Abstract
The genetic code can be expanded to include unnatural amino acids (Uaas) by engineering orthogonal components involved in protein translation. To be compatible with live cells, side chains of Uaas have been limited to either chemically inert or bio-orthogonal (i.e., nonreactive toward biomolecules) functionalities. To introduce bioreactivity into live systems, the genetic code has recently been engineered to encode a new class of Uaas, the bioreactive Uaas. These Uaas, after being incorporated into proteins, specifically react with target natural amino acid residues via proximity-enabled bioreactivity, enabling the selective formation of new covalent linkages within and between proteins both in vitro and in live systems. The new covalent bonding ability has been harnessed within proteins to enhance photostability, increase thermostability, staple proteins recombinantly, and build optical nano-switches, and between proteins to pinpoint ligand-receptor interaction, target native receptors irreversibly, and generate covalent macromolecular inhibitors. These diverse bioreactivities, inaccessible to natural proteins, thus open doors to novel protein engineering and provide new avenues for biological studies, biotherapeutics and synthetic biology.
Keywords: expansion of the genetic code, unnatural amino acid, proximity enabled reactivity, protein crosslinking, protein interaction, protein engineering
Introduction
The essential framework for protein structure and function is provided by covalent peptide bonds in the backbone and noncovalent interactions among amino acid side chains. The disulfide bond formed between two cysteine residues is a unique covalent linkage between amino acid side chains, and plays crucial roles in the folding, stability, and activity of a variety of proteins such as antibodies, cytokines and membrane-associated receptors [1,2]. However, the weak bonding, reversibility, and redox sensitivity of the disulfide bond impose limitations on protein function, engineering, and applications [3]. Isopeptide bonds between Lys and Asn or Asp have also been discovered, but they form in the hydrophobic protein interior only and require an essential Glu or Asp residue to catalyze the reaction [4]. Although protein crosslinking is a common biological mechanism contributing to processes such as blood coagulation and tissue stabilization, the covalent crosslinking requires various enzymes to activate amino acid side chains for reaction [5]. In general, side chains of natural amino acid residues except cysteine rarely form covalent bonds spontaneously in native cellular environments. Therefore, a fundamental limitation of natural proteins is the inadequacy in covalent bonding of their side chains. Additional types of covalent linkages would expand avenues for generating novel protein properties and functions.
The genetic code specifies 20-22 natural amino acids for building proteins, and this code is preserved in virtually all life forms on earth with just minor variations [6,7]. In recent years, the genetic code has been artificially expanded to include unnatural amino acids (Uaas) by introducing into cells an orthogonal tRNA/aminoacyl-tRNA synthetase (aaRS) pair that is specific for the Uaa and compatible with protein translational machinery [8-10]. The orthogonal aaRS charges the desired Uaa onto the orthogonal tRNA, and the aminoacylated-tRNA incorporates the Uaa into proteins in response to a unique codon (the amber stop codon UAG is often used) through protein translation in live cells [11,12]. This methodology was initially established in E. coli [10], and later proven generally applicable in yeast [13,14], mammalian cells [15-17], stem cells [18], plants [19], invertebrates including C. elegans [20,21] and Drosophila [22], even as well as embryonic mouse brain [23]. More than 100 Uaas with a variety of side chains have been genetically incorporated into proteins using this approach [11,24-26], and this number is increasing with efforts from multiple research groups worldwide [27-33].
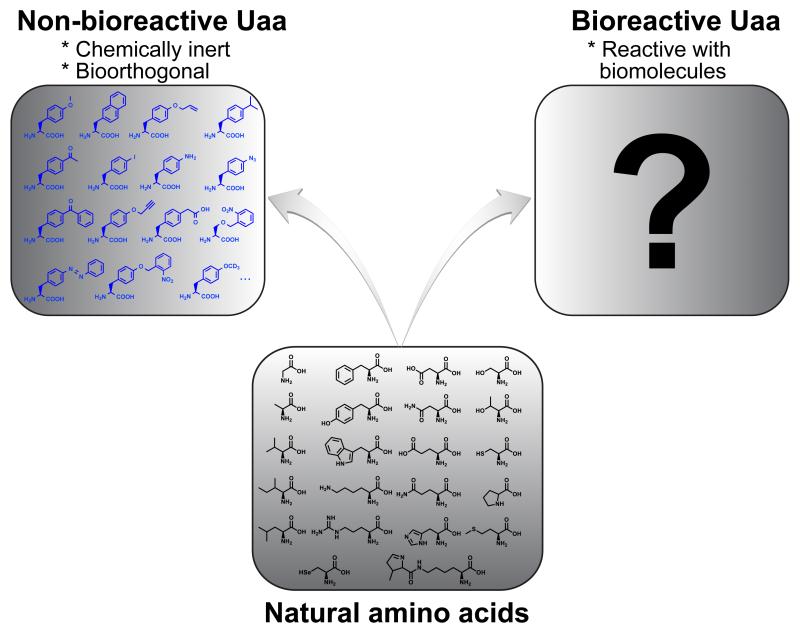
To be compatible with live cells or systems, all Uaas genetically incorporated in the past contain functional groups that are either chemically inert or bio-orthogonal, that is, they are non-bioreactive. To break the natural barrier to protein synthesis by enabling new covalent bonding ability for proteins in live cells, would it be possible to genetically encode bioreactive Uaas, i.e., Uaas with functional groups that can react with biomolecules inside cells (Figure 1)? Here we review how bioreactive Uaas, initially thought infeasible for genetic encoding, became a new class of Uaas specifiable by the genetic code in live cells. Examples are given to illustrate the unique abilities of these bioreactive Uaas affording to proteins via their specific reactivities.
Figure 1.
The genetic code has been expanded to include non-bioreative Uaas. Would it be possible to genetically encode bioreactive Uaas?
Methodology
Genetically encoding a bioreactive Uaa is a conundrum
The method of building new covalent linkages into proteins is to genetically incorporate a Uaa into the target protein and to enable the Uaa to react with a natural amino acid side chain to form a covalent bond selectively. Two conundrums arise for execution: 1) To form a new covalent bond between the side chains of a Uaa and a natural amino acid, the Uaa must be reactive toward the target natural amino acid. However, if the reactivity is uncontrolled, the ubiquitous presence of the natural amino acid in proteins and inside cells will result in undesired nonspecific linkages, which can potentially cause cytotoxicity as well. 2) The required bioreactivity may make the Uaa react with proteins involved in translation, thereby blocking the addition of the Uaa to the nascent protein via translation.
Proximity-enabled bioreactivity
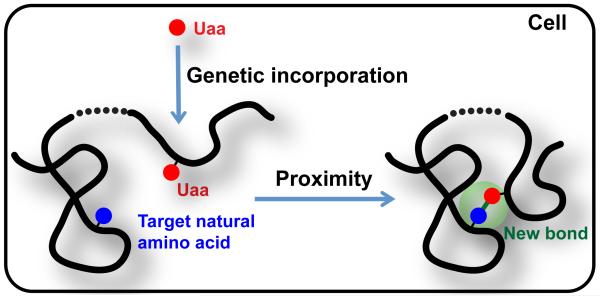
We hypothesize that this impasse of bioreactivity, genetic encoding, and selectivity can be overcome via the concept of Proximity-Enabled Bioreactivity (Figure 2) [34]. Specifically, we propose to tune the reactivity of the Uaa so that it does not react with free natural amino acids and other biomolecules under physiological conditions, thereby permitting genetic incorporation in vivo via endogenous translational machinery. When the Uaa comes into proximity to its target natural amino acid residue in proteins with appropriate orientation, the increased local effective concentration then facilitates the Uaa to selectively react with the side chain of the target natural residue to create a covalent bond. The bond can be built intramolecularly within a protein or intermolecularly between two proteins.
Figure 2.
Build new covalent bonds in proteins via proximity-enabled bioreactivity. Broken line indicates that proximity can be brought by intramolecular protein folding or conformational change, or by intermolecular protein interactions.
Feasibility of this hypothesis is supported by previous research on proximity effects on other systems. When small molecules are brought close by a DNA template, their reactivities can be enhanced several hundred folds [35]. Reactivities between proteins and small molecules can also be enhanced by proximity effect, which have been discovered in natural products [36] and utilized in developing irreversible ligands [37], small molecule inhibitors, and activity-based protein profiling [38]. Amino acid side chains readily get close to each other either within proteins or at the interface of interacting proteins, so we reasoned that harnessing the proximity of a designed Uaa and a target natural amino acid would enable the selective formation of a new covalent bond.
Existing chemistries for protein labeling and modification may not be directly applied for proximity-enabled bioreactivity in cells. Protein modification [39] generally target one type of residue, so the reactivity cannot be limited to a single residue and to the target protein alone in a mixture of proteins in cells. Innovative chemistries developed in activity-based protein profiling [38,40] usually target catalytic residues and are accelerated by enzyme catalytic mechanisms. For proximity-enabled bioreactivity, the covalent linkage can be desired anywhere inside or at the surface of protein, which might lack a catalytic active site. Bioorthogonal click reactions proceed with fast rates under mild biocompatible conditions [41], yet they require two non-natural reaction partners (e.g., azide and alkyne) and are designed inert to biological functionalities. Photocrosslinking is not specific to a particular amino acid residue.
The chemistry for proximity-enabled bioreactivity will involve a functional group incorporated into the protein that is selectively reactive toward a specific natural residue, and the reaction must proceed efficiently under physiological conditions. A single-step reaction is preferred; reactions that require multistep conversion or exogenous catalysts may not be compatible with cellular and in vivo administration. Lastly, the reaction should be non-toxic and should not negatively interfere with other endogenous biological processes.
First success with Uaa Ffact targeting Cys
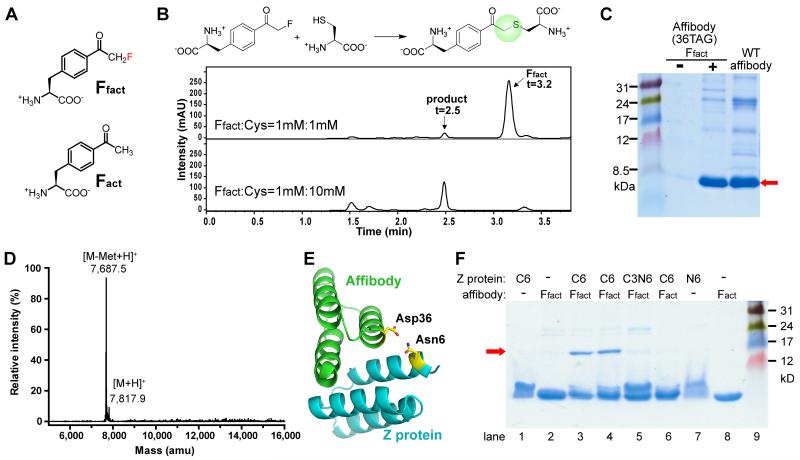
We designed p-2′-fluoroacetyl-phenylalanine 1 (Ffact, Figure 3A) as the Uaa to react with Cys [34]. The sulfhydryl group of Cys has the highest nucleophilicity among chemical groups occurring in natural amino acid side chains, so we expected that a weak electrophilic group, only when close to a Cys residue, would selectively react with it. C-F bond is strong and F is not a great leaving group in SN2 reactions, but the carbonyl group of Ffact would increase its reactivity making the C a weak electrophile. In fact, small molecule or peptidyl fluoromethyl ketones have been shown to react irreversibly with Cys in the active site of a cysteine proteinase [42] and the S6 kinase [40].
Figure 3.
Genetic encoding of the bioreactive Uaa Ffact and its reaction with Cys via proximity-enabled bioreactivity to covalently crosslink two proteins. (A) Structure of Ffact and the isosteric control Fact. (B) LC-MS analysis indicate that 1 mM Ffact reacted completely with Cys at 10 mM but < 5% at 1 mM in PBS pH 7.4 at 37°C for 24 h. (C) Site-specific incorporation of Ffact into affibody, shown by SDS-PAGE of mutant affibody expressed in E. coli with or without Ffact. (D) ESI-MS analysis of the mutant affibody from C confirmed that only Ffact was incorporated at site 36 and unmodified. (E) Structure of the affibody-Z complex (PDB 1LP1) with proximal Asp36 and Asn6 highlighted. (F) Incubation of the affibody(Ffact36) with the Z(Cys6) resulted in formation of the affibody-Z covalent complex (red arrow, lane 3 & 4), as shown in SDS_PAGE analysis. The Z protein contained Cys or Asn at residue 6 (C3 indicates Cys at residue 3); the affibody had Ffact or Fact at residue 36. Minus indicates no protein added. Lane 4 & 8: Tris buffer, pH 8.0; other lanes: PBS, pH 7.4. t = 1 h.
The reactivity of Ffact toward Cys was initially tested in vitro using amino acids. Because intracellular concentrations of amino acids (except Glu) are usually <1 mM and average protein concentration is 3 mM inside cells [43], we used 1 mM of Uaa to react with the target natural amino acid Cys at 0.05, 1 and 10 mM, respectively, in phosphate buffer (pH 7.4) at 37 °C for 24 hr to mimic the mild intracellular conditions. In reactions containing 1 mM Ffact and 50 μM Cys, no Ffact was found to react with Cys. A small percentage (< 5%) of Ffact reacted with Cys when Cys concentration was increased to 1 mM. In contrast, in reactions containing 1 mM Ffact and 10 mM Cys, Ffact was completely converted to the product (Figure 3B). A major effect of proximity is to increase the effective concentration of the reactants, with effective molarity reaching as high as 105 M [37]. These concentration-dependent results thus suggest that the reactivity of Ffact with Cys could be enhanced by proximity.
Given the presumption that bioreactive Uaas will not be able to go through translation, it is crucial to convincingly prove that the Uaa can be genetically incorporated into proteins in live cells. In addition, it is important to determine if the Uaa, after incorporation into the protein, remains intact or is modified by other biomolecules inside the cell. Since Ffact is structurally similar to p-acetyl-L-phenylalanine (Fact, Figure 3A) with only a hydrogen atom replaced by fluorine, we attempted to incorporate Ffact into an affibody protein at site 36 (codon for Asp36 mutated to the amber stop codon TAG) using the orthogonal tRNA-synthetase pair evolved for Fact [44]. After expressing in E. coli, full-length affibody production was markedly increased (43 fold) when Ffact was added in the growth media (Figure 3C). Electrospray ionization mass spectrometric (ESI-MS) analysis of the purified affibody showed that only Ffact was specifically incorporated at the 36TAG site (Figure 3D). More importantly, there were no MS peaks corresponding to the affibody with Ffact reacted with Cys, glutathione, imidazole, or any amino acid residue of the affibody, indicating that Ffact was stable and unmodified during protein biosynthesis and purification.
To determine if Ffact could react with Cys via proximity-enabled bioreactivity in protein context, we used the affibody-Z protein pair as the model system and introduced Ffact and Cys at their binding interface (Figure 3E). The affibody binds to its substrate Z protein with a Kd of 6 μM, and the structure of the complex is available to guide site selection [45]. We introduced Cys at site 6 of the Z protein, and incubated the purified affibody(Ffact36) with the purified Z(Cys6) in PBS buffer (pH 7.4) at 37 °C. A band with molecular weight corresponding to the affibody-Z complex was detected on denaturing SDS-PAGE (Figure 3F), indicating that the two proteins were covalently crosslinked. The yield of covalent complex formation was 63 ± 3% (n = 3). MS analysis of the reaction product confirmed that the complex was covalently linked by Ffact reacting with Cys. This complex band did not form when Ffact36 was replaced by the nonfluorinated Fact in the affibody or when Cys6 was replaced by Asn in the Z protein (Figure 3F). In addition, when we placed Cys at Z protein Asn3 instead of Asn6, no covalent affibody-Z complex was formed (Figure 3F, lane 5), indicating the reaction is proximity-dependent.
These results demonstrate that a bioreactive Uaa can be genetically encoded in E. coli cells and remains intact. Only when in proximity to a target natural amino acid residue, the incorporated bioreactive Uaa then selectively reacts with its target residue via proximity-enabled bioreactivity, forming a covalent linkage in high specificity.
Diverse Uaas targeting different natural amino acid residues
Targeting Cys
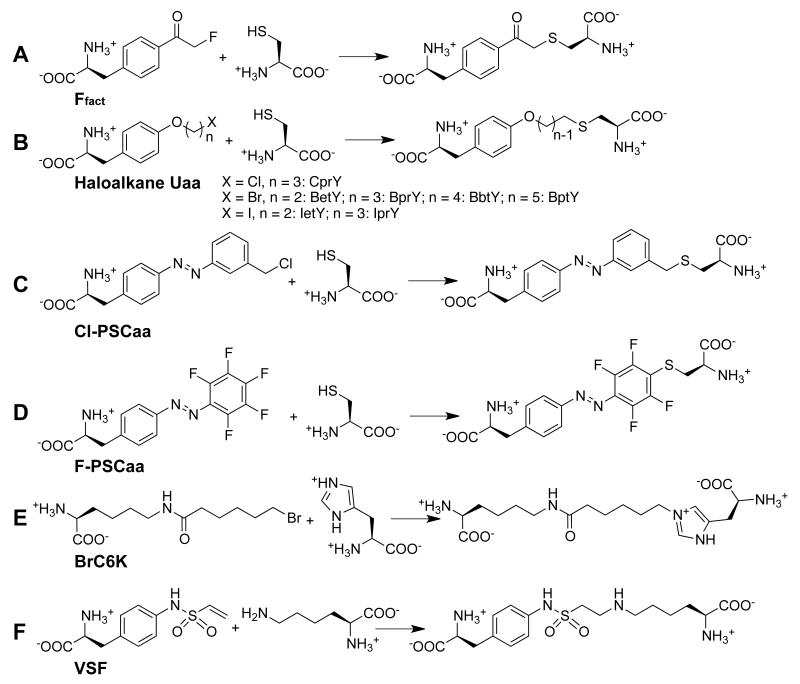
In addition to fluoromethyl ketone embedded in Uaa Ffact [34], other functional groups installed on diverse Uaas have been subsequently demonstrated to be suitable for genetic encoding and reacting with Cys via proximity-enabled bioreactivity. A series of haloalkane Uaas were designed and synthesized containing different halogen atoms linked with aliphatic chains of varying length (Figure 4B) [46]. The M. mazei pair was evolved to incorporate these Phe-scaffold Uaas [32] into myoglobin in E. coli and mammalian cells. ESI-FTMS analysis of mutant myoglobin showed that the Uaas were specifically incorporated and no side reaction of the Uaa with Cys, glutathione or any other amino acid residue was observed, indicating that the Uaa did not undergo modification or nonspecific self-reaction. These haloalkane Uaas, after incorporation into the affibody, react with a proximal Cys of the Z protein at the interface, crosslinking the two proteins irreversibly in the efficiency of ~45% in 1h [46]. Similarly, the Cl-PSCaa, 2-amino-3-(4-((3-(chloromethyl)phenyl)diazenyl)phenyl)propanoic acid, with benzyl chloride installed on an azobenzene was shown to be incorporated by the evolved pair in E. coli and mammalian cells, and the incorporated Cl-PSCaa reacts with a proximal Cys to form an intramolecular photoswitchable bridge (Figure 4C) [47]. Besides the above nucleophilic substitution reactions, nucleophilic aromatic substitution (SNAr) was also successfully exploited for proximity-enabled bioreactivity. The F-PSCaa, (S,E)-2-amino-3-(4-((perfluorophenyl)diazenyl)phenyl)propanoic acid, has been genetically incorporated into proteins by the pair, and reacts with a proximal Cys within the protein to build a photoswitchable bridge in situ (Figure 4D) [48]. Altogether these results suggest the proximity-enabled bioreactivity can be generally applied to a range of Uaas with different functional groups and side chain scaffolds.
Figure 4.
Structures and reactions of bioreactive Uaas that have been genetically encoded.
Targeting His and Lys
Initial efforts in developing proximity-enabled bioreactivity have been focused on targeting the Cys residue [34]. Although Cys plays a crucial role in disulfide bond formation and catalysis in a variety of proteins, other amino acids containing hydrophilic side chains are often found at protein surfaces and interfaces where a Cys may be absent [49]. Cys mutants can be generated to fulfill the requirement sometimes, yet in many situations such as addressing a therapeutic target in vivo it is infeasible to modify the target protein genetically or chemically. Therefore, the ability to target residues other than Cys for covalent bond generation would dramatically expand the diversity of proteins applicable by proximity-enabled bioreactivity.
Chen et al. found that alkyl bromide, when installed on a long linear alkyl side chain, is able to react with His and Lys via proximity-enabled bioreactivity [50]. The Uaa (S)-2-amino-6-(6-bromohexanamido)hexanoic acid (BrC6K) (Figure 4E) was synthesized and incorporated into proteins by the evolved pair. By incorporating BrC6K at site 30 and placing Cys/His/Lys at the proximal site 47 in the affibody, they analyzed the mutant proteins with MS. The results showed that BrC6K formed the covalent bond with Cys at pH 7.4 in 100% yield, with His at pH 8.0 (23% yield) or pH 8.8 (50% yield), and with Lys at pH 8.8 (12% yield). In another study, Furman et al. reported that Uaa carrying vinylsulfonamide (Figure 4F) can be genetically incorporated into proteins in E. coli. They demonstrated that the incorporated Uaa rapidly reacts with Lys in 86% efficiency at pH 7.4 in vitro, enabling the covalent crosslinking of Herceptin Fab to the HER2 receptor [51].
Intramolecular PEPC
The ability to genetically encode a bioreactive Uaa selectively reacting with a natural amino acid residue under mild conditions now enables the specific introduction of covalent bonds for proteins in vitro and in live cells, which we call Proximity Enabled Protein Crosslinking (PEPC) [34,46]. PEPC can occur within or between proteins to afford new opportunities for protein research and engineering.
Enhance photostability of fluorescent proteins
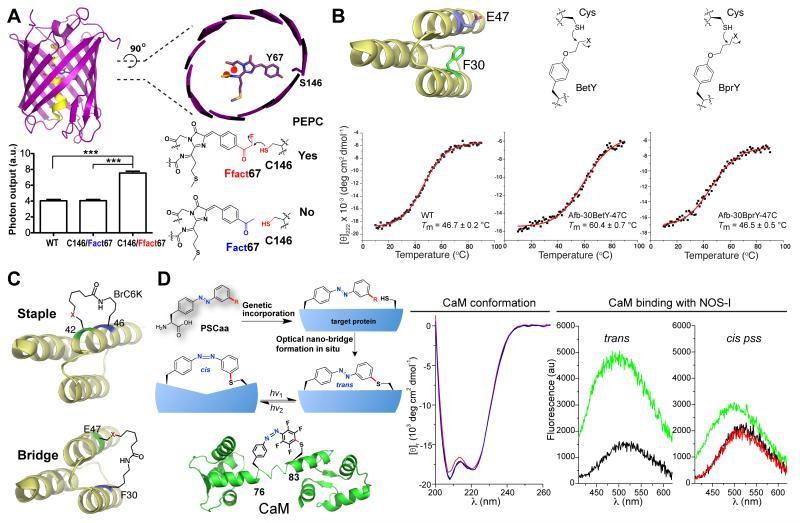
The spatial and temporal resolution of single molecule imaging is determined by photon output of the fluorophore [52,53], which is limited by its photostability. Fluorescent proteins have played an important role in biological imaging with conventional and super resolution [52]. However, it has been challenging to improve the photostability of fluorescent proteins. The fluorophore of all fluorescent proteins has one end covalently linked to the central α-helix with the other end dangling (Figure 5A). We envisioned that if the other end could also be covalently linked to the β-barrel, the fluorophore would become more rigid to increase the photostability. The crystal structure of the red fluorescent protein mPlum shows that Ser146 is 2.6 Å (distance of O-O) away from the Tyr67 of the fluorophore [54]. We thus replaced Tyr67 with Ffact and Ser146 with Cys to facilitate bond formation for covalently attaching the fluorophore [34]. After expressing mPlum S146C/Y67Ffact mutant in E. coli, MS analysis indicate that Ffact67-Cys146 bond has already formed in almost 100% efficiency without any further treatment. Single molecule analysis showed that mutant S146C/Y67Ffact had 2.24 fold longer lifespan and a marked 1.86 fold increase in photon output than the mutant S146C/Y67Fact (Fact is an isosteric control for Ffact and cannot form covalent bond with Cys, Figure 3A). Even when compared to the wild type (WT) mPlum, mutant S146C/Y67Ffact showed 1.92 fold increase in lifespan and 1.86 fold increase in photon output (Figure 5A). Photobleaching of bulk mPlum proteins results in a t1/2 of 7.0 s, while bulk mutant S146C-Y67Ffact yields t1/2 of 14.1 s showing enhanced photostability.
Figure 5.
Intramolecular PEPC for protein engineering and research. (A) A single new covalent bond linking the fluorophore to β-barrel increases photon output of red fluorescent proteins. Structure of red fluorescent protein mPlum is shown, and photon output of WT and mutant mPlum were measured on single molecule level. *** P < 0.0001, n > 156. (B) Increase protein thermal stability by intramolecular PEPC of protein residues. (C) Recombinantly build staples or bridges onto proteins. (D) Build optical nano-switch in situ to modulate protein structure and function. A nano-switch built on CaM via F-PSCaa reacting with Cys is shown. Circular dichroism spectra were recorded for the trans form (black), the cis pss (red) and the trans pss (blue) of the nano-switch CaM in PBS buffer. Binding of NOS-I peptide to CaM was measured by fluorescence. Left panel: the trans nano-switch CaM before (black line) and after (green line) addition of the NOS-I peptide. Right panel: nano-switch CaM in the trans state (black line) was illuminated with green light generating the cis pss state (red line). Subsequent addition of NOS-I peptide to the cis pss state resulted in 50% increase in fluorescence (green line).
When Ffact-Cys were introduced at the corresponding positions into mKate2, another red fluorescent protein with photostability optimized through non-covalent interactions [55], single molecule analysis indicated that the lifespan of mKate2(S143C/Y64Ffact) increased 1.49 fold and the photon output increased 2.31 fold relative to mKate2(S143C/Y64Fact). In comparison to WT mKate2, mKate2(S143C/Y64Ffact) also had 2.04 fold increase in lifespan and 2.46 fold increase in photon output [34]. Altogether these results demonstrate that the Ffact-Cys covalent bond is spontaneously formed within the two fluorescent proteins after protein translation, which is impossible to achieve with other means. This single covalent bond improves the photostability and photon output of fluorescent proteins.
Increase protein thermal stability
Haloalkane Uaa and Cys were introduced into the same Afb protein to build a covalent protein bridge crosslinking its two α–helices (Figure 5B) [46]. After protein expression in E. coli followed by purification, mutant proteins of Afb-30BetY-47C and Afb-30BprY-47C clearly showed strong peaks with monoisotopic masses corresponding to the Afb containing a covalent Cys-Uaa bond at the introduced positions, indicating successful intra-molecular crosslinking. The thermal stabilities of the WT Afb, Afb-30BetY-47C and Afb-30BprY-47C were measured using circular dichroism (CD) [56]. The WT Afb and mutant Afb-30BprY-47C had similar melting temperature (Tm) of 46.7 °C and 46.5 °C, respectively. In contrast, mutant Afb-30BetY-47C showed a markedly higher Tm of 60.4 °C. These results suggest that an intramolecular covalent protein bridge of an appropriate length improves the thermal stability of Afb.
Recombinant protein stapling and protein bridging
Stapled peptides have strong potential for biological therapeutics for their increased interacting surface, resistance to degradation, and cell permeability [57-59]. The flexibility of the Uaa BrC6K side chain was exploited to build covalent linkages on α-helix in cells as a new way to recombinantly staple α-helix (Figure 5C) [50]. Sites 42 and 46 on one α-helix of the affibody [45] were chosen for incorporating Cys and BrC6K, respectively. The affibody was expressed in E. coli and purified for ESI-FTMS analysis. Monoisotopic masses clearly indicate that the covalent staple was built at the introduced sites in 100% efficiency without any further treatment. In addition to staple one α-helix, one can also readily bridge two α-helices by placing the BrC6K and Cys at two different α-helices (similar to the approach shown in the section above). The Stapled peptides could only be chemically synthesized before using toxic catalysts. Bioreactive Uaas now allow recombinant expression of stapled peptides and stapled proteins, which will facilitate the development of this class of biotherapeutics (e.g., library generation, production).
Build optical nano-switch for molecular opto-biology
The ability to control protein function with light provides excellent temporal and spatial resolution for precise investigation in situ, and thus is having significant impact on neuroscience and expanding to general biology [60-64]. Two major barriers of existing optogenetic methods are that 1) they cannot be readily applied to any protein in general, and 2) they cannot provide high specificity and flexibility in selecting modulation site, thus limiting the precision of studies. To address these issues, we genetically built a light-sensitive nano-switch onto proteins to reversibly modulate protein secondary structures or domains (Figure 5D). PhotoSwitchable Click amino acids (PSCaa) are designed to contain the azobenzene photoswitch and an additional click chemical group, which selectively reacts with an appropriately positioned natural amino acid residue to form a covalent nano-switch co-translationally via proximity-enabled bioreactivity.
Two PSCaas, the Cl-PSCaa (Figure 4C) [47] isomerized by light of 365/405 nm and the F-PSCaa (Figure 4D) [48] isomerized by visible light of 405/540 nm, were synthesized and genetically incorporated into proteins in E. coli and mammalian cells by the tRNAPyl/MmPSCaaRS pair. To build a nano-switch, the F-PSCaa was incorporated into the central helix of calmodulin (CaM) at residue 76 and a Cys was placed at the i+7 position (residue 83) (Figure 5D). After expressing in E. coli, the CaM mutant protein was purified and analyzed by high-resolution mass spectrometry. The data indicate that only F-PSCaa was incorporated at site 76 and an intramolecular bond was formed between the F-PSCaa and the Cys, forming the nano-switch in 100% efficiency. Similar results were also obtained for the Cl-PSCaa [47].
The isomerization process of the nano-switch containing CaM was monitored using UV-vis spectroscopy [48]. Green light (540 nm) drives a clear trans to cis photoisomerization of the nano-switch, and subsequent illumination with blue light (405 nm) results in cis to trans photoisomerization yielding the photostationary state (pss) of the trans state. Successive illumination with either green or blue light allows for reversible transformation between the two states without showing fatigue of the nano-switch. Moreover, photoisomerization of the nano-switch drives CaM conformational change reversibly as detected by circular dichroism (Figure 5D). Upon trans to cis photosiomerization the intensity of the n-π* transition band at 208 nm decreased (red line), with magnitude comparable to that of wild-type CaM before and after Ca2+ binding [65], suggesting a clear conformational change of the CaM. Notably, the conformation of the ground trans state was restored by cis to trans photoisomerization using blue light subsequently (blue line).
The binding function of CaM could also be photo-modulated by the nano-switch. The binding of CaM to the CaM-binding domain of the neuronal nitric oxide synthase (NOS-I) was measured [48]. Upon binding, CaM wraps around the NOS-I peptide, which results in fluorescence change of dansyl chloride labeled onto CaM [66]. After addition of NOS-I peptide to the trans form of the nano-switch-CaM, the fluorescence intensity markedly increased by 215%, which is indicative of the NOS-I peptide binding to CaM (green line, Figure 5D). In contrast, addition of NOS-I to the cis pss of the nano-switch-CaM increased the fluorescence by only 50% (green line), indicating that photoswitching the nano-switch to cis form decreased CaM binding.
The nano-switch can be a versatile optical controller to modulate various positions and features of proteins, without the need to know the function of the protein in advance. This new mechanism of modulating the structure, rather than modulating the function that is used in existing optogenetic methods, will be particularly valuable for investigating unknown protein function.
Intermolecular PEPC
Pinpoint ligand-receptor interaction in live mammalian cells
Class B GPCRs are peptide receptors of high pharmacological relevance to widespread diseases, yet molecular determinants regulating their activation by peptide agonists are largely unknown [67]. To investigate the interaction between the corticotropin releasing factor receptor type 1 (CRF1R) and its native peptide ligand Urocortin-I (Ucn1) directly in mammalian cells, photocrosslinking Uaa Azi was first incorporated in CRF1R expressed in mammalian cells at various positions to crosslink the ligand Ucn1 upon UV light activation (Figure 6A) [68,69]. A covalent complex of CRF1R-Ucn1 detected by Western analysis of cell lysate indicates that Ucn1 interacts with CRF1R at the Azi incorporation site, resulting in a map of the interaction sites on the receptor. However, photocrosslinking reaction is not specific to amino acid residues, and therefore how the ligand is positioned in the receptor remains unclear. The bioreactive Uaa Ffact is then introduced into CRF1R and Cys positioned at different sites of the ligand Ucn1. Since Ffact reacts with Cys in residue-specific and distance-dependent manner, the detection of CRF1R-Ucn1 covalent complex resulting from the Ffact-Cys reaction thus provides reciprocal spatial constrains of the receptor-ligand interaction. These data were analyzed in the context of separate structures of CRF1R transmembrane domain and extracellular domain, yielding the first complete conformational model for the peptide-receptor complex. Structural features of the complex yield molecular insights on the mechanism of receptor activation and the basis for discrimination between agonist and antagonist function [69].
Figure 6.
Intermolecular PEPC for protein research and applications. (A) Pinpoint ligand-receptor interaction. Photocrosslinking probe Azi was incorporated into CRF1R to reveal the binding pocket (in purple) for its native peptide agonist Ucn1; Bioreactive probe Ffact was incorporated into CRF1R together with Cys in Ucn1 to determine reciprocal spatial constrains (circle with same colors) of the receptor-ligand complex. Obtained from intact receptor expressed in live cells, these data were used to build a complete conformation model for the CRF1R complexed with Ucn1. (B) Target native receptor irreversibly. The BrC6K-ZHER2 affibody (with BrC6K incorporated at Asp37) covalently bound to the endogenous HER2 receptor on SKBR3 breast cancer cells, while the WT-ZHER2 could not. Cells were incubated with FITC-labeled WT- or BrC6K-ZHER2, washed to disrupt non-covalent interactions, and then imaged for FITC signal and western blotted for the His6 tag appended to ZHER2. (C) Develop covalent peptide inhibitor. Installation of sulfonyl fluoride on SAHp53-8 peptide increases its inhibition of p53-Mdm4 interaction by 10-fold.
Target native receptors irreversibly
The efficacy of protein therapeutics is dependent upon the association and dissociation rates between the therapeutic agent and the target protein [70,71]. Although antibodies can have high affinities for their targets, in many cases such as cancer and infectious diseases, it is crucial to completely remove the pathogenic cells or the microbial pathogen. Antibody dissociation is also an obstacle in imaging [70,71]. These processes would be significantly enhanced if the therapeutic can covalently crosslink to its target. Through intermolecular PEPC, a covalent affibody has been generated to irreversibly crosslink the endogenous human epidermal growth factor receptor 2 (HER2) on breast cancer cells (Figure 6B) [50]. Based on the crystal structure of the HER2 extracellular domain in complex with the HER2-specific affibody ZHER2 [72,73], the bioreactive Uaa BrC6K was incorporated at site 37 into the ZHER2 affibody to target the His490 of HER2. Incubation of this BrC6K-ZHER2 with HER2 positive breast cancer cells results in covalent binding of the BrC6K-ZHER2 to cells, which cannot be washed away using stringent conditions that disrupt noncovalent binding of WT ZHER2 [50]. In another independent study [51], Uaa p-vinylsulfonamido-(S)-phenylalanine (Figure 4F) was incorporated into the Herceptin Fab at site Tyr92, and the resultant mutant Fab was incubated with HER2 positive cancer cells. Fluorescence imaging and western analysis showed that the Fab covalently binds with HER2 receptor on cell surface through the Uaa reacting with Lys569 of HER2. The vinylsulfonamide group shows high crosslinking efficiency, but its high reactivity also results in undesired glutathione modification of the Uaa when the Uaa was incorporated in the mutant Fab in E. coli. Together these two studies prove the feasibility of genetically encoding bioreactive Uaas in therapeutic proteins to covalently bind native receptors without the need to modify the target receptor genetically or chemically.
Develop covalent peptide inhibitors
Small molecules can be covalently attached to proteins via either reacting with the catalytic residues or through ligand-directed chemistry; such covalent reactivities have been harnessed to improve the drug potency [74] and to selectively label proteins [75,76]. In contrast, it has been difficult to design and implement macromolecular covalent inhibitors. Macromolecules such as peptide inhibitors have increased molecular weight and bulk, making them difficult to reach the active site of target proteins. They usually gain access to protein surface or protein-protein interacting interface, but catalytic residues are often lacking there for covalent bond formation. Through intermolecular PEPC, covalent peptide inhibitors have been generated (Figure 6C) [77]. A sulfonyl fluoride containing Uaa was inserted into the stapled peptide SAHp53-8, and the resultant peptide binds Mdm4 covalently through the Uaa reacting with His or Lys of Mdm4. Such irreversible binding dramatically increases the inhibition activity of the SAHp53-8 peptide to p53-Mdm4 interaction by 10-fold. In this study the sulfonyl fluoride group was conveniently installed on the peptide chemically; for lager proteins, genetically encoding PEPC-capable bioreactive Uaas in live cells will readily generate macromolecular covalent inhibitors.
Conclusions
The concept of proximity-enabled bioreactivity now enables the genetic encoding of a new class of Uaas, the bioreactive Uaas, in live cells. Distinct from natural amino acids and Uaas incorporated in the past, the bioreactive Uaas are able to selectively form covalent linkages with a target natural amino acid residue in proximity in proteins. These genetically introduced new covalent linkages for proteins break the natural barrier in protein biosynthesis, and are affording novel avenues for researching protein functions and engineering new protein properties. In the future we expect more bioreactive Uaas will be designed and encoded to target various natural amino acid residues, and the proximity-enabled bioreactivity concept may be further expanded to target other biomolecules inside cells. These explorations will generally impact biological studies, biotechnology, biotherapeutics and synthetic biology.
Acknowledgements
This review is based on the author’s lecture given at the symposium ‘Fifty Years of the Genetic Code: A Symposium to Honor the Legacy of Marshall Nirenberg’ presented by the IUBMB and the New York Academy of Sciences in 2014, for which Dr. Brian Clark was one of the organizers. The author would like dedicate this review to Dr. Brian Clark for his generous encouragement and support. US National Institutes of Health (grant 1R01GM118384-01) is acknowledged for financial support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Sevier CS, Kaiser CA. Formation and transfer of disulphide bonds in living cells. Nat Rev Mol Cell Biol. 2002;3:836–847. doi: 10.1038/nrm954. [DOI] [PubMed] [Google Scholar]
- [2].Liu H, May K. Disulfide bond structures of IgG molecules: structural variations, chemical modifications and possible impacts to stability and biological function. mAbs. 2012;4:17–23. doi: 10.4161/mabs.4.1.18347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Berkmen M. Production of disulfide-bonded proteins in Escherichia coli. Protein Expr Purif. 2012;82:240–251. doi: 10.1016/j.pep.2011.10.009. [DOI] [PubMed] [Google Scholar]
- [4].Kang HJ, Baker EN. Intramolecular isopeptide bonds: protein crosslinks built for stress? Trends Biochem Sci. 2011;36:229–237. doi: 10.1016/j.tibs.2010.09.007. [DOI] [PubMed] [Google Scholar]
- [5].Heck T, Faccio G, Richter M, Thony-Meyer L. Enzyme-catalyzed protein crosslinking. Appl Microbiol Biotechnol. 2013;97:461–475. doi: 10.1007/s00253-012-4569-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Ling J, O'Donoghue P, Soll D. Genetic code flexibility in microorganisms: novel mechanisms and impact on physiology. Nat Rev Microbiol. 2015;13:707–721. doi: 10.1038/nrmicro3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Knight RD, Freeland SJ, Landweber LF. Rewiring the keyboard: evolvability of the genetic code. Nat Rev Genet. 2001;2:49–58. doi: 10.1038/35047500. [DOI] [PubMed] [Google Scholar]
- [8].Wang L, Magliery TJ, Liu DR, Schultz PG. A new functional suppressor tRNA/aminoacyl-tRNA synthetase pair for the in vivo incorporation of unnatural amino acids into proteins. J Am Chem Soc. 2000;122:5010–5011. [Google Scholar]
- [9].Wang L, Schultz PG. A general approach for the generation of orthogonal tRNAs. Chem Biol. 2001;8:883–890. doi: 10.1016/s1074-5521(01)00063-1. [DOI] [PubMed] [Google Scholar]
- [10].Wang L, Brock A, Herberich B, Schultz PG. Expanding the genetic code of Escherichia coli. Science. 2001;292:498–500. doi: 10.1126/science.1060077. [DOI] [PubMed] [Google Scholar]
- [11].Wang L, Schultz PG. Expanding the genetic code. Chem Commun. 2002:1–11. doi: 10.1039/b108185n. [DOI] [PubMed] [Google Scholar]
- [12].Wang L, Schultz PG. Expanding the genetic code. Angew Chem Int Ed Engl. 2005;44:34–66. doi: 10.1002/anie.200460627. [DOI] [PubMed] [Google Scholar]
- [13].Chin JW, Cropp TA, Anderson JC, Mukherji M, Zhang Z, Schultz PG. An expanded eukaryotic genetic code. Science. 2003;301:964–967. doi: 10.1126/science.1084772. [DOI] [PubMed] [Google Scholar]
- [14].Wang Q, Wang L. New methods enabling efficient incorporation of unnatural amino acids in yeast. J Am Chem Soc. 2008;130:6066–6067. doi: 10.1021/ja800894n. [DOI] [PubMed] [Google Scholar]
- [15].Sakamoto K, Hayashi A, Sakamoto A, Kiga D, Nakayama H, Soma A, Kobayashi T, Kitabatake M, Takio K, Saito K, Shirouzu M, Hirao I, Yokoyama S. Site-specific incorporation of an unnatural amino acid into proteins in mammalian cells. Nucleic Acids Res. 2002;30:4692–4699. doi: 10.1093/nar/gkf589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wang W, Takimoto JK, Louie GV, Baiga TJ, Noel JP, Lee KF, Slesinger PA, Wang L. Genetically encoding unnatural amino acids for cellular and neuronal studies. Nat Neurosci. 2007;10:1063–1072. doi: 10.1038/nn1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Liu W, Brock A, Chen S, Chen S, Schultz PG. Genetic incorporation of unnatural amino acids into proteins in mammalian cells. Nat Methods. 2007;4:239–244. doi: 10.1038/nmeth1016. [DOI] [PubMed] [Google Scholar]
- [18].Shen B, Xiang Z, Miller B, Louie G, Wang W, Noel JP, Gage FH, Wang L. Genetically encoding unnatural amino acids in neural stem cells and optically reporting voltage-sensitive domain changes in differentiated neurons. Stem Cells. 2011;29:1231–1240. doi: 10.1002/stem.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Li F, Zhang H, Sun Y, Pan Y, Zhou J, Wang J. Expanding the genetic code for photoclick chemistry in E. coli, mammalian cells, and A. thaliana. Angew Chem Int Ed Engl. 2013;52:9700–9704. doi: 10.1002/anie.201303477. [DOI] [PubMed] [Google Scholar]
- [20].Parrish AR, She X, Xiang Z, Coin I, Shen Z, Briggs SP, Dillin A, Wang L. Expanding the Genetic Code of Caenorhabditis elegans Using Bacterial Aminoacyl-tRNA Synthetase/tRNA Pairs. ACS Chem Biol. 2012;7:1292–1302. doi: 10.1021/cb200542j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Greiss S, Chin JW. Expanding the Genetic Code of an Animal. J Am Chem Soc. 2011;133:14196–14199. doi: 10.1021/ja2054034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Bianco A, Townsley FM, Greiss S, Lang K, Chin JW. Expanding the genetic code of Drosophila melanogaster. Nat Chem Biol. 2012;8:748–750. doi: 10.1038/nchembio.1043. [DOI] [PubMed] [Google Scholar]
- [23].Kang JY, Kawaguchi D, Coin I, Xiang Z, O'Leary DD, Slesinger PA, Wang L. In vivo expression of a light-activatable potassium channel using unnatural amino acids. Neuron. 2013;80:358–370. doi: 10.1016/j.neuron.2013.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Liu CC, Schultz PG. Adding new chemistries to the genetic code. Annu Rev Biochem. 2010;79:413–444. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- [25].Wang L, Xie J, Schultz PG. Expanding the genetic code. Annu Rev Biophys Biomol Struct. 2006;35:225–249. doi: 10.1146/annurev.biophys.35.101105.121507. [DOI] [PubMed] [Google Scholar]
- [26].Wang Q, Parrish AR, Wang L. Expanding the genetic code for biological studies. Chem Biol. 2009;16:323–336. doi: 10.1016/j.chembiol.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Zhu S, Riou M, Yao CA, Carvalho S, Rodriguez PC, Bensaude O, Paoletti P, Ye S. Genetically encoding a light switch in an ionotropic glutamate receptor reveals subunit-specific interfaces. Proc Natl Acad Sci U S A. 2014;111:6081–6086. doi: 10.1073/pnas.1318808111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Hoffmann JE, Plass T, Nikic I, Aramburu IV, Koehler C, Gillandt H, Lemke EA, Schultz C. Highly Stable trans-Cyclooctene Amino Acids for Live-Cell Labeling. Chemistry. 2015;21:12266–12270. doi: 10.1002/chem.201501647. [DOI] [PubMed] [Google Scholar]
- [29].Schmidt MJ, Fedoseev A, Bucker D, Borbas J, Peter C, Drescher M, Summerer D. EPR Distance Measurements in Native Proteins with Genetically Encoded Spin Labels. ACS Chem Biol. 2015;10:2764–2771. doi: 10.1021/acschembio.5b00512. [DOI] [PubMed] [Google Scholar]
- [30].Yang Y, Zhou Q, Wang L, Liu X, Zhang W, Hu M, Dong J, Li J, Xiaoxuan L, Ouyang H, Li H, Gao F, Gong W, Lu Y, Wang J. Significant Improvement of Oxidase Activity through the Genetic Incorporation of a Redox-active Unnatural Amino Acid. Chem Sci. 2015;6:3881–3885. doi: 10.1039/c5sc01126d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Li J, Jia S, Chen PR. Diels-Alder reaction-triggered bioorthogonal protein decaging in living cells. Nat Chem Biol. 2014;10:1003–1005. doi: 10.1038/nchembio.1656. [DOI] [PubMed] [Google Scholar]
- [32].Takimoto JK, Dellas N, Noel JP, Wang L. Stereochemical basis for engineered pyrrolysyltRNA synthetase and the efficient in vivo incorporation of structurally divergent non-native amino acids. ACS Chem Biol. 2011;6:733–743. doi: 10.1021/cb200057a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Lee YJ, Kurra Y, Liu WR. Phospha-Michael Addition as a New Click Reaction for Protein Functionalization. ChemBioChem. 2016;17:456–461. doi: 10.1002/cbic.201500697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Xiang Z, Ren H, Hu YS, Coin I, Wei J, Cang H, Wang L. Adding an unnatural covalent bond to proteins through proximity-enhanced bioreactivity. Nat Methods. 2013;10:885–888. doi: 10.1038/nmeth.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Li X, Liu DR. DNA-templated organic synthesis: nature's strategy for controlling chemical reactivity applied to synthetic molecules. Angew Chem Int Ed Engl. 2004;43:4848–4870. doi: 10.1002/anie.200400656. [DOI] [PubMed] [Google Scholar]
- [36].Drahl C, Cravatt BF, Sorensen EJ. Protein-reactive natural products. Angew Chem Int Ed Engl. 2005;44:5788–5809. doi: 10.1002/anie.200500900. [DOI] [PubMed] [Google Scholar]
- [37].Chmura AJ, Orton MS, Meares CF. Antibodies with infinite affinity. Proc Natl Acad Sci U S A. 2001;98:8480–8484. doi: 10.1073/pnas.151260298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Cravatt BF, Wright AT, Kozarich JW. Activity-based protein profiling: from enzyme chemistry to proteomic chemistry. Annu Rev Biochem. 2008;77:383–414. doi: 10.1146/annurev.biochem.75.101304.124125. [DOI] [PubMed] [Google Scholar]
- [39].Basle E, Joubert N, Pucheault M. Protein chemical modification on endogenous amino acids. Chem Biol. 2010;17:213–227. doi: 10.1016/j.chembiol.2010.02.008. [DOI] [PubMed] [Google Scholar]
- [40].Cohen MS, Zhang C, Shokat KM, Taunton J. Structural bioinformatics-based design of selective, irreversible kinase inhibitors. Science. 2005;308:1318–1321. doi: 10.1126/science1108367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Sletten EM, Bertozzi CR. Bioorthogonal chemistry: fishing for selectivity in a sea of functionality. Angew Chem Int Ed Engl. 2009;48:6974–6998. doi: 10.1002/anie.200900942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Ahmed NK, Martin LA, Watts LM, Palmer J, Thornburg L, Prior J, Esser RE. Peptidyl fluoromethyl ketones as inhibitors of cathepsin B. Implication for treatment of rheumatoid arthritis. Biochem Pharmacol. 1992;44:1201–1207. doi: 10.1016/0006-2952(92)90385-v. [DOI] [PubMed] [Google Scholar]
- [43].Neidhardt FC. Escherichia coli and Salmonella: Cellular and Molecular Biology. ASM Press; 1996. [Google Scholar]
- [44].Wang L, Zhang Z, Brock A, Schultz PG. Addition of the keto functional group to the genetic code of Escherichia coli. Proc Natl Acad Sci U S A. 2003;100:56–61. doi: 10.1073/pnas.0234824100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Hogbom M, Eklund M, Nygren PA, Nordlund P. Structural basis for recognition by an in vitro evolved affibody. Proc Natl Acad Sci U S A. 2003;100:3191–3196. doi: 10.1073/pnas.0436100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Xiang Z, Lacey VK, Ren H, Xu J, Burban DJ, Jennings PA, Wang L. Proximity-Enabled Protein Crosslinking through Genetically Encoding Haloalkane Unnatural Amino Acids. Angew Chem Int Ed Engl. 2014;53:2190–2193. doi: 10.1002/anie.201308794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Hoppmann C, Lacey VK, Louie GV, Wei J, Noel JP, Wang L. Genetically Encoding Photoswitchable Click Amino Acids in Escherichia coli and Mammalian Cells. Angew Chem Int Ed Engl. 2014;53:3932–3936. doi: 10.1002/anie.201400001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Hoppmann C, Maslennikov I, Choe S, Wang L. In Situ Formation of an Azo Bridge on Proteins Controllable by Visible Light. J Am Chem Soc. 2015;137:11218–11221. doi: 10.1021/jacs.5b06234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Lins L, Thomas A, Brasseur R. Analysis of accessible surface of residues in proteins. Protein Sci. 2003;12:1406–1417. doi: 10.1110/ps.0304803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Chen XH, Xiang Z, Hu YS, Lacey VK, Cang H, Wang L. Genetically encoding an electrophilic amino Acid for protein stapling and covalent binding to native receptors. ACS Chem Biol. 2014;9:1956–1961. doi: 10.1021/cb500453a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Furman JL, Kang M, Choi S, Cao Y, Wold ED, Sun SB, Smider VV, Schultz PG, Kim CH. A genetically encoded aza-Michael acceptor for covalent cross-linking of protein-receptor complexes. J Am Chem Soc. 2014;136:8411–8417. doi: 10.1021/ja502851h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Patterson G, Davidson M, Manley S, Lippincott-Schwartz J. Superresolution imaging using single-molecule localization. Annu Rev Phys Chem. 2010;61:345–367. doi: 10.1146/annurev.physchem.012809.103444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Thompson RE, Larson DR, Webb WW. Precise Nanometer Localization Analysis for Individual Fluorescent Probes. Biophys J. 2002;82:2775–2783. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Shu X, Wang L, Colip L, Kallio K, Remington SJ. Unique interactions between the chromophore and glutamate 16 lead to far-red emission in a red fluorescent protein. Protein Sci. 2009;18:460–466. doi: 10.1002/pro.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Shcherbo D, Murphy CS, Ermakova GV, Solovieva EA, Chepurnykh TV, Shcheglov AS, Verkhusha VV, Pletnev VZ, Hazelwood KL, Roche PM, Lukyanov S, Zaraisky AG, Davidson MW, Chudakov DM. Far-red fluorescent tags for protein imaging in living tissues. Biochem J. 2009;418:567–574. doi: 10.1042/BJ20081949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Greenfield NJ. Using circular dichroism collected as a function of temperature to determine the thermodynamics of protein unfolding and binding interactions. Nat Protoc. 2006;1:2527–2535. doi: 10.1038/nprot.2006.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Bock JE, Gavenonis J, Kritzer JA. Getting in shape: controlling peptide bioactivity and bioavailability using conformational constraints. ACS Chem Biol. 2013;8:488–499. doi: 10.1021/cb300515u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Verdine GL, Hilinski GJ. Stapled peptides for intracellular drug targets. Methods Enzymol. 2012;503:3–33. doi: 10.1016/B978-0-12-396962-0.00001-X. [DOI] [PubMed] [Google Scholar]
- [59].Bird GH, Gavathiotis E, LaBelle JL, Katz SG, Walensky LD. Distinct BimBH3 (BimSAHB) stapled peptides for structural and cellular studies. ACS Chem Biol. 2014;9:831–837. doi: 10.1021/cb4003305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Fenno L, Yizhar O, Deisseroth K. The development and application of optogenetics. Annu Rev Neurosci. 2011;34:389–412. doi: 10.1146/annurev-neuro-061010-113817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Szobota S, Isacoff EY. Optical control of neuronal activity. Annu Rev Biophys. 2010;39:329–348. doi: 10.1146/annurev.biophys.093008.131400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Wu YI, Frey D, Lungu OI, Jaehrig A, Schlichting I, Kuhlman B, Hahn KM. A genetically encoded photoactivatable Rac controls the motility of living cells. Nature. 2009;461:104–108. doi: 10.1038/nature08241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Levskaya A, Weiner OD, Lim WA, Voigt CA. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature. 2009;461:997–1001. doi: 10.1038/nature08446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Zhou XX, Chung HK, Lam AJ, Lin MZ. Optical control of protein activity by fluorescent protein domains. Science. 2012;338:810–814. doi: 10.1126/science.1226854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Shifman JM, Choi MH, Mihalas S, Mayo SL, Kennedy MB. Ca2+/calmodulin-dependent protein kinase II (CaMKII) is activated by calmodulin with two bound calciums. Proc Natl Acad Sci U S A. 2006;103:13968–13973. doi: 10.1073/pnas.0606433103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Valentine KG, Ng HL, Schneeweis L, Kranz JK, Frederick KK, Alber T, Wand AJ. PDB code 2O60. 2006.
- [67].Pal K, Melcher K, Xu HE. Structure and mechanism for recognition of peptide hormones by Class B G-protein-coupled receptors. Acta Pharmacol Sin. 2012;33:300–311. doi: 10.1038/aps.2011.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Coin I, Perrin MH, Vale WW, Wang L. Photo-cross-linkers incorporated into G-protein-coupled receptors in mammalian cells: a ligand comparison. Angew Chem Int Ed Engl. 2011;50:8077–8081. doi: 10.1002/anie.201102646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Coin I, Katritch V, Sun T, Xiang Z, Siu FY, Beyermann M, Stevens RC, Wang L. Genetically Encoded Chemical Probes in Cells Reveal the Binding Path of Urocortin-I to CRF Class B GPCR. Cell. 2013;155:1258–1269. doi: 10.1016/j.cell.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Beck A, Wurch T, Bailly C, Corvaia N. Strategies and challenges for the next generation of therapeutic antibodies. Nat Rev Immunol. 2010;10:345–352. doi: 10.1038/nri2747. [DOI] [PubMed] [Google Scholar]
- [71].Tebbutt N, Pedersen MW, Johns TG. Targeting the ERBB family in cancer: couples therapy. Nat Rev Cancer. 2013;13:663–673. doi: 10.1038/nrc3559. [DOI] [PubMed] [Google Scholar]
- [72].Cho HS, Mason K, Ramyar KX, Stanley AM, Gabelli SB, Denney DW, Jr., Leahy DJ. Structure of the extracellular region of HER2 alone and in complex with the Herceptin Fab. Nature. 2003;421:756–760. doi: 10.1038/nature01392. [DOI] [PubMed] [Google Scholar]
- [73].Eigenbrot C, Ultsch M, Dubnovitsky A, Abrahmsen L, Hard T. Structural basis for high-affinity HER2 receptor binding by an engineered protein. Proc Natl Acad Sci U S A. 2010;107:15039–15044. doi: 10.1073/pnas.1005025107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Singh J, Petter RC, Baillie TA, Whitty A. The resurgence of covalent drugs. Nat Rev Drug Discov. 2011;10:307–317. doi: 10.1038/nrd3410. [DOI] [PubMed] [Google Scholar]
- [75].Takaoka Y, Ojida A, Hamachi I. Protein organic chemistry and applications for labeling and engineering in live-cell systems. Angew Chem Int Ed Engl. 2013;52:4088–4106. doi: 10.1002/anie.201207089. [DOI] [PubMed] [Google Scholar]
- [76].Reinhardt U, Lotze J, Zernia S, Morl K, Beck-Sickinger AG, Seitz O. Peptide-templated acyl transfer: a chemical method for the labeling of membrane proteins on live cells. Angew Chem Int Ed Engl. 2014;53:10237–10241. doi: 10.1002/anie.201403214. [DOI] [PubMed] [Google Scholar]
- [77].Hoppmann C, Wang L. Proximity-enabled bioreactivity to generate covalent peptide inhibitors of p53-Mdm4. Chem Commun (Camb) 2016;52:5140–5143. doi: 10.1039/c6cc01226d. [DOI] [PubMed] [Google Scholar]