Abstract
In cereal crops, vegetative and spikelet development play important roles in grain yield and quality, but the genetic mechanisms that control vegetative and spikelet development remain poorly understood in rice. Here, we identified a new rice mutant, defective glume 1 (dg1) mutant from cultivar Zhonghua11 after ethyl methanesulfonate treatment. The dg1 mutant displayed the dwarfism with small, rolled leaves, which resulted from smaller cells and more bulliform cells. The dg1 mutant also had an enlarged leaf angle and defects in brassinosteroid signaling. In the dg1 mutant, both the rudimentary glume and sterile lemma (glumes) were transformed into lemma-like organ and acquired the lemma identity. Additionally, the dg1 mutant produced slender grains. Further analysis revealed that DG1 affects grain size by regulating cell proliferation and expansion. We fine mapped the dg1 locus to a 31-kb region that includes eight open reading frames. We examined the DNA sequence and expression of these loci, but we were not able to identify the DG1 gene. Therefore, more work will be needed for cloning and functional analysis of DG1, which would contribute to our understanding of the molecular mechanisms behind whole-plant development in rice.
Keywords: rice (Oryza sativa L.), dg1 mutant, dwarfism and rolled leaves, leaf angle, BR signaling, glumes, grain size
Introduction
Rice (Oryza sativa L.) is one of the most important cereal crops and feeds more than half of the world’s population as a staple food. The vegetative and floral organs are very important agricultural organs that determine grain yield and quality. In eudicots, the typical flowers have four whorls of floral organs: sepals in whorl 1, petals in whorl 2, stamens in whorl 3, and pistils in whorl 4 from the outer to inner whorls. The classical ABCDE model proposes that how A/B/C/D/E class genes act in combination to specify the identity of each organ and affect floral meristem determinacy (Coen and Meyerowitz, 1991; Jeon et al., 2000; Theissen and Saedler, 2001; Nagasawa et al., 2003; Ditta et al., 2004; Yamaguchi and Hirano, 2006; Dreni et al., 2007; Gao et al., 2010). A- and E-function genes are responsible for the sepal formation (Wang et al., 2010; Kobayashi et al., 2012); A-, B- and E-function genes together determine the petal identity (Nagasawa et al., 2003; Prasad and Vijayraghavan, 2003; Yao et al., 2008; Yun et al., 2013); B-, C-, and E-function genes together modulate the stamen development (Kyozuka and Shimamoto, 2002; Yamaguchi and Hirano, 2006; Dreni et al., 2011; Yun et al., 2013); and C- and E-function genes act together to regulate the pistil identity (Coen and Meyerowitz, 1991; Pelaz et al., 2000; Theissen and Saedler, 2001; Ditta et al., 2004; Gao et al., 2010). D-and E-function genes together specify the placenta and ovule identity (Dreni et al., 2007; Gao et al., 2010; Li et al., 2011). This genetic model is applicable to monocot species including rice and maize (Nagasawa et al., 2003; Dreni et al., 2007), except that the non-MADS-box homeotic gene DROOPINGLEAF (DL) are recruited and mainly determines the pistil identity in rice (Yamaguchi et al., 2004; Yamaguchi and Hirano, 2006; Sang et al., 2012).
Vegetative and reproductive development plays important roles in high yield because plant architecture is the foundation of reproductive development, and the development of spikelet which directly produced seeds determines the final grain quantity and quality. Ongoing genetic research has identified several genes that pleiotropic affect vegetative growth and spikelet development in rice. Rice Aberrant Panicle Organization 1 (APO1), the ortholog of Arabidopsis Unusual Floral Organ (UFO), encodes an F-box protein and functions in leaf formation, inflorescence shape and the identity of floral organs (Ikeda et al., 2005, 2007; Ikeda-Kawakatsu et al., 2009). The apo1 mutant produces more leaves, smaller inflorescences, lodicule-like stamens, and extra carpels, suggesting that the mutant affects the identity of floral organs and floral meristem determinacy. Super Apical Dormant1 (SAD1) encodes a Mediator-interacting protein that binds to OsMED4 and controls various aspects of plant development (Duan et al., 2012). The sad1 mutant exhibited a low number of tillers, severe dwarfism, slender and short leaves, and abnormal floral organs. Dwarf and Deformed Flower1 (DDF1) and Abnormal Flower and Dwarf1 (AFD1) encodes an RNA polymerase I and a DUF640 domain protein, respectively (Li et al., 2015; Ren et al., 2016b). The loss-of-function mutants of DDF1 and AFD1 displayed variable defects such as dwarfism, low seed-setting rate, and defective floral organs, suggesting that DDF1 and AFD1 are involved in the regulation of cell expansion proliferation, and floral organ identity. More work is required to better understand the molecular mechanism that controls vegetative growth and reproductive development in rice.
In this study, we characterized a new recessive mutant, defective glume 1 (dg1) from an M2 population of the cultivar Zhonghua11 (ZH11) treated with ethyl methanesulfonate (EMS). The dg1 mutant displayed a wide range of defects, including dwarfism, increased leaf angle, small and rolled leaves, bushy stigmas.
Our observations of the elongated glumes (rudimentary glumes and sterile lemmas) with lemma-like cellular patterns in the dg1 mutant indicated that the rudimentary glume, sterile lemma, and lemma may be homologous organs and DG1 plays important roles in the determination of floral organ identity. In the dg1 mutant, we observed slender grains, which were caused by abnormalities in the number and size of the hull (i.e., lemma and palea) cells. Our results also showed that dg1 mutant was a brassinosteroid (BR)-hypersensitive and DG1 was involved in regulating BR signaling. Further, we investigated the phenotypes of the dg1 mutant and fine mapped the dg1 locus. DNA sequencing and gene expression were used in an attempt to clone the DG1 gene.
Materials and Methods
Plant Materials
The dg1 mutant of rice (O. sativa) was obtained from the EMS treatment of the japonica cultivar ZH11. ZH11 plants were used as the wild-type strain for phenotypic observation. The dg1 mutant was crossed with indica cultivars Nanjing 6 (NJ6) and Taichung Native 1 (TN1) and the F1 plants were self-pollinated to generate the F2 population. Rice plants were cultivated at the experimental paddy in China National Rice Research Institute, Hangzhou from May to October, and in Lingshui, Hainan Province from January to May.
Identification of DG1
The dg1 mutation was primary mapped in the F2 population of a cross between the dg1 mutant and the indica cultivar NJ6, with which six simple sequence repeat (SSR) markers from publicly available rice databases flanking the target region. For fine mapping of the dg1 mutation, more F2 plants were screened and new insertion or deletion (InDel) molecular markers were used. The sequences of primers are listed in Supplementary Table 1.
Microscopic Examination
Fresh spikelet specimens were acquired from the same position of ZH11 and the dg1 mutant grown under the same conditions. Specimens were fixed at 4°C overnight in 50% FAA solution (50% ethanol, 10% formaldehyde, 5% glacial acetic acid, and 35% sterile water). Spikelet specimens were placed under vacuumed for about 30 min prior to fixation to ensure the materials were completely immersed in the FAA solution. The fixed samples were then treated with an ethanol gradient (50, 70, 80, 90, 100, 100, and 100%). Spikelet specimens were infiltrated in xylene and embedded in Paraplast Plus (Sigma). Using a microtome (HM340E), samples were cut into approximately 8-μm thick sections and flattened on glass slides coated with poly-lysine. After deparaffinating the samples in a xylene series and dehydrating them with an ethanol series, the samples were stained using 1% safranine and 1% Fast Green, then dehydrated in an ethanol series and cleared in a xylene series. For microscopy, sections were covered with neutral resins at 42°C for 48 h. Optical microscopy was conducted with a Nikon Eclipse 90i microscope. For electron microscopy, fresh spikelet hull specimens were observed using a HITACHI S-3500 scanning electron microscope that had a –30°C cool stage. We examined cell size, number, and area using the measurement tools of the Nikon Eclipse 90i microscope, and six biological repeats were examined for cell size, cell area, and cell number to obtain the final results.
RNA Extraction and Quantitative Real-Time PCR (qRT-PCR) Analysis
The total RNA was isolated from floral organs and inflorescences of the wild type and dg1 mutant using the AxyPrepTM total RNA Miniprep Kit (Axygen) according to the manufacturer’s instructions. The first strand of complementary DNA (cDNA) was reverse transcribed from 500 ng total RNA in a 50 uL reaction volume using the ReverTra Ace® quantitative PCR RT Master Mix Kit with gDNA remover (Toyobo) and qRT-PCR was conducted with a CFX96 TouchTM Real-time PCR Detection System and the 2x SsoFastTM EvaGreen® SuperMix (Bio-Rad) to amplify the cDNA of all tested genes. All target genes were normalized to the rice internal control gene Actin to detect the relative expression levels. Three biological repeats were conducted to obtain the final results. The primers for the qRT-PCR assays are listed in Supplementary Table 1.
BR Response Test
Germinated seeds from ZH11 and dg1 mutant were grown in standard culture solutions of 0, 0.01, 0.1, and 1 uM 24-Epibrassinolide (24-eBL) under darkness for a week, respectively. The angle of leaf inclination was then measured and photographed.
Results
Defects in Vegetative Development in the dg1 Mutant
In the vegetative phase, the dg1 mutant was shorter and thinner than the wild type (Figures 1A,B and Supplementary Figure 1). The first internode was distinctly shorter in the dg1 mutant, but the second, third, fourth, and fifth internodes showed no differences when compared with the wild type (Figure 1B and Supplementary Figure 1). The culms and leaves of dg1 mutant were thinner than those of the wild type (Figures 1C,D,F,G and Supplementary Figure 1) and the base of the flag leaves was rolled (Figure 1D). In addition, the leaf angle was enlarged in the dg1 mutant compared with the wild type; the angle of leaf inclination was increased by 51% in the dg1 mutant (Figure 1E and Supplementary Figure 1). Additionally, the average collar length of the adaxial surfaces in the lamina joint of the dg1 mutant was increased by 40%, while the average collar length in the abaxial side was indistinguishable from the wild type (Supplementary Figure 1).
FIGURE 1.

Morphological comparison of the wild type and dg1 mutant. (A) Wild type (left) and dg1 plants (right) at heading stage. (B) Lengths of internodes in the dg1 mutant (left) and wild type (right); fifth internode of the wild type, fourth internode of the wild type, third internode of the wild type, second internode of the wild type, first internode of the wild type, fifth internode of the dg1 mutant, fourth internode of the dg1 mutant, third internode of the dg1 mutant, second internode of the dg1 mutant, and first internode of the dg1 mutant from left to right. (C) Comparison of leaf morphology in the wild type (left) and dg1 mutant (right). (D) Base of flag leaves in the wild type (left) and dg1 mutant (right). (E) Comparison of flag leaf angle. (F) Culm of the wild type. (G) Culm of dg1 mutant. (H) Longitudinal section of the first internode in the wild type. (I) Longitudinal section of the first internode in the dg1 mutant. (J) Cross section of the middle part in the wild type leaf. (K) Cross section of the middle part in the dg1 leaf. (L) Partial magnification in (J). (M) Partial magnification in (K). (N) Cross section of the base of the flag leaves in the wild type. (O) Cross section of the base of the flag leaves in the dg1 mutant. (P) Magnification of red box region in (N). (Q) Magnification of red box region in (O). (R) Cross section of the culm in the wild type. (S) Cross section of the culm in the the dg1 mutant. BV, big vascular bundle. SV, small vascular bundle. BC, bulliform cell. Red arrows represent big vascular bundles. Bars = 20 cm in (A); 10 cm in (B,C); 2 cm in (D); 1 cm in (E); 5 mm in (F,G); 100 μm in (H,I,P,Q); 2000 μm in (J,K); 500 μm in (L,M,R,S). 1000 μm in (N,Q).
We also performed histocytological analysis to study the cell structures. The cells were shorter in the first internodes of the dg1 mutant (Figures 1H,I and Supplementary Figure 1). Analysis of cross sections of the culms and leaves showed that the total cell numbers were reduced and the number of small vascular bundles was markedly decreased in the dg1 mutant compared with the wild type (Figures 1J,K,L,M,R,S and Supplementary Figure 1). However, the number of big vascular bundles remained unchanged between the wild type and the dg1 mutant (Figures 1J,K,R,S and Supplementary Figure 1). These results suggested that DG1 affects the diameter of culms and the width of leaves mainly by regulating the number of small vascular bundles. In the dg1 leaves, the number and area of bulliform cells were also increased (Figures 1N,O,P,Q and Supplementary Figure 1), resulting in adaxial leaf rolling.
The dg1 Mutant is Hypersensitive to BR and Has Altered Expression of BR Response and Biosynthesis Genes
Brassinosteroid plays a vital role in plant growth and development, especially in leaf morphology and leaf angle (Sakamoto et al., 2013; Zhang et al., 2015). Thus, to test whether the dg1 mutant has defects in BR signaling, we conducted a BR sensitivity experiment using different concentrations of 24-eBL and measured the degree of leaf inclination. Notably, the dg1 mutant showed a larger lamina joint angle than that of the wild type after treatment with 0.1 μM 24-eBL (Figures 2A–E). This showed that the dg1 mutant was more hypersensitive to BR than the wild type and suggested that dg1 mutant may be related to BR signaling. We examined the expressions of related genes on BR response and biosynthesis in the BR-treated and untreated wild type and dg1 plants by qRT-PCR. In the untreated plants, the expression of the BR response genes OsBRI1 (brassinosteroid insensitive 1) and OsBZR1 (Brassinazole-resistant 1), was higher in the dg1 mutant than in the wild type (Figure 2F). Similarly, the mRNA levels of BR biosynthetic genes D2 (CYP90D2) and BRD1 (BR-deficient dwarf1) were significantly up-regulated in the dg1 mutant compared with those of the wild type under normal growth condition (Figure 2F). In the treated plants, the expression levels of OsBRI1, OsBZR1, D2 and BRD1 were reduced in the wild type and dg1 mutant.
FIGURE 2.

Morphological response to brassinosteroid (BR) treatment. (A–D) Phenotype of the wild type (left) and dg1 mutant (right) after BR treatment. (E) Statistical analysis of the leaf angle in the wild type and dg1 mutant after BR treatment. (F) expression analysis of BR biosynthetic and signaling genes. 0, 0.01, 0.1, and 1 μM represent a series of BR concentrations. Total RNA was isolated from the leaves of the untreated wild type, the untreated dg1 mutant, the wild type after BR treatment and the dg1 mutant after BR treatment at the seedling stage. WT, wild type. Black asterisk indicates the significant difference of the expression levels of D2, BRD1, OsBRI1, and OsBZR1 between the wild type and dg1 mutant. Red asterisk indicates the significant difference of the decreased range of expressions levels of D2, BRD1, and OsBZR1 between the wild type group and dg1 group. Error bars indicate SD. ∗∗Significant difference at P < 0.01 compared with the wild type by Student’s t-test. ∗Significant difference at P < 0.05 compared with the wild type by Student’s t-test. Bars = 0.5 cm.
Compared with the wild type without BR treatment, the expression of D2, BRD1 and OsBZR1 in the wild type under BR treatment was reduced by 70, 73, and 23%, respectively. By contrast, the expression of D2, BRD1 and OsBZR1 in the dg1 mutant under BR treatment was reduced by 75, 77, and 31%, respectively. The decreased ranges of expressions of D2, BRD1 and OsBZR1 were higher in the dg1 mutant than that in the wild type under BR treatment, further revealing that the dg1 mutant was hypersensitive to exogenous BR. These findings indicated that OsBRI1, OsBZR1, D2 and BRD1 were modulated by DG1 under normal growth condition, and confirmed that DG1 is involved in regulating BR signaling.
Phenotypic Defects of Spikelet in the dg1 Mutant
The wild type spikelet in rice is composed of two pairs of vestigial glumes: rudimentary glumes and sterile lemmas, and one terminal floret comprising a lemma, a palea, two lodicules, six stamens and a central pistil (Figures 3A–E). At heading stage, we found that about 39% spikelets showed longer rudimentary glumes in the dg1 mutant (Figures 3A,B,F,G). The average size of wild type rudimentary glumes was about 0.5 mm length, while the size of rudimentary glumes was variable, from 1.1 to 4.0 mm in length in the 39% spikelets of the dg1 mutant (Supplementary Table 2). About 94% spikelets of the dg1 mutant produced larger sterile lemmas that were similar to the lemmas of the wild type or dg1 mutant (Figures 3A,B,F,G). The length of the dg1 sterile lemmas was varied from 6.0 to 7.9 mm (Supplementary Table 2).
FIGURE 3.

Phenotypes of the spikelets in the wild type and the dg1 mutant. (A) Wild-type spikelet. (B) Rudimentary glume and sterile lemma in the wild type. (C) Wild type floret. (D) Stamen of the wild type floret. (E) Pistil of the wild type floret. (F) dg1 spikelet. (G) Rudimentary glume and sterile lemma in the dg1 mutant. (H) dg1 floret. (I) Stamen of the dg1 floret. (J) Pistil of the dg1 floret. rg, rudimentary glume; sl, sterile lemma; le, lemma; pa, palea; sti, stigma; lo, lodicule; st, stamen; pi, pistil. Bars = 1000 μm in (A,F); 500 μm in (B–E,G–J).
We next investigated the floral organs of four whorls in the dg1 flowers. Interestingly, the dg1 mutant showed slightly whiter and smaller stamens, and highly bushy stigmas compared with the wild type (Figures 3C,D,E,H,I,J). However, the number of floral organs was not changed in the dg1 mutant (Figures 3A,C,F,H). To investigate the cellular morphology, we performed paraffin section and scanning electron microscopy (SEM). In the wild type, the lemma had five vascular bundles and two unique inward hook-like structures (Supplementary Figures 2A,C). The palea had three vascular bundles and exhibited a uniquely smooth epidermis that lacked outer silicified epicuticular cells (Figures 4A–C and Supplementary Figures 2A,C). The sterile lemma developed one vascular bundle (Supplementary Figure 2D) and had a smooth epidermis with regularly arranged cells and rare trichomes (Figures 4A,D). The epidermis of the rudimentary glumes displayed irregularly arranged cells and bore numerous small protrusions and trichomes (Figures 4A,E). No obvious vascular bundles were observed in the rudimentary glumes of the wild type (Supplementary Figure 2E). We also found that the stigmas of the wild-type pistil were sparse and the pistils had normal ovaries (Figures 4F,G and Supplementary Figures 2B,F,G). By contrast, the sterile lemmas and rudimentary glumes were elongated in the dg1 mutant (Figures 4H–J). The sterile lemmas and rudimentary glumes had more vascular bundles, big protrusions and trichomes, and inward hook-like structures (Figures 4K,L and Supplementary Figures 2H–K), which was similar with that of the lemma in the wild type (Figure 4B and Supplementary Figures 2A,C). In addition, the dg1 mutant displayed denser stigmas and an abnormal ovary (Figures 4M,N and Supplementary Figures 2B,G).
FIGURE 4.
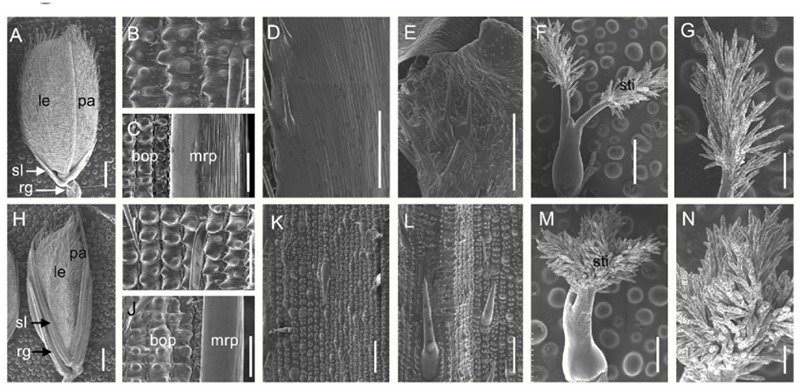
Scanning electron microscopy (SEM) analysis of floral organs in the wild type and the dg1 mutant at heading stage. (A) Wild-type spikelet. (B) Epidermal surface of the lemma in the wild type. (C) Epidermal surface of the palea in the wild type. (D) Epidermal surface of the sterile lemma in the wild type. (E) Epidermal surface of the rudimentary glume in the wild type. (F) Pistil in the wild-type floret. (G) Stigma of pistil in the wild-type floret. (H) dg1 spikelet. (I) Epidermal surface of the lemma in the dg1 mutant. (J) Epidermal surface of the palea in the dg1 mutant. (K) Epidermal surface of sterile lemma in the dg1 mutant. (L) Epidermal surface of the rudimentary glume in the dg1 mutant. (M) Pistil in the dg1 floret. (N) Stigma of pistil in the dg1 floret. rg, rudimentary glume; sl, sterile lemma; le, lemma; pa, palea; bop, body of palea; mrp, marginal region of palea; sti, stigma. Bars = 1000 μm in (A,H); 500 μm in (F,M); 200 μm in (G,N); and 100 μm in (B–E,I–L).
To clarify the identities of the floral organs, we examined the expression of related marker genes in the dg1 spikelets. The expressions of OsMADS1, OsMADS14, OsMADS15, and DROOPING LEAF (DL) was detected in the sterile lemmas and rudimentary glumes of the dg1 spikelets, implying that the sterile lemmas and rudimentary glumes were converted to lemma-like organs (Figure 5A). No OsMADS6 transcript was found in the sterile lemmas and rudimentary glumes of the wild type and dg1 spikelets, suggesting that the sterile lemmas and rudimentary glumes did not acquire the palea identity (Figure 5A). The expression of OsMADS2, OsMADS4, and OsMADS16 was lower in the dg1 stamens (Figure 5B), and the expression of OsMADS3, OsMADS58, and DL was lower in the dg1 pistils (Figure 5C), which was consistent with the phenotype defects in the dg1 mutant. These results revealed that the glumes of the dg1 mutant partly acquired the lemma identity, and the identities of stamens and pistils were altered.
FIGURE 5.
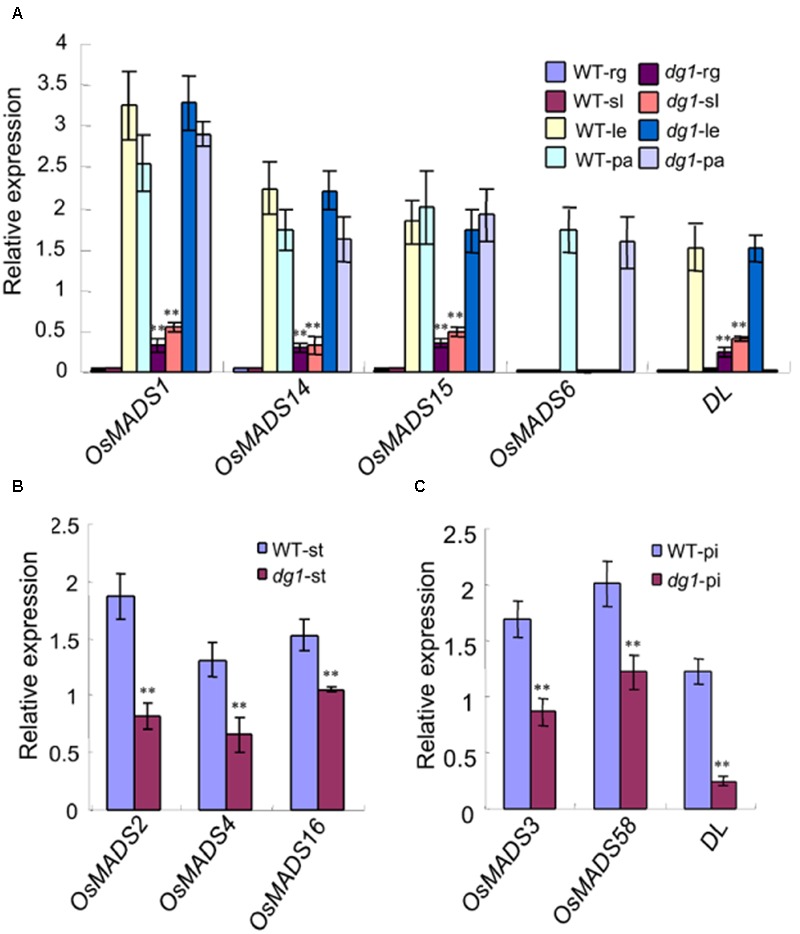
Expression analysis of genes determining floral organ identity in the wild type and dg1 mutant. rg, rudimentary glume; sl, sterile lemma; le, lemma; pa, palea; st, stamen; pi, pistil. (A) The expression of genes in the rudimentary glume and sterile lemma. (B) The expression of genes in the stamen. (C) The expression of genes in the pistil. Total RNA was isolated from spikelets at heading stage. WT, wild type. Error bars indicate SD. ∗∗Significant difference at P < 0.01 compared with the wild type by Student’s t-test.
Abnormal Early Spikelet Development in the dg1 Mutant
We examined spikelet development in the dg1 mutant by SEM. The wild-type spikelets formed sterile lemma and rudimentary glume primordia, and the palea and lemma primordia were growing at the spikelet 4 stage (Sp4) (Figure 6A). At the Sp5 and Sp6 stages, six stamen primordia were observed in the wild-type spikelet and the growth of the stamen primordium on the lemma side was delayed (Figure 6B). Also at this stage the rudimentary glume ceased growing and the sterile lemma continued growing (Figures 6B,C). During the Sp7 and Sp8 stages in the wild-type spikelet, the pistil primordium was observed and the sterile lemma was further enlarged and was much longer than the rudimentary glume (Figures 6C,D). By contrast, no obvious differences were detected in the rudimentary glume and sterile lemma of the dg1 spikelets at the Sp4–Sp6 stages (Figures 6E,F). At the Sp7 and Sp8 stages, the rudimentary glume appeared normal in the dg1 spikelets, while the sterile lemma was longer (Figures 6G,H). We concluded that the dg1 glumes continue growing after the Sp8 stage, thus producing further elongated glumes at heading stage, and the rudimentary glume develops at later stages. Additionally, the development of the dg1 stamens was not synchronous and other floral organs appeared normal in the dg1 florets. These results revealed that the enlargement of glumes was strongly affected and the development of stamens was disrupted in the dg1 mutant.
FIGURE 6.
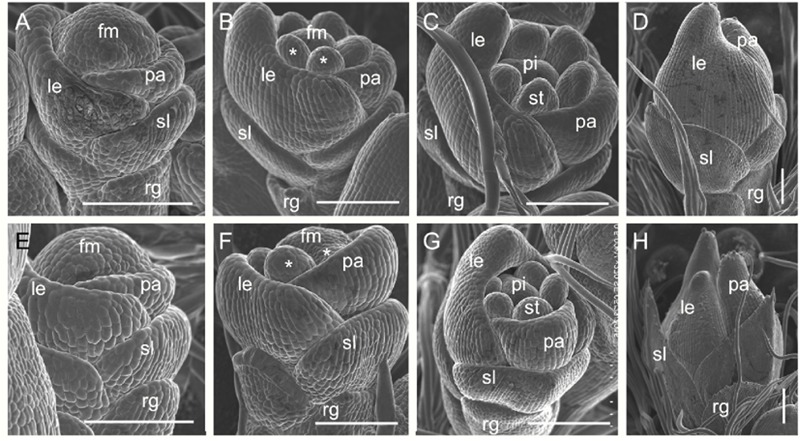
Early spikelet development in the wild type and the dg1 mutant. (A–D) Wild-type spikelet. (A) Sp4, (B) Sp5-6, (C) Sp7, (D) Sp8. (E–H) dg1 spikelet. (E) Sp4, (F) Sp5-6, (G) Sp7, (H) Sp8. fm, floral meristem; le, lemma; pa, palea; st, stamen; pi, pistil, rg, rudimentary glume; sl, sterile lemma. Bars = 100 μm.
The dg1 Mutant Has Reduced Grain Yield
At maturity, the dg1 mutant had shorter and smaller panicles (Supplementary Figure 3). Compared with the wild type, the panicles were 33% shorter in the dg1 mutant (Supplementary Figure 3). The number of spikelets from the dg1 mutant was only 66% of that of the wild type (Supplementary Figure 3). The pollen viability of the stamens was 52% and the seed-setting rate was 45% in the dg1 mutant (Supplementary Figures 3, 4), compared with 96 and 85% in the wild type, respectively (Supplementary Figures 3, 4). This suggested that the low seed-setting rate in the dg1 mutant may be due to defective pollen grains. The wild-type grains and brown rice averaged 7.1 and 5.4 mm long, and 3.5 and 3.0 mm wide, respectively (Figures 7A–E and Supplementary Figure 3). The dg1 grains were longer and thinner than those of the wild type (Figures 7A,D,E and Supplementary Figure 3). The dg1 grains and brown rice averaged 7.9 and 5.6 mm long and 3.3 and 2.8 mm wide, respectively (Supplementary Figure 3). Compared with the wild type, the 1,000-grain weight was not affected in the dg1 mutant, but the weight of 1,000-brown rice from the dg1 mutant was decreased markedly (Supplementary Figure 3), suggesting that the proportion of hulls in the dg1 grains was higher than that of the wild type, which supports the ideal that the hulls can affect grain yield. We also examined the expression of genes involved in the regulation of grain size. Compared with the wild type, BG1, BG2, GS3, GW2, GW8, and TGW6 were down-regulated in the dg1 mutant (Figure 7F). These data supported the phenotypic observations and the ideal that DG1 may affect grain size by negatively regulating the related genes expression.
FIGURE 7.
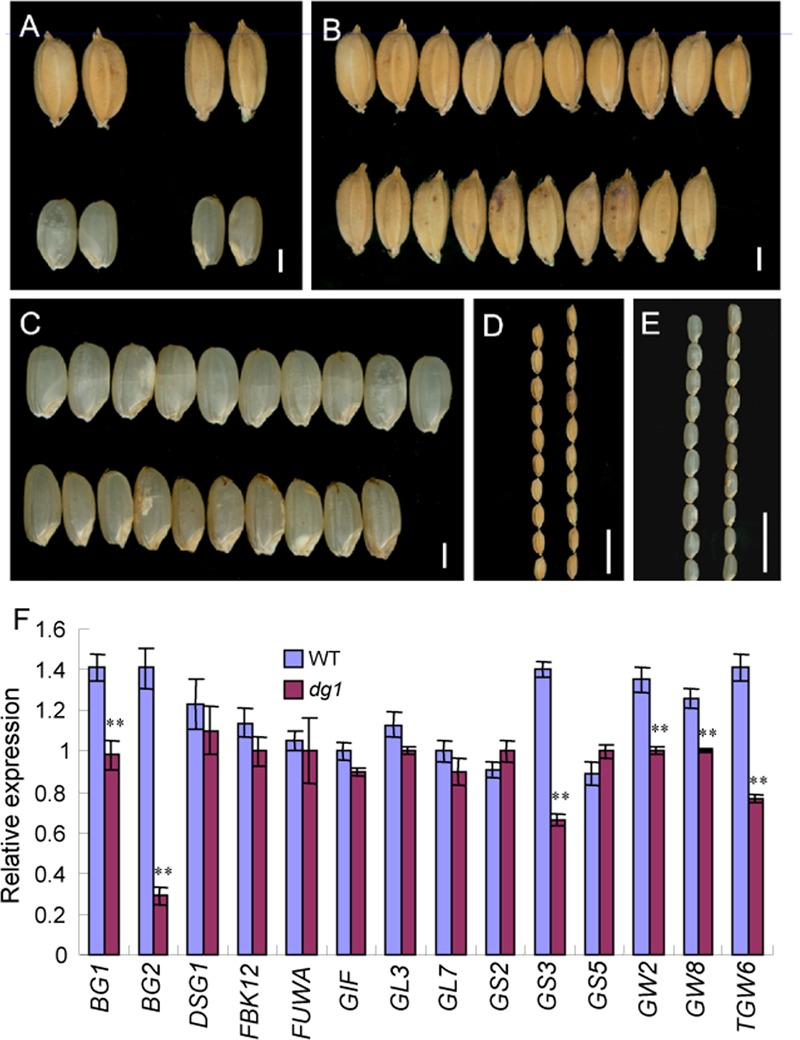
Phenotypic observations of grain and expression analysis of related genes controlling grain size in the wild type and dg1 mutant. (A) Grains phenotype of the wild type (left) and dg1 mutant (right). (B) Grains of the wild type (upper) and dg1 mutant (lower). (C) Brown rice of the wild type (upper) and dg1 mutant (lower). (D) Grains of the wild type (left) and dg1 mutant (right). (E) Brown rice of the wild type (left) and dg1 mutant (right). (F) Expression analysis of related genes controlling grain size in the wild type and dg1 mutant. Total RNA was isolated from the endosperms at 5 days after fertilization. Bars = 2000 μm in (A–C); 1 cm in (D, E). Error bars indicate SD. ∗∗Significant difference at P < 0.01 compared with the wild type by Student’s t-test.
To investigate the abnormal grains in the dg1 mutant at the cellular level, we performed histocytological analysis, including SEM and paraffin section. The observations of the outer epidermal cells of the lemmas showed that the cells were compressed longitudinally in the dg1 mutant (Figures 8A,B,E,F). However, the total cell number and the number of cells per millimeter along the longitudinal axis of the lemmas from the dg1 mutant were significantly increased compared to the wild type (Figures 8B,F,I,J), resulting in longer grains in the dg1 mutant. Cross sections of central parts of the hulls revealed that the dg1 spikelet contained shorter length of total cells and smaller cells than the corresponding region in the wild type (Figures 8C,D,G,H,K,L); this differences likely were responsible for the slender grains observed in the dg1 mutant. Taken together, our results implied that DG1 controls grain size by regulating cell number and size.
FIGURE 8.
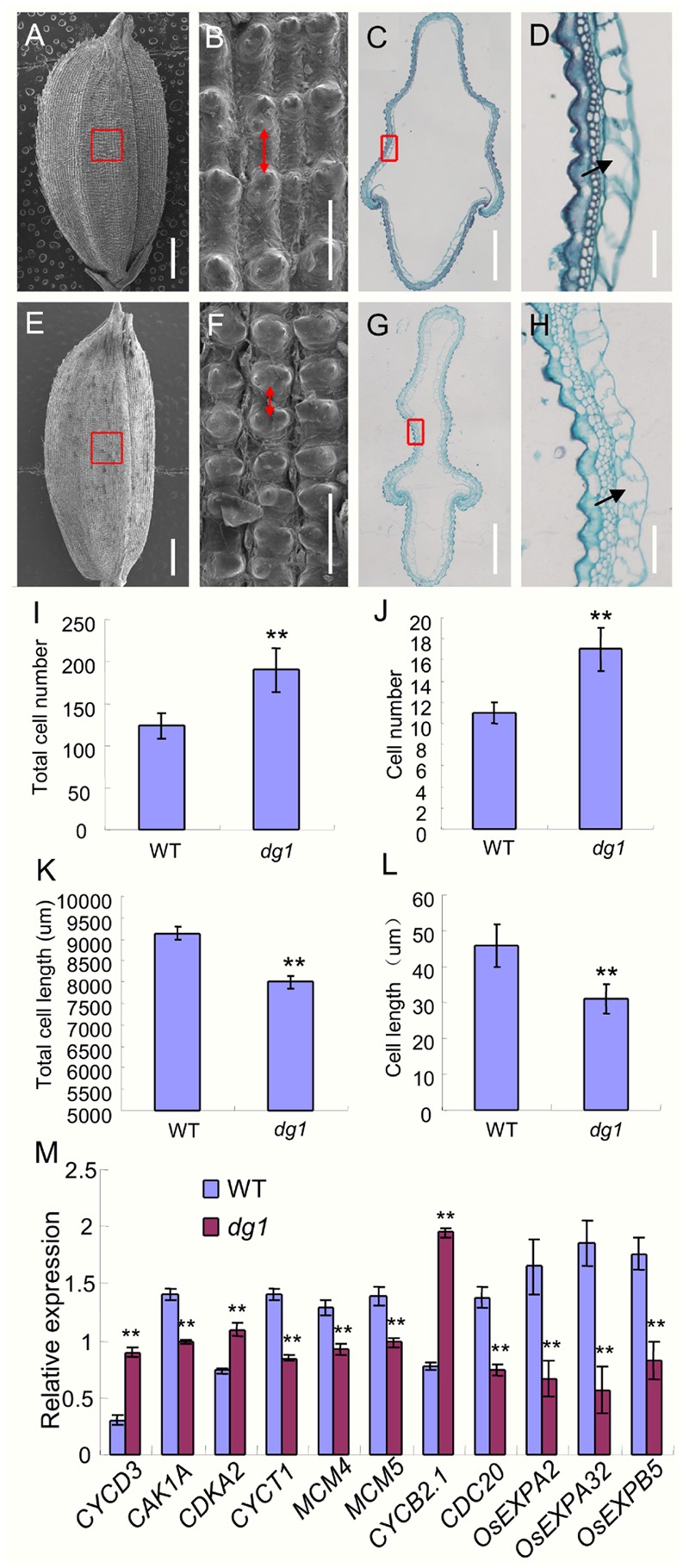
Histocytological analysis of grain hull in the wild type and dg1 mutant. (A) Grain of the wild type. (B) Red box region in (A). (C) Cross section of the hull in the wild type. (D) Partial magnification of red box region in (C). (E) dg1 grain. (F) Red box region in (E). (G) Cross section of the hull in the dg1 mutant. (H) Partial magnification of red box region in (G). (I) Total cell number along the longitudinal axis of the lemma. (J) Cell number per millimeter along the longitudinal axis of the lemma. (K) Total cell length in the middle part of the hull. (L) Cell length. (M) Expression analysis of cell cycle and expansion-related genes in panicles of the wild type and dg1 mutant. Red arrows represent cell length in the epidermal surface of the hull. Black arrows represent cell length in the cross section of the hull. Total RNA was isolated from the spikelets at the heading stage. Error bars indicate SD. ∗∗Significant difference at P < 0.01 compared with the wild type by Student’s t-test.
Because cell number and size were altered in the dg1 mutant, we next investigated the expression of several genes that regulate cell cycle and cell expansion in rice. The expressions of eight cell cycle-related genes was altered. Compared with the wild type, CYCD3, CDKA2, and CYCB2.1 were up-regulated in the dg1 mutant, and CAK1A, CYCT1, MCM4, MCM5, and CDC20 were down-regulated in the dg1 mutant (Figure 8M). The expression of three cell expansion-related genes (OsEXPA2, OsEXPA32, and OsEXPB5) was reduced in the dg1 mutant compared to the wild type (Figure 8M). These results were consistent with the phenotypic observations and implied that DG1 mainly affects grain size by regulating cell proliferation and cell expansion.
Genetic Analysis of the dg1 Mutant
To survey whether the dg1 mutant phenotype is controlled by multiple genes or a single gene and whether it is dominant or recessive, we performed the reciprocal crosses between the dg1 mutant and two indica cultivars, NJ6 and TN1 to obtain F1 and F2 plants (Table 1). The F1 offspring of all combinations showed a normal phenotype (Table 1), indicating that the dg1 mutation is recessive. In the four F2 populations, the segregation ratio of the wild type to dg1 phenotype was 3:1 (Table 1), indicating that the mutated phenotype is controlled by a single, recessive nuclear gene.
Table 1.
Genetic analysis of the phenotype of the dg1 mutant.
| F2 |
||||||
|---|---|---|---|---|---|---|
| Phenotype of F1 | No. of wild type plants | No. of dg1 plants | Total plants | X2 (3:1) | P-value | |
| dg1/NJ06 | Normal | 1612 | 518 | 2130 | 0.526 | 0.468 |
| NJ06/dg1 | Normal | 1165 | 400 | 1565 | 0.261 | 0.610 |
| dg1/TN1 | Normal | 1209 | 381 | 1590 | 0.913 | 0.339 |
| TN1/dg1 | Normal | 961 | 342 | 1303 | 1.081 | 0.299 |
Fine Mapping and Candidate Genes Analysis of DG1
To map the locus responsible for the dg1 mutant phenotype, we selected the cross of dg1 mutant and NJ6. We then tested 312 pairs of SSR primers evenly distributed on the 12 rice chromosomes and found that 165 of these markers showed polymorphisms between the two parental lines. Using the polymorphic markers, we screened DNA pools from 15 wild type individuals and 15 recessive dg1 individuals and compared the pools with the parental lines. Two markers, C2 and C21 on chromosome 8, had the same amplified fragment length in the recessive DNA pool and the dg1 mutant, and the fragment length differed from that of the wild-type DNA pool. Using this small F2 population (70 plants), the mutated locus was primarily mapped to chromosome 8 between markers C2 and C21 (Figure 9A).
FIGURE 9.
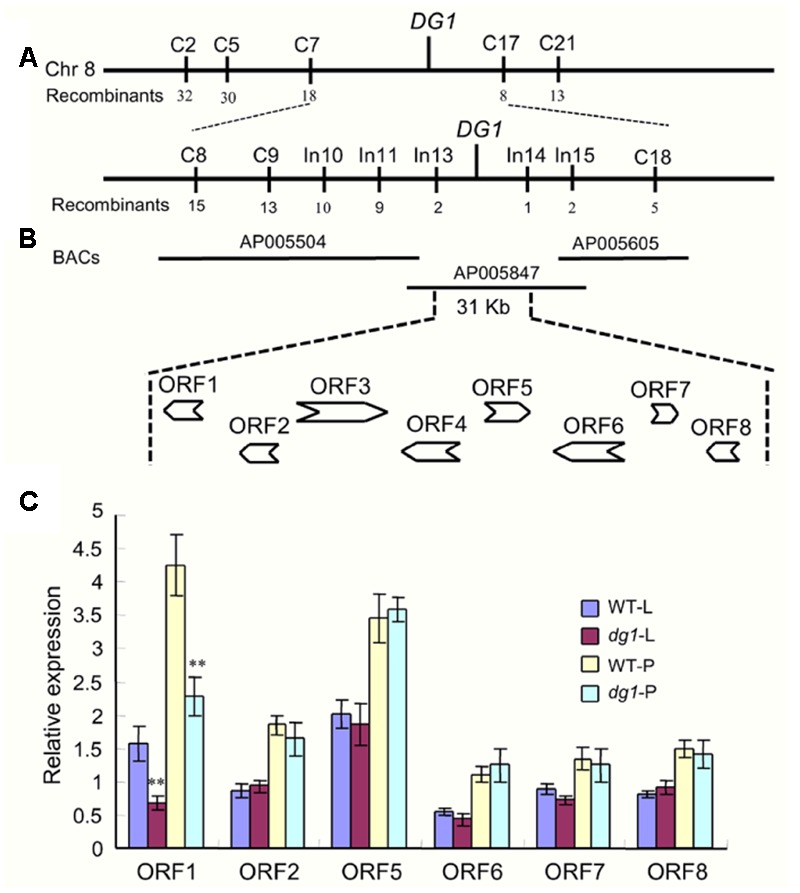
Fine mapping of the DG1 gene and analysis of candidate genes. (A,B) Fine mapping of the DG1 gene. (C) Expression analysis of candidate genes. WT-L, leaf of the wild type at heading stage; dg1-L, leaf of the dg1 mutant at heading stage; WT-P, panicles of the wild type at heading stage; dg1-P, panicles of the dg1 mutant at heading stage. Error bars indicate SD. ∗∗Significant difference at P < 0.01 compared with the wild type by Student’s t-test.
To further access the dg1 locus, we designed a large number of SSR and InDel markers flanking C2 and C21. The markers C5, C7, C8, C9, C17, C18, In10, In11, In13, In14, and In15 exhibited polymorphisms between the two parents (Figure 9A). Using 518 recessive individuals, the DG1 gene was finally delimited between InDel markers In13 and In14, an approximately 31-kb physical distance in Nipponbare (Figure 9B). In the 31-kb target region, there were eight predicted opening reading frames (ORFs) (Figure 9B). ORF1 encodes an acyl CoA binding protein that was reported to be related to plant growth and stress tolerance. ORF2 encodes a ribosomal protein and ORF3 and ORF4 encode retrotransposon proteins. ORF5 encodes an expression protein and ORF6 encodes a nuf2 family protein. ORF7 and ORF8 encode an Mps one binder kinase activator-like 1A and a DNA polymerase, respectively. We next sequenced all the annotated genes, but no nucleotide changes were detected between the wild type and dg1 mutant. We also investigated the expression of all annotated genes in the leaf and panicles by qRT-PCR. Compared with the wild type, only LOC_Os08g06550 (ORF1) showed a lower level of expression in the dg1 mutant; no differences were detected in the other genes between the wild type and dg1 mutant (Figure 9C). More work is needed to identify the gene responsible for the dg1 mutant phenotype.
The Expression of Genes Related to Floral Development in the dg1 Mutant
Since the dg1 mutant has defects in spikelet development, we examined whether DG1 is involved in the regulation of genes associated with floral development in rice. We detected several MADS-box and non-MADS-box genes including A-class genes (OsMADS14, and OsMADS15), B-class genes (OsMADS2, OsMADS4, and OsMADS16), C-class genes (OsMADS3 and OsMADS58), E-class genes (OsMADS1, OsMADS6, and OsMADS34), and non-MADS-box genes (DL, G1, SNB, IDS1, and MFS1). Except for OsMADS6 and OsMADS34, most of the tested MADS-box genes showed lower expression levels in the dg1 mutant (Figure 10A). Lower expression of OsMADS1, OsMADS14, and OsMADS15 may be attributed to abnormal heading and short panicles in the dg1 mutant. Abnormal expression of OsMADS2, OsMADS4, OsMADS16, OsMADS3, OsMADS58, and DL likely caused the defective stamens and pistils in the dg1 mutant.
FIGURE 10.
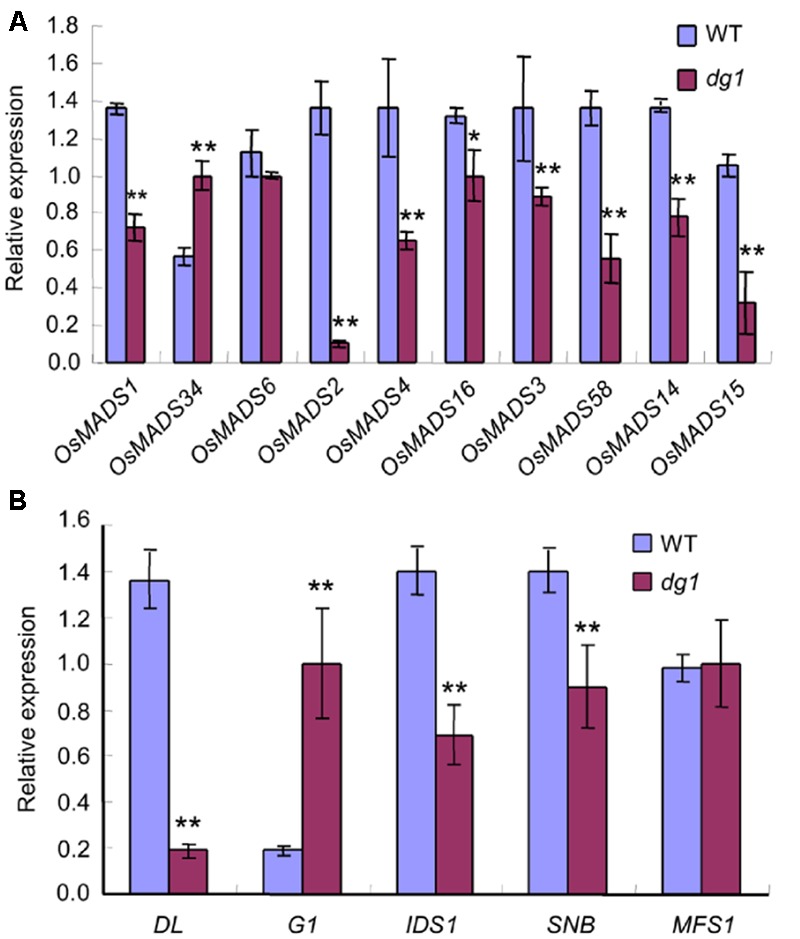
Expression analysis of related floral development genes. (A) The expression of MADS-box genes. (B) The expression of other related genes. The total RNA was isolated from young panicles at the heading stage. ∗∗Significant difference at P < 0.01 compared with the wild type by Student’s t-test.
We also observed more abundant transcripts of G1 and OsMADS34 which restrained the elongation of glumes (rudimentary glume and sterile lemma) (Figure 10B) and fewer transcripts of SNB and IDS1 which positively regulated fate of glumes in the dg1 mutant (Figure 10B). These results supported the phenotypic observations and suggested that DG1 negatively regulated the identities of glumes, resembling the functions of G1 and OsMADS34. Taken together, our findings revealed pleiotropic roles of DG1 in the floral development and DG1 regulated the expression of these genes associated with floral development.
Discussion
Many rice mutants associated with the vegetative development and floral organ identity have been obtained such as apo1, sad1, opb, afd1, and ddf1 (Ikeda et al., 2007; Horigome et al., 2009; Duan et al., 2012; Ikeda-Kawakatsu et al., 2012; Li et al., 2015; Ren et al., 2016b). However, our understanding of vegetative and reproductive development in rice remains limited, and the discovery of more rice mutants will further our knowledge of the regulatory mechanism behind these processes. The apo1 mutant rapidly produced more leaves than wild type, the stamens were transformed into lodicules, and extra pistils emerged (Ikeda et al., 2007). The sad1 mutant exhibited a variety of defects including few tillers, severe dwarfism, poor root, and abnormal floral organs (Li et al., 2015). In the opb mutant, leaf blades were malformed and the sheath cells partly invaded the blade region. Additionally, lateral growth of the lemma and palea was suppressed and the resulting structure was unable to enclose the inner floral organs (Horigome et al., 2009). Dwarfism occurred in the afd1 and ddf1 mutants due to defects in cell proliferation and expansion and the lodicules and stamens were transformed into glume-like organs (Duan et al., 2012; Ren et al., 2016b). The genes responsible for these phenotypes are expressed in all tissues and organs, suggesting that these genes are not tissue-specific and play important roles in the regulation of whole-plant development. In our study, the dg1 mutant had defects in plant height, leaf morphology, and leaf angle. The dg1 mutant had slender culms with small and semi-rolled leaves. Further analysis revealed that these defects could attributed to abnormal cell number and cell size in those organs, including fewer vascular bundles, smaller cells, and more bulliform cells.
In our study, the dg1 mutant had an enlarged leaf angle and BR was a key regulator of leaf angle (Sakamoto et al., 2013; Chen et al., 2015; Zhang et al., 2015). The 24-eBL treatments at concentrations over 0.1 μM induced a lamina joint angle of above 90° in the wild type and dg1 mutant. However, the dg1 mutant produced a larger leaf angle than that of the wild type at 24-eBL treatment of 0.1 and 1 μM, and no differences were observed between the wild type and dg1 mutant at 24-eBL treatment of 0 and 0.01 μM. The expression of BR response and biosynthetic genes OsBRI1, OsBZR1, CYP90D2, and BRD1 were markedly increased in the dg1 mutant compared with the wild type under normal growth conditions and following BR treatment. However, the decreased ranges of expressions of D2, BRD1, and OsBZR1 were higher in the dg1 mutant than that in the wild type under BR treatment. These findings suggested that the dg1 mutant was hypersensitive to BR and may be had defects in BR signaling.
The glumes (i.e., rudimentary glume and sterile lemma) are a unique structure in grass spikelets (Zanis, 2007; Li et al., 2009; Ren et al., 2013, 2016a) and the dg1 mutation also affected the development of glumes. In the dg1 mutant, some spikelets exhibited larger rudimentary glumes and sterile lemmas. The rudimentary glume and sterile lemma in the dg1 mutant exhibited similar protrusions, trichomes, and cell layers, resembling the wild-type lemma. Also, in the dg1 glumes, OsMADS1, OsMADS14, OsMADS15, and DL were expressed ectopically in the glumes, but no OsMADS6 transcript was detected. These results revealed that the glumes of dg1 mutant partly acquired the lemma identity and DG1 may be an important regulator of the glume identity.
There are two prevailing hypotheses on the origin and evolution of the glumes. One suggests that the rudimentary glume and sterile lemma are severely reduced bract-like organs (Schmidt and Ambrose, 1998; Terrell et al., 2001; Hong et al., 2010; Ren et al., 2013). The other suggests that the sterile lemma may be derived from a morphological modification of the remaining lemma (Yoshida et al., 2009; Kobayashi et al., 2010; Ren et al., 2016a). FRIZZY PANICLE (FZP), SUPERNUMERARY BRACT (SNB), Oryza sativa INDETERMINATE SPIKELET1 (OsIDS1), and MULTI-FLORET SPIKELET1 (MFS1) encode APETALA2/ethyleneresponsive (AP2/ERF) proteins that determine the identity of sterile lemma and/or the rudimentary glume. Loss of function of FZP and SNB resulted in extra rudimentary glumes, but no sterile lemmas were found in the corresponding position (Komatsu et al., 2003; Lee et al., 2007). A mutation of OsIDS1 and MFS1 caused the sterile lemmas to be converted into bract-like organs resembling the rudimentary glumes. The bract-like glume organs of maize and wheat are severely reduced and equivalent to the rudimentary glume in rice, and are similar to the lemma in size and structure (Kellogg, 2001; Yoshida et al., 2009; Hong et al., 2010). The sterile lemmas were elongated and partly acquired the lemma identity in the dg1, g1/ele, osmads34, and eg1 mutants (Li et al., 2009; Yoshida et al., 2009; Hong et al., 2010; Gao et al., 2010; Lin et al., 2014; Ren et al., 2016a), which supports the second hypothesis. Therefore, these findings reveal that the rudimentary glume, sterile lemma and the lemma may have been homologous organs during evolution. We also observed slender grains in the dg1 mutant. Histocytological observation found that the number and size of hull cells were altered in the dg1 mutant. The qRT-PCR analyses showed that DG1 affected the expression of cell cycle and expansion related genes, indicating that DG1 influences grain size by regulating cell proliferation and expansion.
Using the SSR and InDel markers, we mapped the DG1 locus between InDel markers In13 and In14 on chromosome 8 within a 31-kb physical region. This region contains eight annotated genes, including an acyl CoA binding protein. Although the acyl CoA binding proteins have been associated with floral development, no differences were detected in the DNA sequences between the wild type and dg1 mutant. However, the expression level of LOC_Os08g06550 (predicted acyl CoA binding protein) was dramatically decreased in the dg1 mutant. We also investigated other annotated genes, and the DNA sequences and transcripts levels were not altered between the wild type and dg1 mutant. Thus, LOC_Os08g06550 may contribute to the dg1 mutant phenotypes. However, these findings did not provide valid evidences to identify the DG1 gene. Other factors besides DNA sequence changes, such as epigenetic alterations can lead to altered phenotypes. A previous study showed that FERTILIZATION-INDEPENDENT ENDOSPERM 1 (FIE1) repressed the expression of the H3K27me3-mediated gene and interacted with rice enhancer of Zeste homologs (Zhang et al., 2012). The fie1 allele had no nucleotide sequence changes but was hypomethylated in the promoter region of FIE1, and fie1 mutant had dwarfism and various floral defects. Thus, more work is needed to isolate the target gene, such as the analysis of promoter regions and untranslational region sequences of the candidate gene, intergenic enhancers or repressors, and epigenetic regulation in the dg1 mutant (Luo et al., 2008).
Taken together, cloning and functional analysis of the DG1 gene would facilitate a better understanding of the molecular mechanisms involved in vegetative and spikelet development in rice, and may provide a new opportunity to improve rice grain yield. Additionally, the dg1 mutant is an ideal material for further studies of BR signaling.
Author Contributions
Experimental design: QQ, DR, and WC; Experiments: HY, DR, BR, ZW, YZ, YL, JH, and LZ; Data analysis: DZ, GZ, ZG, GC, and LG; Manuscript preparation: DR and QQ.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31401464 and 31521064). This work was supported by the National Key Research and Development Program of China (2016YFD0100902-07), and the National Science and Technology Major Project (2016ZX08009003-003-008).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00486/full#supplementary-material
References
- Chen Q., Xie Q., Gao J., Wang W., Sun B., Liu B., et al. (2015). Characterization of rolled and erect leaf 1 in regulating leave morphology in rice. J. Exp. Bot. 66 6047–6058. 10.1111/pce.12397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen E., Meyerowitz E. (1991). The war of the whorls: genetic interactions controlling flower development. Nature 353 31–37. 10.1038/353031a0 [DOI] [PubMed] [Google Scholar]
- Ditta G., Pinyopich A., Robles P., Pelaz S., Yanofsky M. (2004). The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr. Biol. 14 1935–1940. 10.1016/j.cub.2004.10.028 [DOI] [PubMed] [Google Scholar]
- Dreni L., Jacchia S., Fornara F., Fornari M., Ouwerkerk P., An G., et al. (2007). The D-lineage MADS-box gene OsMADS13 controls ovule identity in rice. Plant J. 52 690–699. 10.1111/j.1365-313X.2007.03272.x [DOI] [PubMed] [Google Scholar]
- Dreni L., Pilatone A., Yun D., Erreni S., Pajoro A., Caporali E., et al. (2011). Functional analysis of all AGAMOUS subfamily members in rice reveals their roles in reproductive organ identity determination and meristem determinacy. Plant Cell 23 2850–2863. 10.1105/tpc.111.087007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Y., Li S., Chen Z., Zheng L., Diao Z., Zhou Y., et al. (2012). Dwarf and deformed flower 1, encoding an F-box protein, is critical for vegetative and floral development in rice (Oryza sativa L.). Plant J. 72 829–842. 10.1111/j.1365-313X.2012.05126.x [DOI] [PubMed] [Google Scholar]
- Gao X., Liang W., Yin C., Ji S., Wang H., Su X., et al. (2010). The SEPALLATA-like gene OsMADS34 is required for rice inflorescence and spikelet development. Plant Physiol. 153 728–740. 10.1104/pp.110.156711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong L., Qian Q., Zhu K., Tang D., Huang Z., Gao L., et al. (2010). ELE restrains empty glumes from developing into lemmas. J. Genet. Genomics 37 101–115. 10.1016/S1673-8527(09)60029-1 [DOI] [PubMed] [Google Scholar]
- Horigome A., Nagasawa N., Ikeda K., Ito M., Itoh J., Nagato Y. (2009). Rice OPEN BEAK is a negative regulator of class 1 knox genes and a positive regulator of class B floral homeotic gene. Plant J. 58 724–736. 10.1111/j.1365-313X.2009.03823.x [DOI] [PubMed] [Google Scholar]
- Ikeda K., Ito M., Nagasawa N., Kyozuka J., Nagato Y. (2007). Rice ABERRANT PANICLE ORGANIZATION 1, encoding an F-box protein, regulates meristem fate. Plant J. 51 1030–1040. 10.1111/j.1365-313X.2007.03200.x [DOI] [PubMed] [Google Scholar]
- Ikeda K., Nagasawa N., Yasuo N. (2005). ABERRANT PANICLE ORGANIZATION 1 temporally regulates meristem identity in rice. Dev. Biol. 282 349–360. 10.1016/j.ydbio.2005.03.016 [DOI] [PubMed] [Google Scholar]
- Ikeda-Kawakatsu K., Maekawa M., Izawa T., Itoh J., Nagato Y. (2012). ABERRANT PANICLE ORGANIZATION 2/RFL, the rice ortholog of Arabidopsis LEAFY, suppresses the transition from inflorescence meristem to floral meristem through interaction with APO1. Plant J. 69 168–180. 10.1111/j.1365-313X.2011.04781.x [DOI] [PubMed] [Google Scholar]
- Ikeda-Kawakatsu K., Yasuno N., Oikawa T., Iida S., Nagato Y., Maekawa M., et al. (2009). Expression Level of ABERRANT PANICLE ORGANIZATION1 determines rice inflorescence form through control of cell proliferation in the meristem. Plant Physiol. 150 736–747. 10.1104/pp.109.136739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon J., Jang S., Lee S., Nam J., Kim C., Lee S., et al. (2000). leafy hull sterile1 Is a homeotic mutation in a rice MADS box gene affecting rice flower development. Plant Cell 12 871–884. 10.1105/tpc.12.6.871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg E. (2001). Evolutionary history of the grasses. Plant Physiol. 125 1198–1205. 10.1104/pp.125.3.1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K., Maekawa M., Miyao A., Hirochika H., Kyozuka J. (2010). PANICLE PHYTOMER2 (PAP2), encoding a SEPALLATA subfamily MADS-box protein, positively controls spikelet meristem identity in rice. Plant Cell Physiol. 51 47–57. 10.1093/pcp/pcp166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K., Yasuno N., Sato Y., Yoda M., Yamazaki R., Kimizu M., et al. (2012). Inflorescence meristem identity in rice is specified by overlapping functions of three AP1/FUL-like MADS box genes and PAP2, a SEPALLATA MADS box gene. Plant Cell 24 1848–1859. 10.1105/tpc.112.097105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M., Chujo A., Nagato Y., Shimamoto K., Kyozuka J. (2003). FRIZZY PANICLE is required to prevent the formation of axillary meristems and to establish floral meristem identity in rice spikelets. Development 130 3841–3850. 10.1242/dev.00564 [DOI] [PubMed] [Google Scholar]
- Kyozuka J., Shimamoto K. (2002). Ectopic expression of OsMADS3, a rice ortholog of AGAMOUS, caused a homeotic transformation of lodicules to stamens in transgenic rice plants. Plant Cell Physiol. 43 130–135. 10.1093/pcp/pcf010 [DOI] [PubMed] [Google Scholar]
- Lee D., Lee J., Moon S., Park S., An G. (2007). The rice heterochronic gene SUPERNUMERARY BRACT regulates the transition from spikelet meristem to floral meristem. Plant J. 49 64–78. 10.1111/j.1365-313X.2006.02941.x [DOI] [PubMed] [Google Scholar]
- Li H., Liang W., Hua Y., Zhu L., Yin C., Xu J., et al. (2011). Rice MADS6 interacts with the floral homeotic genes SUPERWOMAN1, MADS3, MADS58, MADS13, and DROOPING LEAF in specifying floral organ identities and meristem fate. Plant Cell 23 2536–2552. 10.1105/tpc.111.087262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Xue D., Gao Z., Yan M., Xu W., Xin Z., et al. (2009). A putative lipase gene EXTRA GLUME1 regulates both empty-glume fate and spikelet development in rice. Plant J. 57 593–605. 10.1111/j.1365-313X.2008.03710.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Yoshida A., Takahashi M., Maekawa M., Kojima M., Sakakibara H., et al. (2015). SAD1, an RNA polymerase I subunit A34.5 of rice, interacts with Mediator and controls various aspects of plant development. Plant J. 81 282–291. 10.1111/tpj.12725 [DOI] [PubMed] [Google Scholar]
- Lin X., Wu F., Du X., Shi X., Liu Y., Liu S., et al. (2014). The pleiotropic SEPALLATA-like gene OsMADS34 reveals that the ‘empty glumes’ of rice (Oryza sativa) spikelets are in fact rudimentary lemmas. New Phytol. 202 689–702. 10.1111/nph.12657 [DOI] [PubMed] [Google Scholar]
- Luo J., Hao W., Jin J., Gao J., Lin H. (2008). Fine mapping of Spr3, a locus for spreading panicle from African cultivated rice (Oryza glaberrima Steud.). Mol. Plant 1 830–838. 10.1093/mp/ssn045 [DOI] [PubMed] [Google Scholar]
- Nagasawa N., Miyoshi M., Sano Y., Satoh H., Hirano H., Sakai H., et al. (2003). SUPERWOMAN1 and DROOPING LEAF genes control floral organ identity in rice. Development 130 705–718. 10.1242/dev.00294 [DOI] [PubMed] [Google Scholar]
- Pelaz S., Ditta G., Baumann E., Wisman E., Yanofsky M. (2000). B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405 200–203. 10.1038/35012103 [DOI] [PubMed] [Google Scholar]
- Prasad K., Vijayraghavan U. (2003). Double-stranded RNA interference of a rice PI/GLO paralog, OsMADS2, uncovers its second-whorl-specific function in floral organ patterning. Genetics 165 2301–2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D., Li Y., Zhao F., Sang X., Shi J., Wang N., et al. (2013). MULTI-FLORET SPIKELET1, which encodes an AP2/ERF protein, determines spikelet meristem fate and sterile lemma identity in rice. Plant Physiol. 162 872–884. 10.1104/pp.113.216044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D., Rao Y., Leng Y., Li Z., Xu Q., Wu L., et al. (2016a). Regulatory role of OsMADS34 in the determination of glumes fate, grain yield, and quality in rice. Front. Plant Sci. 7:1853 10.3389/fpls.2016.01853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D., Rao Y., Wu L., Xu Q., Li Z., Yu H., et al. (2016b). 2016 The pleiotropic ABNORMAL FLOWER AND DWARF1 affects plant height, floral development and grain yield in rice. J. Integr. Plant Biol. 58 529–539. 10.1111/jipb.12441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T., Morinaka Y., Inukai Y., Kitano H., Fujioka S. (2013). Auxin signal transcription factor regulates expression of brassinosteroid receptor gene in rice. Plant J. 73 676–688. 10.1111/tpj.12071 [DOI] [PubMed] [Google Scholar]
- Sang X., Li Y., Luo Z., Ren D., Fang L., Wang N., et al. (2012). CHIMERIC FLORAL ORGANS1, encoding a monocot-specific MADS box protein, regulates floral organ identity in rice. Plant Physiol. 160 788–807. 10.1104/pp.112.200980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt R., Ambrose B. (1998). The blooming of grass flower development. Curr. Opin. Plant Biol. 1 60–67. 10.1016/S1369-5266(98)80129-5 [DOI] [PubMed] [Google Scholar]
- Terrell E. E., Peterson P. P., Wergin W. P. (2001). Epidermal features and spikelet micromorphology in Oryza and related genera (Poaceae: Oryzeae). Smithsonian Contr. Bot. 91 1–50. 10.5479/si.0081024X.91 [DOI] [Google Scholar]
- Theissen G., Saedler H. (2001). Plant biology. Floral quartets. Nature 409 469–471. 10.1038/35054172 [DOI] [PubMed] [Google Scholar]
- Wang K., Tang D., Hong L., Xu W., Huang J., Li M., et al. (2010). DEP and AFO regulate reproductive habit in rice. PLoS Genet. 6:e1000818 10.1371/journal.pgen.1000818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T., Hirano H. (2006). Function and diversification of MADS-box genes in rice. Sci. World J. 6 1923–1932. 10.1100/tsw.2006.320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T., Nagasawa N., Kawasaki S., Matsuoka M., Nagato Y., Hirano H. (2004). The YABBY gene DROOPING LEAF regulates carpel specification and midrib development in Oryza sativa. Plant Cell 16 500–509. 10.1105/tpc.018044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao S., Ohmori S., Kimizu M., Yoshida H. (2008). Unequal genetic redundancy of rice PISTILLATA orthologs, OsMADS2 and OsMADS4, in lodicule and stamen development. Plant Cell Physiol. 49 853–857. 10.1093/pcp/pcn050 [DOI] [PubMed] [Google Scholar]
- Yoshida A., Suzaki T., Tanaka W., Hirano H. (2009). The homeotic gene long sterile lemma (G1) specifies sterile lemma identity in the rice spikelet. Proc. Natl. Aacd. Sci. U.S.A. 106 20103–20108. 10.1073/pnas.0907896106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun D., Liang W., Dreni L., Yin C., Zhou Z., Kater M., et al. (2013). OsMADS16 genetically interacts with OsMADS3 and OsMADS58 in specifying floral patterning in rice. Mol. Plant 6 743–756. 10.1093/mp/sst003 [DOI] [PubMed] [Google Scholar]
- Zanis M. (2007). Grass spikelet genetics and duplicate gene comparisons. Int. J. Plant Sci. 168 93–110. 10.1086/509787 [DOI] [Google Scholar]
- Zhang L., Cheng Z., Qin R., Qiu Y., Wang J., Cui X., et al. (2012). Identification and characterization of an epi-allele of FIE1 reveals a regulatory linkage between two epigenetic marks in rice. Plant Cell 24 4407–4421. 10.1105/tpc.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Wang S., Xu Y., Yu C., Shen C., Qian Q., et al. (2015). The auxin response factor, OsARF19 controls rice leaf angles through positively regulating OsGH3-5 and OsBRI1. Plant Cell Environ. 38 638–654. 10.1111/pce.12397 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


