Abstract
Human papillomaviruses (HPVs) target PML nuclear bodies (NBs) during infectious entry and PML protein is important for efficient transcription of incoming viral genome. However, the transcriptional down regulation was shown to be promoter-independent in that heterologous promoters delivered by papillomavirus particles were also affected. To further investigate the role of PML protein in HPV entry, we used shRNA to knockdown PML protein in HaCaT keratinocytes. Confirming previous findings, PML knockdown in HaCaT cells reduced HPV16 transcript levels significantly following infectious entry without impairing binding and trafficking. However, when we quantified steady-state levels of pseudogenomes in interphase cells, we found strongly reduced genome levels compared to parental HaCaT cells. Since nuclear delivery was comparable in both cell lines, we conclude that viral pseudogenome must be removed after successful nuclear delivery. Transcriptome analysis by gene array revealed that PML knockdown in clonal HaCaT cells was associated with a constitutive interferon (IFN) response. Abrogation of JAK1/2 signaling prevented genome loss, however, did not restore viral transcription. In contrast, knockdown of PML protein in HeLa cells did not affect HPV genome delivery and transcription. HeLa cells are transformed by HPV18 oncogenes E6 and E7, which have been shown to interfere with the JAK/Stat signaling pathway. Our data imply that PML NBs protect incoming HPV genomes. Furthermore, they provide evidence that PML NBs are key regulators of the innate immune response in keratinocytes.
INTRODUCTION
Promyelocytic leukemia (PML) nuclear bodies (NBs) are distinct subnuclear structures that are variable in number and size and have been implicated in a variety of cellular processes including: transcriptional regulation, growth suppression, innate immune response, and apoptosis (1). PML protein is the main structural component of PML NBs (2). Seven isoforms of PML protein have been identified, all of which can reside in PML NBs with the exception of PMLVII, which lacks a nuclear localization signal (3, 4). A cell in which PML has been knocked out fails to assemble these structures (2). In addition to other proteins permanently residing in PML NBs, such as Sp100 and Daxx, many proteins have been shown to transiently localize to these bodies (5, 6). PML NBs are highly dynamic and vary during cell cycle progression (7-9). They disassemble during mitosis to form large cytoplasmic aggregates and reassemble after mitosis and nuclear envelope reformation have been completed (10). Despite many attempts, the exact cellular function of PML NBs has not been uncovered yet. Because of the transient nature of protein association with PML NBs, they were suggested to serve as intracellular storage compartment for excess protein that allows quick access if needed (5). In support of this, post-translational modifications, such as SUMOylation, regulate association of proteins with PML NBs (11, 12).
Many DNA viruses, including members of the herpesviridae family, target PML NBs during primary infection and induce a reorganization of these subnuclear structures (13-20). To achieve reorganization, these viruses encode proteins, either expressed as immediate early proteins and/or delivered to host cells as a virion component, that target specific PML isoforms for degradation. Failure to induce reorganization of PML NBs prevents efficient establishment of infection (16, 21-27). Conversely, knockdown of PML alleviates the requirement for the PML-targeting viral proteins (for review see ref. (28). Evidence has been presented that supports an antiviral function for PML NBs, specifically PML protein, and suggests that the reorganization allows the virus to escape this innate immune response (for review see ref. (3).
While PML NBs have been shown to restrict viral infection for most viruses unless they are reorganized, papillomaviruses (PVs), including bovine papillomavirus 1 (BPV1) and human papillomavirus 16 (HPV16) and HPV18, require PML protein for efficient establishment of infection (29, 30). PVs transiently target PML NBs during infectious entry. Prior to accumulating at PML NBs, HPV16 attaches to host cells by interaction of the major capsid protein, L1, with basement membrane- and cell surface-resident receptors (31-37). The interaction with heparan sulfate moieties induces conformational changes affecting both capsid proteins (34, 36, 38). Conformational changes reduce the affinity to the primary receptor, which in turn allows transfer to less-well defined uptake receptors (39-43), endocytosis (44-46), and uncoating in acidified endocytic vesicles (45, 47-49). The viral genome in association with the minor capsid protein, L2, is rescued from lysosomal degradation by retromer complexes. Retromer complexes mediate trafficking of the L2/genome complex to the trans-Golgi network (TGN) (50-52). The L2 protein likely mediates the interaction with the transport machinery, since it partially penetrates the endocytic membrane following uncoating (53-55). The L2/genome complex gains access to the nuclei during mitosis, requiring nuclear envelope breakdown rather than active nuclear import via nuclear pores (56, 57). During these processes, the viral genome is still present in membrane bound transport vesicles and requires microtubules for transport (58). The L2/DNA complex associates with PML NBs after reformation of the nuclei (29). HPV16 L2 protein can be SUMOylated and harbors a sumo interaction motif (SIM), suggesting that L2 protein may mediate association with PML NBs (59, 60). This is supported by the observation that L2 over-expression in mammalian cells results in its accumulation at PML NBs (61-63). Furthermore, L2 protein may transiently localize to PML NBs in differentiated keratinocytes of naturally infected tissue (64).
Triggered by our recent finding that Epstein Barr Virus (EBV)-harboring epithelial cells, which have lower numbers of PML NBs compared to parental cells (65, 66) and displayed reduced levels of HPV16 genome following infectious entry, we revisited the role of PML NBs in HPV16 infection using knockdown approaches. Herein, we report that knockdown of PML protein in HaCaT keratinocytes reduces nuclear genome levels following infectious entry, probably due to degradation after successful delivery. This points to a protective role of PML NBs in HPV16 infection. We also observed that PML knockdown constitutively activated an interferon (IFN) response.
MATERIALS AND METHODS
Cell lines
293TT and HeLa cells were cultured in DMEM supplemented with 10% FBS, non-essential amino acids, antibiotics, and L-Glutamax. HaCaT cells were grown in low glucose DMEM containing 5% FBS and antibiotics. AGS-R and AGS-R-BX (infected with EBV) cells, a kind gift of Lindsey Hutt-Fletcher (Shreveport, USA), were propagated in F12 medium supplemented with 10% FBS, antibiotics, and L-Glutamax. AGS-R-BX cells were grown under Neomycin selection to retain the EBV genome. Lentivirus-transduced cells were maintained with continuous antibiotic selection, as appropriate.
Antibodies and reagents
Antibodies used for the study were as follows: PML (BETHYL; catalog number (#) A301-167A), lamin A/C mouse (Sigma; #SAB4200263), Nup153 (Covance; #MMS-102P), pStat1 and Stat1 (Cell Signaling; #9167S and 9176S, respectively), and β-actin (Santa Cruz; #sc-47778). L1-specific mouse monoclonal antibody 33L1-7 and rabbit polyclonal K-75 were described previously (42, 67). Click-iT® EdU Imaging Kit (Molecular Probes; C10338), AlexaFluor (AF)-labeled secondary antibodies (Life Technologies; #A11029, A11034, A21236, and A21245) and phalloidin were purchased from Life Technologies. Peroxidase-conjugated AffiniPure goat anti-mouse and anti-rabbit antibodies were purchased from Jackson ImmunoResearch.
Generation of HPV16 pseudo- and quasivirions
The pSheLL16 L1/L2 packaging plasmid and pfwB plasmid, expressing enhanced green fluorescent protein (GFP) were a kind gift from John Schiller, Bethesda, MA. pCMV-LoxP-HPV16-LoxP-eGFP plasmid and pBCre plasmid were provided by Jason Bodily (Shreveport, USA) (68). Quasivirions were generated using 293TT cells following the improved protocol of Buck and Thompson (69) with minor modifications. Briefly, 293TT cells were first cotransfected with pSheLL16 L1/L2 packaging plasmid and pCMV-LoxP-HPV16-LoxP-eGFP plasmid and 24 hours later transfected with pBCre plasmid. Two days later, cells were harvested and viral particles were purified as described previously (69, 70). Because activity of the cre recombinase generates two circular plasmids of packable size (pEGFPN1 and HPV16 genome), isolated virions comprise a mixture of pseudovirions (pEGFPN1 plasmid) and quasivirions (HPV16 genome). Pseudovirions harboring GFP were also generated in 293TT cells as described by Buck et al. (69, 70). For pseudogenome detection by fluorescence microscopy, pseudogenomes were labeled with EdU (5-ethynyl-2’-deoxyuridine) by supplementing the growth medium with 100 μM EdU at 6 hours post transfection as described (71) during generation of pseudovirions. To determine the viral genome equivalent (vge) by real time quantitative PCR (RT-qPCR), encapsidated DNA was isolated using the NucleoSpin® Blood QuickPure (Macherey-Nagel; #740569.250). To determine the level of cellular DNA contamination in our quasivirion preparations, sequencing libraries were prepared with the Illumina Nextera XT kit from 2 independent isolations of encapsidated viral DNA. Paired-end sequencing (2×300 cycles) was performed on an Illumina MiSeq. 508,500 and 611,402 sequencing reads were obtained, respectively. Reads were aligned to the human hg19 reference genome using the Illumina MiSeq Reporter v2.5.1.
Lentivirus transduction
Lentivirus vector plasmid expressing anti-PML (pLKO-shPML1) was a kind gift from Dr. Roger Everett, MRC University of Glasgow Centre for Virus Research (72). Lentivirus supernatants were prepared after co-transfection into HEK-293TT cells of a lentivirus vector plasmid with pMD2.G and psPAX2. HaCaT or HeLa cells were transduced with lentiviruses expressing shRNAs directed against PML (shPML) or scrambled control (ctrl). Stable cell lines were then selected with puromycin (10 μg/ml)(Calbiochem; #540222). A PML-deficient single-cell clone was established and used for most experiments. However, similar results were achieved using the bulk population prior to subcloning. PML knockdown was confirmed by immunofluorescent staining and Western Blot.
Immunofluorescence staining and microscopy
Cells were grown on cover slips at approximately 50% confluency and infected with HPV16 pseudovirus. EdU staining was performed according to the manufacturer's directions. In brief, at the indicated times post infection, cells were washed with PBS and fixed with 4% paraformaldehyde for 15 min at room temperature, washed, permeabilized with 0.5% Triton X-100 in PBS for 10 min, washed, and blocked with 5% goat serum in PBS for 30 min followed by a 30 min incubation with Click-iT® reaction cocktail containing AlexaFluor 555 for EdU-labeled pesudogenome detection. After extensive washing, cells were incubated for 30 min with primary antibodies at room temperature, washed again extensively, and subsequently incubated with AlexaFluor-tagged secondary antibodies for 30 min. After extensive washing with PBS, cells were mounted in ‘Gold Antifade’ containing DAPI (Life Technologies; #P3693). All IF images were captured by confocal microscopy with a 63x objective using a Leica TCS SP5 Spectral Confocal Microscope and processed with Adobe Photoshop software. Approximately 5-10×105 viral genome equivalents per cover slip were used. Number of viral pseudogenome in the nuclei was quantified in z-stacks spanning the whole nucleus, unless mentioned. Results are expressed as a number of EdU or an average percent of EdU-labeled viral pseudogenome in the nucleus of control cells ± SEM.
Cell-binding assay
For the binding analysis, cells were incubated with equal amounts of PsV for one hour at 37°C. Unbound particles were removed by several washes with PBS. Cells were then fixed and stained with L1-specific polyclonal rabbit antibody K-75 as described above without EdU detection. Assays were quantified and expressed as the pixel sum ratio of the L1 to ROI.
Infection in presence of drugs and immunofluorescence
HaCaT cells were grown on cover slips at approximately 50% confluency and infected with HPV16 pseudovirus in the presence of 0.4 μM INBC (Ruxolitinib; Cayman Chemical; #11609). At 30 hpi, samples were fixed with 4% paraformaldehyde and stained as described above for EdU, PML, lamin A/C, and DAPI using primary and appropriate AF tagged secondary antibody.
For immunofluorescence in the presence of Leptomycin B (LMB), 10ng/ml LMB (Sigma; #L2913) was added at 24 hpi and incubated for an additional two hours. At 26 hpi, samples were fixed with 4% paraformaldehyde and stained as described above for EdU, PML, lamin A/C and DAPI using primary and appropriate AF tagged secondary antibody.
Western blot
Whole-cell extracts were obtained from cell pellets lysed in 1x Laemmli sample buffer supplemented with 2-mercaptoethanol and 1× phosphatase inhibitor Cocktail 2 and 3 (Sigma; #P5726 and P0044, respectively). Proteins were resolved on SDS-PAGE and transferred to nitrocellulose membrane. Membranes were blocked 1 hour in 5% Blotting-Grade Blocker (BIO-RAD; #170-6404) in 1× TBST and incubated at 4°C overnight with primary antibodies. The primary antibodies and dilutions were as follows: PML (1:2000), phospho-Stat1 (Tyr701) (1:1000), and Stat1 (1:1000). After primary antibody incubation, membranes were washed 3 × 15 minutes in 1× TBST wash buffer. Membranes were then incubated with horseradish peroxidase–tagged goat anti-mouse and goat anti-rabbit secondary antibodies (1:2,500) at room temperature for 1 hour, washed 3 × 15 minutes in 1× TBST. Signals were detected by enhanced chemiluminescence (Thermo Scientific; #1859701 and 1859698)). Equal protein loading was confirmed by probing with β-actin monoclonal antibody (1:2,000).
RNA isolation, cDNA synthesis, real-time qPCR
Total RNA was extracted using the RNeasy Plus Mini RNA Isolation Kit (Qiagen; #74236). RNA samples from cells infected with quasivirions in the absence or presence of INBC or IFN β (Calbiochem; #407318) along with untreated controls were treated with DNAse I (BioLabs) prior reverse transcription. 0.5 μg total RNA was used to reverse-transcribe into cDNA using ImProm-II Reverse Transcriptase kit (Promega). Equal amounts of cDNA were quantified by RT-qPCR using the IQ SYBR Green Supermix (BIO-RAD) and a CFX96 Real-Time System (BIO-RAD). PCR reactions were carried out in triplicate, and transcript levels were normalized to β-actin. Mock reverse-transcribed samples were included as negative control. A list of oligonucleotide sequences used is provided in Table 1. The BIO-RAD CFX Manager 3.1 software was used to analyze the data.
Table 1.
List of oligonucleotides used in the study
| Name | 5′ Sequence | 3′ Sequence | Reference |
|---|---|---|---|
| MX1 | ctttccagtccagctcggca | agctgctggccgtacgtctg | (94) |
| MX2 | aaactgttcagagcacgattgaag | accatctgctccattctgaactg | (95) |
| OAS2 | aggtggctcctatggacggaa | ggcttctcttctgatcctggaattg | (94) |
| IFI27 | ggcagccttgtggctactct | atggagcccaggatgaacttg | (95) |
| IFI6 | gttctcactatattgtccaggctagagt | agtttattctgttgtcacatctaggttgtt | (96) |
| CXCL10 | caaatctgctttttaaagaatgctc | aagaatttgggccccttg | (97) |
| IRF7 | taccatctacctgggcttcg | agggttccagcttcacca | (95) |
| BST2 | acgcgtctgcagaggtggag | gcagcggagctggagtcct | (94) |
| HPV16E7 | atgagcaattaaatgacagctcagag | gcagcggagctggagtcct | (98) |
| HPV16E6 | ctgcaatgtttcaggaccc | tcacgtcgcagtaactgttg | |
| HPV16E1^E4 | cagaaaccataatctaccatggctg | tgtgtttcttcggtgccca | |
| IFNα | gcctgggaggttgtcagagcagaa | gtgcctgcacaggtatacaccaag | |
| IFNβ | gcctttgctctggcacaacaggt | tgccacaggagcttctgacactga | |
| IFNγ | caggtcattcagatgtagcggat | actctcctctttccaattcttcaaaa | |
| IFNκ | gccccaagagtttctgcaatac | ggcctgtagggacatttcataga | |
| IFNλ | ggaattgggacctgaggctt | ctaggacgtcctccagggct | |
| β-actin | ggcatcctcaccctgaagta | cagaggcgtacagggatagc |
Gene Array
RNA samples were analyzed by microarray using the GeneChip Human Genome U133 2.0 plus array (Affymetrix). RNA integrity was assessed with an Agilent TapeStation RNA Assay (Agilent Technologies, Palo Alto, CA). Microarray hybridization and fluorescence detection were performed as described in the Affymetrix GeneChip Expression Technical Manual. Pixel intensities were measured, expression signals were analyzed and features extracted using the commercial software package Expression Console 1.4 (Affymetrix). Arrays were globally scaled to a target intensity value of 500 in order to compare individual experiments. The absolute call (present, marginal, absent) of each gene expression in each sample was identified using the above-mentioned software. The data files generated by the Affymetrix microarray hybridization platform were analyzed using GeneSifter Analysis Edition by Geospiza software. The analysis was carried out on RNA samples isolated from vector control and shPML stably transduced HaCaT cells, each in duplicate. Signals that changed by >2-fold, with differential P values of ≤0.05 in shPML HaCaT compared to control cells, were analyzed for differential gene expression. Data deposited in Gene Expression Omnibus (accession number pending).
RESULTS
PML knock down reduces HPV pseudogenome levels in HPV16-infected cells
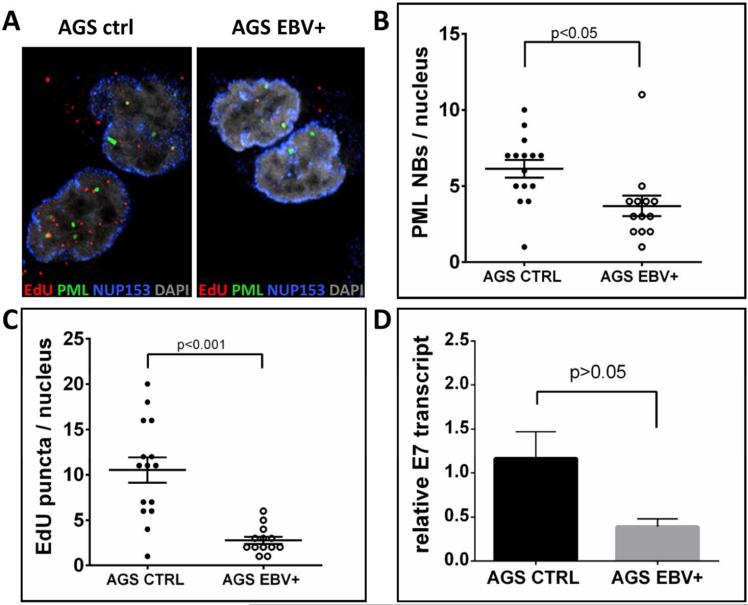
Using mouse cells deficient for PML protein, it was reported that PML protein is important for efficient transcription of incoming BPV1. Similarly, transcription driven by the heterologous CMV major immediate early promoter was impaired 10 fold in the absence of PML protein (29). To explain activation of homologous and heterologous promoters, it was concluded that PML NBs provide an environment favoring transcription. While investigating a possible synergy between EBV and HPV16, we observed that the amount of EdU-labeled nuclear DNA delivered by HPV16 particles was reduced approximately three-fold in EBV-harboring epithelial AGS cells as compared to parental AGS cells (Fig. 1A and C). Furthermore, the number of PML NBs was reduced in cells infected with EBV as well (Fig. 1B). We also observed reduced HPV16 transcript levels in EBV-containing compared to parental AGS cells (Fig. 1D)).
Fig 1. HPV16 pseudogenome delivery to the nucleus of EBV-harboring epithelial AGS cells.
(A) Cells were grown on glass coverslips and infected with EdU-labeled HPV16 PsV. At 24 hpi cells were fixed and processed for the detection of EdU-labeled DNA (red), PML (green), NUP153 (blue) and DAPI (gray). (B) PML NBs and (C) EdU-labeled viral pseudogenome number was counted manually in z-stacks spanning the whole nucleus for each cell. 15 AGS cells and 13 EBV-harboring AGS cells were included in the count. P value was determined using Student's t-test. (D) RT-qPCR for viral transcription at 48 hpi with HPV16 quasivirions. cDNA samples were analyzed for HPV16E7 and the data shown are fold changes relative to AGS cells. Error bars represent SEM of five independent experiments.
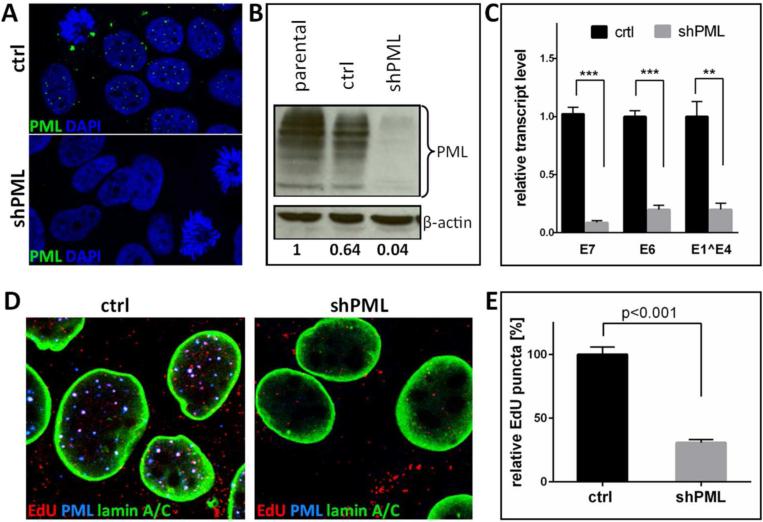
Since EBV has been shown to induce the degradation of PML isoforms (73) and PML protein was recently indirectly implicated in nuclear delivery of HPV16 (74), we revisited the role of PML protein in the establishment of HPV16 infection. To this end, we generated stable HaCaT-derived cell lines deficient for all isoforms of PML protein using shRNA delivered by lentivirus transduction using constructs kindly provided by R. Everett (72). Various groups have used this shRNA in a number of studies and no off-target effects have been reported (72, 75-78). As control, HaCaT cells were also stably transduced with a lentivirus expressing a scrambled shRNA. Knockdown was confirmed by IF and Western blot analysis (Fig. 2A and B). Confirming previous findings, transcription of HPV16 genome delivered by HPV16 quasivirions, which contain an authentic HPV16 genome with a loxP element inserted downstream of the L1 open reading frame (68) and were assembled using 293TT cells, was strongly reduced in PML protein-deficient cells compared to vector control-transduced HaCaT cells (Fig. 2C). In line with our observations with EBV-harboring AGS cells, we found that the amount of EdU-labeled DNA delivered by HPV16 particles present in nuclei of HPV16 infected cells at 30 hours post infection (hpi) was reduced by at least three-fold in PML protein-deficient HaCaT (Fig. 2D and E).
Fig 2. Knock down of PML protein in HaCaT cells.
(A) Immunofluorescent staining of HaCaT cells stably transduced with lentiviruses expressing scrambled or shRNA directed against PML. Cells were seeded onto glass coverslips and stained with PML antibody (green) and DAPI (blue). (B) Western blot analysis of PML protein in HaCaT cells stably transduced with vector control or shPML. Cells lysates were analyzed with anti-PML specific antibody. The levels of β-actin served as a loading control. Differences in PML protein levels were calculated by densitometry of the blots. Results shown are a representative of two independent experiments in which the repeat experiment also yielded similar results. (C) RT-qPCR for viral transcription at 72 hpi with HPV16 quasivirions. cDNA samples were analyzed for HPV16 E6, E7 and E1^E4 and the data shown are fold changes relative to vector control-transduced HaCaT cells samples. Error bars represent SEM of three (E6, E1^E4) and ten (E7) independent experiments, respectively. (D) Cells were grown on glass coverslips and infected with EdU-labeled PsV and processed at 24 hpi for the detection of EdU-labeled DNA (red), PML (blue), lamin A/C (green) and DAPI (not shown). (E) Quantification of EdU-labeled viral pseudogenome detected in the nuclei of vector control- and shPML-transduced HaCaT cells. The data shown are average percent of viral pseudogenome present in the nuclei of control cells. EdU-labeled viral pseudogenome number was counted manually in z-stacks spanning the whole nucleus for each cell. Results were calculated from 55 cells collected in three independent experiments. Error bars represent SEM. P value was determined using Student's t-test. ***: p<0.001; **: p<0.005
Reduced nuclear HPV16 genome levels in PML knockout cells are due to genome loss following successful nuclear delivery
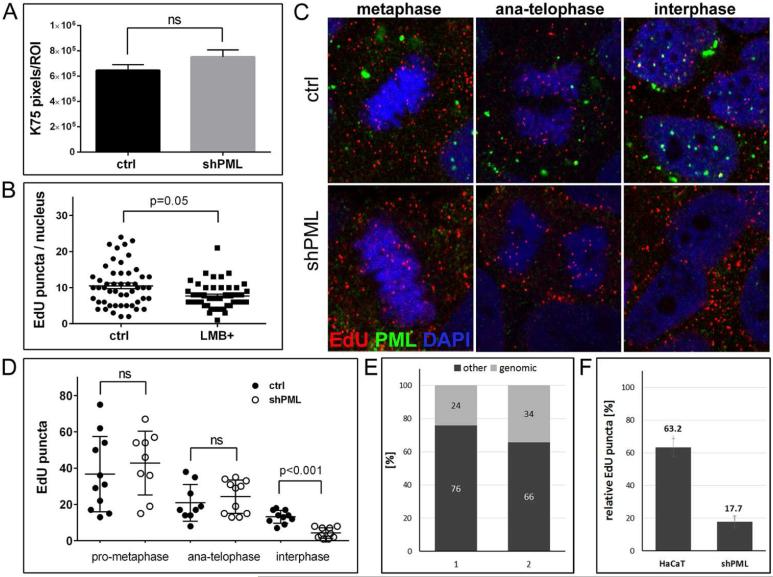
To rule out that the reduced nuclear level of viral pseudogenome was due to early events, we quantified virion binding in vector control and PML protein-deficient HaCaT cells. As shown in Fig. 3A, binding was not significantly affected by PML knockdown. L2 protein, which accompanies viral genome to the nucleus, harbors a nuclear export signal recognized by the nuclear export factor CRM1 (79). To test the possibility that DNA delivered by HPV16 particles is exported from the nucleus in absence of PML protein, we measured EdU-stained puncta in infected cells treated with the CRM1 inhibitor LMB during the last two hours of infection. LMB treatment did not restore levels of EdU-positive puncta in PML protein-deficient cells (Fig. 3B). LMB treatment resulted in CRM1 accumulation in the nucleus (data not shown). During these analyses, we noticed similar numbers of EdU puncta associated with mitotic chromosomes in PML-expressing and -deficient HaCaT cells. As a reminder, it was shown that HPV infection requires nuclear envelope breakdown for nuclear access of the viral genome and the viral genome is still in a vesicle during mitosis (56-58). Therefore, we quantified association of EdU-labeled encapsidated DNA with mitotic chromosomes at different stages of mitosis in both vector control and PML-deficient HaCaT cells. Reduced levels of EdU-labeled puncta in interphase of PML-deficient cells compared to vector control cells were again confirmed. However, we found no significant difference between these cells from metaphase through telophase (Fig. 3C and D) providing further support that the observed difference in interphase is not due to early events of infectious entry. The rather low degree association of viral genomes with mitotic chromosomes is best explained by the asynchronous entry with reported half times of up to 12 hours (42). We would like to point out that we did not observe co-localization of PML protein with viral pseudogenome on mitotic chromosomes, suggesting that interaction between PML protein and viral genome may not be important for efficient nuclear genome delivery (Fig. 3C) in line with observations by Day et al. (29). Although co-localization of individual viral genomes with large cytoplasmic PML aggregates was observed in rare cases (74). When we compared EdU-labeled puncta in pro-metaphase and ana-/telophase of vector control cells, we observed approximately a 50 % decrease in the number of puncta associated with each of the newly forming nuclei (Fig. 3D). This is best explained by the segregation of sister chromatids rather than being due to dissociation from the mitotic chromosomes. An additional reduction in EdU puncta is seen from ana-/telophase to interphase and approximately 63 % are retained in interphase cells compared to late mitotic stages (Fig. 3F). In PML protein-deficient cells, we again see a reduction by approximately 50 % of EdU puncta in late mitotic stages compared to pro-metaphase due to sister chromatid segregation. However, only 17 % of EdU puncta are retained in interphase nuclei of PML protein-deficient cells (Fig. 3F). Based on signal size and intensity, which is similar in viral particles bound to ECM and present intracellularly (data not shown), we assume that on average each EdU puncta associated with mitotic chromosomes or present in interphase cells represents a single DNA molecule delivered by one viral particle rather than representing an aggregate of viral particles. Therefore, loss of EdU puncta cannot be explained by diffusion of aggregated DNA.
Fig 3. Binding, uncoating, and delivery of HPV16 pseudogenome to the nucleus of PML-deficient HaCaT cells.
(A) Cell-binding assay. HPV16 pseudoviruses were bound to of HaCaT cells stably transduced with lentiviruses expressing scrambled or shRNA directed against PML for 1 h and L1 was detected using K-75 antibody. (B) Quantification of EdU-labeled viral pseudogenome in the nuclei of PML-deficient HaCaT cells after inhibition of nuclear export. Cells were grown on glass coverslips and infected with EdU-labeled PsV and processed at 26 hpi. Leptomycin B (10ng/ml) was added 2 h prior to fixation. Cells were stained for EdU, PML, lamin A/C and DAPI. Number of EdU-labeled viral pseudogenome was quantified in z-stacks spanning the whole nucleus in 51 control and 47 LMB-treated cells. Student's t-test was used to determine statistical significance of the data. (C) EdU-labeled viral pseudogenome in different stages of mitosis. Cells were grown on glass coverslips and infected with EdU-labeled PsV and processed at 24 hpi for the detection of EdU-labeled DNA (red), PML (green), lamin A/C (not shown) and DAPI (blue). A single slice from a z stack is shown. (D) Number of EdU-labeled viral genome bound to cellular chromatin or in the nucleus was scored in cells captured in different stages of mitosis. An example representative of two independent experiments is shown. Results were calculated from at least 30 cells per experiment. Student's t-test was used to determine statistical significance of the data. (E) Composition of encapsidated DNA in quasivirion preparations according to MiSeq Reporter. Two samples of DNA isolated from independent quasivirion preparations were sequenced using Illumina NextSeq 500. Results of alignment to the human genome are shown as percent of total sequenced encapsidated DNA. (F) Quantification of EdU-labeled viral genome bound to cellular chromatin in cells at interphase normalized to anaphasetelophase (shown in (D)).
Taken together, these findings suggested that DNA delivered by viral particles is lost in PML protein-deficient cells. In an attempt to confirm these findings, we isolated nuclei of HaCaT cells after infection to measure nuclear viral genome by qPCR. Unfortunately, we were not able to completely remove viral genome still present on the ECM, the cell surface or in the cytoplasm despite employing various means of cell fractionation. Nuclear fractions obtained from cells infected with L1 only or mutant HPV16 particles harboring 16L2-R302/5A, which are both defective for delivering genome to the nucleus, contained similar levels of viral genomes as nuclei from wild type infected cells (51). To rule out that reduced levels of EdU puncta in PML protein-deficient cells are due to loss of encapsidated linear cellular DNA, which always contaminates pseudovirus preparations, we subjected DNA isolated from two HPV16 quasivirion preparations and aligned the reads to the human genome. We found that 24 and 34 % of total encapsidated DNA was of human origin (Fig. 3E). Therefore, the 83 % reduction of EdU puncta in PML protein-deficient HaCaT cannot solely be explained by degradation of encapsidated cellular DNA, suggesting that the loss is mostly due to removal of encapsidated pseudogenome. However, the level of encapsidated cellular DNA correlates well with the observed 37 % loss of EdU puncta in parental HaCaT cells. Taken together, these data suggest that the loss of viral genome following nuclear delivery may be partially responsible for the PML knockdown phenotype. The data furthermore indicate that viral genome is likely lost by degradation if not associated with PML NBs.
PML knockdown does not impair HPV16 infection of HeLa cells
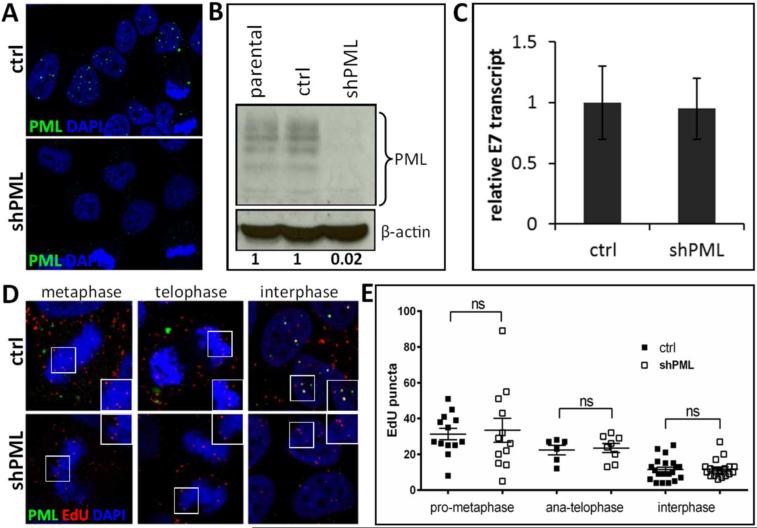
Next, we wanted to confirm our findings using HeLa cells, a cervical cancer-derived cell line commonly used in HPV entry studies. PML protein-deficient HeLa derivatives were established together with vector control lines (Fig. 4A and B) and analyzed for HPV16 transcription and pseudogenome delivery as described above for HaCaT cell lines. To our surprise, PML knockdown neither impaired transcription of HPV16 genomes and pseudogenomes nor did it affect levels of nuclear DNA delivered by viral particles at 24 hpi in interphase cells (Fig. 4C-E). However, we again observed a drop in genome levels similar to HaCaT when early mitotic stages were compared to later stages, which is due to sister chromatid segregation.
Fig 4. Knock down of PML protein in HeLa cells.
(A) Immunofluorescent staining of HeLa cells stably transduced with lentiviruses expressing scrambled (ctrl) or shRNA directed against PML (shPML). Cells were seeded onto glass coverslips and stained with PML antibody (green) and DAPI (blue). (B) Western blot analysis of PML protein in HeLa cells stably transduced with vector control or shPML lentiviruses. Cell lysates were analyzed with anti-PML specific antibody. The levels of β-actin served as a loading control. Differences in PML protein levels were calculated by densitometry of the blots. Results shown are a representative of two independent experiments in which the repeat experiment also yielded similar results. (C) RT-qPCR for viral transcription at 72 hpi with HPV16 quasivirions. cDNA samples were analyzed for HPV16E7 and the data shown are fold changes relative to vector control transduced HeLa cells samples. Error bars represent SEM of two independent experiments. (D) Detection of EdU-labeled viral pseudogenome in different stages of mitosis. Cells were grown on glass coverslips and infected with EdU-labeled PsV and processed at 24 hpi for the detection of EdU-labeled DNA, PML, lamin A/C (not shown) and DAPI. Colors are coded as indicated by the labels on the figure. (E) Number of EdU-labeled viral genome bound to cellular chromatin or in the nucleus was scored in cells captured in different stages of mitosis. Results were calculated from at least 30 cells per experiment. Student's t-test was used to determine statistical significance of the data.
PML protein knockdown induces an IFN response in HaCaT cells
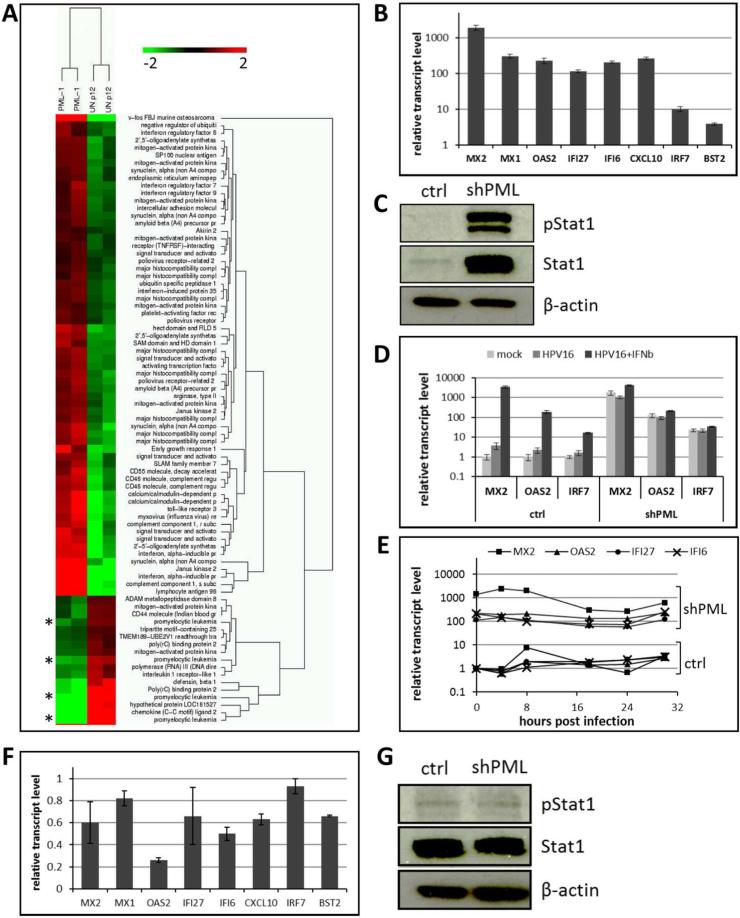
To start addressing possible mechanisms responsible for the unexpected loss of HPV genome, we compared the transcriptome of PML protein-deficient with vector control-transduced HaCaT cells using gene array analysis. Strikingly, factors typically induced by IFN signaling were highly upregulated in PML protein-deficient cells (Fig. 5A). This analysis also confirmed the knockdown of PML protein. We chose eight genes and confirmed their increased expression in PML knockdown cells by RT-qPCR (Fig. 5B). Interferon-stimulated gene (ISG) expression levels increased as a function of passage number (data not shown), suggesting that the constitutive IFN response is not an immediate response after PML knockdown. Among the upregulated genes was Stat1, which is a key signaling factor in the IFN pathways. Western blot analysis confirmed an increase in the amount and phosphorylation of Stat1 protein (Fig. 5C). Surprisingly, the common type I IFNs are not expressed at appreciable levels in either cell line (Table 2). The exception is IFN κ, a keratinocyte-specific type I IFN, which is expressed at very low levels, however, not affected by PML knockdown. The type II IFN γ is also not expressed in HaCaT cells. In contrast, the type III IFN λ is expressed, albeit at low levels, and expression is increased approximately 10 fold in PML protein-deficient cells. We then confirmed that the type I IFN pathway is functional. Addition of recombinant interferon β (IFN β) induces a strong interferon response in vector control and further increases by two- to four-fold the already elevated ISG expression levels in PML protein-deficient HaCaT cells (Fig. 5D).
Fig 5. Expression of interferon-stimulated genes (ISGs) in PML-deficient HaCaT and HeLa cells.
(A) Heat map of innate immune response genes generated based on microarray analysis of PML protein-deficient cells (PML) compared to vector control HaCaT cells (UN-p12). Asterisks highlight down regulation of PML gene expression. (B) RT-qPCR analysis of ISGs in PML-deficient HaCaT cells. The data shown are fold changes relative to vector control-transduced HaCaT cells. Error bars represent SEM of three independent experiments. (C) Immunoblot analysis of pStat1 and Stat1 in lysates from control and PML-deficient HaCaT cells. β-actin was used as a loading control. (D) Expression of ISGs in cells infected with HPV16 quasivirions in the presence or absence of IFNβ. Cells were treated with 1000 U/ml of IFNβ 1 h prior to infection. Samples were collected 48 hpi and analyzed by RT-qPCR. The data shown are fold changes relative to mock-treated vector control HaCaT cells. Error bars represent SEM of four independent experiments. P value was determined using Student's t-test. * P<0.02. (E) RT-qPCR analysis of ISGs in PML deficient and control HaCaT cells at indicated times post infection with HPV16 pseudovirus. The data shown are fold changes relative to uninfected vector control HaCaT cells. (F) RT-qPCR analysis of ISGs in PML-deficient HeLa cells. The data shown are fold changes relative to vector control-transduced cells. Error bars represent SEM of two independent experiments. (G) Western blot analysis of pStat1 and Stat1 in lysates from control and shPML HeLa cells. The levels of β-actin served as a loading control. Results shown are a representative of two independent experiments.
Table 2.
interferon mRNA levels in HaCaT cells
| Gene | Ct value | Fold change (±SEM) | |
|---|---|---|---|
| Ctrl (ave) | shPML (ave) | shPML | |
| Actin | 20.07 | 20.97 | - |
| IFN α | 40* | 40* | - |
| IFN β | 40* | 40* | - |
| IFN γ | 40* | 40* | - |
| IFN κ | 35.95 | 36.04 | 1.75 (±0.59) |
| IFN λ | 35.62 | 33.33 | 9.36 (±1.86) |
no amplification observed
The upregulation of ISGs was observed in uninfected PML protein-deficient HaCaT cells. To determine whether HPV16 infection alters the IFN response, we infected PML protein-deficient and control HaCaT cells with HPV16 pseudovirus and followed expression of a subset of ISGs over time. While we observed a slight fluctuation of ISGs in PML knock down cells, HPV16 infection did not consistently modulate ISG expression (Fig. 5E). In contrast, a slight increase of ISG expression was observed in control cells over time that never reached the levels of PML-deficient HaCaT, though. Control HeLa cells had comparable base line levels of ISG transcript. However, PML protein-deficient HeLa cells did neither display increased expression of ISGs nor Stat1 activation (Fig. 5F and G). This is not surprising since HeLa cells express HPV18 E6 and E7 oncogenes at high levels and E6 expression has been demonstrated to interfere with type I IFN signaling (80, 81).
Blocking the interferon response prevents loss of DNA delivered by HPV16
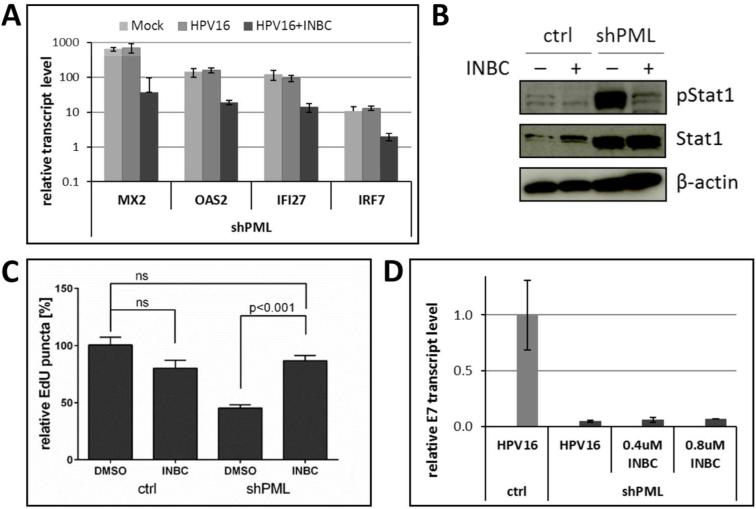
Next, we chose to determine whether the JAK/Stat signaling was causally linked to impaired HPV16 infection. To this end, PML protein-deficient HaCaT cells were infected with HPV16 quasivirions in the presence of INBC, a JAK1/2 kinase inhibitor. All IFN receptors are known to signal through this pathway. INBC treatment significantly decreased ISG expression levels and the phosphorylation of Stat1, suggesting that kinase inhibition was successful (Fig. 6A and B). At the same time, INBC treatment did restore EdU-stained puncta levels following infection with HPV16 pseudovirions (Fig. 6C). These data suggest that signaling through the JAK/Stat pathway is required for the loss of DNA delivered by viral particles to cells deficient for PML protein. However, HPV16 early transcription was not restored in the presence of INBC (Fig. 6D), suggesting that PML protein positively contributes to viral transcription as previously suggested, in addition to protecting incoming viral genome.
Fig 6. Inhibition of the JAK/Stat signaling pathway in PML-deficient HaCaT cells.
(A) Expression of ISGs in cells infected with HPV16 quasivirions in the presence or absence of Jak1/Jak2 kinase inhibitor. Cells were treated with 0.4 μM of INBC 1 h prior to infection. Samples were collected at 48 hpi and analyzed using RT-qPCR. The data shown are fold changes relative to mock-treated vector control HaCaT cells. Error bars represent SEM of two independent experiments. Student's t-test was used to determine P value. * P<0.01. (B) Immunoblot analysis of pStat1 and Stat1 in lysates from control and shPML HaCaT cells treated for 8 h with 0.8 μM INBC. β-actin was used as a loading control. (C) Quantification of EdU-labeled viral pseudogenome in the nuclei of cells infected with HPV16 pseudovirions in the presence of INBC. Stably transduced HaCaT cells were grown on coverslips infected with HPV16 pseudovirus in absence or presence of INBC (0.4 μM). At 30 hpi, samples were fixed and stained for EdU, PML, lamin A/C and DAPI. Number of EdU-labeled viral genome in the nuclei was counted in z-stacks spanning the whole nucleus for each cell. The data shown are average percent of viral pseudogenome present in the nuclei of vector control cells. Results were calculated in total from 60 to 116 cells collected from three independent experiments. Error bars represent the SEM, P value was determined using Student's t-test. (D) Indicated HaCaT cell derivatives were infected with HPV16 quasivirions in the absence or presence of INBC for 48 h. Relative E7 transcript levels were determined by RT-qPCR.
DISCUSSION
PML NBs have long been believed to serve important functions in antiviral defense. They are targeted and reorganized by many DNA viruses and the expression of PML protein is induced by treatment with IFN. Knockdown of PML facilitates infections of many DNA viruses and alleviates the need for viral proteins responsible for reorganization of PML NBs. In contrast to these observations, PML protein was shown to be important for efficient transcription from incoming PV genomes, which was interpreted as PML NBs providing an environment favoring transcription. The findings presented herein support the notion that PML protein has a protective role in HPV16 infection in addition. Our data suggest that PML NBs prevent loss of incoming viral genome after successful delivery to the nucleus. Since treatment with inhibitors of the nuclear export factor CRM1 did not restore genome levels in PML protein-deficient cells, we propose that genome loss is due to degradation. Our data also suggest that PML protein is not directly involved in nuclear genome delivery as recently suggested (74), which confirms previous findings by Day et al. (29). We only see rare events of co-localization of viral genome in large cytoplasmic PML aggregates, never on mitotic chromosomes. Furthermore, association of viral genome with chromosomes during mitosis is not altered by PML knockdown. One possible explanation for the loss of viral genome in HaCaT but not in HeLa cells is that it is being recognized as foreign by nuclear innate immune sensors. This is in line with our observation that PML protein-deficient HaCaT but not HeLa cells display elevated transcript levels of interferon responsive genes. The encapsidated genome is mostly chromatinized and as such similar to the host cell DNA. It will be interesting to investigate in future, which sensors are involved. A possible trigger may be the presence of viral capsid proteins or the circular form of incoming viral genome. However, the current experimental system for pseudovirus production and labeling of the genome is based on transfecting plasmids derived from bacteria into the packaging cell line. Therefore, we cannot exclude that virus-delivered encapsidated DNA is partially derived from bacteria and recognized as such.
Our data suggest that PML protein is required for efficient HPV transcriptional activity, in addition to ensuring the integrity of incoming HPV genome. While treatment with the inhibitor of the JAK/Stat signaling pathway INBC restored genome levels, it did not rescue transcription. This is in line with a previous report using a rather artificial experimental system of PML deficient mouse embryonic stem cells and BPV1 (29). The findings were recently confirmed in a study using primary human foreskin keratinocytes, in which the authors showed that transient knockdown of PML protein reduced HPV18 transcription and impaired the establishment of immortalized cell lines upon transfection of HPV18 genome (30). However, the underlying mechanism was not investigated. The same study described that another marker of PML NBs, Sp100, restricts transcription from HPV18 genomes. Taken together with our results, the data suggest that, in addition to providing a protective environment for HPV genomes upon infectious entry, PML NBs regulate transcription as well. An attractive interpretation is that the initial stimulation of viral transcription in the presence of PML protein allows the establishment of infection and recruitment of Sp100 may help to silence viral promoters at later times. Since PML NBs are completely disassembled during mitosis and are reassembled after nuclear envelope reformation (10), it is conceivable that Sp100 recruitment to these sites is slightly delayed allowing establishment of infection before silencing viral genomes to aid immune evasion. Further studies are required to establish the exact timing of Sp100 recruitment to PML NBs following infectious entry. It will also be of interest to delineate the timing of PML protein recruitment to the L2/viral genome complex, which may occur prior to or after release of the viral genome from the nuclear transport vesicles (58).
The herein described protective function of PML NBs may also explain why other DNA viruses, such as members of the herpesviridae family, reorganize PML NBs rather than disassembling them completely. One previous study indirectly implied that PML NBs might be required for stabilizing the herpes simplex virus type 1 (HSV-1) genome. Using HSV-1 virions that packaged an amplicon, which was comprised of the HSV-1 origin of replication, the “a” sequence for cleavage and packaging, and a reporter, it was observed that exclusively amplicon sequences located at PML NBs were transcriptionally active (82). The protective function of PML NBs might have gone unnoticed in other studies because viral factors counteracting the host cell innate immune response might have prevented degradation of viral genome. Indeed, using HeLa cells we provide some evidence that viral proteins E6 and E7 may prevent HPV genome degradation by interfering with innate immunity. E6 protein is known to interfere with JAK/Stat signaling (80, 81). Unfortunately, HeLa cells are addicted to the viral oncogenes excluding a knockdown approach to directly test this assumption. Our attempts to express E6 and E7 at levels that would allow detection of the viral proteins in HaCaT cells were unsuccessful. It is well known in the field that HaCaT cells do not easily express foreign DNA as is evident from the low expression level of reporters delivered by HPV particles despite efficient delivery of viral pseudogenome.
Our findings also imply that the PML protein-deficient host cell can sense incoming PV DNA or the viral capsid protein(s) and target the DNA for degradation. While cytosolic and nuclear sensors of foreign DNA have been identified previously, these sensors detect DNA that is bacterial, artificial or naked, i.e. not packaged into chromatin (83-89). In contrast, the HPV genome is organized into chromatin composed of the four core histones and does not contain any viral protein other than the capsid proteins that help deliver the viral genome (90). Currently, we don't know the nature of this sensor and where it might be located. According to current models, incoming viral DNA escapes the endocytic compartment at the TGN or endoplasmic reticulum prior to mitosis (55). Thus, both cytosolic and nuclear sensors should have access to viral genome. However, only IFI16, a sensor present in both the nuclei and cytosol, is expressed in HaCaT cells, and preliminary data suggest that knockdown of IFI16 does not prevent genome loss. In turn, this suggests that other sensors must be present that have not been identified as of yet. Since APOBEC3 induced by IFN α has recently been shown to degrade hepatitis B virus DNA, it may be an attractive additional candidate (91).
The results presented herein also suggest that PML NBs play an important role in regulating innate immune responses in keratinocytes. Knockdown of PML protein induces a strong and constitutive IFN response. Since the JAK kinase inhibitor INBC significantly reduces ISG expression in PML protein-deficient cells, it seems likely that autocrine signaling is involved. However, since only INF λ is upregulated and expressed at rather low levels, it is conceivable that an unknown intracellular activation is driving ISG expression. Further experimentation is required to distinguish between these possibilities. A similar activation of IFN signaling upon PML knockdown, in this case IFN γ signaling, has been reported once before in mouse embryonic stem cells even though these results are disputed (92, 93). It might have gone unnoticed by others because it is not found in all cell lineages and many transformed cell lines express viral oncogenes, which are able to interfere with innate immune responses. Indeed, HeLa cells do not display a similar activation upon PML knockdown. Other transformed cell lines of various origins that we tested also showed no increased ISG expression (data not shown). Furthermore, the slow build up of the IFN response might have contributed to it not being more often reported.
In summary, we have identified a novel function of PML NBs; protecting incoming HPV16 genomes and thus allowing transcription to be initiated. Our data also suggest that IFN responses can restrict HPV infection and it will be interesting to test whether the constitutively active IFN pathway may be responsible for our difficulty to infect primary keratinocytes. We have also presented evidence that PML NBs are key regulators of the innate immune response, at least in HaCaT keratinocytes. Whether this also applies to other keratinocytes, including primary cell lines, and to cells of other lineages is under current investigation.
IMPORTANCE.
Promyelocytic leukemia nuclear bodies (PML NBs) are important for antiviral defense. Many DNA viruses target these subnuclear structures and reorganize them. Reorganization of PML NBs by viral proteins is important for establishment of infection. In contrast, human papillomaviruses (HPVs) require the presence of PML protein for efficient transcription of incoming viral genome. Our finding that PML protein prevents the loss of HPV genome following infection implies that the host cell may be able to recognize chromatinized HPV genome or the associated capsid proteins. A constitutively active interferon response in absence of PML protein suggests that PML NBs are key regulators of the innate immune response in keratinocytes.
ACKNOWLEDGMENTS
We are grateful to Roger Everett, MRC University of Glasgow Centre for Virus Research, for providing the lentiviral vectors for PML knockdown. The project described was supported by R01AI081809 from the National Institute of Allergy and Infectious diseases, by R01DE01669-08S1 from the National Institute of Dental and Craniofacial Research, and in part by grants from the Feist-Weiller Cancer Center and the National Institute of General Medical Sciences (P30GM110703).
REFERENCES
- 1.Bernardi R, Pandolfi PP. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat Rev Mol Cell Biol. 2007;8:1006–1016. doi: 10.1038/nrm2277. [DOI] [PubMed] [Google Scholar]
- 2.Ishov AM, Sotnikov AG, Negorev D, Vladimirova OV, Neff N, Kamitani T, Yeh ET, Strauss JF, 3rd, Maul GG. PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J Cell Biol. 1999;147:221–234. doi: 10.1083/jcb.147.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maarifi G, Chelbi-Alix MK, Nisole S. PML control of cytokine signaling. Cytokine Growth Factor Rev. 2014;25:551–561. doi: 10.1016/j.cytogfr.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 4.Nisole S, Maroui MA, Mascle XH, Aubry M, Chelbi-Alix MK. Differential Roles of PML Isoforms. Front Oncol. 2013;3:125. doi: 10.3389/fonc.2013.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Negorev D, Maul GG. Cellular proteins localized at and interacting within ND10/PML nuclear bodies/PODs suggest functions of a nuclear depot. Oncogene. 2001;20:7234–7242. doi: 10.1038/sj.onc.1204764. [DOI] [PubMed] [Google Scholar]
- 6.Van Damme E, Laukens K, Dang TH, Van Ostade X. A manually curated network of the PML nuclear body interactome reveals an important role for PML-NBs in SUMOylation dynamics. Int J Biol Sci. 2010;6:51–67. doi: 10.7150/ijbs.6.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dellaire G, Eskiw CH, Dehghani H, Ching RW, Bazett-Jones DP. Mitotic accumulations of PML protein contribute to the re-establishment of PML nuclear bodies in G1. J Cell Sci. 2006;119:1034–1042. doi: 10.1242/jcs.02817. [DOI] [PubMed] [Google Scholar]
- 8.Everett RD, Lomonte P, Sternsdorf T, van Driel R, Orr A. Cell cycle regulation of PML modification and ND10 composition. J Cell Sci. 1999;112:4581–4588. doi: 10.1242/jcs.112.24.4581. [DOI] [PubMed] [Google Scholar]
- 9.Jul-Larsen A, Grudic A, Bjerkvig R, Boe SO. Cell-cycle regulation and dynamics of cytoplasmic compartments containing the promyelocytic leukemia protein and nucleoporins. J Cell Sci. 2009;122:1201–1210. doi: 10.1242/jcs.040840. [DOI] [PubMed] [Google Scholar]
- 10.Palibrk V, Lang E, Lang A, Schink KO, Rowe AD, Boe SO. Promyelocytic leukemia bodies tether to early endosomes during mitosis. Cell Cycle. 2014;13:1749–1755. doi: 10.4161/cc.28653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng X, Kao HY. Post-translational modifications of PML: consequences and implications. Front Oncol. 2012;2:210. doi: 10.3389/fonc.2012.00210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmitz ML, Grishina I. Regulation of the tumor suppressor PML by sequential post-translational modifications. Front Oncol. 2012;2:204. doi: 10.3389/fonc.2012.00204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adamson AL, Kenney S. Epstein-barr virus immediate-early protein BZLF1 is SUMO-1 modified and disrupts promyelocytic leukemia bodies. J Virol. 2001;75:2388–2399. doi: 10.1128/JVI.75.5.2388-2399.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berscheminski J, Groitl P, Dobner T, Wimmer P, Schreiner S. The adenoviral oncogene E1A-13S interacts with a specific isoform of the tumor suppressor PML to enhance viral transcription. J Virol. 2013;87:965–977. doi: 10.1128/JVI.02023-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Everett RD. DNA viruses and viral proteins that interact with PML nuclear bodies. Oncogene. 2001;20:7266–7273. doi: 10.1038/sj.onc.1204759. [DOI] [PubMed] [Google Scholar]
- 16.Everett RD, Freemont P, Saitoh H, Dasso M, Orr A, Kathoria M, Parkinson J. The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110- and proteasome-dependent loss of several PML isoforms. J Virol. 1998;72:6581–6591. doi: 10.1128/jvi.72.8.6581-6591.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ishov AM, Maul GG. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J Cell Biol. 1996;134:815–826. doi: 10.1083/jcb.134.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maul GG, Negorev D. Differences between mouse and human cytomegalovirus interactions with their respective hosts at immediate early times of the replication cycle. Med Microbiol Immunol. 2008;197:241–249. doi: 10.1007/s00430-008-0078-1. [DOI] [PubMed] [Google Scholar]
- 19.Sternsdorf T, Grotzinger T, Jensen K, Will H. Nuclear dots: actors on many stages. Immunobiology. 1997;198:307–331. doi: 10.1016/S0171-2985(97)80051-4. [DOI] [PubMed] [Google Scholar]
- 20.Tavalai N, Adler M, Scherer M, Riedl Y, Stamminger T. Evidence for a dual antiviral role of the major nuclear domain 10 component Sp100 during the immediate-early and late phases of the human cytomegalovirus replication cycle. J Virol. 2011;85:9447–9458. doi: 10.1128/JVI.00870-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahn JH, Brignole EJ, 3rd, Hayward GS. Disruption of PML subnuclear domains by the acidic IE1 protein of human cytomegalovirus is mediated through interaction with PML and may modulate a RING finger-dependent cryptic transactivator function of PML. Mol Cell Biol. 1998;18:4899–4913. doi: 10.1128/mcb.18.8.4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahn JH, Hayward GS. Disruption of PML-associated nuclear bodies by IE1 correlates with efficient early stages of viral gene expression and DNA replication in human cytomegalovirus infection. Virology. 2000;274:39–55. doi: 10.1006/viro.2000.0448. [DOI] [PubMed] [Google Scholar]
- 23.Boutell C, Orr A, Everett RD. PML residue lysine 160 is required for the degradation of PML induced by herpes simplex virus type 1 regulatory protein ICP0. J Virol. 2003;77:8686–8694. doi: 10.1128/JVI.77.16.8686-8694.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Everett RD, Maul GG. HSV-1 IE protein Vmw110 causes redistribution of PML. EMBO J. 1994;13:5062–5069. doi: 10.1002/j.1460-2075.1994.tb06835.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Korioth F, Maul GG, Plachter B, Stamminger T, Frey J. The nuclear domain 10 (ND10) is disrupted by the human cytomegalovirus gene product IE1. Exp Cell Res. 1996;229:155–158. doi: 10.1006/excr.1996.0353. [DOI] [PubMed] [Google Scholar]
- 26.Muller S, Dejean A. Viral immediate-early proteins abrogate the modification by SUMO-1 of PML and Sp100 proteins, correlating with nuclear body disruption. J Virol. 1999;73:5137–5143. doi: 10.1128/jvi.73.6.5137-5143.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parkinson J, Everett RD. Alphaherpesvirus proteins related to herpes simplex virus type 1 ICP0 affect cellular structures and proteins. J Virol. 2000;74:10006–10017. doi: 10.1128/jvi.74.21.10006-10017.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saffert RT, Kalejta RF. Promyelocytic leukemia-nuclear body proteins: herpesvirus enemies, accomplices, or both? Future Virol. 2008;3:265–277. doi: 10.2217/17460794.3.3.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Day PM, Baker CC, Lowy DR, Schiller JT. Establishment of papillomavirus infection is enhanced by promyelocytic leukemia protein (PML) expression. Proc Natl Acad Sci U S A. 2004;101:14252–14257. doi: 10.1073/pnas.0404229101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stepp WH, Meyers JM, McBride AA. Sp100 provides intrinsic immunity against human papillomavirus infection. MBio. 2013;4:e00845–00813. doi: 10.1128/mBio.00845-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Culp TD, Budgeon LR, Marinkovich MP, Meneguzzi G, Christensen ND. Keratinocyte-secreted laminin 5 can function as a transient receptor for human papillomaviruses by binding virions and transferring them to adjacent cells. J Virol. 2006;80:8940–8950. doi: 10.1128/JVI.00724-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giroglou T, Florin L, Schäfer F, Streeck RE, Sapp M. Human papillomavirus infection requires cell surface heparan sulfate. J Virol. 2001;75:1565–1570. doi: 10.1128/JVI.75.3.1565-1570.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson KM, Kines RC, Roberts JN, Lowy DR, Schiller JT, Day PM. Role of heparan sulfate in attachment to and infection of the murine female genital tract by human papillomavirus. J Virol. 2009;83:2067–2074. doi: 10.1128/JVI.02190-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richards KF, Bienkowska-Haba M, Dasgupta J, Chen XS, Sapp M. Multiple heparan sulfate binding site engagements are required for the infectious entry of human papillomavirus type 16. J Virol. 2013;87:11426–11437. doi: 10.1128/JVI.01721-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Richards KF, Mukherjee S, Bienkowska-Haba M, Pang J, Sapp M. Human papillomavirus species-specific interaction with the basement membrane-resident non-heparan sulfate receptor. Viruses. 2014;6:4856–4879. doi: 10.3390/v6124856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Richards RM, Lowy DR, Schiller JT, Day PM. Cleavage of the papillomavirus minor capsid protein, L2, at a furin consensus site is necessary for infection. Proc Natl Acad Sci U S A. 2006;103:1522–1527. doi: 10.1073/pnas.0508815103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roberts JN, Buck CB, Thompson CD, Kines R, Bernardo M, Choyke PL, Lowy DR, Schiller JT. Genital transmission of HPV in a mouse model is potentiated by nonoxynol-9 and inhibited by carrageenan. Nat Med. 2007;13:857–861. doi: 10.1038/nm1598. [DOI] [PubMed] [Google Scholar]
- 38.Bienkowska-Haba M, Patel HD, Sapp M. Target cell cyclophilins facilitate human papillomavirus type 16 infection. PLoS Pathog. 2009;5:e1000524. doi: 10.1371/journal.ppat.1000524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dziduszko A, Ozbun MA. Annexin A2 and S100A10 regulate human papillomavirus type 16 entry and intracellular trafficking in human keratinocytes. J Virol. 2013;87:7502–7515. doi: 10.1128/JVI.00519-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Evander M, Frazer IH, Payne E, Qi YM, Hengst K, McMillan NA. Identification of the alpha6 integrin as a candidate receptor for papillomaviruses. J Virol. 1997;71:2449–2456. doi: 10.1128/jvi.71.3.2449-2456.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scheffer KD, Gawlitza A, Spoden GA, Zhang XA, Lambert C, Berditchevski F, Florin L. Tetraspanin CD151 mediates papillomavirus type 16 endocytosis. J Virol. 2013;87:3435–3446. doi: 10.1128/JVI.02906-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Selinka HC, Florin L, Patel HD, Freitag K, Schmidtke M, Makarov VA, Sapp M. Inhibition of Transfer to Secondary Receptors by Heparan Sulfate-Binding Drug or Antibody Induces Non-Infectious Uptake of Human Papillomavirus. J Virol. 2007;81:10970–10980. doi: 10.1128/JVI.00998-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Woodham AW, Da Silva DM, Skeate JG, Raff AB, Ambroso MR, Brand HE, Isas JM, Langen R, Kast WM. The S100A10 subunit of the annexin A2 heterotetramer facilitates L2-mediated human papillomavirus infection. PLoS One. 2012;7:e43519. doi: 10.1371/journal.pone.0043519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schelhaas M, Shah B, Holzer M, Blattmann P, Kuhling L, Day PM, Schiller JT, Helenius A. Entry of human papillomavirus type 16 by actin-dependent, clathrin- and lipid raft-independent endocytosis. PLoS Pathog. 2012;8:e1002657. doi: 10.1371/journal.ppat.1002657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spoden G, Freitag K, Husmann M, Boller K, Sapp M, Lambert C, Florin L. Clathrin- and Caveolin-Independent Entry of Human Papillomavirus Type 16 - Involvement of Tetraspanin-Enriched Microdomains (TEMs). PLoS ONE. 2008;3:e3313. doi: 10.1371/journal.pone.0003313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spoden G, Kuhling L, Cordes N, Frenzel B, Sapp M, Boller K, Florin L, Schelhaas M. Human papillomavirus types 16, 18, and 31 share similar endocytic requirements for entry. J Virol. 2013;87:7765–7773. doi: 10.1128/JVI.00370-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bienkowska-Haba M, Williams C, Kim SM, Garcea RL, Sapp M. Cyclophilins Facilitate Dissociation of the Human Papillomavirus Type 16 Capsid Protein L1 from the L2/DNA Complex following Virus Entry. J Virol. 2012;86:9875–9887. doi: 10.1128/JVI.00980-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Day PM, Lowy DR, Schiller JT. Papillomaviruses infect cells via a clathrin-dependent pathway. Virology. 2003;307:1–11. doi: 10.1016/s0042-6822(02)00143-5. [DOI] [PubMed] [Google Scholar]
- 49.Selinka HC, Giroglou T, Sapp M. Analysis of the infectious entry pathway of human papillomavirus type 33 pseudovirions. Virology. 2002;299:279–287. doi: 10.1006/viro.2001.1493. [DOI] [PubMed] [Google Scholar]
- 50.Day PM, Thompson CD, Schowalter RM, Lowy DR, Schiller JT. Identification of a Role for the trans-Golgi Network in Human Papillomavirus 16 Pseudovirus Infection. J Virol. 2013;87:3862–3870. doi: 10.1128/JVI.03222-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.DiGiuseppe S, Bienkowska-Haba M, Hilbig L, Sapp M. The nuclear retention signal of HPV16 L2 protein is essential for incoming viral genome to transverse the trans-Golgi network. Virology. 2014;458-459:93–105. doi: 10.1016/j.virol.2014.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lipovsky A, Popa A, Pimienta G, Wyler M, Bhan A, Kuruvilla L, Guie MA, Poffenberger AC, Nelson CD, Atwood WJ, DiMaio D. Genome-wide siRNA screen identifies the retromer as a cellular entry factor for human papillomavirus. Proc Natl Acad Sci U S A. 2013;110:7452–7457. doi: 10.1073/pnas.1302164110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.DiGiuseppe S, Keiffer TR, Bienkowska-Haba M, Luszczek W, Guion LG, Muller M, Sapp M. Topography of the human papillomavirus minor capsid protein L2 during vesicular trafficking of infectious entry. J Virol. 2015 doi: 10.1128/JVI.01588-15. doi:10.1128/jvi.01588-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Popa A, Zhang W, Harrison MS, Goodner K, Kazakov T, Goodwin EC, Lipovsky A, Burd CG, DiMaio D. Direct binding of retromer to human papillomavirus type 16 minor capsid protein L2 mediates endosome exit during viral infection. PLoS Pathog. 2015;11:e1004699. doi: 10.1371/journal.ppat.1004699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang W, Kazakov T, Popa A, DiMaio D. Vesicular trafficking of incoming human papillomavirus 16 to the Golgi apparatus and endoplasmic reticulum requires gamma-secretase activity. MBio. 2014;5:e01777–01714. doi: 10.1128/mBio.01777-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aydin I, Weber S, Snijder B, Samperio Ventayol P, Kuhbacher A, Becker M, Day PM, Schiller JT, Kann M, Pelkmans L, Helenius A, Schelhaas M. Large scale RNAi reveals the requirement of nuclear envelope breakdown for nuclear import of human papillomaviruses. PLoS Pathog. 2014;10:e1004162. doi: 10.1371/journal.ppat.1004162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pyeon D, Pearce SM, Lank SM, Ahlquist P, Lambert PF. Establishment of human papillomavirus infection requires cell cycle progression. PLoS Pathog. 2009;5:e1000318. doi: 10.1371/journal.ppat.1000318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.DiGiuseppe S, Luszczek W, Keiffer TR, Bienkowska-Haba M, Guion LG, Sapp MJ. Incoming human papillomavirus type 16 genome resides in a vesicular compartment throughout mitosis. Proc Natl Acad Sci U S A. 2016;113:6289–6294. doi: 10.1073/pnas.1600638113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bund T, Spoden GA, Koynov K, Hellmann N, Boukhallouk F, Arnold P, Hinderberger D, Florin L. An L2 SUMO interacting motif is important for PML localization and infection of human papillomavirus type 16. Cell Microbiol. 2014;16:1179–1200. doi: 10.1111/cmi.12271. [DOI] [PubMed] [Google Scholar]
- 60.Marusic MB, Mencin N, Licen M, Banks L, Grm HS. Modification of human papillomavirus minor capsid protein L2 by sumoylation. J Virol. 2010;84:11585–11589. doi: 10.1128/JVI.01269-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Becker KA, Florin L, Sapp C, Maul GG, Sapp M. Nuclear localization but not PML protein is required for incorporation of the papillomavirus minor capsid protein L2 into virus-like particles. J Virol. 2004;78:1121–1128. doi: 10.1128/JVI.78.3.1121-1128.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Becker KA, Florin L, Sapp C, Sapp M. Dissection of human papillomavirus type 33 L2 domains involved in nuclear domain (ND) 10 homing and reorganization. Virology. 2003;314:161–167. doi: 10.1016/s0042-6822(03)00447-1. [DOI] [PubMed] [Google Scholar]
- 63.Day PM, Roden RB, Lowy DR, Schiller JT. The papillomavirus minor capsid protein, L2, induces localization of the major capsid protein, L1, and the viral transcription/replication protein, E2, to PML oncogenic domains. J Virol. 1998;72:142–150. doi: 10.1128/jvi.72.1.142-150.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Florin L, Schäfer F, Sotlar K, Streeck RE, Sapp M. Reorganization of nuclear domain 10 induced by papillomavirus capsid protein L2. Virology. 2002;295:97–107. doi: 10.1006/viro.2002.1360. [DOI] [PubMed] [Google Scholar]
- 65.Sivachandran N, Cao JY, Frappier L. Epstein-Barr virus nuclear antigen 1 Hijacks the host kinase CK2 to disrupt PML nuclear bodies. J Virol. 2010;84:11113–11123. doi: 10.1128/JVI.01183-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sivachandran N, Sarkari F, Frappier L. Epstein-Barr nuclear antigen 1 contributes to nasopharyngeal carcinoma through disruption of PML nuclear bodies. PLoS Pathog. 2008;4:e1000170. doi: 10.1371/journal.ppat.1000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sapp M, Kraus U, Volpers C, Snijders PJ, Walboomers JM, Streeck RE. Analysis of type-restricted and cross-reactive epitopes on virus-like particles of human papillomavirus type 33 and in infected tissues using monoclonal antibodies to the major capsid protein. J Gen Virol. 1994;75:3375–3383. doi: 10.1099/0022-1317-75-12-3375. [DOI] [PubMed] [Google Scholar]
- 68.Lee JH, Yi SM, Anderson ME, Berger KL, Welsh MJ, Klingelhutz AJ, Ozbun MA. Propagation of infectious human papillomavirus type 16 by using an adenovirus and Cre/LoxP mechanism. Proc Natl Acad Sci U S A. 2004;101:2094–2099. doi: 10.1073/pnas.0308615100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Buck CB, Thompson CD. Production of papillomavirus-based gene transfer vectors. Curr Protoc Cell Biol. 2007 doi: 10.1002/0471143030.cb2601s37. Chapter 26:Unit 26.21. [DOI] [PubMed] [Google Scholar]
- 70.Buck CB, Thompson CD, Pang YYS, Lowy DR, Schiller JT. Maturation of papillomavirus capsids. J Virol. 2005;79:2839–2846. doi: 10.1128/JVI.79.5.2839-2846.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ishii Y, Tanaka K, Kondo K, Takeuchi T, Mori S, Kanda T. Inhibition of nuclear entry of HPV16 pseudovirus-packaged DNA by an anti-HPV16 L2 neutralizing antibody. Virology. 2010;406:181–188. doi: 10.1016/j.virol.2010.07.019. [DOI] [PubMed] [Google Scholar]
- 72.Everett RD, Parada C, Gripon P, Sirma H, Orr A. Replication of ICP0- null mutant herpes simplex virus type 1 is restricted by both PML and Sp100. J Virol. 2008;82:2661–2672. doi: 10.1128/JVI.02308-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sarkari F, Wang X, Nguyen T, Frappier L. The herpesvirus associated ubiquitin specific protease, USP7, is a negative regulator of PML proteins and PML nuclear bodies. PLoS ONE. 6:e16598. doi: 10.1371/journal.pone.0016598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Broniarczyk J, Massimi P, Bergant M, Banks L. Human Papillomavirus Infectious Entry and Trafficking Is a Rapid Process. J Virol. 2015;89:8727–8732. doi: 10.1128/JVI.00722-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Everett RD, Murray J, Orr A, Preston CM. Herpes simplex virus type 1 genomes are associated with ND10 nuclear substructures in quiescently infected human fibroblasts. J Virol. 2007;81:10991–11004. doi: 10.1128/JVI.00705-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sarkari F, Wang X, Nguyen T, Frappier L. The herpesvirus associated ubiquitin specific protease, USP7, is a negative regulator of PML proteins and PML nuclear bodies. PLoS One. 2011;6:e16598. doi: 10.1371/journal.pone.0016598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sivachandran N, Wang X, Frappier L. Functions of the Epstein-Barr virus EBNA1 protein in viral reactivation and lytic infection. J Virol. 2012;86:6146–6158. doi: 10.1128/JVI.00013-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Smith MC, Box AC, Haug JS, Lane WS, Davido DJ. A Phospho-SIM in the Antiviral Protein PML is Required for Its Recruitment to HSV-1 Genomes. Cells. 2014;3:1131–1158. doi: 10.3390/cells3041131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mamoor S, Onder Z, Karanam B, Kwak K, Bordeaux J, Crosby L, Roden RB, Moroianu J. The high risk HPV16 L2 minor capsid protein has multiple transport signals that mediate its nucleocytoplasmic traffic. Virology. 2012;422:413–424. doi: 10.1016/j.virol.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hong S, Mehta KP, Laimins LA. Suppression of STAT-1 expression by human papillomaviruses is necessary for differentiation-dependent genome amplification and plasmid maintenance. J Virol. 2011;85:9486–9494. doi: 10.1128/JVI.05007-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Reiser J, Hurst J, Voges M, Krauss P, Munch P, Iftner T, Stubenrauch F. High-risk human papillomaviruses repress constitutive kappa interferon transcription via E6 to prevent pathogen recognition receptor and antiviral-gene expression. J Virol. 2011;85:11372–11380. doi: 10.1128/JVI.05279-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tang Q, Li L, Ishov AM, Revol V, Epstein AL, Maul GG. Determination of minimum herpes simplex virus type 1 components necessary to localize transcriptionally active DNA to ND10. J Virol. 2003;77:5821–5828. doi: 10.1128/JVI.77.10.5821-5828.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Diner BA, Li T, Greco TM, Crow MS, Fuesler JA, Wang J, Cristea IM. The functional interactome of PYHIN immune regulators reveals IFIX is a sensor of viral DNA. Mol Syst Biol. 2015;11:787. doi: 10.15252/msb.20145808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, Fitzgerald KA. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kerur N, Veettil MV, Sharma-Walia N, Bottero V, Sadagopan S, Otageri P, Chandran B. IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi Sarcoma-associated herpesvirus infection. Cell Host Microbe. 2011;9:363–375. doi: 10.1016/j.chom.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Orzalli MH, DeLuca NA, Knipe DM. Nuclear IFI16 induction of IRF-3 signaling during herpesviral infection and degradation of IFI16 by the viral ICP0 protein. Proc Natl Acad Sci U S A. 2012;109:E3008–3017. doi: 10.1073/pnas.1211302109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rathinam VA, Jiang Z, Waggoner SN, Sharma S, Cole LE, Waggoner L, Vanaja SK, Monks BG, Ganesan S, Latz E, Hornung V, Vogel SN, Szomolanyi-Tsuda E, Fitzgerald KA. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat Immunol. 2010;11:395–402. doi: 10.1038/ni.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, Fitzgerald KA, Paludan SR, Bowie AG. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Favre M, Breitburd F, Croissant O, Orth G. Chromatin-like structures obtained after alkaline disruption of bovine and human papillomaviruses. J Virol. 1977;21:1205–1209. doi: 10.1128/jvi.21.3.1205-1209.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lucifora J, Xia Y, Reisinger F, Zhang K, Stadler D, Cheng X, Sprinzl MF, Koppensteiner H, Makowska Z, Volz T, Remouchamps C, Chou WM, Thasler WE, Huser N, Durantel D, Liang TJ, Munk C, Heim MH, Browning JL, Dejardin E, Dandri M, Schindler M, Heikenwalder M, Protzer U. Specific and nonhepatotoxic degradation of nuclear hepatitis B virus cccDNA. Science. 2014;343:1221–1228. doi: 10.1126/science.1243462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Choi YH, Bernardi R, Pandolfi PP, Benveniste EN. The promyelocytic leukemia protein functions as a negative regulator of IFN-gamma signaling. Proc Natl Acad Sci U S A. 2006;103:18715–18720. doi: 10.1073/pnas.0604800103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.El Bougrini J, Dianoux L, Chelbi-Alix MK. PML positively regulates interferon gamma signaling. Biochimie. 2011;93:389–398. doi: 10.1016/j.biochi.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 94.Cheon H, Holvey-Bates EG, Schoggins JW, Forster S, Hertzog P, Imanaka N, Rice CM, Jackson MW, Junk DJ, Stark GR. IFNbeta-dependent increases in STAT1, STAT2, and IRF9 mediate resistance to viruses and DNA damage. Embo j. 2013;32:2751–2763. doi: 10.1038/emboj.2013.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zahoor MA, Xue G, Sato H, Murakami T, Takeshima SN, Aida Y. HIV- 1 Vpr induces interferon-stimulated genes in human monocyte-derived macrophages. PLoS One. 2014;9:e106418. doi: 10.1371/journal.pone.0106418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tanaka S, Honda Y, Honda M. Identification of differentially expressed genes in blood cells of narcolepsy patients. Sleep. 2007;30:974–979. doi: 10.1093/sleep/30.8.974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Li C, Wang Z, Liu F, Zhu J, Yang L, Cai G, Zhang Z, Huang W, Cai S, Xu Y. CXCL10 mRNA expression predicts response to neoadjuvant chemoradiotherapy in rectal cancer patients. Tumour Biol. 2014;35:9683–9691. doi: 10.1007/s13277-014-2234-0. [DOI] [PubMed] [Google Scholar]
- 98.Donaldson MM, Mackintosh LJ, Bodily JM, Dornan ES, Laimins LA, Morgan IM. An interaction between human papillomavirus 16 E2 and TopBP1 is required for optimum viral DNA replication and episomal genome establishment. J Virol. 2012;86:12806–12815. doi: 10.1128/JVI.01002-12. [DOI] [PMC free article] [PubMed] [Google Scholar]