Abstract
Cooperation is abundant in nature, occurring at all levels of biological complexity. Yet cooperation is continually threatened by subversion from non-cooperating cheaters. Previous studies have shown that cooperation can nevertheless be maintained when the benefits that cooperation provides to relatives outweigh the associated costs. These fitness costs and benefits are not fixed properties, but can be affected by the environment in which populations reside. Here, we describe how one environmental factor, resource abundance, decisively affects the evolution of cooperative public goods production in two independent evolving systems. In the Avida digital evolution platform, populations evolved in environments with different levels of a required resource, while populations of Vibrio cholerae evolved in the presence of different nutrient concentrations. In both systems, cooperators and cheaters co-existed stably in resource-rich environments, while cheaters dominated in resource-poor environments. These two outcomes were separated by a sharp transition that occurred at a critical level of resource. These results offer new insights into how the environment affects the evolution of cooperation and highlight the challenges that populations of cooperators face when they experience environmental change.
Keywords: Public goods, biofilms, Vibrio cholerae, digital evolution, experimental evolution, cooperation, Avida
Introduction
Cooperation takes many forms in nature, and each instance presents an evolutionary opportunity for exploitation by those that take advantage of the costly behavior but do not contribute. Over time, these cheaters can drive cooperators from the population and lead to a tragedy of the commons, whereby the entire population declines (Hardin, 1968). How cooperators persist despite these formidable challenges is often attributed to inclusive fitness (Hamilton, 1964a, Hamilton, 1964b), where an individual’s fitness is determined not only by its own activities, but also by the effects that cooperation has on its relatives. With this perspective, cooperative behaviors can flourish when the benefits they provide to close relatives outweigh the associated costs (West et al., 2007).
Quantifying these fitness costs and benefits is often quite difficult. Compounding this difficulty is the fact that evolution does not occur in a vacuum. In natural settings, a myriad of biotic and abiotic factors combines to continually alter the costs and benefits of a cooperative act as well as the relatedness of the individuals involved. These fluctuations in fitness effects over both time and space make the maintenance of cooperation over evolutionary timescales even more daunting.
Here, we examine how one such environmental factor, resource abundance, affects the evolution of biofilm formation, a prevalent form of cooperation within microbial communities (Hall-Stoodley et al., 2004). Biofilms are spatially structured communities of microorganisms connected through a matrix of extracellular polymeric substance (EPS), which is produced by individuals at a cost and placed into the surrounding environment. The biofilm lifestyle often affords its constituents a wide variety of benefits, including adhesion to surfaces in fluid environments, protection from predation, and increased resistance to antibiotic treatments (Hall-Stoodley & Stoodley, 2009, Mah et al., 2003, Mah & O’Toole, 2001). Since EPS is a public good that can be utilized by the surrounding community, biofilms present an opportunity for exploitation by cheaters, which take advantage of the biofilm without contributing to its formation (Xavier & Foster, 2007, Brockhurst et al., 2008). The presence of cheaters can significantly weaken biofilms, thereby reducing the potential benefits and threatening the long-term stability of cooperation (Rainey & Rainey, 2003, Popat et al., 2012).
In this article, we employ two experimental systems, one digital and the other biological, to investigate how resource abundance affects the evolution of cooperative biofilm formation. First, using the Avida digital evolution platform (Ofria & Wilke, 2004), populations of “digital organisms” evolved in spatially-structured environments containing different levels of a resource. This resource was required to complete logic-based tasks that conferred fitness rewards. One task, however, yielded no direct reward when completed, but instead produced a diffusible public good. Sufficient levels of this public good allowed patches to survive a periodic environmental disturbance.
To further explore the relationship between resource abundance and cooperation in a living system, we performed additional experiments in which populations of the bacterium Vibrio cholerae grew and evolved in environments with different nutrient concentrations. In these experiments, the formation of biofilms enhanced survival of periodic population bottlenecks. The amount of biofilm produced and the composition of each population were measured to determine the success of evolved cooperators in each environment.
Although the microbial and digital systems used in this study were quite distinct, we observed the same evolutionary response to resource abundance. Specifically, we found that cooperation does not simply increase as resource becomes more abundant; rather, that it can be maintained only above a critical level of resource.
Materials and Methods
Computational Model
Avida (Ofria & Wilke, 2004) is a computational platform that has been used to study complex ecological and evolutionary dynamics in a wide variety of contexts (see e.g., Chow et al., 2004, Clune et al., 2012, Lenski et al., 2003, Wilke et al., 2001). This section provides a brief overview of Avida and describes how it was used to model the cooperative formation of biofilms.
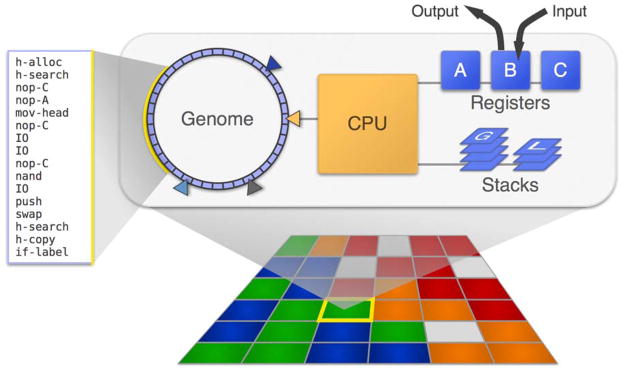
Experiments in Avida act on populations of self-replicating digital organisms (small computer programs), each of which resides independently in a cell on a lattice. As depicted in Figure 1, each organism is “embodied” by a circular list of instructions (its genome), which are executed on virtual computer hardware allocated to that organism. This virtual hardware includes registers and stacks for storing data values and a central processing unit (CPU) for executing instructions. Instructions perform arithmetic and logical operations (e.g., addition, multiplication, and bit-shifts), control the flow of execution, perform input and output, and execute behaviors associated with replication. Instructions are typically executed sequentially, but some instructions enable execution to jump to a different location in the genome. An organism’s environment includes a small set of random numbers that can be input and stored in registers; an organism can also output the value of a register to the environment.
Figure 1. Anatomy of an Avidian.
Within populations (below), each individual organism (top) possesses a circular genome of instructions, which are executed on “virtual computer hardware” to produce that individual’s phenotype. Organisms compete for resources and space in their environment by completing logic tasks and can produce and excrete products into their surrounding environment.
In this study, populations were seeded using a single ancestor organism capable only of replication. Any additional behaviors, including the completion of logic tasks described below, arose through mutations that occurred during asexual replication. An organism replicated by first allocating additional space in its genome, into which it copied instructions from its genome line by line. During this process, mutations substituted one instruction with another with probability 0.75%. Mutations also deleted or inserted a randomly chosen instruction from the set of 26 instructions with probability 5% as the organism divided. Once this process completed, the genome was cleaved, and the resulting daughter organism was placed into a randomly chosen adjacent cell, replacing any organism occupying that cell. This process created positive assortment of types. Since genomes are circular, an organism continued to execute until it was replaced, typically replicating with each pass through the genome.
Individuals competed for space in a 100×100-cell lattice by completing a set of binary logic tasks, listed in Table 1. Whenever an organism output a number to its environment, Avida determined whether that value matched the result of any of the logic tasks when applied to numbers in the organism’s initial environment. If so, Avida recorded that the organism had completed the task and conferred on the organism a merit bonus proportional to that task’s complexity (for a discussion of task complexity, see Lenski et al., 2003). These merit bonuses, awarded once for each unique task completed, were given to both the parent and offspring immediately following replication. During each update, Avida’s unit of time, each organism executed an average of 30 instructions. An organism with more merit than its competitors executed proportionately more instructions from its genome per update. Therefore, by completing these tasks, an organism replicated more quickly, thus increasing its fitness.
Table 1.
Binary Logic Tasks in the Avida Biofilm Model
| Task | Logic Operation | Output* | Merit Bonus |
|---|---|---|---|
| NOT | ¬A, ¬B | 0110, 0101 | 2 |
| NAND | ¬(A ∧ B) | 0111 | 2 |
| AND | A ∧ B | 1000 | 4 |
| OR NOT | A ∨ ¬B, ¬A ∨ B | 1101, 1110 | 0** |
| OR | A ∨ B | 1011 | 8 |
| AND NOT | A ∧ ¬B, ¬A ∧ B | 0001, 0010 | 8 |
| NOR | ¬(A ∨ B) | 0100 | 16 |
| XOR | (A ∧ ¬B) ∨ (¬A ∧ B) | 0011 | 16 |
| EQU | (A ∧ B) ∨ (¬A ∧ ¬B) | 1100 | 32 |
The symbol ¬ denotes negation.
Outputs are listed for the example input values A: 1001, B: 1010. Note that Avida typically uses 32-bit integers.
The cooperative OR NOT task did not confer a merit bonus.
In addition to random numbers, the Avida environment also contains resources. In this study, populations evolved in environments with different levels of a required resource R, which diffused throughout the lattice. One unit of R was required to complete any task. As described by Equation 1, the level of R was determined for each cell c at the beginning of each update t, where RI represents the total inflow of resource, C is the set of all lattice cells, Nc is the set of cells adjacent to cell c, φ is the rate of resource diffusion into neighboring cells, and δ is the rate of resource decay. For all experiments reported, |C|=10000, φ=0.01, and δ=0.01. The inflow rate RI and initial resource level Rc(0) were adjusted to create environments with equilibrium R levels ranging f rom 2 to 64 units per cell.
| (1) |
The environment was defined by eight binary logic tasks (Table 1), each of which was rewarded proportionally to its complexity. A ninth task of intermediate complexity was defined that did not confer a merit reward. Instead, the completion of this OR NOT task created one unit of a public good P, which was placed into the environment at that location. P also diffused and decayed at a rate of 1% per update. This public good, akin to EPS produced by biofilm-forming bacteria, enabled organisms to survive a periodic environmental disturbance. During each update, this disturbance event randomly selected one lattice cell and examined each neighboring cell within a 5-cell distance, a region totaling 121 cells. If the average level of P in these cells was below 3 units, all organisms residing in that region were killed. Otherwise, all organisms were spared. These environmental disturbances presented an opportunity for non-producers to exploit the public good produced by neighboring cooperators. Importantly, the formation of these “biofilms” did not affect the flow of R or P in the environment (see Nadell et al., 2010 and Julou et al., 2013 for studies where flow is affected, leading to different biofilm structures). Individuals could not directly sense the level of resource in their surrounding or the presence or phenotypes of neighboring individuals.
For each of the equilibrium R levels studied, 30 replicate populations evolved for 100,000 updates, or an average of 16,700 generations. Each simulation started with a different seed for the pseudorandom number generator, which allowed each population to follow a unique evolutionary trajectory.
To allow populations to first reach sufficient densities, the environmental disturbance began at update 1,000. Otherwise, populations tended to die out. However, we note that the formation of biofilms was not necessary for survival. Most populations persisted throughout the duration of the simulations, regardless of the level of R. By forming biofilms, however, populations could maintain greater densities.
In all resource environments, the periodic disturbances drove a subset of the replicate populations to extinction. This typically occurred early in the simulations, when populations had not yet reached sufficient densities. We found no relationship between resource abundance and the proportion of populations that met extinction. Data for populations driven to extinction are not included in our results.
Ancestral Strain and Growth Media
Our wild type strain of Vibrio cholerae was derived from the wild-type El Tor biotype strain C6706str2, a streptomycin-resistant isolate of C6706 (Thelin & Taylor, 1996). Cultures were grown in Miller LB broth (Neogen Corp. Acumedia Product #7279). To provide different resource environments, LB concentrations of 0.25x, 0.5x, 0.75x, 1x, 1.25x, 1.5x, 1.75x, and 2x were created by serially diluting a 2x LB stock with a 2x NaCl solution (20 g L−1), thereby ensuring an equivalent salt concentration at each resource level.
Experimental Evolution of Biofilm Formation
To start cultures, cells were placed from frozen glycerol stock into 2 mL 1x LB and incubated overnight with vigorous (220 rpm) shaking at 35 °C. Cultures were then diluted 100-fold in fresh medium for each LB concentration studied, vortexed, and 160 μL aliquots were placed into wells in a sterile MBEC assay plate (Innovotech Inc.; Harrison et al., 2005). For each concentration, five replicate populations were grown for 24 hours at 35 °C on a fixed-speed nutating mixer (Fisher Scientific Inc. Model 22-363-152). To remove cells from the resulting biofilms, the lid was placed into a sterile microtiter plate containing 150 μL 1x LB per well and sonicated on a suspended tray for 45 minutes at 40 kHz (Bransonic 2510 bath sonicator, Branson Ultrasonics Corp.). The resulting product in each well was then diluted 100-fold using the appropriate media, and 150 μL was used to inoculate the same well in a sterile MBEC plate. The remainder was mixed with 30 μL 80% glycerol per well and stored at −80 °C. This process was repeated for 7 days.
Biofilm Growth Measurement
Biofilms were measured by crystal violet staining. Cultures were started and grown as previously described. 5 μL samples were then vortexed with 145 μL of the appropriate media concentration and used to inoculate wells in an MBEC assay plate. After 12 hours of growth in conditions identical to those described above, plates were washed with 150 μL phosphate buffered saline (PBS), fixed in 150 μL 95% ethanol (EtOH), and stained with 150 μL of 0.41% crystal violet solution (w/v in 12% EtOH). Following three additional washes with 150 μL PBS, the crystal violet was diluted in 300 μL 95% EtOH. The absorbance of the diluted crystal violet was then measured via spectroscopy at 595 nm (SpectraMax M5, Molecular Devices, LLC).
Estimating Population Composition
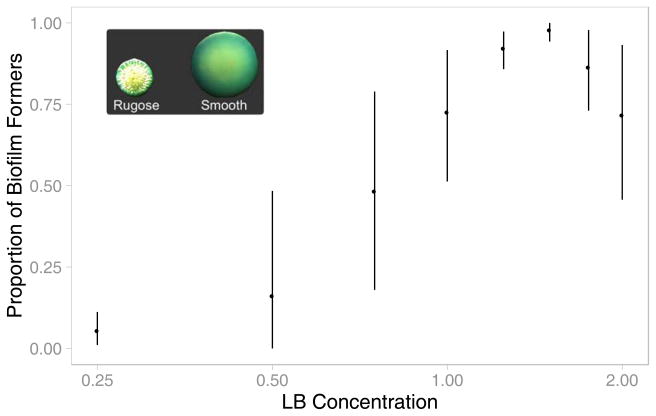
In V. cholerae, biofilm formers display a rugose colony morphology, while non-formers display a smooth morphology when plated on LB agar (see Figure 6 inset; Yildiz et al., 2004). To estimate the proportions of cooperative biofilm formers and non-forming cheaters present in populations, we compared the abundances of these two morphotypes. Biofilms were first removed from the pegs on which they were grown by sonication, as previously described. The resulting solutions were serially diluted in 1x LB, plated on LB agar, and incubated at 35 °C. After 24 hours, the number of rugose and smooth colony forming units (CFUs) were counted by visual examination.
Figure 6. Relative Frequency of Biofilm Formers Increases with Resource Richness.
The average proportion of biofilm-forming types within five replicate populations is shown with the LB concentrations in which they evolved. Evolved populations were plated, and the number of biofilm-forming rugose- and non-forming smooth colony forming units (CFUs) were counted. Error bars indicate 95% confidence intervals. Inset: Rugose and smooth colony morphologies exhibited by V. cholerae. Images are to scale.
Fitness of Evolved Strains in Biofilm and Planktonic Growth
One evolved rugose type and one evolved smooth type were isolated from one replicate evolved population (1x LB) and frozen as previously described. To compare these isolates, cultures were grown overnight in 1xLB. Competitions were initiated by combining equal volumes of the two overnight cultures after dilution to equal densities (absorbances at 595 nm). Rugose, smooth, and mixed cultures were grown in 1x LB overnight at 35 °C in an MBEC assay plate. Biofilms were sonicated from the pegs into 0.86% saline, diluted, and plated on LB agar. For planktonic growth, the cultures were extracted from the wells, diluted, and plated on LB agar. Rugose and smooth CFUs were then counted after incubation. These experiments were repeated using absorbance at 595 nm as a proxy for density. We used the change in frequency of rugose types during growth as a measure of its relative fitness (v). Specifically, v = p24(1 − p0)/p0 (1 − p24), where po is the initial proportion of rugose CFUs per mL, and p24 is their proportion after 24 hours (Ross-Gillespie et al., 2007).
Software, Data Analysis, and Archival
Avida 2.12.4 was used for the simulations described. Data analysis was done using R 3.2.3 (R Core Team, 2015). Reported confidence intervals were estimated by bootstrapping with 1000 resamples. The model, configurations, data, and analysis scripts will be archived and made publicly available prior to publication.
Results
In the Avida model, populations were grown and evolved in environments with different levels of a resource. Although this resource was not necessary for growth, it was required for completing self-benefitting tasks. This resource was also required to complete a cooperative task that produced a public good. As described above, a periodic environmental disturbance killed all organisms in a localized region when the amount of this public good present in that region was below a configured threshold. In this study, cooperators are defined as individuals that completed the public-good-producing task at least once during their lifetime, while cheaters did not. This classification is irrespective of the self-benefitting tasks performed, so many different cooperator and cheater phenotypes were possible. Cheaters could increase their survival rates under the protection of neighboring cooperators. A snapshot of a representative population in this system is shown in Figure S1 and Supplemental Video 11.
Cooperation is Maintained Above a Critical Resource Threshold
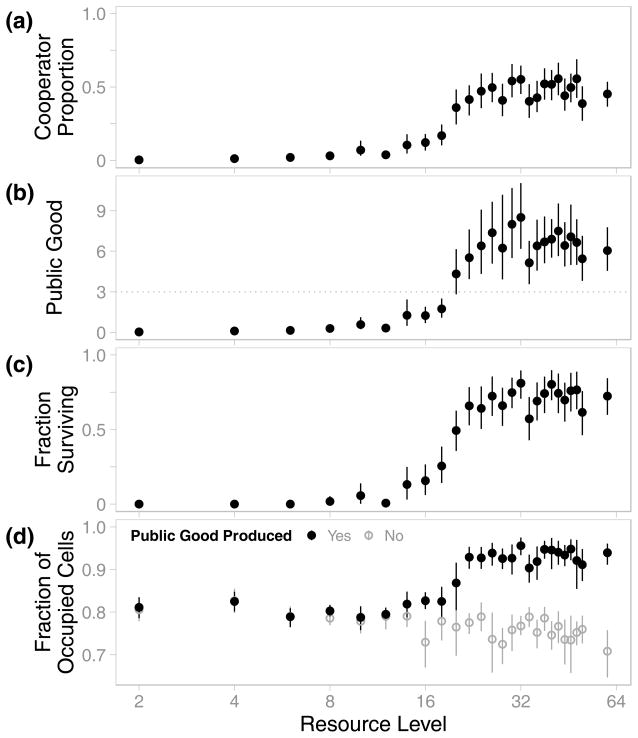
The relative frequency of cooperators at the end of simulations depended on the abundance of resource in the environments in which populations evolved (Figure 2a). In resource-rich environments (i.e., those with 20 units of resource per cell and greater), cooperators uniformly represented 46.8% (95% CI [44.2%, 49.6%]) of their populations (Figure 2a; see also Figure S2). Conversely, cooperator frequency was significantly lower in environments with14 units of resource per cell or less. In these resource-poor environments, individuals that completed the cooperative task were either absent or exceedingly rare (approx. 2.9%; 95% CI [2.09%, 4.03%]). Importantly, the transition between these two regimes occurred rapidly at intermediate levels of resource (approx. 14–20 units per cell).
Figure 2. Cooperation and Survival at Different Resource Levels.
(a) Although cooperation arose in all environments, it was only maintained at high proportions when resource was abundant. (b) In environments where cooperation was maintained, the average level of public good exceeded the threshold for survival imposed by the environmental disturbance events (3 units, dotted line). (c) These high levels of public good in resource-rich environments enabled populations to survive the majority of environmental disturbances. (d) As a result, populations in resource-rich environments maintained greater densities. When the cooperative task did not produce public good (open grey points), density did not change with resource abundance. For all panels, data shown are the averages among replicate populations at the end of simulations, and disturbance data are from the final 100 updates. Error bars indicate 95% confidence intervals.
As a result of increased cooperator abundances, populations in resource-rich environments produced more public good than populations in resource-poor environments (Figure 2b). In fact, the average per-cell level of public good produced by these evolved populations was substantially higher than the amount required to survive the environmental disturbance. As shown in Figure 2c, this behavior enabled populations in resource-rich environments to survive approximately 70% (95% CI [66.2%, 72.5%]) of the environmental disturbance events, while the survival rate was only 3% (95% CI [1.2%, 5.1%]) among populations in resource-poor environments.
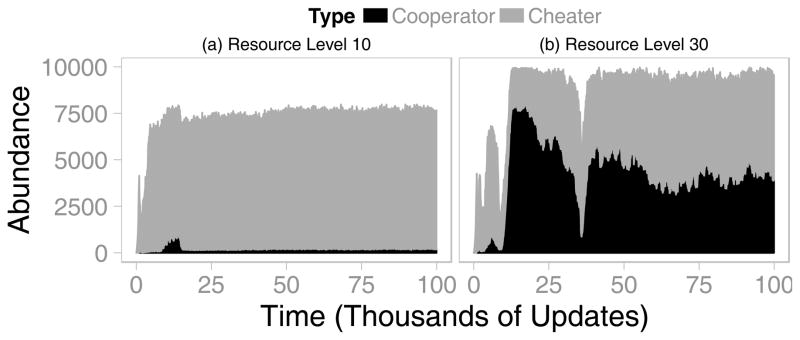
In our model, localized reproduction and environmental disturbances created positive assortment, which can promote cooperation (Kerr et al., 2002, Pepper & Smuts, 2002). As depicted in Figure S1, these processes segregated cooperators and cheaters into different patches. Although these changes continually altered the spatial distribution of phenotypes (Supplemental Video 1), the overall abundances of the cooperator and cheater phenotypes reached stability (Figures 3, S2). In the population shown in Figure 3b, for example, cooperators reached a frequency of approximately 45% shortly after environmental disturbances had nearly driven that population to extinction at around update 35,000.
Figure 3. Population Dynamics of Cooperation in Different Environments.
The number of cooperators and cheaters are shown over time in two representative populations. Note that cheater abundances are shown stacked on top of cooperator abundances. (a) In a resource-poor environment, cheaters maintain their initial dominance throughout the simulation. (b) In a resource-rich environment, cooperators quickly rise in abundance immediately after their population is drastically thinned by environmental disturbance. Cheaters nearly drive cooperators to extinction by approximately 35,000 updates. However, the susceptible population was thinned, which once again allowed cooperators to rebound.
Populations with Cooperators Reach Higher Densities
Populations in resource-rich environments maintained greater densities than those in resource-poor environments (Figure 2d). These increases in population size were not simply due to the addition of resource. In control simulations where completion of the cooperative task did not produce public good, population sizes were not significantly affected by resource level (Figure 2d). Therefore, these larger populations were not the result of environmental enrichment, but rather the increased survival of environmental disturbances provided by the cooperative production of public good (Figures 2b, 2c). This finding indicates that public good production provided a selective advantage in resource-rich environments.
Cooperator Success is Not Due to Increased Mutational Opportunities
Importantly, because populations in resource-rich environments were larger, they experienced more growth, and therefore more mutational opportunities. Because of this, the relative lack of cooperation in resource-poor environments seen in Figure 2a may have occurred because these populations had not yet evolved to complete the cooperative task. However, Figure S3a shows that the cooperative task arose in all environments, and that the rate at which populations acquired this behavior was not affected by resource level. In fact, populations evolved to complete the most complex (and most beneficial) task, EQU, in all environments (Figure S3a). Although the time required for this trait to first arise was not significantly affected by resource level (Figure S3b), the number of populations in which it arose was (Figure S3a).
Changing Environments Affect Cooperation
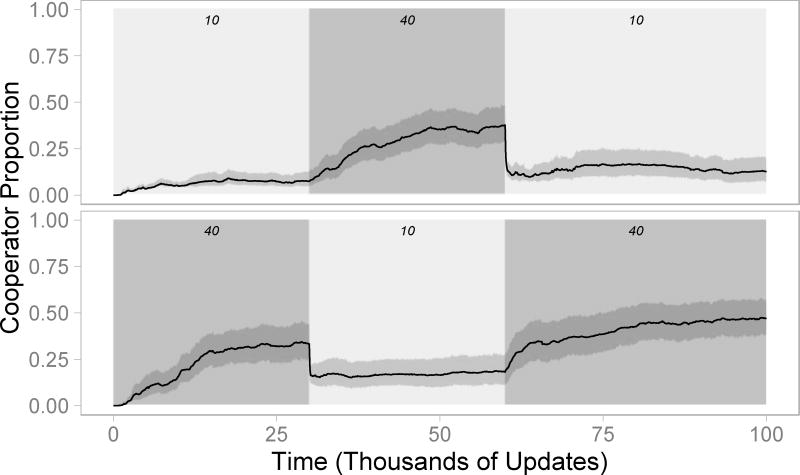
We next subjected populations to changes in resource levels that crossed the critical threshold. Starting in either resource-rich (R=40) or resource-poor (R=10) environments, populations evolved for 30,000 updates before the resource level abruptly changed. During the next 30,000 updates, populations from the resource rich environment experienced the resource poor environment, and vice versa. A second environmental shift then brought resource levels back to their initial values for the remainder of the simulations.
These environmental shifts strongly affected cooperation in both treatments (Figure 4). When populations transitioned from resource poor to resource rich environments, cooperation increased substantially. Conversely, when resource levels shifted from rich to poor, cooperation quickly dropped. These results demonstrate that environmental changes can rapidly alter the types of interactions that occur within populations.
Figure 4. Cooperation in Changing Environments.
Evolving populations experienced an environmental shift at updates 30,000 and 60,000. Numbers above each region indicate the per-cell equilibrium resource level (R). Mean cooperator proportion is shown among replicate populations, while shaded areas indicate 95% confidence bands. (top) When populations shifted from resource-poor to resource-rich environments, mean cooperator proportion increased from 7% to 38%. Upon returning to a resource-poor environment, mean cooperator proportion rapidly fell and remained near 12%. (bottom) Similarly, when populations temporarily experienced resource-poor environments, mean cooperator proportion dropped from 33% to 17%. When populations were returned to resource-rich environments, mean cooperator proportion reached 47%.
Critical Resource Threshold in Vibrio cholerae
To determine whether this critical transition to cooperation is a general feature of the evolution of cooperation in different resource environments, we also conducted experiments using a microbial system. Populations of the bacterial pathogen Vibrio cholerae were grown in different concentrations of LB medium in MBEC assay plates (Harrison et al., 2005), where the formation of biofilms allowed constituent cells to adhere to polystyrene pegs protruding from the lid into each well. Periodically, biofilms were sonicated from the pegs, diluted, and used to inoculate a fresh plate. This process was repeated for 7 days. Importantly, the wild-type, ancestral strain exhibits poor biofilm formation due to repression of genes that synthesize EPS by the density-dependent process of quorum sensing (Hammer & Bassler, 2003, Zhu & Mekalanos, 2003, Waters et al., 2008).
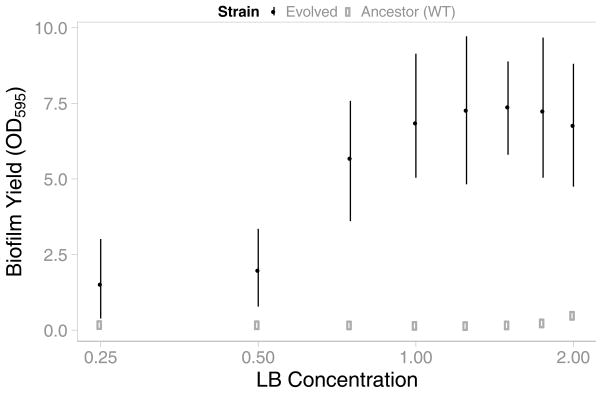
Populations that evolved in resource-rich environments formed significantly more robust biofilms than those that evolved in resource-poor environments (Figure 5). As with the simulated populations, this increase in cooperation occurred rapidly above a critical level of resource. In the environments studied, this transition occurred between 0.5x and 0.75x LB. We refer to environments with levels of resource below this threshold as resource-poor (0.25x and 0.50x LB), and those above it as resource-rich (0.75x LB and greater). Although EPS production was enriched in all environments due to our method of daily passaging, the increases seen in resource-rich environments were significantly greater than those in resource-poor environments (Figure 5). Among the environments within each of these two regimes, no significant differences in biofilm formation were observed, again leading to a bimodal distribution of non-cooperators and cooperators. In contrast, biofilm formation by the ancestral wild type strain was not affected by resource concentration (Figure 5). This indicates that the increases in biofilm formation by evolved populations were not simply due to increased access to resource or nutrient regulation of biofilm formation. Moreover, the lack of biofilm formation at low resource levels was not due to a lack of growth, as V. cholerae was able to reach densities in 0.5x LB equivalent to those observed at 1.75x and higher than those at 2.0x (Figure S4). Therefore, the differences in biofilm formation among evolved populations are not due to adaptive responses to different environments as has been observed in other species (Ramli et al., 2013, Vivas et al., 2008), but rather to selection for increased EPS production.
Figure 5. Biofilm Formation in Different Resource Concentrations.
Biofilm formation increased among all evolved strains (filled black points). However, strains that evolved in resource-rich environments (LB concentrations of approx. 0.75x and above) formed substantially larger biofilms than those that evolved in resource-poor environments (LB concentrations below 0.75x). The wild type ancestral strain formed weak biofilms in all environments (open grey points). Error bars indicate 95% confidence intervals.
EPS production by V. cholerae leads to altered colony morphology (Yildiz et al., 2004). As depicted in Figure 6 (inset), smooth colonies, which are larger and appear more translucent, form poor biofilms, while rugose colonies, which are smaller in size and wrinkly in appearance, form robust biofilms. Consistent with its poor biofilm formation, the wild-type ancestor strain displayed a smooth colony morphology. Figure 6 shows that the proportion of biofilm-forming rugose types increases in resource-rich environments, which corresponds with the observed increases in biofilm formation (Figure 5).
Non-EPS Producers Utilize Public-Goods Production to Increase Fitness
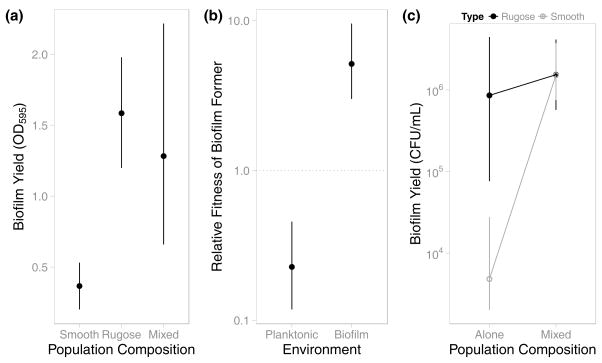
Biofilm formation in V. cholerae has been shown to be a cooperative trait allowing sequestration of public goods (Drescher et al., 2014), while providing mechanisms to exclude conspecific members from the group (Nadell et al., 2015). We therefore sought to determine if biofilm formation in our experimental evolution studies was exploitable. Six unique smooth and rugose clones were isolated from the 1x evolved lines. As expected, EPS production by the rugose isolates significantly enhanced biofilm formation versus smooth cells (Figure 7a). When rugose and smooth cells were co-inoculated at approximately equivalent initial frequencies, smooth isolates dominated the planktonic culture (Figure 7b). After growth in monoculture for 24 hours, approximately 104 smooth cells were measured on the pegs as compared with approximately 106 rugose cells (Figure 7c). In mixed cultures, the number of rugose cells associated with the pegs did not significantly change, however the number of smooth cells increased more than 100-fold to 2×106 cells, nearly the same as the number of rugose cells. Importantly, however, on average 50% of the biofilm consisted of smooth cells (95% CI [36.2%, 57.5%]), indicating these cells were able to access the biofilm formed by rugose cells and survive the passaging event. Therefore, EPS production by rugose cells could serve as a cooperative public good that was utilized by the smooth cells. However, the incorporation of smooth cells did not significantly reduce biofilm formation or the number of rugose cells (Figure 7c). This finding is consistent with previous studies showing that V. cholerae prevented invasion of planktonic cells to the biofilm interior (Nadell et al., 2015, Schluter et al., 2015). Similarly, Irie et al. recently demonstrated that although Pseudomonas aeruginosa socially produce the PSL polysaccharide and its benefits are shared amongst producers, it is not cheatable (Irie et al., 2016).
Figure 7. Competition Between Evolved Types.
Biofilm-forming (rugose) and non-forming (smooth) types were isolated from evolved populations and grown both alone and together. (a) Populations containing the evolved biofilm-forming type produced significantly more robust biofilms than monocultures of the non-forming type. (b) When grown with the evolved non-forming type, the biofilm-forming type was disadvantaged in planktonic growth. However, the biofilm-forming type handily outcompeted the non-former in colonizing a surface. (c) Although the non-forming type produced significantly weaker biofilms in monoculture, cultures initiated using an approx. 1:1 mixture of biofilm-forming and non-forming types produced biofilms that were as robust as those formed by biofilm-forming monocultures. Despite this enrichment for non-forming cells in mixed cultures, growth of the biofilm-forming type was not affected. In all plots, error bars indicate 95% confidence intervals.
Discussion
We have explored how resource abundance affects the evolution and maintenance of biofilm formation, a form of public goods cooperation. In both digital and microbial populations, cooperation was rare when a required resource was sparse (Figures 2a and 5). However, cooperation persisted when resource was abundant. In both systems, the transition between these two evolutionary outcomes occurred rapidly at a critical level of resource, which suggests that selection for biofilm formation was different in these two environments. When simulated populations experienced a shift from resource-rich to resource-poor environments, cooperation rapidly decreased (Figure 4). Conversely, when the environment became resource rich, cooperation increased substantially.
Environmental conditions and other ecological factors drive selection for all traits, and social behaviors are no exception. In a clear demonstration of this relationship, Zhang and Rainey showed that the costly production of pyoverdin, an extracellular metabolite secreted by Pseudomonas fluorescens, is adaptive and cheatable in iron-limiting environments, but maladaptive when iron was more abundant (Zhang & Rainey, 2013). Similarly, several studies have shown that resource concentration can dictate where the interactions of cross-feeding microbial strains fall on the spectrum between mutualism and parasitism (Bull and Harcombe, 2008, Hom and Murray, 2014, Nielsen et al., 2000). Notably, by adjusting amino acid concentrations, Hoek et al. recently altered the relationship between two yeast strains through several distinct outcomes from obligate mutualism to competitive exclusion (Hoek et al., 2016).
The bimodal response that occurs in response to resource abundance suggests that below some critical level of resource, the benefits of biofilm formation do not compensate for the costs. In these environments, selection favors non-production (Figures 2a and 5). Meanwhile, above critical resource levels, cooperation can be beneficial, which also presents opportunities for exploitation by cheaters. The plateau in cooperation that we observe in resource rich environments may indicate that additional investment in biofilm production yields diminishing returns (Foster 2004). Indeed, further production did not enhance survival once subpopulations had produced sufficient public good to survive the environmental disturbance or to adhere to the pegs. A similar response to changes in resource concentration has been shown by cooperative traits that mediate plant infection by Agrobacterium tumefaciens (Platt et al., 2012). In that study, harboring a plasmid encoding those traits was costly when resources were limited, but beneficial in resource rich environments.
Importantly, different cooperative behaviors or species can respond differently to changing environmental conditions. The proportion of P. fluorescens SBW25 wrinkly spreader morphotypes, which cooperatively form a mat at the air-broth interface, increases linearly with resource concentration (Travisano and Rainey, 2000, Brockhurst et al., 2008). When populations of these strains instead face different levels of environmental disturbance, cooperation shows a hump-shaped response, whereby cooperation peaks at intermediate disturbance (Brockhurst et al., 2010).
Natural populations commonly reside in complex environments, where organisms are likely to experience ecological changes within their lifetime. Our work has provided further evidence that cooperative behaviors are occasionally maladaptive. In these instances, types can gain evolutionary stability by cooperating only when doing so is beneficial, and otherwise focusing on individual growth (Cornforth et al., 2012, Heilmann et al., 2015, Darch et al., 2012). Indeed, several mechanisms have been identified that allow organisms to cooperate facultatively.
In Pseudomonas aeruginosa, swarming is enabled by the cooperative secretion of biosurfactants. By producing these public goods only when carbon concentrations exceed what is necessary for growth, wild-type cells are immune to cheating, a phenomenon termed metabolic prudence (Xavier et al., 2011). While our Avida populations were not able to sense their environments, our evolved Vibrio cholerae types may be displaying a similar form of metabolic prudence. Our experiments indicate that the metabolic prudence may not just affect the expression of cooperative functions, but may extend to evolutionary timescales as well. Further experiments exploring how biofilm formers respond to fluctuating environments could highlight whether metabolic prudence leads populations down diverging paths based on environmental conditions.
Facultative cooperation can also be mediated by quorum sensing, a widespread form of signaling that allows microbial populations to respond to changes in population density and environmental conditions through the production and detection of small molecules (Waters and Bassler, 2005). Indeed, many cooperative traits are now understood to be regulated by quorum sensing systems (Bruger and Waters, 2015). Importantly, whether a cooperative trait is stabilized by quorum sensing in changing environments depends critically on the relationship between cooperative benefit and environmental change (Cornforth et al., 2012, Heilmann et al., 2015, Archetti and Scheuring, 2011). For instance, the formation of biofilms, which showed a sigmoidal response to changes in resource abundance in our experiments, is normally under quorum sensing control in V. cholerae. By using quorum sensing to selectively produce EPS only when doing so is beneficial, cooperator populations may gain evolutionary stability in fluctuating environments.
Similarly, P. aeruginosa and Vibrios use quorum sensing to regulate the cooperative production of extracellular proteases, which exhibit a similar bimodal response to changes in carbon and population density (Darch et al., 2012). Indeed, in Vibrio harveyi, quorum sensing control of extracellular proteases prevents defector invasion of the wild type strain, whereas a constitutive cooperator is rapidly driven to extinction (Bruger and Waters, 2016). Conversely, quorum sensing does not regulate the production of iron-scavenging siderophores, which instead shows a linear response to resource abundance (Brockhurst et al., 2008). Understanding how cooperation co-evolves with mechanisms like quorum sensing and metabolic prudence remains an important challenge that requires consideration of how selection for the cooperative behavior in question is affected by the environment in which it occurs
The relatedness of interacting individuals was an important factor in both of our systems. In the computational model, localized replication and a lack of mixing provided a degree of spatial structuring for all individuals. The formation of biofilms, however, allowed patches of producers to maintain persistent interactions, while patches of cheaters were continually fragmented by environmental disturbances. Similarly, the formation of biofilms created structuring within the populations of V. cholerae. As a result, producers were more likely to interact with closely related producers than with others. Kin were therefore more likely to benefit from cooperation, thus lessening the detrimental impact of cheaters. Further, populations can physically exclude cheaters and non-relatives from the biofilm interior (Nadell et al., 2015) and may induce biofilm formation in response to competition (Oliviera et al., 2015). Several previous studies have demonstrated that spatial structuring and other mechanisms that support preferential cooperation among relatives can be crucial for maintaining cooperation (e.g., Strassmann et al., 2011, Kerr et al., 2002, Nadell & Bassler, 2011).
When biofilms enable populations to reach greater densities, cooperation may be further challenged by increased competition among kin (Taylor, 1992, Platt and Bever, 2009). Under these circumstances, cooperation can nevertheless be bolstered by pleiotropy (Mitri and Foster, 2016, Dandekar et al., 2012), policing (Wang et al., 2015; Frank, 2003), or non-social adaptation (Hammarlund et al., 2016, Asfahl et al., 2015). In our systems, the environmental disturbance created opportunities for the population to expand, allowing cooperators to escape competition from non-cooperators (Supplemental Video 1; Alizon and Taylor, 2008). Natural biofilms are often transient, and dispersal may provide a similar means of escape. To fully understand how cooperation evolves in biofilms, future studies therefore must consider all stages of the biofilm life cycle (see e.g., Poltak and Cooper, 2011).
This work has demonstrated that in some environments, cooperation is simply a poor strategy. Determining how environmental change alters selection is crucial for gaining a broader understanding of how cooperation evolves and is maintained in nature. This perspective is also necessary in the development of “anti-infective” treatments that target cooperative virulence factors (Crespi et al., 2014, Petersen et al., 2009), as pathogens are likely to face a large diversity of ecological conditions within their host.
Supplementary Material
Acknowledgments
This work was supported in part by National Science Foundation grants CNS-1059373, CNS-0915855, DBI-0939454, CCF-0820220, CNS-0751155, DBI-1309318, and National Institutes of Health grants R01-GM109259 and R01-GM110444. Computational resources and support were provided by the Institute for Cyber Enabled Research at Michigan State University and A. Pakanati.
Footnotes
doi:10.6084/m9.figshare.2063355.v1
References
- Alizon S, Taylor P. Empty sites can promote altruistic behavior. Evolution. 2008;62:1335–1344. doi: 10.1111/j.1558-5646.2008.00369.x. [DOI] [PubMed] [Google Scholar]
- Asfahl KL, Walsh J, Gilbert K, Schuster M. Non-social adaptation defers a tragedy of the commons in Pseudomonas aeruginosa quorum sensing. ISME J. 2015;9(1):734–1746. doi: 10.1038/ismej.2014.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archetti M, Scheuring I. Coexistence of cooperation and defection in public goods games. Evolution. 2011;65:1140–1148. doi: 10.1111/j.1558-5646.2010.01185.x. [DOI] [PubMed] [Google Scholar]
- Brockhurst MA, Buckling A, Racey D, Gardner A. Resource supply and the evolution of public-goods cooperation in bacteria. BMC Biol. 2008;6:20. doi: 10.1186/1741-7007-6-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockhurst MA, Habets MG, Libberton B, Buckling A, Gardner A. Ecological drivers of the evolution of public-goods cooperation in bacteria. Ecology. 2010;91:334–340. doi: 10.1890/09-0293.1. [DOI] [PubMed] [Google Scholar]
- Bruger E, Waters C. Sharing the sandbox: Evolutionary mechanisms that maintain bacterial cooperation [version 1; referees: 2 approved] F1000Research. 2015;4(1):504. doi: 10.12688/f1000research.7363.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruger E, Waters C. Bacterial quorum sensing stabilizes cooperation by optimizing growth strategies. Appl Environ Microbiol. 2016 doi: 10.1128/AEM.01945-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull JJ, Harcombe WR. Population dynamics constrain the cooperative evolution of cross-feeding. PLoS ONE. 2008;4:e4115. doi: 10.1371/journal.pone.0004115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow SS, Wilke CO, Ofria C, Lenski RE, Adami C. Adaptive radiation from resource competition in digital organisms. Science. 2004;305:84–86. doi: 10.1126/science.1096307. [DOI] [PubMed] [Google Scholar]
- Clune J, Pennock RT, Ofria C, Lenski RE. Ontogeny tends to recapitulate phylogeny in digital organisms. Am Nat. 2012;180:E54–63. doi: 10.1086/666984. [DOI] [PubMed] [Google Scholar]
- Cornforth DM, Sumpter DJ, Brown SP, Brannstrom A. Synergy and group size in microbial cooperation. Am Nat. 2012;180:296–305. doi: 10.1086/667193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespi B, Foster K, Ubeda F. First principles of Hamiltonian medicine. Phil Trans R Soc B. 2014;369(2):0130366. doi: 10.1098/rstb.2013.0366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandekar AA, Chugani S, Greenberg EP. Bacterial quorum sensing and metabolic incentives to cooperate. Science. 2012;338(2):64–266. doi: 10.1126/science.1227289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darch SE, West SA, Winzer K, Diggle SP. Density-dependent fitness benefits in quorum-sensing bacterial populations. PNAS. 2012;109(8):259–8263. doi: 10.1073/pnas.1118131109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drescher K, Nadell CD, Stone HA, Wingreen NS, Bassler BL. Solutions to the public goods dilemma in bacterial biofilms. Curr Biol. 2014;24:50–55. doi: 10.1016/j.cub.2013.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster KR. Diminishing returns in social evolution: the not-so-tragic commons. J Evol Biol. 2004;17:1058–1072. doi: 10.1111/j.1420-9101.2004.00747.x. [DOI] [PubMed] [Google Scholar]
- Frank SA. Repression of competition and the evolution of cooperation. Evolution. 2003;57(6):93–705. doi: 10.1111/j.0014-3820.2003.tb00283.x. [DOI] [PubMed] [Google Scholar]
- Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol. 2004;2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- Hall-Stoodley L, Stoodley P. Evolving concepts in biofilm infections. Cell Microbiol. 2009;11:1034–1043. doi: 10.1111/j.1462-5822.2009.01323.x. [DOI] [PubMed] [Google Scholar]
- Hamilton WD. The genetical evolution of social behaviour. I. J Theor Biol. 1964a;7:1–16. doi: 10.1016/0022-5193(64)90038-4. [DOI] [PubMed] [Google Scholar]
- Hamilton WD. The genetical evolution of social behaviour. II. J Theor Biol. 1964b;7:17–52. doi: 10.1016/0022-5193(64)90039-6. [DOI] [PubMed] [Google Scholar]
- Hammarlund SP, Connelly BD, Dickinson KJ, Kerr B. The evolution of cooperation by the Hankshaw effect. Evolution. 70:1376–1385. doi: 10.1111/evo.12928. [DOI] [PubMed] [Google Scholar]
- Hammer BK, Bassler BL. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol Microbiol. 2003;50:101–104. doi: 10.1046/j.1365-2958.2003.03688.x. [DOI] [PubMed] [Google Scholar]
- Hardin G. The tragedy of the commons. Science. 1968;162:1243–1248. [PubMed] [Google Scholar]
- Harrison JJ, Stremick CA, Turner RJ, Allan ND, Olson ME, Ceri H. Microtiter susceptibility testing of microbes growing on peg lids: a miniaturized biofilm model for high-throughput screening. Nat Protoc. 2005;5:1236–1254. doi: 10.1038/nprot.2010.71. [DOI] [PubMed] [Google Scholar]
- Heilmann S, Krishna S, Kerr B. Why do bacteria regulate public goods by quorum sensing?-How the shapes of cost and benefit functions determine the form of optimal regulation. Front Microbiol. 2015;6:767. doi: 10.3389/fmicb.2015.00767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoek TA, Axelrod K, Biancalani T, Yurtsev EA, Liu J, Gore J. Resource availability modulates the cooperative and competitive nature of a microbial cross-feeding mutualism. PLoS Biol. 2016;14:e1002540. doi: 10.1371/journal.pbio.1002540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hom EFY, Murray AW. Niche engineering demonstrates a latent capacity for fungal-algal mutualism. Science. 2014;345(9):4–98. doi: 10.1126/science.1253320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie Y, Roberts A, Kragh KN, Gordon VD, Hutchinson J, Allen RJ, Melaugh G, Bjarnsholt T, West SA, Diggle SP. The Pseudomonas aeruginosa PSL polysaccharide is a social but non-cheatable trait in biofilms. bioRxiv. 2016 doi: 10.1128/mBio.00374-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julou T, Mora T, Guillon L, Croquette V, Schalk IJ, Bensimon D, Desprat N. Cell-cell contacts confine public goods diffusion inside Pseudomonas aeruginosa clonal microcolonies. Proc Natl Acad Sci U S A. 2013;110:12577–12582. doi: 10.1073/pnas.1301428110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr B, Riley MA, Feldman MW, Bohannan BJ. Local dispersal promotes biodiversity in a real-life game of rock-paper-scissors. Nature. 2002;418:171–174. doi: 10.1038/nature00823. [DOI] [PubMed] [Google Scholar]
- Lenski RE, Ofria C, Pennock RT, Adami C. The evolutionary origin of complex features. Nature. 2003;423:139–144. doi: 10.1038/nature01568. [DOI] [PubMed] [Google Scholar]
- Mah TF, O’Toole GA. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 2001;9:34–39. doi: 10.1016/s0966-842x(00)01913-2. [DOI] [PubMed] [Google Scholar]
- Mah TF, Pitts B, Pellock B, Walker GC, Stewart PS, O’Toole GA. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature. 2003;426:306–310. doi: 10.1038/nature02122. [DOI] [PubMed] [Google Scholar]
- Mitri S, Foster KR. Pleiotropy and the low cost of individual traits promote cooperation. Evolution. 2016;70(4):88–494. doi: 10.1111/evo.12851. [DOI] [PubMed] [Google Scholar]
- Nadell CD, Bassler BL. A fitness trade-off between local competition and dispersal in Vibrio cholerae biofilms. Proc Natl Acad Sci U S A. 2011;108:14181–14185. doi: 10.1073/pnas.1111147108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadell CD, Drescher K, Wingreen NS, Bassler BL. Extracellular matrix structure governs invasion resistance in bacterial biofilms. ISME J. 2015 doi: 10.1038/ismej.2014.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadell CD, Foster KR, Xavier JB. Emergence of spatial structure in cell groups and the evolution of cooperation. PLoS Comput Biol. 2010;6:e1000716. doi: 10.1371/journal.pcbi.1000716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen AT, Tolker-Nielsen T, Barken KB, Molin S. Role of commensal relationships on the spatial structure of a surface-attached microbial consortium. Environ Microbiol. 2000;2(5):9–68. doi: 10.1046/j.1462-2920.2000.00084.x. [DOI] [PubMed] [Google Scholar]
- Ofria C, Wilke CO. Avida: a software platform for research in computational evolutionary biology. Artif Life. 2004;10:191–229. doi: 10.1162/106454604773563612. [DOI] [PubMed] [Google Scholar]
- Oliviera NM, Martinez-Garcia E, Xavier J, Durham WM, Kolter R, Kim W, Foster KR. Biofilm formation as a response to ecological competition. PLoS Biol. 2015;13:e1002191. doi: 10.1371/journal.pbio.1002191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepper JW, Smuts BB. A mechanism for the evolution of altruism among non-kin: Positive assortment through environmental feedback. American Naturalist. 2002;160:205–213. doi: 10.1086/341018. [DOI] [PubMed] [Google Scholar]
- Petersen A, Aarestrup FM, Olsen JE. The in vitro fitness cost of antimicrobial resistance in. Escherichia coli varies with the growth conditions. FEMS Microbiol Lett. 2009;299(5):3–59. doi: 10.1111/j.1574-6968.2009.01734.x. [DOI] [PubMed] [Google Scholar]
- Platt TG, Bever JD. Kin competition and the evolution of cooperation. Trends Ecol Evol. 2009;24:370–377. doi: 10.1016/j.tree.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt TG, Bever JD, Fuqua C. A cooperative virulence plasmid imposes a high fitness cost under conditions that induce pathogenesis. Proc Biol Sci. 2012;279:1691–1699. doi: 10.1098/rspb.2011.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poltak SR, Cooper VR. Ecological succession in long-term experimentally evolved biofilms produces synergistic communities. ISME J. 2011;5(3):69–378. doi: 10.1038/ismej.2010.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popat R, Crusz SA, Messina M, Williams P, West SA, Diggle SP. Quorum-sensing and cheating in bacterial biofilms. Proc Biol Sci. 2012;279:4765–4771. doi: 10.1098/rspb.2012.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainey PB, Rainey K. Evolution of cooperation and conflict in experimental bacterial populations. Nature. 2003;425:72–74. doi: 10.1038/nature01906. [DOI] [PubMed] [Google Scholar]
- R Core Team. R: A language and environment for statistical computing. Vienna, Austria: 2015. [Google Scholar]
- Ramli NS, Eng Guan C, Nathan S, Vadivelu J. The effect of environmental conditions on biofilm formation of Burkholderia pseudomallei clinical isolates. PLoS ONE. 2013;7:e44104. doi: 10.1371/journal.pone.0044104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Gillespie A, Gardner A, West SA, Griffin AS. Frequency dependence and cooperation: theory and a test with bacteria. Am Nat. 2007;170:331–342. doi: 10.1086/519860. [DOI] [PubMed] [Google Scholar]
- Schluter J, Nadell CD, Bassler BL, Foster KR. Adhesion as a weapon in microbial competition. ISME J. 2015;9:139–149. doi: 10.1038/ismej.2014.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strassmann JE, Gilbert OM, Queller DC. Kin discrimination and cooperation in microbes. Annu Rev Microbiol. 2011;65:349–367. doi: 10.1146/annurev.micro.112408.134109. [DOI] [PubMed] [Google Scholar]
- Taylor PD. Altruism in viscous populations-an inclusive fitness model. Evol Ecol. 1992;6(3):52. [Google Scholar]
- Thelin KH, Taylor RK. Toxin-coregulated pilus, but not mannose-sensitive hemagglutinin, is required for colonization by Vibrio cholerae O1 El Tor biotype and O139 strains. Infect Immun. 1996;64:2853–2856. doi: 10.1128/iai.64.7.2853-2856.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travisano M, Rainey PB. Studies of adaptive radiation using model microbial systems. Am Nat. 2000;156:S35–S44. doi: 10.1086/303414. [DOI] [PubMed] [Google Scholar]
- Vivas J, Padilla D, Real F, Bravo J, Grasso V, Acosta F. Influence of environmental conditions on biofilm formation by Hafnia alvei strains. Vet Microbiol. 2008;129:150–155. doi: 10.1016/j.vetmic.2007.11.007. [DOI] [PubMed] [Google Scholar]
- Wang M, Schaefer AL, Dandekar AA, Greenberg EP. Quorum sensing and policing of Pseudomonas aeruginosa social cheaters. PNAS. 2015;112(2):187–2191. doi: 10.1073/pnas.1500704112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters CM, Bassler BL. Quorum Sensing: Cell-to-cell communication in bacteria. Annu Rev Cell Dev Bi. 2005;21(3):19–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- Waters CM, Lu W, Rabinowitz JD, Bassler BL. Quorum Sensing Controls Biofilm Formation in Vibrio cholerae through Modulation of Cyclic Di-GMP Levels and Repression of vpsT. J Bacteriol. 2008;190:2527–2536. doi: 10.1128/JB.01756-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West SA, Griffin AS, Gardner A. Evolutionary explanations for cooperation. Curr Biol. 2007;17:R661–672. doi: 10.1016/j.cub.2007.06.004. [DOI] [PubMed] [Google Scholar]
- Wilke CO, Wang JL, Ofria C, Lenski RE, Adami C. Evolution of digital organisms at high mutation rates leads to survival of the flattest. Nature. 2001;412:331–333. doi: 10.1038/35085569. [DOI] [PubMed] [Google Scholar]
- Xavier JB, Foster KR. Cooperation and conflict in microbial biofilms. Proc Natl Acad Sci U S A. 2007;104:876–881. doi: 10.1073/pnas.0607651104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier JB, Kim W, Foster KR. A molecular mechanism that stabilizes cooperative secretions in Pseudomonas aeruginosa. Mol Microbiol. 2011;79:166–179. doi: 10.1111/j.1365-2958.2010.07436.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz FH, Liu XS, Heydorn A, Schoolnik GK. Molecular analysis of rugosity in a Vibrio cholerae O1 El Tor phase variant. Mol Microbiol. 2004;53:497–515. doi: 10.1111/j.1365-2958.2004.04154.x. [DOI] [PubMed] [Google Scholar]
- Zhang XX, Rainey PB. Exploring the sociobiology of pyoverdin–producing Pseudomonas. Evolution. 2013;67:3161–3174. doi: 10.1111/evo.12183. [DOI] [PubMed] [Google Scholar]
- Zhu J, Mekalanos JJ. Quorum sensing-dependent biofilms enhance colonization in Vibrio cholerae. Dev Cell. 2003;5:647–656. doi: 10.1016/s1534-5807(03)00295-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.