Abstract
Here we report of summary of the characteristics of ‘Collinsella phocaeensis’ strain Marseille-P3245T sp. nov., ‘Clostridium merdae’ strain Marseille-P2953T, ‘Sutterella massiliensis’ strain Marseille-P2435T sp. nov., ‘Sutturella timonensis’ strain Marseille-P3282T sp. nov., ‘Enorma phocaeensis’ Marseille-P3242T sp. nov., ‘Mailhella massiliensis’ strain Marseille-P3199T gen. nov., sp. nov., ‘Mordavella massiliensis’ strain Marseille-P3246T sp. nov. and ‘Massiliprevotella massiliensis’ strain Marseille-P2439T sp. nov. isolated from fresh stool samples of healthy French patients.
Keywords: Culturomics, human gut microbiota, new bacteria, new species, taxonogenomics
In 2015, using the culturomics approach [1], we isolated eight bacteria which could not be identified by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) (Bruker Daltonics, Bremen, Germany) [1], [2]. These species were isolated from the stool samples of healthy French patients. All patients gave informed consent, and this study was approved by the ethics committee of the IFR48 Federative Research Institute under number 09-022. Because the identification of these eight isolates failed, we sequenced their 16S rRNA gene using fD1-rP2 primers as described previously by using a 3130-XL sequencer (Applied Biosciences, Saint Aubin, France) [3].
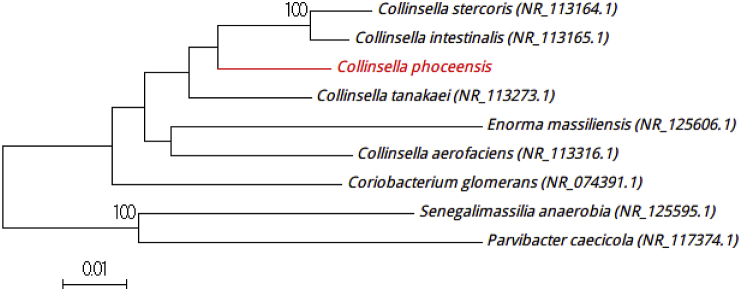
Strain Marseille-P3245T was isolated after 5 days of preincubation at 37°C in an anaerobic blood culture bottle (Becton-Dickinson Diagnostics, Le Pont-de-Claix, France) supplemented with 3 mL of rumen fluid filter-sterilized through a 0.2 μm pore filter (Thermo Fisher Scientific, Villebon-sur-Yvette, France) and 3 mL of sheep's blood (bioMérieux, Marcy l'Etoile, France). Then isolated colonies of the strain Marseille-P3245T were obtained by subculturing on 5% sheep's blood agar (bioMérieux) in anaerobic conditions generated by AnaeroGen (bioMérieux) after 72 hours of incubation. The colonies on sheep's blood agar were whitish with a diameter of 1 mm. Cells were Gram-negative bacilli, with a mean width of 2.5 μm and a length of 3.5 to 6.0 μm under electron microscopy. Bacterial cells were nonmotile, nonhaemolytic, non–endospore forming and obligately anaerobic. This strain did not exhibit catalase or oxidase activities. Strain Marseille-P3245T exhibited a 96.54% sequence similarity with Collinsella tanakaei strain JCM 16071 (NR_113273), the phylogenetically closest species with standing in nomenclature (Fig. 1). Collinsella tanakaei strain JCM 16071 is a Gram-negative bacillus isolated from human faeces in 2010 [4]. The 16S rRNA gene sequence similarity was <98.7% between strain Marseille-P3245T and its phylogenetically closest species with standing in nomenclature, which putatively classifies it as a new member of the Collinsella genus in the Actinobacteria phylum [5]. Thus, we propose the creation of the new species ‘Collinsella phocaeensis’ (pho.cae.en′sis, N.L. adj. fem., from Phocaea, the antic name of Phocea, the Greek city which founded Marseille, France, where the strain was isolated). Marseille-P3245T is the type strain of the species ‘Collinsella phocaeensis.’
Fig. 1.
Phylogenetic tree showing position of ‘Collinsella phocaeensis’ strain Marseille-P3245T (red) relative to other phylogenetically close neighbours. Sequences were aligned using CLUSTALW, and phylogenetic inferences obtained with Kimura-2 parameter models using maximum-likelihood method within MEGA software. Numbers at nodes are percentages of bootstrap values obtained by repeating analysis to generate majority consensus tree 1000 times. Only values >95% are displayed. Scale bar represents 1% nucleotide sequence divergence.
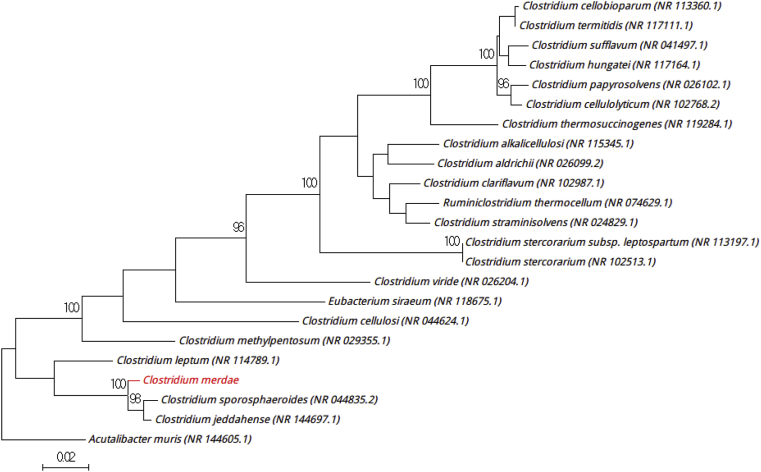
The growth of strain Marseille-P2953T was obtained after a 7-day incubation period in a 5% sheep's blood–enriched anaerobic blood culture bottle enriched with filter-sterilized rumen (Becton-Dickinson Diagnostics) at 37°C. Then subculture was performed on 5% sheep's blood–enriched Columbia agar (bioMérieux) at 37°C under anaerobic atmosphere generated by AnaeroGen (bioMérieux). The colonies were beige, translucent and nonhaemolytic with about 1 to 2 mm in diameter on blood-enriched agar (bioMérieux). Gram staining highlighted rod-shaped Gram-positive bacilli. It is an obligate anaerobic, motile and spore-forming, with a mean diameter and length of 0.5 μm and 3.0 μm respectively observed under electron microscopy. The cells of strain Marseille-P2953T were flagellate rods. Catalase and oxidase tests were negative. Strain Marseille-P2953T exhibited a 97.8% 16S rRNA gene sequence identity with Clostridium sporosphaeroides strain DSM 1294 (GenBank accession no. NZ_KB911066), the phylogenetically closest species with standing in nomenclature (Fig. 2), which putatively classifies it as a member within the family Clostridiaceae in the Firmicutes phylum. Clostridium sporosphaeroides DSM 1294 is a Gram-positive strict anaerobic and was isolated from the environment [6]. The 16S rRNA gene sequencing of strain Marseille-P2953T diverged by more than 2.3% from the other members of the genus Clostridium, which allowed us to delineate the bacterial genera without carrying out DNA-DNA hybridization [5]. From these results, we propose the creation of the new species, ‘Clostridium merdae’ sp. nov. (mer′dae, L. gen. adj. merdae, ‘of faeces,’ referring to the source where the strain Marseille-P2953T was isolated). Marseille-P2953T is the type strain of the new species ‘Clostridium merdae.’
Fig. 2.
Phylogenetic tree highlighting position of ‘Clostridium merdae’ strain Marseille-P2953T (red) relative to other phylogenetically close members of family Clostridiaceae. Alignment and phylogenetic inferences were performed as described in Fig. 1. Scale bar represents 2% nucleotide sequence divergence.
The initial growth of strain Marseille-P2435T was obtained after a 3-day incubation period in a 5% sheep's blood– and filter-sterilized rumen-enriched anaerobic blood culture bottle (Becton-Dickinson Diagnostics) at 37°C. Strain Marseille-P2435T was isolated by subculture in a 5% sheep's blood–enriched Columbia agar (bioMérieux) at 37°C under strict anaerobic conditions generated by AnaeroGen (bioMérieux). Strain Marseille-P2435T is a Gram-negative bacillus, non–spore forming and nonmotile (4.0 × 4–10 μm). It presents neither catalase nor oxidase activities. On the agar plates, the colonies were slightly translucent, greyish and about 0.5 to 1 mm in diameter after 3 days of culture on 5% sheep's blood agar. The strain Marseille-P2435T had a 16S rRNA gene sequence identity of 97.94% with Sutterella stercoricanis strain 5BAC4 (NR_025600), the phylogenetically closest species with standing in nomenclature (Fig. 3), which potentially classifies it as a member of a new genus within the family Sutterellaceae in the Proteobacteria phylum. Sutterella stercoricanis strain 5BAC4 is a Gram-negative, rod-shaped bacterium which was isolated from canine faeces [7]. Strain Marseille-P2435T shows a 16S rRNA gene sequence divergence of >1.3% with the phylogenetically closest species with a validly published name and standing in nomenclature [5]. Therefore, we propose the creation of the new species ‘Sutterella massiliensis’ (ma.ssi.li.en′sis, N.L. adj. fem., from Massili, the antic name of Marseille, France, where the strain was isolated). Marseille-P2435T is the type strain of the species ‘Sutterella massiliensis.’
Fig. 3.
Phylogenetic tree showing positions of ‘Sutterella massiliensis’ strain Marseille-P2435T and ‘Sutturella timonensis’ Marseille-P3282T (red) relative to other phylogenetically close neighbours. Alignment and phylogenetic inferences were performed as described in Fig. 1. Scale bar represents 1% nucleotide sequence divergence.
Strain Marseille-P3282T was isolated under the same conditions as ‘Sutterella massiliensis’ strain Marseille-P2435T. After 72 hours of incubation, the colonies were light grey, nonhaemolytic and nontranslucent, and exhibited a diameter of 1 to 2 mm on 5% blood-enriched agar (bioMérieux). Cells from this strain were Gram-negative bacilli, and were mobile and non–spore forming. Under electron microscopy, bacterial cells exhibited a diameter of 2.5 to 3.0 μm and a length of 3.7 to 6.0 μm. Catalase and oxidase activities were negative. The strain Marseille-P3282T had a 16S rRNA gene sequence identity of 95.34% with Sutterella stercoricanis strain 5BAC4 (NR_025600), the phylogenetically closest species with standing in nomenclature (Fig. 3). This divergence of the 16S rRNA gene sequence >1.3% with its phylogenetically closest species with a validly published name standing in nomenclature classifies Marseille-P3282 as a second new Sutterella species in the Sutterellaceae family [5]. Consequently, we propose the creation of the new species ‘Sutterella timonensis’ (ti.mo.nen′sis, N.L. adj. fem., from Timone, the name of the main hospital in Marseille, France, where the strain was isolated). Marseille-P3282T is the type strain of the species ‘Sutterella timonensis.’
Strain Marseille-P3242T was isolated by directly seeding the fresh stool on 5% sheep's blood–enriched Columbia agar under anaerobic conditions generated by AnaeroGen after 72 hours of incubation at 37°C. Strain Marseille-P3242T presented light nonhemolytic grey colonies, with a diameter of 0.5 to 1 mm on agar plates. The bacteria appeared as bacilli arranged in pairs or long chains with a length and diameter between 2.0 to 3.0 μm and 5.0 to 6.5 μm respectively under electron microscopy. Strain Marseille-P3242T is an obligately anaerobic bacterium that is nonmotile, non–spore forming and Gram-negative. Cells are negative for oxidase and catalase. The strain Marseille-P3242T had a 16S rRNA sequence identity of 96.17% with ‘Enorma massiliensis’ strain ph1 (NR_125606), the phylogenetically closest species with standing in nomenclature (Fig. 4). ‘Enorma massiliensis’ strain phI is a Gram-positive bacillus, obligately anaerobic and nonmotile, and was isolated from the human faecal flora [8]. Given the 96.17% similarity level of the 16S r RNA gene sequence of strain Marseille-P3242T with ‘Enorma massiliensis’ of <98.7%, we propose that it may be a representative strain of a new species within the Enorma genus in the Actinobacteria phylum [5]. Thus, we propose the creation of the new species ‘Enorma phocaeensis’ (pho.cae.en′sis, N.L. adj. fem., from Phocaea, the antic name of Phocea, the Greek city which founded Marseille, France, where the strain was isolated). Marseille-P3242T is the type strain of the species ‘Enorma phocaeensis.’
Fig. 4.
Phylogenetic tree showing position of Enorma phocaeensis Marseille-P3242T (red) relative to other phylogenetically close neighbours. Alignment and phylogenetic inferences were performed as described in Fig. 1. Scale bar represents 1% nucleotide sequence divergence.
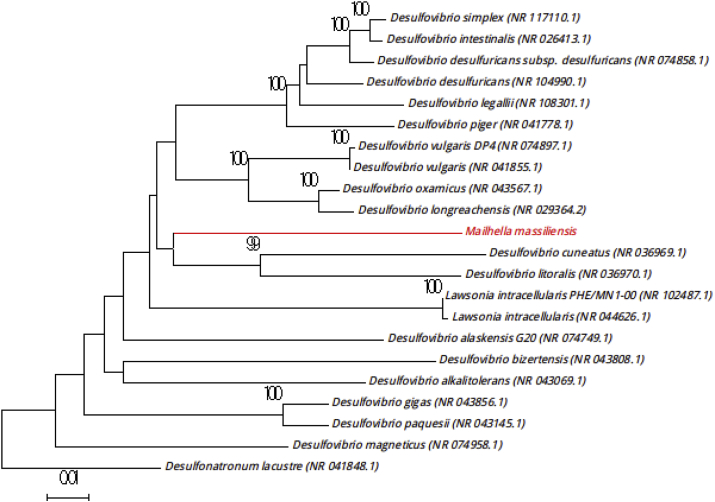
The first isolation of the strain Marseille-P3199T occurred after direct inoculation of fresh stool on 5% sheep's blood–enriched Columbia agar, without enrichment, into blood bottles. Surface colonies on sheep's blood agar after 72 hours of incubation under anaerobic atmosphere were circular, translucent and grayish, and approximately 1 mm in diameter. Strain Marseille-P3199T is a Gram-negative bacillus with a mean diameter and length of 3.5 μm and 15 to 30 μm respectively. It is strictly anaerobic, motile and non–spore forming. Cells were negative for catalase and oxidase. The test of sulfate reducing was positive. Furthermore, the strain Marseille-P3199T had a 16S rRNA gene sequence identity of 89.13% with Desulfovibrio oxamicus strain DSM 1925 (NR_043567) [9], the phylogenetically closest species with standing in nomenclature (Fig. 5). A similarity of 16S rRNA gene sequence <95% between our strain and the phylogenetically closest species with standing in nomenclature lead us to putatively classify Marseille-P3199 as a new member of the Desulfovibrionaceae family in the Proteobacteria phylum [5]. Thus, we propose the creation of the new genus Mailhella (mail.hel′la, L. gen. fem., from Mailhe, for the infectious disease resident Morgane Mailhe, who works in Timone Hospital, Marseille, France). Marseille-P3199T is the type strain of the species ‘Mailhella massiliensis’ (ma.ssi.li.en′sis, N.L. adj. fem., from Massili, the antic name of Marseille, France, where the strain was isolated).
Fig. 5.
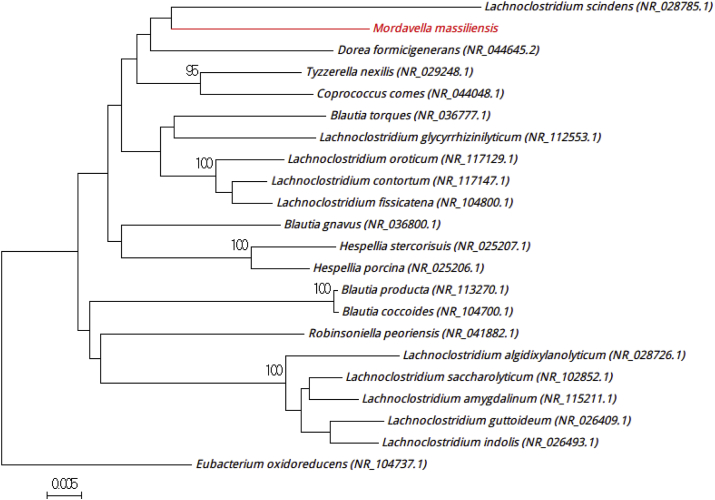
Phylogenetic tree showing position of ‘Mordavella massiliensis’ strain Marseille-P3246T (red) relative to other phylogenetically close neighbours. Alignment and phylogenetic inferences were performed as described in Fig. 1. Scale bar represents 0.5% nucleotide sequence divergence.
Strain Marseille- P3246T was isolated under the same conditions as ‘Mordavella massiliensis’ strain Marseille-P3199T, from fresh stools seeded within 5 minutes after specimen collection. After 72 hours of incubation on petri dishes of 5% sheep's blood–enriched Columbia agar, the colonies were whitish, circular and nonhaemolytic, with an average diameter of 0.5 mm. Gram staining showed Gram-negative colonies, occurring singly or in long chains of rods. Observed under the electron microscope, the cells presented a mean diameter and length of 2 μm and 6 to 12 μm respectively. Catalase and oxidase activities were negative. Strain Marseille-P3246T exhibited a 16S rRNA gene sequence identity of 94.27% with Clostridium oroticum strain DSM 1287 (NR_117130), the phylogenetically closest species with standing in nomenclature (Fig. 6). Clostridium oroticum strain DSM 1287 is a Gram-positive rod-shaped bacteria that is anaerobic and nonmotile, as described by Cato et al. [10]. This phylogenetic analysis, based on 16S rRNA, showed that strain Marseille-P3246 presented a similarity <98.7% with the other phylogenetically closest species, which lead us to putatively classify Marseille-P3246 as a new genus of the Lachnospiraceae family in the Firmicutes phylum [5]. The name proposed for this new genus is Mordavella (mor.da.ve′lla, L. gen. fem., from Mordav, for the infectious diseases residents Morgane Mailhe and Davide Ricaboni, who worked in Timone Hospital, Marseille, France). Marseille-P3246T is the type strain of the species ‘Mordavella massiliensis’ (ma.ssi.li.en′sis, N.L. adj. fem., from Massili, the antic name of Marseille, France, where the strain was isolated).
Fig. 6.
Phylogenetic tree showing position of ‘Mailhella massiliensis’ strain Marseille-P3199T (red) relative to other phylogenetically close neighbours. Alignment and phylogenetic inferences were performed as described in Fig. 1. Scale bar represents 1% nucleotide sequence divergence.
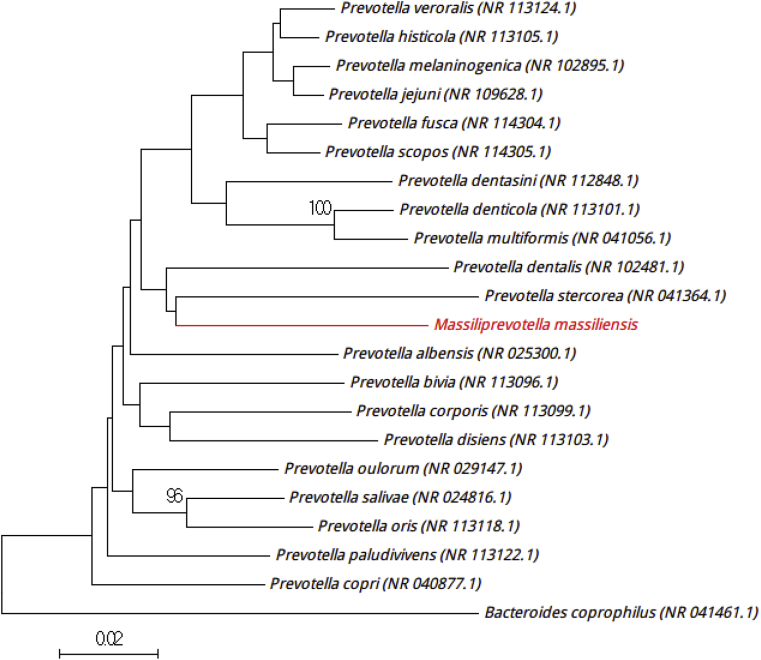
‘Massiliprevotella massiliensis’ strain Marseille-P2439T was isolated by culture on 5% sheep's blood agar (bioMérieux) under anaerobic conditions after 48 hours, preceding a preincubation of the stool specimens in a blood culture bottle, with an addition of 5 mL of sheep rumen for 7 days. Surface colonies on sheep's blood agar were beige, circular and transparent, 1 to 1.5 mm in diameter. Strain Marseille-P2439T is a Gram-negative bacillus, motile and non–spore forming. The catalase and oxidase tests were negative. Observed under electron microscopy, the bacterial cells were polymorphic. Strain Marseille-P2439T (LT223577) showed a 16S rRNA sequence identity of 91% with Prevotella fusca strain JCM17724 (NZ_CP012074), the phylogenetically closest species with standing in nomenclature (Fig. 7), which putatively classifies it as a member of a new genus within the family Prevotellaceae in the Firmicutes phylum. Prevotella fusca strain JCM17724 is a Gram-negative bacillus, obligately anaerobic and nonmotile, and was isolated from the human oral cavity [11]. Strain Marseille-P2439T exhibits a 16S rRNA gene sequence divergence >5% with its phylogenetically closest species with a validly published name with standing in nomenclature [5]. Consequently, we formally propose the creation of the new genus ‘Massiliprevotella’ (by similarity with the Prevotella genus), and strain Marseille-P2439T is the type strain of the new species ‘Massiliprevotella massiliensis’ gen. nov., sp. nov. (ma.si.li.e′n.sis, L. gen. masc. n., from massiliensis, ‘of Massilia,’ the Latin name of Marseille where the strain was first isolated).
Fig. 7.
Phylogenetic tree showing position of ‘Massiliprevotella massiliensis’ strain Marseille- P2439T (red) relative to other phylogenetically close members of family Prevotellaceae. Scale bar represents 2% nucleotide sequence divergence.
MALDI-TOF MS spectra
The MALDI-TOF MS spectra of these species are available at online (http://mediterranee-infection.com/article.php?laref=256&titre=urms-database).
Nucleotide sequence accession number
The 16s r RNA gene sequence was deposited in GenBank under the following accession numbers: ‘Collinsella phocaeensis’ strain Marseille-P3245T (LT598576), ‘Clostridium merdae’ strain Marseille-P2953T (LT732642), ‘Sutterella massiliensis’ strain Marseille-P2435T (LT223579), ‘Sutturella timonensis’ Marseille-P3282T (LT623892), ‘Enorma phocaeensis’ Marseille-P3242T (LT598579), ‘Mailhella massiliensis’ strain Marseille-P3199T (LT598584), ‘Mordavella massiliensis’ strain Marseille-P3246T (LT615363) and ‘Massiliprevotella massiliensis’ strain Marseille-P2439T (LT223577).
Deposit in a culture collection
The strains were deposited in the Collection de Souches de l’Unité des Rickettsies (CSUR, WDCM 875) under numbers P3245 (‘Collinsella phocaeensis’ strain Marseille-P3245T), P2953 (Clostridium merdae’ strain Marseille-P2953), P2435 (‘Sutterella massiliensis’ strain Marseille-P2435T), P3282 (‘Sutturella timonensis’ Marseille-P3282T), P3242 (‘Enorma phocaeensis’ Marseille-P3242T), P3246 ‘Mordavella massiliensis’ strain Marseille-P3246T), P3199 (‘Mailhella massiliensis’ strain Marseille-P3199T) and P2439 (‘Massiliprevotella massiliensis’ strain Marseille-P2439T).
Acknowledgement
This study was funded by the Fondation Méditerranée Infection.
Conflict of Interest
None declared.
References
- 1.Lagier J.C., Hugon P., Khelaifia S., Fournier P.E., La Scola B., Raoult D. The rebirth of culture in microbiology through the example of culturomics to study human gut microbiota. Clin Microbiol Rev. 2015;28:237–264. doi: 10.1128/CMR.00014-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lagier J.C., Khelaifia S., Alou M.T., Ndongo S., Dione N., Hugon P. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat Microbiol. 2016;1:16–203. doi: 10.1038/nmicrobiol.2016.203. [DOI] [PubMed] [Google Scholar]
- 3.Drancourt M., Bollet C., Carlioz A., Martelin R., Gayral J.P., Raoult D. 16S ribosomal DNA sequence analysis of a large collection of environmental and clinical unidentifiable bacterial isolates. J Clin Microbiol. 2000;38:3623–3630. doi: 10.1128/jcm.38.10.3623-3630.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nagai F., Watanabe Y., Morotomi M. Slackia piriformis sp. nov. and Collinsella tanakaei sp. nov., new members of the family Coriobacteriaceae, isolated from human faeces. Int J Syst Evol Microbiol. 2010;60:2639–2646. doi: 10.1099/ijs.0.017533-0. [DOI] [PubMed] [Google Scholar]
- 5.Kim M., Oh H.S., Park S.C., Chun J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol. 2014;64:346–351. doi: 10.1099/ijs.0.059774-0. [DOI] [PubMed] [Google Scholar]
- 6.Logan N.A., De Vos P., Genus I. Clostiridum Prazmowski 1880. In: De Vos P., Garrity D., Jones D., Krieg N.R., Ludwig W., Rainey F.A., editors. Bergey’s manual of systematic bacteriology. vol. 3. the Firmicutes; New York: Springer: 2009. pp. 738–828. [Google Scholar]
- 7.Greetham H.L., Collins M.D., Gibson G.R., Giffard C., Falsen E., Lawson P.A. Sutterella stercoricanis sp. nov., isolated from canine faeces. Int J Syst Evol Microbiol. 2004;54:1581–1584. doi: 10.1099/ijs.0.63098-0. [DOI] [PubMed] [Google Scholar]
- 8.Mishra A.K., Hugon P., Lagier J.C., Nguyen T.T., Couderc C., Raoult D. Non contiguous-finished genome sequence and description of Enorma massiliensis gen. nov., sp. nov., a new member of the family Coriobacteriaceae. Stand Genomic Sci. 2013;8:290–305. doi: 10.4056/sigs.3426906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.López-Cortés A., Fardeau M.L., Fauque G., Joulian C., Ollivier B. Reclassification of the sulfate- and nitrate-reducing bacterium Desulfovibrio vulgaris subsp. oxamicus as Desulfovibrio oxamicus sp. nov., comb. nov. Int J Syst Evol Microbiol. 2006;56:1495–1499. doi: 10.1099/ijs.0.64074-0. [DOI] [PubMed] [Google Scholar]
- 10.Cato E.P., Moore W.E.C., Holdeman L.V. Clostridium oroticum comb. nov. amended description. Int J Syst Evol Microbiol. 1968;18:9–13. [Google Scholar]
- 11.Downes J., Wade W.G. Prevotella fusca sp. nov. and Prevotella scopos sp. nov., isolated from the human oral cavity. Int J Syst Evol Microbiol. 2011;61(pt 4):854–858. doi: 10.1099/ijs.0.023861-0. [DOI] [PubMed] [Google Scholar]