers in introns 7 and 8 and attention/deficit hyperactivity disorder (ADHD) in a separate sample [Couto et al., 2009]. Thus, this acetylated region in intron 7 remains of interest because of its proximity to positive markers in several samples. Based on this proximity, and the acetylation in mouse tissues (lung and liver) and human Y79 cells (Fig. 3) we examined the region for conservation using the ECR browser (http://ecrbrowser.dcode.org/), a threshold of 100 bp, and 70% identity, empirically shown to provide high sensitivity for human/ mouse conservation profiles [Loots et al., 2000]. A 244 bp region with 72.5% conservation was observed at mm9 chr13: 25,207,393–25,207,629 corresponding to chr6: 24,391,816–24,392,059 in humans, and a second 192 bp conserved region with 70.3% conservation at mm9 chr13: 25,206,990–25,208,357 corresponding to chr6: 24,391,011–24,392,479 was also observed. The acetylated peak observed in mouse lung spans this conserved region and the acetylated peak in mouse liver is located just outside the region, pointing to this 244 bp subregion of the intron 7 peak as a likely position for the regulatory region. Using the Mulan tool within the ECR browser conserved transcription factor binding sites for CUT homeodomain proteins were noted (data not shown), both of which are expressed in the developing cortex [Nieto et al., 2004; Ferrere et al., 2006].
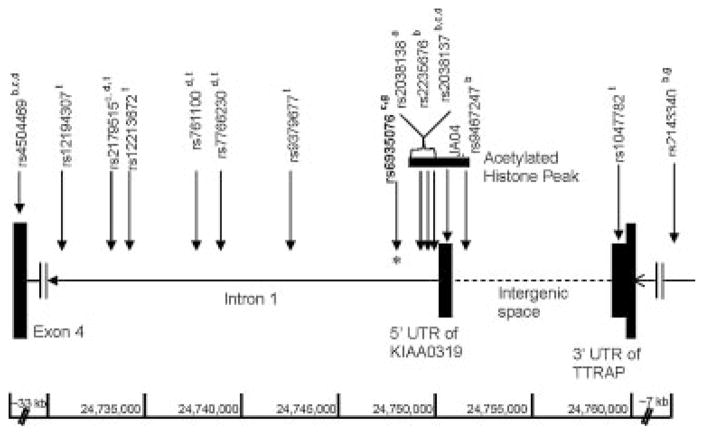
The putative alternative KIAA0319 promoter discussed above (Fig. 3D) and marked with H3ac in the Y79 cell line (chr6: 24,690,380–24,693,734; hg18), is also of interest due to its proximity to association signals. This site is located 3.1 kb from the marker rs4504469 (chr6: 24,696,863) that was associated with RD in several samples [Francks et al., 2004; Cope et al., 2005; Harold et al., 2006] but not others [Schumacher et al., 2006]. We found no significant evidence of association for either this marker or for rs2817200, located under the peak at position chr6: 24,692,345 in intron 4 (Table I), in our sample. The region adjacent (5′) to the KIAA0319 locus is of highest interest because of replicated association findings for markers in this region, and the location of our most significant marker and haplotypes. Within the 70 kb KIAA0319 haplotype [Francks et al., 2004], the only acetylated region was a 2.5 kb stretch in mouse brain tissue spanning ~800 bp 5′ to the first untranslated exon and into intron 1 (Fig. 3B). In the human Y79 cell line, the 5′ acetylated region was similar, spanning a ~2.7 kb region (chr6: 24,752,922–24,755,657) across the first untranslated exon (Figs. 3E and 4). Interestingly, five markers previously associated with RD were located within this 2.7 kb acetylated region (Fig. 4). These were the microsatellite marker, JA04 [Kaplan et al., 2002-], rs2038138 [Deffenbacher et al., 2004], rs2235676 (same as rs17491230), rs2038137 [Francks et al., 2004; Harold et al., 2006], and rs9467247 [Francks et al., 2004]. Furthermore, rs6935076 (chr6: 24,752,301) the marker with the strongest association in our sample was located ~600 bp downstream of the 2.7 kb region marked by acetylation in Y79 (Fig. 4). Two regions within the 2.7 kb acetylated region were conserved between humans and mice, one 169 bp region just 5′ to the first exon at mm9 chr13: 24,936,321–24,938,641 (74.6% similarity) and a second 189 bp region with 69.3% similarity located at mm9 chr13: 24,936,321–24,938,641.
FIG. 4.
Schematic showing part of intron 1, the 5′ untranslated exon (5′ UTR) of KIAA0319, the 3′ UTR of TTRAP, and the markers rs4504469 and rs2143340. The acetylation peak (H3ac) identified in mouse brain and Y79 is marked by the thick black line and spans ~2.7 kb. Within this region, one microsatellite (JA04) and four SNP markers showed evidence for association to RD in at least two out of three independent samples [Kaplan et al., 2002; Deffenbacher et al., 2004; Francks et al., 2004; Harold et al., 2006]. The marker rs6935076 (*) showed the most significant evidence for association in our sample. Significant marker associations from previous studies both within and beyond the H3ac peak are denoted by: (a) Deffenbacher et al. [2004], (b) Francks et al. [2004], (c) Cope et al. [2005], (d) Harold et al. [2006], (f) Kaplan et al. [2002], (g) Luciano et al. [2007]. The tagging SNPs are marked by a (t). The 22 kb LD block extends from (and includes) rs12194307 to rs1047782.
Based on the convergence of association findings from multiple independent samples in the H3ac marked region, we investigated it in more detail. Analysis of the LD pattern across the region of this H3ac peak and KIAA0319 was conducted using the CEPH Northern and Western European (CEU) genotype data from the HapMap project [The International HapMap Consortium, 2005]. These data showed that part of intron 1 of KIAA0319, the H3ac region, and some 5′, upstream sequence of KIAA0319 were all contained in one 22 kb haplotype block (Fig. 4). This haplotype block contains the associated markers within and just outside the acetylated region discussed above, and an additional three markers (rs7766230, rs761100, rs2179515) associated in the study of Harold et al. [2006]. The 22 kb block is also flanked by two additional associated markers (rs4504469, rs2143340) from five studies [Francks et al., 2004; Cope et al., 2005; Harold et al., 2006; Luciano et al., 2007; Paracchini et al., 2008] (Fig. 4). We found that haplotype variation greater than 1% could be tagged by seven SNPs (tagSNPs) including rs761100 and rs2179515, the two markers that showed evidence for association with RD in the combined analysis of two independent UK samples in a recent study [Harold et al., 2006]. We genotyped the seven additional tagSNPs markers and TDT analysis revealed no significant evidence for association, however a weak trend was observed for rs12194307 (χ2 =2.000; P =0.157) and rs761100 (χ2 =2.032; P =0.154) (Table IV). A stepwise regression was conducted to identify a model containing markers that best explained variance contributed by affection status in our sample [Cordell et al., 2004]. The analysis was conducted using the seven tagSNPs, as well as four significant SNPs from the haplotypes of Francks et al. [2004] and Cope et al. [2005] genotyped in our sample. Of these 11 SNPs, rs4504469, rs12194307, rs12213672, rs6935076, rs2038137, and rs2143340 made up the most favorable model. These six markers were used to construct haplotypes that were tested for evidence of association with RD. One haplotype was modestly under transmitted to the affected subjects (χ2 =3.965; P =0.046) in our sample and a number of other low frequency haplotypes also showed evidence for biased transmission.
TABLE IV.
TDT Analysis of tagSNPs for the 22 kb LD block* Across KIAA0319 and TTRAP on 6p
| Name | Allele | Frequency | Transmitted | Un-transmitted | χ2 | P |
|---|---|---|---|---|---|---|
| rs12194307 | T | 0.85 | 42 | 30 | 2.000 | 0.157 |
| A | 0.15 | 30 | 42 | |||
| rs2179515 | T | 0.35 | 62 | 52 | 0.877 | 0.349 |
| C | 0.65 | 52 | 62 | |||
| rs12213672 | G | 0.05 | 13 | 9 | 0.727 | 0.394 |
| T | 0.95 | 9 | 13 | |||
| rs761100 | C | 0.41 | 71 | 55 | 2.032 | 0.154 |
| A | 0.59 | 55 | 17 | |||
| rs7766230 | A | 0.21 | 46 | 41 | 0.287 | 0.592 |
| G | 0.79 | 41 | 46 | |||
| rs9379677 | G | 0.77 | 44 | 43 | 0.011 | 0.917 |
| A | 0.23 | 43 | 44 | |||
| rs1047782 | T | 0.29 | 60 | 52 | 0.571 | 0.450 |
| G | 0.71 | 52 | 60 |
This block extends from a portion of intron 1, exon 1 (untranslated), and 5′ region with the acetylation peak in KIAA0319 and part of the 3′ UTR of TTRAP (Fig. 3).
DISCUSSION
In this study we investigated the association of five candidate genes in a narrowed region on chromosome 6p with RD. After investigating 37 markers across VMP (NRSN1), DCDC2, KIAA0319, TTRAP, and THEM2, the single marker analysis revealed evidence for association with polymorphisms in VMP (NRSN1), KIAA0319, and TTRAP. Of these, the marker rs6935076 in KIAA0319 was also associated in two recent analyses of RD [Cope et al., 2005; Luciano et al., 2007]. However, our finding was for the alternate allele of this marker than previously reported. No association was observed with markers in DCDC2 or a marker in THEM2.
Previous studies have found some evidence for association of VMP (NRSN1) with RD, however the results across studies showed stronger support for either DCDC2 or KIAA0319 [Francks et al., 2004; Cope et al., 2005; Meng et al., 2005; Harold et al., 2006; Schumacher et al., 2006]. Of the markers identified that were associated with VMP (NRSN1), there was no overlap across studies. Therefore, although markers in VMP (NRSN1) have shown some evidence for association with RD in previous studies, overall, the neighboring genes DCDC2 and KIAA0319 provided much stronger association, and focus converged on these two genes. As a result, VMP (NRSN1) was considered the less likely candidate for RD. However there is strong LD between VMP (NRSN1) and DCDC2 as well as punctate LD between the positive markers across the 6p region, including between VMP (NRSN1) and TTRAP in this sample (Fig. 2). This complex LD pattern complicates the determination of the risk gene as well as risk alleles within these genes.
The locus on 6p has been linked and associated with single-word reading, phonological decoding, spelling, and orthographic processing by previous studies [Grigorenko et al., 1997, 2000; Fisher et al., 1999; Gayan et al., 1999; Kaplan et al., 2002; Deffenbacher et al., 2004; Francks et al., 2004; Harold et al., 2006]. Association of spelling, single-word reading, and phonological decoding was found by Deffenbacher et al. [2004], Francks et al., [2004], and Harold et al. [2006] with markers in VMP (NRSN1), DCDC2, KIAA0319, TTRAP, and THEM2. In our sample, we observed some evidence for association of markers in KIAA0319 and VMP (NRSN1) with the spelling, decoding, and word identification phenotypes using quantitative analyses. Given the stronger evidence of association for some of these makers using a categorical definition, it was surprising that we did not see a more robust finding across markers and phenotypes for the quantitative analysis. Previous studies indicate that the 6p locus may be more significantly associated with a more severe form of RD [Deffenbacher et al., 2004; Francks et al., 2004; Bates et al., 2007]. However two recent studies using unselected samples identified association with a marker in KIAA0319 and TTRAP, as well as haplotypes of these markers, indicating that this locus also contributes to the spectrum of reading skills in the population [Luciano et al., 2007; Paracchini et al., 2008].
Haplotypes associated in previous studies [Francks et al., 2004; Cope et al., 2005; Luciano et al., 2007] were also investigated for association in our sample. Of the three sets of haplotypes tested, significant association was observed for haplotypes of two markers in KIAA0319 first associated with RD in a study from the UK [Cope et al., 2005]. The 1-2 (G-A) haplotype was significantly under-transmitted in our sample (Table III). This 1-2 haplotype showed marginally significant evidence for association in the study of Cope et al. [2005] in both their case/control and trio samples, however, it was over-transmitted in their trio sample and appeared more frequently in their cases, compared to their controls. Association of their under-transmitted haplotype 2-1 was much more significant in both their samples, which lead to the suggestion that this haplotype conferred a protective effect. Therefore, although under-transmitted haplotypes of rs4504469 and rs6935076 are significantly associated with RD in the sample from Cardiff and our sample from Toronto, they are made up of opposite alleles of these markers in each sample (2-1 in Cardiff and 1-2 in Toronto). The study of Luciano et al. [2007] did not find an association with haplotypes of these two markers. Instead, in their study, the haplotypes of Francks et al. [2004] (rs4504469–rs2038137–rs2143340) were significantly associated with the measures of reading tested. These haplotypes were not associated with RD in our sample. This disparity between studies is somewhat surprising given that the two sets of haplotypes overlap in both their location (Fig. 4) and their first component marker (rs4504469). One possible explanation is that this locus contains multiple risk alleles that could have occurred on different haplotype backgrounds that segregate differently in subsets of the European population. Our sample is made up of individuals of mixed European ancestry from a large urban center, and thus more ethnically diverse compared to the samples of Cope et al. [2005] and Francks et al. [2004] from the UK and the predominantly “Anglo-Celtic” sample of Luciano et al. [2007] from Brisbane, Australia. Ethnicity could contribute to differences in inter-marker LD in this region across samples resulting in differences in association results between the studies.
Evidence for association in both our sample and previous studies indicates KIAA0319 to be a strong candidate for RD. The 12% “risk” haplotype identified in the study of Francks et al. [2004] has been associated with lower levels of KIAA0319 transcripts in vitro, suggesting that the component markers, or yet unknown polymorphisms in LD with these markers, could affect expression of this gene [Paracchini et al., 2006]. This haplotype spans an approximately 70 kb region, containing a considerable amount of non-coding sequence in which causal variants for RD could be located. Our ChIP-chip assays identified a 2.7 kb H3ac peak (chr6: 24,752,922–24,755,657) in Y79 within this 70 kb region that included the proximal promoter, first untranslated exon, and part of intron 1, supporting this region of KIAA0319 as the possible location of genetic variation contributing to differential expression. Five markers previously associated with RD are situated within this acetylated region. Furthermore, this H3ac peak is located in a LD block with rs6935076, our most significant marker, and additional markers previously associated with RD in independent studies [Deffenbacher et al., 2004; Francks et al., 2004; Harold et al., 2006; Luciano et al., 2007]. Although haplotype analyses of six markers across this LD block only showed weak evidence for association with RD in our study (Table V), this region has been supported by more robust results from multiple studies, pointing to this region as a strong candidate for the location of RD risk alleles.
TABLE V.
Haplotype Analysis of Markers in the Narrowed Region on Chromosome 6p
| Marker 1 | Marker 2 | Marker 3 | Marker 4 | Marker 5 | Marker 6 | Frequency | Observed | Expected | Var (O-E) | χ2 (1 df) | P-value |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs4504469 | rs12194307 | rs12213672 | rs6935076 | rs2038137 | rs2143340 | ||||||
| A | T | T | G | T | A | 0.29 | 94.495 | 85.971 | 28.777 | 2.525 | 0.112 |
| G | T | T | A | G | A | 0.16 | 41.679 | 51.082 | 22.297 | 3.965 | 0.046 |
| G | T | T | G | G | G | 0.11 | 38.764 | 38.149 | 15.282 | 0.025 | |
| G | A | T | A | G | A | 0.11 | 29.852 | 31.420 | 9.514 | 0.258 | |
| G | T | T | G | G | A | 0.09 | 34.771 | 28.367 | 13.313 | 3.080 | |
| G | T | T | G | T | A | 0.05 | 13.748 | 15.117 | 7.294 | 0.257 | |
| A | T | T | A | G | A | 0.05 | 14.915 | 14.322 | 5.557 | 0.063 | |
| G | T | G | G | G | A | 0.04 | 13.000 | 11.000 | 6.015 | 0.665 | |
| A | A | T | A | G | A | 0.02 | 6.342 | 8.195 | 3.321 | 1.034 | |
| G | A | T | G | G | G | 0.01 | 0.573 | 3.286 | 1.520 | 4.846 | |
| G | A | T | G | G | A | 0.01 | 4.591 | 3.783 | 1.307 | 0.499 | |
| G | T | T | A | G | G | 0.01 | 0.000 | 1.059 | 0.508 | 2.207 | |
| A | T | T | G | G | G | 0.01 | 5.111 | 4.067 | 1.517 | 0.718 |
Global test 24 df χ2 = 35.467, P = 0.0618.
Global test grouping haplotypes with frequency <10%, 4 df χ2 =5.2071, P = 0.267 haplotypes of frequency <1% not shown.
The 2.7 kb H3ac region at the 5′ end of KIAA0319 is the only acetylated region within the 22 kb haplotype associated with reduced expression. A distinct H3ac site across exon 5 is 3.5 kb distal to the exon 4 marker rs4504469 that was associated in previous studies [Francks et al., 2004; Cope et al., 2005; Harold et al., 2006], and is the most distal marker included in the haplotype correlated with reduced expression [Paracchini et al., 2006]. However, aside from rs4504469 this exon 5 region has not received nearly as much support from previous studies as the 5′ region of KIAA0319. Previous screening of the KIAA0319 promoter region identified 12 DNA variants in the region and the 3 of those tested were associated with RD [Francks et al., 2004]. Our acetylation results now indicate that this region should be a focus of continued study for this gene. This 2.7 kb region likely regulates transcription of KIAA0319, and thus could contain causal variant(s) for RD that alter expression. This region is highly polymorphic and includes both microsatellites, including the marker JA04, and several insertion/deletion and SNP markers. Therefore a detailed screen of this known variation, along with a search for new variation is a logical step to identify causal variants for RD. Of note is that there may be more than one DNA change on a haplotype that changes gene function [Drysdale et al., 2000]. In addition, the change in function may require the combination of alleles to produce the phenotype because of the modularity of the transcription factor binding sites [Wasserman and Sandelin, 2004]. Parallel in vitro studies of the relationship of this variation to KIAA0319 function will also be needed. A limiting factor is that this gene is not expressed in lymphocytes [Paracchini et al., 2006], and can therefore not be studied directly in patient cell lines. However, promising work has been previously done in neuroblastoma cell lines [Paracchini et al., 2006], providing an effective alternative for initial functional studies.
Within DCDC2, the peak located in intron 7, is also the possible position of risk alleles. Although we found no evidence for association with RD and markers in DCDC2 in this study, the proximity of the remote site in intron 7 to the location of positive markers in several samples [Deffenbacher et al., 2004; Meng et al., 2005; Schumacher et al., 2006] points to this peak as a region of interest for future studies.
While our association studies also supported the gene for VMP (NRSN1), the only significant H3ac site was located in the promoter. The markers with evidence for association in our sample were located in introns 1 and 2, and the 3′ UTR of the gene. Thus far the coding region of this gene has not been screened for DNA changes in any study, thus this would be a priority for further study of VMP (NRSN1) followed by screening of the promoter.
In total, this study combined with previous studies provides evidence in support of KIAA0319 as the susceptibility locus for RD on chromosome 6p, although in this sample, other genes including VMP (NRSN1) as a candidate cannot be ruled out. Using ChIP-chip we have highlighted a 2.7 kb subsection within the 70 kb candidate regulatory region of KIAA0319 marked by the haplotype of Paracchini et al. [2006] and Francks et al. [2004]. This region will now be prioritized in screens for causal variant(s) for RD and directed in vitro studies.
Acknowledgments
We thank Jennifer Archibald for help with assessment of families. This work was supported by a Canadian Institute of Health Research grant (MOP-36358, CLB), a grant from the Krembil Foundation, and a graduate student fellowship from the Hospital for Sick Children Research Training Centre (JMC).
Footnotes
Additional Supporting Information may be found in the online version of this article.
References
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin—Rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30(1):97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Bates TC, Luciano M, Castles A, Coltheart M, Wright MJ, Martin NG. Replication of reported linkages for dyslexia and spelling and suggestive evidence for novel regions on chromosomes 4 and 17. Eur J Hum Genet. 2007;15(2):194–203. doi: 10.1038/sj.ejhg.5201739. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ, III, Gingeras TR, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120(2):169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Boffelli D, Nobrega MA, Rubin EM. Comparative genomics at the vertebrate extremes. Nat Rev Genet. 2004;5(6):456–465. doi: 10.1038/nrg1350. [DOI] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19(2):185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Boyle MH, Offord DR, Racine Y, Fleming JE, Szatmari P, Sanford M. Evaluation of the revised Ontario Child Health Study scales. J Child Psychol Psychiatry. 1993;34(2):189–213. doi: 10.1111/j.1469-7610.1993.tb00979.x. [DOI] [PubMed] [Google Scholar]
- Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, Furey TS, Crawford GE. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132(2):311–322. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardon LR, Smith SD, Fulker DW, Kimberling WJ, Pennington BF, DeFries JC. Quantitative trait locus for reading disability on chromosome 6. Science. 1994;266(5183):276–279. doi: 10.1126/science.7939663. [DOI] [PubMed] [Google Scholar]
- Clayton D, Jones H. Transmission/disequilibrium tests for extended marker haplotypes. Am J Hum Genet. 1999;65(4):1161–1169. doi: 10.1086/302566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conners CK. Conners’ Rating Scales—Revised. Toronto, Canada: Multi-Health Systems Inc; 1997. [Google Scholar]
- Cope N, Harold D, Hill G, Moskvina V, Stevenson J, Holmans P, Owen MJ, O’Donovan MC, Williams J. Strong evidence that KIAA0319 on chromosome 6p is a susceptibility gene for developmental dyslexia. Am J Hum Genet. 2005;76(4):581–591. doi: 10.1086/429131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordell HJ, Clayton DG. A unified stepwise regression procedure for evaluating the relative effects of polymorphisms within a gene using case/ control or family data: Application to HLA in type 1 diabetes. Am J Hum Genet. 2002;70(1):124–141. doi: 10.1086/338007. Epub 2001 Nov 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordell HJ, Barratt BJ, Clayton DG. Case/pseudocontrol analysis in genetic association studies: A unified framework for detection of genotype and haplotype associations, gene-gene and gene-environment interactions, and parent-of-origin effects. Genet Epidemiol. 2004;26(3):167–185. doi: 10.1002/gepi.10307. [DOI] [PubMed] [Google Scholar]
- Couto JM, Gomez L, Wigg K, Cate-Carter T, Archibald J, Anderson B, Tannock R, Kerr EN, Lovett MW, Humphries T, et al. The KIAA0319-like (KIAA0319L) gene on chromosome 1p34 as a candidate for reading disabilities. J Neurogenet. 2008;22(4):295–313. doi: 10.1080/01677060802354328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couto JM, Gomez L, Wigg K, Ickowicz A, Pathare T, Malone M, Kennedy JL, Schachar R, Barr CL. Association of attention-deficit/hyperactivity disorder with a candidate region for reading disabilities on chromosome 6p. Biol Psychiatry. 2009 doi: 10.1016/j.biopsych.2009.02.016. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kovel CG, Hol FA, Heister JG, Willemen JJ, Sandkuijl LA, Franke B, Padberg GW. Genomewide scan identifies susceptibility locus for dyslexia on Xq27 in an extended Dutch family. J Med Genet. 2004;41(9):652–657. doi: 10.1136/jmg.2003.012294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz X, Lois S, Sanchez-Molina S, Martinez-Balbas MA. Do protein motifs read the histone code? Bioessays. 2005;27(2):164–175. doi: 10.1002/bies.20176. [DOI] [PubMed] [Google Scholar]
- Deffenbacher KE, Kenyon JB, Hoover DM, Olson RK, Pennington BF, DeFries JC, Smith SD. Refinement of the 6p21.3 quantitative trait locus influencing dyslexia: Linkage and association analyses. Hum Genet. 2004;115(2):128–138. doi: 10.1007/s00439-004-1126-6. [DOI] [PubMed] [Google Scholar]
- DeFries JC, Gillis JJ. Genetics of reading disability. In: Plomin R, McClearn GE, editors. Nature, nurture, and psychology. Washington, D.C: American Psychological Association; 1993. pp. 121–145. [Google Scholar]
- Drysdale CM, McGraw DW, Stack CB, Stephens JC, Judson RS, Nandabalan K, Arnold K, Ruano G, Liggett SB. Complex promoter and coding region beta 2-adrenergic receptor haplotypes alter receptor expression and predict in vivo responsiveness. Proc Natl Acad Sci USA. 2000;97(19):10483–10488. doi: 10.1073/pnas.97.19.10483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberharter A, Becker PB. Histone acetylation: A switch between repressive and permissive chromatin. Second in review series on chromatin dynamics. EMBO Rep. 2002;3(3):224–229. doi: 10.1093/embo-reports/kvf053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagerheim T, Raeymaekers P, Tonnessen FE, Pedersen M, Tranebjaerg L, Lubs HA. A new gene (DYX3) for dyslexia is located on chromosome 2. J Med Genet. 1999;36(9):664–669. [PMC free article] [PubMed] [Google Scholar]
- Ferrere A, Vitalis T, Gingras H, Gaspar P, Cases O. Expression of Cux-1 and Cux-2 in the developing somatosensory cortex of normal and barrel-defective mice. Anat Rec A Discov Mol Cell Evol Biol. 2006;288(2):158–165. doi: 10.1002/ar.a.20284. [DOI] [PubMed] [Google Scholar]
- Fisher SE, Marlow AJ, Lamb J, Maestrini E, Williams DF, Richardson AJ, Weeks DE, Stein JF, Monaco AP. A quantitative-trait locus on chromosome 6p influences different aspects of developmental dyslexia. Am J Hum Genet. 1999;64(1):146–156. doi: 10.1086/302190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher SE, Francks C, Marlow AJ, MacPhie IL, Newbury DF, Cardon LR, Ishikawa-Brush Y, Richardson AJ, Talcott JB, Gayan J, et al. Independent genome-wide scans identify a chromosome 18 quantitative-trait locus influencing dyslexia. Nat Genet. 2002;30(1):86–91. doi: 10.1038/ng792. [DOI] [PubMed] [Google Scholar]
- Forsberg EC, Bresnick EH. Histone acetylation beyond promoters: Long-range acetylation patterns in the chromatin world. Bioessays. 2001;23(9):820–830. doi: 10.1002/bies.1117. [DOI] [PubMed] [Google Scholar]
- Francks C, Fisher SE, Olson RK, Pennington BF, Smith SD, DeFries JC, Monaco AP. Fine mapping of the chromosome 2p.12–16 dyslexia susceptibility locus: Quantitative association analysis and positional candidate genes SEMA4F and OTX1. Psychiatr Genet. 2002;12(1):35–41. doi: 10.1097/00041444-200203000-00005. [DOI] [PubMed] [Google Scholar]
- Francks C, Paracchini S, Smith SD, Richardson AJ, Scerri TS, Cardon LR, Marlow AJ, MacPhie IL, Walter J, Pennington BF, et al. A 77-kilobase region of chromosome 6p22.2 is associated with dyslexia in families from the United Kingdom and from the United States. Am J Hum Genet. 2004;75(6):1046–1058. doi: 10.1086/426404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, et al. The structure of haplotype blocks in the human genome. Science. 2002;296(5576):2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- Gayan J, Olson RK. Reading disability: Evidence for a genetic etiology. Eur Child Adolesc Psychiatry. 1999;8(Suppl3):52–55. doi: 10.1007/pl00010695. [DOI] [PubMed] [Google Scholar]
- Gayan J, Olson RK. Genetic and environmental influences on orthographic and phonological skills in children with reading disabilities. Dev Neuropsychol. 2001;20(2):483–507. doi: 10.1207/S15326942DN2002_3. [DOI] [PubMed] [Google Scholar]
- Gayan J, Smith SD, Cherny SS, Cardon LR, Fulker DW, Brower AM, Olson RK, Pennington BF, DeFries JC. Quantitative-trait locus for specific language and reading deficits on chromosome 6p. Am J Hum Genet. 1999;64(1):157–164. doi: 10.1086/302191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigorenko EL. Developmental dyslexia: An update on genes, brains, and environments. J Child Psychol Psychiatry. 2001;42(1):91–125. [PubMed] [Google Scholar]
- Grigorenko EL, Wood FB, Meyer MS, Hart LA, Speed WC, Shuster A, Pauls DL. Susceptibility loci for distinct components of developmental dyslexia on chromosomes 6 and 15. Am J Hum Genet. 1997;60(1):27–39. [PMC free article] [PubMed] [Google Scholar]
- Grigorenko EL, Wood FB, Meyer MS, Pauls DL. Chromosome 6p influences on different dyslexia-related cognitive processes: Further confirmation. Am J Hum Genet. 2000;66:715–723. doi: 10.1086/302755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigorenko EL, Wood FB, Meyer MS, Pauls JE, Hart LA, Pauls DL. Linkage studies suggest a possible locus for developmental dyslexia on chromosome 1p. Am J Med Genet. 2001;105(1):120–129. [PubMed] [Google Scholar]
- Grigorenko EL, Wood FB, Golovyan L, Meyer M, Romano C, Pauls D. Continuing the search for dyslexia genes on 6p. Am J Med Genet Part B. 2003;118B(1):89–98. doi: 10.1002/ajmg.b.10032. [DOI] [PubMed] [Google Scholar]
- Habib M. The neurological basis of developmental dyslexia: An overview and working hypothesis. Brain. 2000;123(Pt12):2373–2399. doi: 10.1093/brain/123.12.2373. [DOI] [PubMed] [Google Scholar]
- Harlaar N, Spinath FM, Dale PS, Plomin R. Genetic influences on early word recognition abilities and disabilities: A study of 7-year-old twins. J Child Psychol Psychiatry. 2005;46(4):373–384. doi: 10.1111/j.1469-7610.2004.00358.x. [DOI] [PubMed] [Google Scholar]
- Harold D, Paracchini S, Scerri T, Dennis M, Cope N, Hill G, Moskvina V, Walter J, Richardson AJ, Owen MJ, et al. Further evidence that the KIAA0319 gene confers susceptibility to developmental dyslexia. Mol Psychiatry. 2006;11(12):1085–1091. 1061. doi: 10.1038/sj.mp.4001904. [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39(3):311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Hsiung GY, Kaplan BJ, Petryshen TL, Lu S, Field LL. A dyslexia susceptibility locus (DYX7) linked to dopamine D4 receptor (DRD4) region on chromosome 11p15.5. Am J Med Genet Part B. 2004;125(1):112–119. doi: 10.1002/ajmg.b.20082. [DOI] [PubMed] [Google Scholar]
- Jacobson RH, Ladurner AG, King DS, Tjian R. Structure and function of a human TAFII250 double bromodomain module. Science. 2000;288(5470):1422–1425. doi: 10.1126/science.288.5470.1422. [DOI] [PubMed] [Google Scholar]
- Kadam S, Emerson BM. Mechanisms of chromatin assembly and transcription. Curr Opin Cell Biol. 2002;14(3):262–268. doi: 10.1016/s0955-0674(02)00330-7. [DOI] [PubMed] [Google Scholar]
- Kaminen N, Hannula-Jouppi K, Kestila M, Lahermo P, Muller K, Kaaranen M, Myllyluoma B, Voutilainen A, Lyytinen H, Nopola-Hemmi J, et al. A genome scan for developmental dyslexia confirms linkage to chromosome 2p11 and suggests a new locus on 7q32. J Med Genet. 2003;40(5):340–345. doi: 10.1136/jmg.40.5.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan DE, Gayan J, Ahn J, Won TW, Pauls D, Olson RK, DeFries JC, Wood F, Pennington BF, Page GP, et al. Evidence for linkage and association with reading disability on 6p21.3-22. Am J Hum Genet. 2002;70(5):1287–1298. doi: 10.1086/340449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinjan DA, van Heyningen V. Long-range control of gene expression: Emerging mechanisms and disruption in disease. Am J Hum Genet. 2005;76(1):8–32. doi: 10.1086/426833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurdistani SK, Tavazoie S, Grunstein M. Mapping global histone acetylation patterns to gene expression. Cell. 2004;117(6):721–733. doi: 10.1016/j.cell.2004.05.023. [DOI] [PubMed] [Google Scholar]
- Laird NM, Horvath S, Xu X. Implementing a unified approach to family-based tests of association. Genet Epidemiol. 2000;19(Suppl1):S36–S42. doi: 10.1002/1098-2272(2000)19:1+<::AID-GEPI6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Londin ER, Meng H, Gruen JR. A transcription map of the 6p22.3 reading disability locus identifying candidate genes. BMC Genomics. 2003;4(1):25. doi: 10.1186/1471-2164-4-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loots GG, Locksley RM, Blankespoor CM, Wang ZE, Miller W, Rubin EM, Frazer KA. Identification of a coordinate regulator of interleukins 4,13 and 5 by cross-species sequence comparisons. Science. 2000;288(5463):136–140. doi: 10.1126/science.288.5463.136. [DOI] [PubMed] [Google Scholar]
- Luca P, Laurin N, Misener VL, Wigg KG, Anderson B, Cate-Carter T, Tannock R, Humphries T, Lovett MW, Barr CL. Association of the dopamine receptor D1 gene, DRD1, with inattention symptoms in families selected for reading problems. Mol Psychiatry. 2007;12:776–785. doi: 10.1038/sj.mp.4001972. [DOI] [PubMed] [Google Scholar]
- Luciano M, Lind PA, Duffy DL, Castles A, Wright MJ, Montgomery GW, Martin NG, Bates TC. A haplotype spanning KIAA0319 and TTRAP is associated with normal variation in reading and spelling ability. Biol Psychiatry. 2007;26:26. doi: 10.1016/j.biopsych.2007.03.007. [DOI] [PubMed] [Google Scholar]
- Ludwig MZ, Bergman C, Patel NH, Kreitman M. Evidence for stabilizing selection in a eukaryotic enhancer element. Nature. 2000;403(6769):564–567. doi: 10.1038/35000615. [DOI] [PubMed] [Google Scholar]
- Lyon GR. Part I defining dyslexia, comorbidity, teachers’ knowledge of language and reading. Ann Dyslexia. 2003;53:1–14. [Google Scholar]
- Marlow AJ, Fisher SE, Francks C, MacPhie IL, Cherny SS, Richardson AJ, Talcott JB, Stein JF, Monaco AP, Cardon LR. Use of multivariate linkage analysis for dissection of a complex cognitive trait. Am J Hum Genet. 2003;72(3):561–570. doi: 10.1086/368201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng H, Smith SD, Hager K, Held M, Liu J, Olson RK, Pennington BF, DeFries JC, Gelernter J, O’Reilly-Pol T, et al. DCDC2 is associated with reading disability and modulates neuronal development in the brain. Proc Natl Acad Sci USA. 2005;102(47):17053–17058. doi: 10.1073/pnas.0508591102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16(3):1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris DW, Robinson L, Turic D, Duke M, Webb V, Milham C, Hopkin E, Pound K, Fernando S, Easton M, et al. Family-based association mapping provides evidence for a gene for reading disability on chromosome 15q. Hum Mol Genet. 2000;9(5):843–848. doi: 10.1093/hmg/9.5.843. [DOI] [PubMed] [Google Scholar]
- Ni Z, Bremner R. Brahma-related gene 1-dependent STAT3 recruitment at IL-6-inducible genes. J Immunol. 2007;178(1):345–351. doi: 10.4049/jimmunol.178.1.345. [DOI] [PubMed] [Google Scholar]
- Nieto M, Monuki ES, Tang H, Imitola J, Haubst N, Khoury SJ, Cunningham J, Gotz M, Walsh CA. Expression of Cux-1 and Cux-2 in the subventricular zone and upper layers II-IV of the cerebral cortex. J Comp Neurol. 2004;479(2):168–180. doi: 10.1002/cne.20322. [DOI] [PubMed] [Google Scholar]
- Nopola-Hemmi J, Myllyluoma B, Haltia T, Taipale M, Ollikainen V, Ahonen T, Voutilainen A, Kere J, Widen E. A dominant gene for developmental dyslexia on chromosome 3. J Med Genet. 2001;38(10):658–664. doi: 10.1136/jmg.38.10.658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson RK. Dyslexia: Nature and nurture. Dyslexia. 2002;8(3):143–159. doi: 10.1002/dys.228. [DOI] [PubMed] [Google Scholar]
- Olson R, Forsberg H, Wise B, Rack J. Measurement of word recognition, orthographic, and phonological skills. In: Lyon GR, editor. Frames of reference for the assessment of learning disabilities. Baltimore: Paul H. Brookes; 1994. pp. 243–277. [Google Scholar]
- Paracchini S, Thomas A, Castro S, Lai C, Paramasivam M, Wang Y, Keating BJ, Taylor JM, Hacking DF, Scerri T, et al. The chromosome 6p22 haplotype associated with dyslexia reduces the expression of KIAA0319, a novel gene involved in neuronal migration. Hum Mol Genet. 2006;15(10):1659–1666. doi: 10.1093/hmg/ddl089. Epub 2006 Apr 6. [DOI] [PubMed] [Google Scholar]
- Paracchini S, Steer CD, Buckingham LL, Morris AP, Ring S, Scerri T, Stein J, Pembrey ME, Ragoussis J, Golding J, et al. Association of the KIAA0319 dyslexia susceptibility gene with reading skills in the general population. Am J Psychiatry. 2008;165(12):1576–1584. doi: 10.1176/appi.ajp.2008.07121872. [DOI] [PubMed] [Google Scholar]
- Pennington BF. The genetics of dyslexia. J Child Psychol Psychiatry. 1990;31(2):193–201. doi: 10.1111/j.1469-7610.1990.tb01561.x. [DOI] [PubMed] [Google Scholar]
- Pennington BF. Using genetics to dissect cognition. Am J Hum Genet. 1997;60(1):13–16. [PMC free article] [PubMed] [Google Scholar]
- Petryshen TL, Kaplan BJ, Liu MF, Schmill de French N, Tobias R, Hughes ML, Field LL. Evidence for a susceptibility locus on chromosome 6q influencing phonological coding dyslexia. Am J Med Genet (Neuropsychiatr Genet) 2001;105:507–517. doi: 10.1002/ajmg.1475. [DOI] [PubMed] [Google Scholar]
- Petryshen TL, Kaplan BJ, Hughes ML, Tzenova J, Field LL. Supportive evidence for the DYX3 dyslexia susceptibility gene in Canadian families. J Med Genet. 2002;39(2):125–126. doi: 10.1136/jmg.39.2.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabin M, Wen XL, Hepburn M, Lubs HA, Feldman E, Duara R. Suggestive linkage of developmental dyslexia to chromosome 1p34-p36. Lancet. 1993;342(8864):178. doi: 10.1016/0140-6736(93)91384-x. [DOI] [PubMed] [Google Scholar]
- Roh TY, Cuddapah S, Zhao K. Active chromatin domains are defined by acetylation islands revealed by genome-wide mapping. Genes Dev. 2005;19(5):542–552. doi: 10.1101/gad.1272505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh TY, Wei G, Farrell CM, Zhao K. Genome-wide prediction of conserved and nonconserved enhancers by histone acetylation patterns. Genome Res. 2007;17(1):74–81. doi: 10.1101/gr.5767907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher J, Anthoni H, Dahdouh F, Konig IR, Hillmer AM, Kluck N, Manthey M, Plume E, Warnke A, Remschmidt H, et al. Strong genetic evidence of DCDC2 as a susceptibility gene for dyslexia. Am J Hum Genet. 2006;78(1):52–62. doi: 10.1086/498992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seigel GM, Hackam AS, Ganguly A, Mandell LM, Gonzalez-Fernandez F. Human embryonic and neuronal stem cell markers in retinoblastoma. Mol Vis. 2007;13:823–832. [PMC free article] [PubMed] [Google Scholar]
- Sham PC, Curtis D. An extended transmission/disequilibrium test (TDT) for multi-allele marker loci. Ann Hum Genet. 1995;59(Pt3):323–336. doi: 10.1111/j.1469-1809.1995.tb00751.x. [DOI] [PubMed] [Google Scholar]
- Smith SD, Kimberling WJ. Screening for multiple genes influencing dyslexia. In: Pennington B, editor. Reading disabilities: genetic and neurological influences. Boston: Kluwer Academic; 1991. pp. 97–111. [Google Scholar]
- Smith SD, Kimberling WJ, Pennington BF, Lubs HA. Specific reading disability: Identification of an inherited form through linkage analysis. Science. 1983;219(4590):1345–1347. doi: 10.1126/science.6828864. [DOI] [PubMed] [Google Scholar]
- Smith SD, Kimberling WJ, Pennington BF. Screening for multiple genes influencing dyslexia. Reading Writing: An Interdiscip J. 1991;3:285–298. [Google Scholar]
- Song JS, Johnson WE, Zhu X, Zhang X, Li W, Manrai AK, Liu JS, Chen R, Liu XS. Model-based analysis of two-color arrays (MA2C) Genome Biol. 2007;8(8):R178. doi: 10.1186/gb-2007-8-8-r178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein CM, Schick JH, Gerry TH, Shriberg LD, Millard C, Kundtz-Kluge A, Russo K, Minich N, Hansen A, Freebairn LA, et al. Pleiotropic effects of a chromosome 3 locus on speech-sound disorder and reading. Am J Hum Genet. 2004;74(2):283–297. doi: 10.1086/381562. Epub 2004 Jan 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannock R, Hum M, Masellis M, Humphries T, Schachar R. Teacher telephone interview for children’s academic performance, attention, behavior and learning: DSM-IV Version (TTI-IV) Toronto, Canada: The Hospital for Sick Children; 2002. unpublished document. [Google Scholar]
- The International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437(7063):1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torgesen JK, Wagner RK, Rashotte CA. TOWRE: Test of Word Reading Efficiency. Austin, Texas: Pro-Ed; 1999. [Google Scholar]
- Turic D, Robinson L, Duke M, Morris DW, Webb V, Hamshere M, Milham C, Hopkin E, Pound K, Fernando S, et al. Linkage disequilibrium mapping provides further evidence of a gene for reading disability on chromosome 6p21.3-22. Mol Psychiatry. 2003;8(2):176–185. doi: 10.1038/sj.mp.4001216. [DOI] [PubMed] [Google Scholar]
- Tzenova J, Kaplan BJ, Petryshen TL, Field LL. Confirmation of a dyslexia susceptibility locus on chromosome 1p34-p36 in a set of 100 Canadian families. Am J Med Genet Part B. 2004;127B(1):117–124. doi: 10.1002/ajmg.b.20139. [DOI] [PubMed] [Google Scholar]
- Wasserman WW, Sandelin A. Applied bioinformatics for the identification of regulatory elements. Nat Rev Genet. 2004;5(4):276–287. doi: 10.1038/nrg1315. [DOI] [PubMed] [Google Scholar]
- Wechsler DI. Examiner’s Manual: Wechsler Intelligence Scale for Children—3rd edition. New York, NY: Psychological Corporation; 1991. [Google Scholar]
- Weller EB, Weller RA, Fristad MA, Rooney MT, Schecter J. Children’s Interview for Psychiatric Syndromes (ChIPS) J Am Acad Child Adolesc Psychiatry. 2000;39(1):76–84. doi: 10.1097/00004583-200001000-00019. [DOI] [PubMed] [Google Scholar]
- Wigg KG, Couto JM, Feng Y, Anderson B, Cate-Carter TD, Macciardi F, Tannock R, Lovett MW, Humphries TW, Barr CL. Support for EKN1 as the susceptibility locus for dyslexia on 15q21. Mol Psychiatry. 2004;9(12):1111–1121. doi: 10.1038/sj.mp.4001543. [DOI] [PubMed] [Google Scholar]
- Wilkinson GS. Wide Range Achievement Test 3—Revision 3. Wilmington, DE: Jastak Associates; 1993. [Google Scholar]
- Woodcock RW. Woodcock Reading Mastery Tests—Revised. San Antonio, TX: Pearson Assessments; 1987. [Google Scholar]