Abstract
Staphylococcus xylosus is used as a starter culture in fermented meat products and contributes to color formation by the reduction of nitrate to nitrite. Nitrite is a food additive that is chemically turned to nitric oxide (NO) in meat but its safety has been questioned. The objective of this study was to determine the ability of NO synthase (NOS) of S. xylosus C2a to produce NO. For this purpose, a nos deletion mutant (Δnos) in S. xylosus was constructed and NO production was evaluated in a test based on its ability to form nitrosomyoglobin and nitrosoheme. Production of NO was abrogated in the Δnos mutant under aerobic conditions and reduced about 35-40% comparing to the wild type C2a under limited oxygenation. This mutant was sensitive to oxidative stress. The expression of genes encoding catalase was modulated in the mutant with an up-regulation of katA and a down-regulation of katB and katC. The Δnos mutant displayed high colony pigmentation after prolonged growth on agar medium. Finally, the Δnos mutant showed no growth in minimal medium. Growth was not restored in the minimal medium by complementation with nos, but was restored by either addition of phenylalanine or complementation with pdt, a gene that encodes a prephenate dehydratase involved in phenylalanine biosynthesis and co-transcribed with nos. Our findings clearly demonstrate NOS-mediated NO production in S. xylosus, a meat-associated coagulase-negative Staphylococcus.
Keywords: nitric oxide synthase, nitric oxide, nitrosoheme, Staphylococcus xylosus, coagulase-negative Staphylococcus, oxidative stress
Introduction
Staphylococcus xylosus, a coagulase-negative Staphylococcus (CNS), is commonly used as starter culture to boost color development of cured meat products such as dried fermented sausages (Talon and Leroy, 2006). The typical reddish color relies on the presence of the curing salts, nitrate and nitrite. Nitrate salts are reduced to nitrite by nitrate reductase activity of S. xylosus (Talon et al., 1999; Gøtterup et al., 2008). Nitrite is subsequently converted by chemical reactions to nitric oxide (NO), which is able to bind to the ferrous heme-iron to form the stable red nitrosomyoglobin pigment (Gøtterup et al., 2007, 2008).
The use of the curing agents nitrate and nitrite is regulated by law, with specific indications in the United States (Code of Federal Regulations, 2016) and Europe (Commission Regulation [EU], 2011). Nowadays, the safety regarding the use of such additives on meat products is questioned. Nitrites can react with secondary amine groups of muscle proteins as well as compounds present in the gastrointestinal tract to form undesired N-nitroso compounds such as nitrosamines, which could pose a health threat (Honikel, 2008). The meat industry is therefore looking for alternatives to decrease nitrate/nitrite in the production of meat products.
Staphylococcus xylosus has been shown to convert metmyoglobin to nitrosomyoglobin in culture medium, in salami (Morita et al., 1998) and in raw meat batter (Li et al., 2013, 2016), without addition of nitrate or nitrite. NO production has been suggested to be linked to NO synthase (NOS) activity. NOS catalyzes the production of NO from l-arginine and was initially described in mammals (Alderton et al., 2001). Homologs of the oxygenase domain of mammalian NOS were identified in Gram-positive bacteria. NOS is involved in the production of NO by Bacillus subtilis (Gusarov and Nudler, 2005; Gusarov et al., 2008), Bacillus anthracis (Shatalin et al., 2008), Bacillus cereus (Montgomery et al., 2010), Geobacillus stearothermophilus (Kabir et al., 2008), Streptomyces turgidiscabies (Kers et al., 2004), Deinococcus radiodurans (Patel et al., 2009), and Staphylococcus aureus (van Sorge et al., 2013). S. aureus NOS is involved in resistance to oxidative stress and antibiotics, and virulence (Vaish and Singh, 2013; van Sorge et al., 2013; Sapp et al., 2014). In all sequenced S. aureus genomes, the nos gene is part of a cluster containing another gene encoding a prephenate dehydratase, which is involved in phenylalanine biosynthesis. This genetic organization appears to be unique to staphylococcal genomes (Sapp et al., 2014). The nos genes have been identified in many staphylococci (van Sorge et al., 2013; Sánchez-Mainar et al., 2014; Sapp et al., 2014), but NOS-mediated NO production has only been characterized in the pathogenic S. aureus. The contribution of the NOS activity of S. xylosus to formation of nitrosomyoglobin remains to be demonstrated.
In an attempt to determine the ability of S. xylosus NOS to produce NO, we generated a nos deletion mutant in the S. xylosus C2a strain. NO production in S. xylosus C2a and mutants was investigated using an assay based on the conversion of metmyoglobin to red pigment derivatives followed by an extraction of nitrosoheme. We also assessed the sensitivity of the mutants to oxidative stress.
Materials and Methods
Bacterial Strains and Growth Conditions
The bacterial strains and plasmids used in this study are listed in Table 1. Experiments were performed using S. xylosus strain C2a or its isogenic mutants. S. xylosus was routinely cultured in Tryptic Soy Broth (TSB, Difco) under aerobic conditions (1:10 volume to flask ratio, 150 rpm) or on Tryptic Soy Agar (TSA, Difco) at 30°C. When required, strains were grown in a minimal medium (MX) as already described (Fiegler and Brückner, 1997). The MX medium, when needed, was supplemented with amino acids (100 mg/L). For genetic manipulation, Escherichia coli was cultured in Luria–Bertani (LB, Difco) broth with shaking aeration (150 rpm) or on LB agar (Difco) at 37°C. Antibiotics, when required to maintain plasmids or select recombinants, were added at the following concentrations: for E. coli, 100 μg/mL ampicillin; for S. xylosus, 20 μg/mL chloramphenicol, or 2.5 to 10 μg/mL erythromycin. All chemicals were from Sigma–Aldrich.
Table 1.
List of strains and vectors used in this study.
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Staphylococcus xylosus | ||
| C2a | Derived from the type strain DSM20267 and genetically transformable strain | Götz et al., 1983, LN554884 |
| Δnos | Isogenic mutant of C2a deleted of the SXYL_00923 gene, Δnos::ermB | This study |
| ΔnospRBnos | Δnos complemented with nos | This study |
| ΔnospRBpdt | Δnos complemented with pdt (SXYL_00922) | This study |
| ΔnospRBnospdt | Δnos complemented with nospdt | This study |
| C2apRB473 | C2a containing empty vector pRB473 | This study |
| C2apRB474 | C2a containing empty vector pRB474 | This study |
| ΔnospRB473 | Δnos containing empty vector pRB473 | This study |
| ΔnospRB474 | Δnos containing empty vector pRB474 | This study |
| Escherichia coli | ||
| TOP10 | Competent strain for plasmid transformation | Invitrogen |
| Plasmids | ||
| pBT2 | E. coli-S. xylosus thermosensitive shuttle vector, ApR CmR | Brückner, 1997 |
| pEC4 | pBluescript KS + derivative. Source of ermB gene (EmR). ApR | Brückner, 1997 |
| pRB473 | Shuttle vector ApR and CmR | Brückner, 1992 |
| pRB474 | Shuttle vector ApR and CmR | Brückner, 1992 |
| pBTΔnos | pBT2, SXYL_00923 [13-317]::ermB shuttle vector | This study |
| pRBnos | pRB473 derivate for expression of nos | This study |
| pRBpdt | pRB474 derivate for expression of pdt | This study |
| pRBnospdt | pRB473 derivate for expression of nos and pdt | This study |
To compare growth kinetics under microaerobic conditions, overnight cultures of S. xylosus were diluted to an optical density (OD) of 0.05 to 0.10 at 600 nm and incubated in 100-well microtiter plates with shaking at 30°C in a Bioscreen C plate reader (Labsystems France) while the turbidity was monitored every 30 min for 24 h. Three independent experiments were done for each set of conditions.
DNA Manipulation
Genomic DNA from S. xylosus was prepared from overnight cultures. Briefly, cells were resuspended in Tris-EDTA-sucrose buffer containing 0.1 mg/mL lysostaphin (Sigma-Aldrich) and incubated for 30 min at 37°C. Cells were lysed with sodium dodecyl sulfate and treated with RNase A. Following extraction with phenol-chloroform-isoamyl alcohol (25/24/1) and chloroform, DNA was precipitated with ethanol and resuspended in Tris-EDTA buffer (pH 8.0). Plasmid DNA from E. coli was isolated using the NucleoSpin Plasmid Quick-Pure kit (Macherey-Nagel). Restriction digests were performed using high-fidelity restriction enzymes (New England Biolabs). Ligations were performed using T4 DNA ligase (Roche). Amplifications were performed using Phusion High-Fidelity DNA Polymerase (New England Biolabs) for cloning and GO Taq DNA polymerase (Promega) for standard purposes. PCR products were visualized by 0.8% agarose gel electrophoresis and ethidium bromide staining, and imaged on a Gel Doc 2000 (Bio-Rad). As required, they were purified using the QIAquick Gel Extraction kit (Qiagen). DNA sequencing was performed by GATC Biotech (Mulhouse, France).
Construction of S. xylosus NO Synthase Mutant
The oligonucleotides used in this study were designed based on the genome sequence of S. xylosus C2a (GenBank accession no. LN554884) and are listed in Table 2. A chromosomal nos disruption mutant was constructed by deletion of the nos coding sequence corresponding to amino acids 11 to 345 (out of 354) and insertion of an erythromycin resistance cassette (ermB) using the temperature-sensitive vector pBT2. Briefly, a 756-bp upstream fragment and a 783-bp downstream fragment were separately amplified by using the genomic DNA of S. xylosus C2a as template. The ermB gene was amplified from the pEC4 vector generating an amplicon of 1,366-bp. The three purified PCR products were annealed by overlapping PCR using the outside primers and inserted into pBT2 using appropriate restriction enzymes. The resulting plasmid was constructed in competent E. coli TOP10 and introduced into S. xylosus C2a by electroporation as described (Götz et al., 1983; Brückner, 1997). Gene replacement was allowed to take place by incubating the temperature-sensitive plasmid at 42°C as described (Brückner, 1997). Successful C2aΔnos mutant was screened for resistance to erythromycin and sensitivity to chloramphenicol, indicating loss of the pBT2 backbone, and this was confirmed by PCR and sequencing.
Table 2.
Oligonucleotides used in this study.
| Name | Primer Sequence (5′ to 3′)a,b | Used for |
|---|---|---|
| Up923F1-EcoRI Up923R1-ErmF | CGGAATTCGACGCGTACCλTTCCATT CACAATAGAGAGATGTCACCTGTCGATλGGATTTCGCTTC | Amplification of sequence upstream nos |
| Dn923F2-ErmR Dn923R2-BamHI | GGTATACTACTGACAGCTTCCAACAGAAGCTACTGGTTGTCCA CGGGATCCGGTGTGTCTGCTTGGACλ | Amplification of sequence downstream nos |
| ErmF-Up923R1 ErmR-Dn923F2 | GAAGCGλTCCTTTATCGACAGGTGACATCTCTCTATTGTG TGGACAACCAGTAGCTTCTGTTGGAAGCTGTCAGTAGTATACC | Amplification of ermB |
| Up923F3 Dn923R3 | TCCTGCTCGCACATTACTTG CGTCAGGTATCTTGTTGCTCA | Control of nos::erm construction |
| Up923PF5-SalI Dn923R5-EcoRI | ACGCGTCGACGTGCTTGATAGCACATGλAAGGA CGGAATTCGGGTCGTTAATGλTGGACA | Complementation of nos in pRB473 |
| Up923PF5-SalI Dn922-EcoRI | ACGCGTCGACGTGCTTGATAGCACATGλAAGGA CGGAATTCTTGCTCATTAGGTTGTGCTAATTC | Complementation of nos-pdt in pRB473 |
| Up922-SalI Dn922-EcoRI | ACGCGTCGACGTAAGGGGTAACGTCλTGλ CGGAATTCTTGCTCATTAGGTTGTGCTAATTC | Complementation of pdt in pRB474 |
| 923F4 922R1 | AAGCGλTCCTTTATCGACAC CTAATGGCAGGCCCAATAGA | Co-transcription nos-pdt |
| 923F2 923R2 | TGCAGAAGCGTTTGAATTTG GCTTCGATGCAGTGAGATGA | qRT-PCR of nos (SXYL_00923) |
| 2505F 2505R | CGTCATCTTCACGλGTCATATTC CGCTAGTACACATTATTATCCAATAG | qRT-PCR of katA (SXYL_2505) |
| 1551F 1551R | ATTCGTGGATTCGCATλAG AGCTTCTGGTAGTGACGT | qRT-PCR of katB (SXYL_1551) |
| 2533F 2533R | TTCGATCATGAACGTATACCA GTGTCTGGTGAACCTTTAGAG | qRT-PCR of katC (SXYL_2533) |
| 1303F 1303R | CGCAGCAGTAGAAGGAACTG ATGTCCACCGCCATTATTGC | qRT-PCR of the housekeeping gene sod (SXYL_1303) and control of DNA contamination in RNA extracts |
aUnderlined regions represent restriction enzyme sequences.
bPrimer containing overlapping end (in bold).
Creation of Complementation Plasmids
Complementation of the nos mutation was performed by amplifying the nos gene with its putative promoter. The corresponding amplicon was cloned into the shuttle vector pRB473 using appropriate enzymes, generating pRBnos. Similarly, an nos-pdt complementation vector (pRBnospdt) was constructed. A complementation vector pRBpdt was constructed, placing the expression of pdt under the constitutive promoter of pRB474. All PCR-amplified fragments were verified by sequencing. The resulting plasmids were constructed in competent E. coli TOP10 and introduced into S. xylosus C2aΔnos by electroporation according to an established protocol (Brückner, 1997).
Metmyoglobin Conversion and Assessment of Nitrosoheme Formation
A 20 mg/mL metmyoglobin solution was freshly prepared from equine heart myoglobin (Sigma-Aldrich) as described (Gündoğdu et al., 2006). Overnight S. xylosus cultures were inoculated in TSB supplemented with 2 mg/mL metmyoglobin to a final OD600 nm of 0.5 (8 log CFU/mL). Cultures were incubated either under limited oxygenation (covered with mineral oil and without stirring) or under aerobic conditions as described above. After 24 h of incubation at 30°C, OD600 nm and pH of the cultures were measured and cells were serial-diluted and plated on TSA for enumeration. Cultures were centrifuged and supernatants were used to measure the absorbance spectrum between 500 and 600 nm (BioMate 3, Thermo Fisher Scientific) using as control a solution of sterile TSB supplemented with metmyoglobin and incubated in the same conditions. Finally, supernatants were treated with acetone 1:4 (v/v) to extract the nitrosoheme from nitrosomyoglobin, and the absorbance spectrum was acquired between 450 and 640 nm (V-770 UV/visible, Jasco). Sterile TSB supplemented with metmyoglobin and extracted with acetone was used as control. Experiments were carried out in three independent biological replicates.
Analysis of colony pigmentation
After overnight growth in TSB, S. xylosus strains were plated on TSA. After 2 days of growth (when pigmentation is strongest), 100 mg of each strain was collected from plates and pigments were extracted and quantified according the method of Morikawa et al. (2001). Briefly, cells were washed once in water, centrifuged for 5 min at 15,000 g, re-suspended in 1 mL of methanol and heated for 5 min at 55°C. Cells were centrifuged and the supernatant containing pigment was retrieved. The absorbance spectrum of methanol-extracted pigment was measured between 400 and 600 nm to evaluate carotenoid content. A peak at λ = 460 nm was detected and results are expressed as relative optical density at 460 nm with normalization to C2a. Experiments were performed with three independent biological replicates.
Survival Experiments after Exposure to Different Stresses
The S. xylosus strains were grown in TSB under aerobic conditions. To assess sensitivity to oxidative stress, overnight cultures were diluted to an OD600 nm of 0.05 with fresh medium and incubated until OD600 nm was approximately 1. One milliliter of each culture was collected for serial dilutions and determination of colony-forming units (CFU)/mL on TSA plates. The remaining culture was treated with 150 mM hydrogen peroxide (Sigma-Aldrich) and incubated for 1 h at 30°C. 4,000 U/mL of catalase (Sigma–Aldrich) was added to quench residual H2O2 as described (Barrière et al., 2002). Cells were serial-diluted and plated in duplicate on TSA for enumeration of CFU from surviving cells.
To evaluate the impact of saline and acid stresses, overnight cultures of S. xylosus strains were freshly diluted (1/100) in TSB and grown under aerobic conditions at 30°C until OD600 nm was approximately 0.3. Strains were diluted in NaCl-supplemented TSB (10 and 20% NaCl) or diluted in TSB with adjusted pH (5.0 and 6.0) to an OD600 nm of 0.1 and incubated in 100-well microtiter plates with shaking at 30°C in a Bioscreen C plate reader, where turbidity was monitored every 30 min for 20 hours. Assays were performed with two independent cultures for each strain.
Total RNA Extraction and Reverse Transcription
Staphylococcus xylosus strains were grown in TSB under aerobic conditions for 6 h and 24 h. Cell pellets were immediately frozen in liquid nitrogen to stabilize the bacterial RNA and stored at –80°C. For RNA extraction, cell pellets were thawed on ice and resuspended in 500 μL of cold Tris-EDTA buffer. Samples were transferred into tubes containing 600 mg zirconia-silica beads (0.1 mm diameter), 50 μL of sodium dodecyl sulfate (10%), 500 μL of acid phenol and 3.5 μL of β-mercaptoethanol. Cells were disrupted in a FastPrepTM machine (MP Biomedicals). After addition of 200 μL of chloroform and centrifugation, the aqueous phase containing RNA was purified with the Nucleospin RNAII kit (Macherey Nagel) according to the manufacturer’s instructions. A supplementary treatment was performed with Turbo DNAse (Ambion) to remove residual DNA. Absence of contaminating DNA was checked by PCR targeting the sod gene (Table 2). Total RNA isolated was quantified using a Nanodrop 1000 (Thermo Fisher Scientific). RNA samples were stored at –80°C. RNA isolated from three independent biological replicates for each strain or condition (wild-type and its mutants at 6 and 24 h) was reverse-transcribed to cDNA with a SuperScript Reverse Transcriptase kit following the manufacturer’s instructions (Invitrogen).
Evaluation of nos-pdt Co-transcription
To determine if the two adjacent genes, nos and pdt, were co-transcribed, cDNA from the mRNA species transcribed from C2a 24-h cultures was subject to PCR using a pair of primers targeting the nos-pdt junction region (Table 2). The conditions for the amplification were 5 min at 95°C, followed by 25 cycles of 30 s at 95°C, 30 s at 60°C and 120 s at 72°C and finally 5 min at 72°C. As controls, this amplification was also performed on C2a genomic template DNA and no template sample.
Quantitative Real-time PCR
Expression of genes of interest was carried out by quantitative real-time PCR (qRT-PCR) using iQTM SYBR® Green Supermix (Bio-Rad) and the MasterCycler RealPlex (Eppendorf). Thermal cycling consisted of 30 s at 95°C, followed by 40 cycles of 15 s at 95°C and 60 s at 60°C. The primers used are listed in Table 2. The relative fold change of gene expression, using measured sod housekeeping gene expression, was determined by the Livak (2-ΔΔCt) method (Livak and Schmittgen, 2001).
Statistics
The data were analyzed by using GraphPad Prism software (version 5.01). The significance of experimental differences in bacterial growth, formation of myoglobin derivatives, and colony pigment production were analyzed by one-way analysis of variance (ANOVA) with Tukey’s multiple comparison.
Results
NOS Contributes to Growth under Aerobic Conditions
To define the role of NOS activity in S. xylosus C2a, a mutant strain (Δnos) was created by insertion of an erythromycin resistance gene cassette into the nos coding sequence. Deletion of the nos gene was confirmed by PCR analysis and sequencing (data not shown). When grown in TSB under aerobic conditions, the Δnos mutant displayed a slight growth defect compared with the wild type (Figure 1A). Complementation of the Δnos mutant with a plasmid expressing the nos gene (ΔnospRBnos) showed restored growth (Figure 1B). The wild type and the Δnos and nos complemented mutants did not differ in growth in microtiter plates under microaerobic conditions (Supplementary Figure S1).
FIGURE 1.
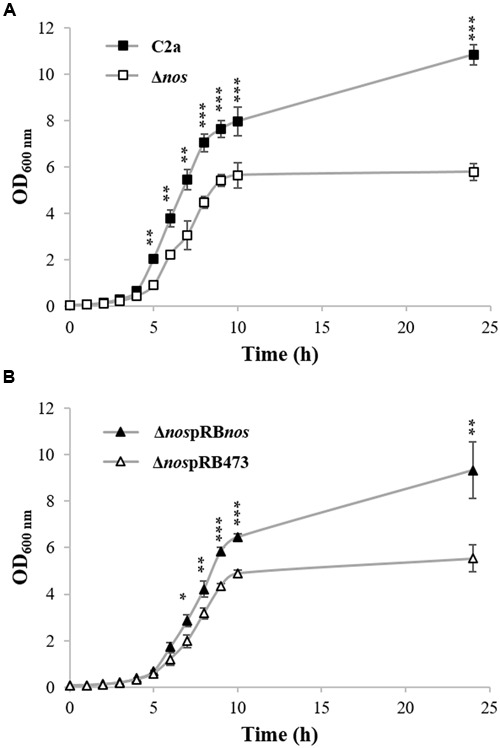
Growth of Staphylococcus xylosus strains in TSB under aerobic conditions. (A) S. xylosus C2a and Δnos. (B) ΔnospRBnos and ΔnospRB473 with chloramphenicol. Data represent means ± SD from n = 3 independent biological replicates. *p < 0.05; **p < 0.01; ***p < 0.001.
PDT But Not NOS Is Required for Growth in Minimal Medium
Growth was measured in minimal medium under microaerobic conditions for the wild-type C2a, the Δnos mutant and the nos complemented Δnos mutant (Figure 2A). The Δnos mutant was not able to grow in this minimal medium and growth was not restored by plasmid complementation with the nos gene (Figure 2A). Since the nos gene is in a cluster with the pdt gene, we tested the hypothesis that both genes were co-transcribed (Supplementary Figure S2). RT-PCR experiments were carried out using a primer annealing specifically with nos and a primer annealing specifically with pdt (Table 2). Amplification of a 1,421-bp fragment indicated a co-transcription of both nos and pdt (data not shown). We therefore speculated that the deletion of nos and the insertion of ermB had a polar effect on the downstream gene pdt, leading to a frameshift in the pdt open reading frame (ORF). The pdt gene encodes a prephenate dehydratase (EC4.2.1.51), which catalyzes the penultimate reaction in the phenylalanine biosynthesis. Supplementation with phenylalanine restored the growth of the Δnos and the nos complemented (ΔnospRBnos) mutants (Figure 2B), while no restoration of growth was observed with other amino acids such as tryptophan or tyrosine, which, like phenylalanine, are derived from chorismate (data not shown). Moreover, complementation of the Δnos mutant with pdt (ΔnospRBpdt) or with the nos-pdt operon (ΔnospRBnospdt) restored growth to the wild-type C2a level (Figure 2C).
FIGURE 2.
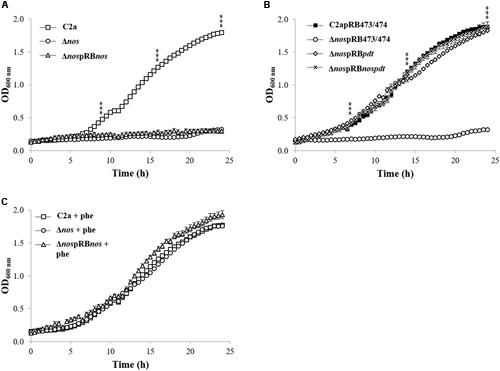
Growth of S. xylosus strains in chemically defined minimal medium in the Bioscreen Assay. (A) Growth of S. xylosus C2a, Δnos, and ΔnospRBnos, (B) Growth of C2a, Δnos and ΔnospRBnos with phenylalanine (phe), (C) Growth of C2apRB473 (identical growth as C2apRB474), ΔnospRB473 (identical growth as ΔnospRB474), ΔnospRBpdt and ΔnospRBnospdt. Data represent means ± SD from n = 3 independent biological replicates. ***p < 0.001.
NOS Contributes to NO Production
The formation of the red myoglobin derivatives occurred for S. xylosus C2a under limited oxygenation (Figure 3A) and aerobic conditions (Figure 3B), where different forms of myoglobin came from the conversion of metmyoglobin (brown) to some red derivatives, such as oxymyoglobin and nitrosomyoglobin. The red pigment content was sharply decreased, but not completely abolished, in the Δnos mutant compared with the wild type, and was restored to the wild-type level in the nos complemented Δnos mutant under both conditions (Figure 3). Under limited oxygenation after 24 h of incubation, the cellular population was close to initial level, 8 log CFU/mL, for C2a (7.8 ± 0.04), Δnos mutant (7.6 ± 0.40) and nos complemented Δnos mutant (7.2 ± 0.01). Under aerobic conditions, the red pigment level was higher (Figure 3B) and the cellular population reached 9 log CFU/mL for C2a (9.1 ± 0.04), Δnos mutant (9.2 ± 0.20) and Δnos complemented mutant (9.2 ± 0.10).
FIGURE 3.
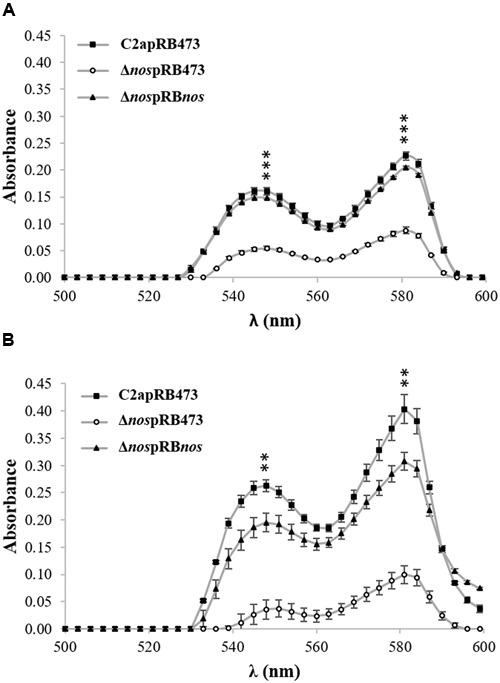
Formation of red myoglobin derivatives. S. xylosus C2apRB473, ΔnospRB473 and ΔnospRBnos were incubated in TSB supplemented with metmyoglobin (2 mg/mL) under (A) limited oxygenation (no stirring and reaction mixture covered with mineral oil) and (B) aerobic conditions (150 rpm). Formation of red pigment was evaluated by measuring absorbance. Data represent means ± SD from n = 3 independent biological replicates. **p < 0.01; ***p < 0.001 at λ548 nm and λ581 nm.
To estimate the production of nitrosomyoglobin, the nitrosoheme of culture supernatants from S. xylosus C2a and its mutants was extracted and measured (Figure 4). Under limited oxygenation (Figure 4A), where the pH of cultures was about 6.0, the nitrosoheme formation of the Δnos mutant was reduced (A540nm = 0.060 ± 1.1E-03) compared with the wild type (A540 nm = 0.090 ± 7.1E-04) and the nos complemented Δnos mutant (A540 nm = 0.090 ± 1.1E-03). Under aerobic conditions (Figure 4B), where the pH of the cultures was about 8.0, maximum absorption shifted, as described (Yu et al., 2016), to λ580 nm instead of λ540 nm. In these conditions, no nitrosoheme was formed for the Δnos mutant (A580 nm < 0.005) compared with the wild type (A580 nm = 0.060 ± 2.8E-03) and the nos complemented Δnos mutant (A580nm = 0.080 ± 1.4E-03).
FIGURE 4.
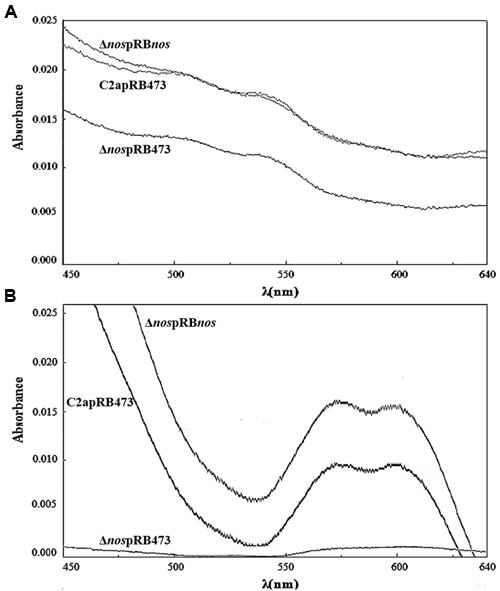
Evidence of nitrosoheme. Nitrosoheme was extracted from nitrosomyoglobin with 80% acetone from supernatants of strains grown in TSB supplemented with metmyoglobin and inoculated under (A) limited oxygenation (no stirring and reaction mixture covered with mineral oil) and (B) aerobic conditions (150 rpm). Curves show spectra representative of three biological replicates.
The Δnos mutant and the pdt complemented Δnos mutant had the same activity, while for the nospdt complemented Δnos mutant the formation of red myoglobin derivatives was restored to the wild-type level (data not shown). These results clearly demonstrated the contribution of NOS and not PDT to the production of NO in S. xylosus C2a.
NOS Affects Colony Pigmentation
When grown on TSA plates, the Δnos mutant displayed enhanced pigmentation relative to the wild type. This pigmentation returned to wild-type level upon complementation with the nos and nos-pdt genes, but not with the pdt gene alone (Figure 5A). Pigments of the wild type and the Δnos and ΔnospRBnos mutants were extracted and absorbance at 460 nm was measured. Production of carotenoid pigment in the Δnos mutant was significantly higher than in the wild type, and returned to the wild-type level in the nos complemented mutant (Figure 5B). These results clearly demonstrated that solely NOS was involved in the pigmentation phenotype. This difference in carotenoid pigment production between the wild type and the Δnos mutant was, however, not observed with the bacterial pellets after liquid cultures in TSB, even after prolonged growth for several days (data not shown).
FIGURE 5.
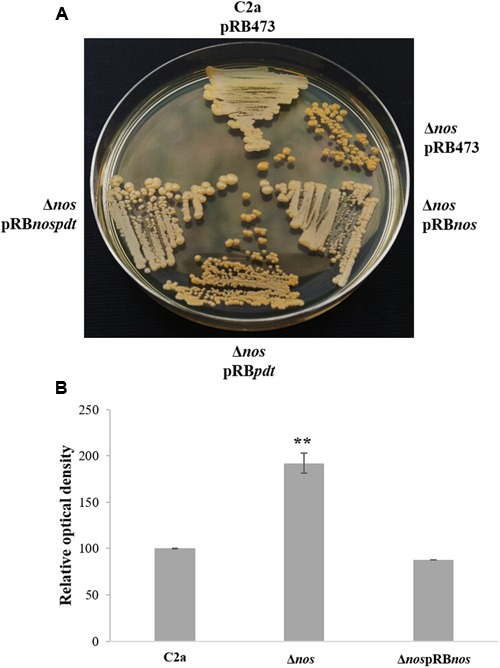
Colony pigment production by S. xylosus strains. (A) S. xylosus C2apRB473, ΔnospRB473, ΔnospRBnos, ΔnospRBpdt and ΔnospRBnospdt streaked on Tryptic Soy Agar (TSA) plates containing chloramphenicol for three days. (B) Measurement of pigmentation after methanol extraction at λ460 nm. The relative optical density is normalized to C2a, which was set at 100. Data represent means ± SD from n = 3 independent biological replicates. **p < 0.01.
NOS Protects Specifically against Peroxide Stress
Stress challenge assays with 150 mM H2O2 were performed on the wild-type C2a and the Δnos and nos complemented Δnos mutants to investigate a possible oxidative stress-sensitive phenotype in the Δnos mutant. After 1-h treatment, the Δnos mutant presented a significant reduction in cell viability (5 log) compared with the wild type (Figure 6). The phenotype was restored to the wild-type level in the nos complemented mutant (Figure 6). To evaluate the specificity of NOS in protecting against oxidative stress, stress challenge assays in saline and acid conditions were performed on the wild type and the Δnos and nos complemented Δnos mutants (Supplementary Figure S3). As the salt concentration increased (Supplementary Figure S3A) or the pH decreased (Supplementary Figure S3B), bacterial growth was affected and decreased for all strains with no significant differences between strains. These results demonstrated that NOS was not required to overcome those stresses.
FIGURE 6.
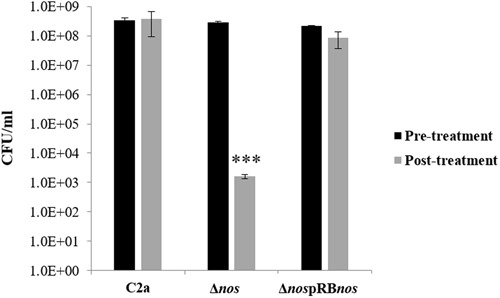
Effect of nos deletion on sensitivity to oxidative stress. S. xylosus C2a, Δnos, and ΔnospRBnos strains were grown under aerobic conditions up to OD = 1 and treated for 1 h with 150 mM hydrogen peroxide (H2O2). Cells, before and after treatment, were enumerated. Data represent means ± SD from n = 2 independent biological replicates. *p < 0.05.
NOS Modulates Catalase Gene Expression
To determine whether increased H2O2-sensitive phenotype and defects in growth under aerobic conditions of the Δnos mutant were due to differential expression of catalase genes, we evaluated expression of the katA, katB, and katC genes by qRT-PCR. The katA gene was overexpressed 6- and 10-fold at 6 h and 24 h, respectively in the Δnos mutant related to the wild type (Figure 7). Expression of the two genes katB and katC was not modified in the Δnos mutant at 6 h and was sharply downregulated at 24 h by comparison with the wild type (Figure 7). In the nos complemented mutant, only the nos gene was highly overexpressed at the two times of incubation, while the expression of the three catalase genes was not modified by comparison with the wild type. However, there was no difference in survival between the wild type and the nos complemented Δnos mutant when treated with 200 mM H2O2 (4 log reduction) and 250 mM (6 log reduction).
FIGURE 7.
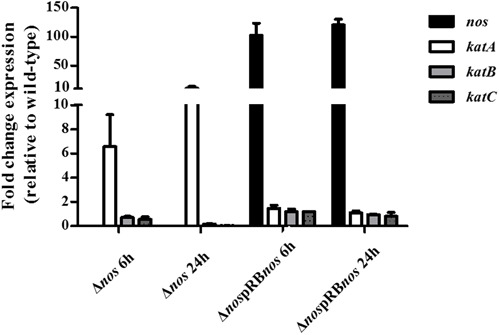
Expression of nos and kat genes. S. xylosus C2a, Δnos, and ΔnospRBnos strains were grown in TSB under aerobic conditions for 6 and 24 h. Expression of nos and kat genes was evaluated by qRT-PCR. The Livak method (2-ΔΔCt) was applied to analyze expression, using sod as reference gene. Results are expressed as means ± SD from n = 3 independent biological replicates.
Discussion
Staphylococcus xylosus is commonly used as a starter culture for fermented meat products, mainly because it reduces nitrate to nitrite, which undergoes chemical reactions leading to NO, which is responsible for color development (Talon and Leroy, 2006). As the safety of nitrite as a food additive is questioned, we studied the ability of S. xylosus to produce NO via a pathway catalyzed by NOS. To demonstrate the production of NO by S. xylosus, we used the property of NO to form a NO complex of Fe(II) myoglobin, called nitrosomyoglobin. In previous studies S. xylosus FAX-1 and S. xylosus A1 were able to convert metmyoglobin to nitrosomyoglobin in vitro and in raw meat batter (Morita et al., 1998; Li et al., 2013, 2016). However, the mechanism of such NO production was not established. Here we demonstrate that NOS activity of S. xylosus led to production of NO. Under limited oxygenation, the Δnos mutant remained able to form nitrosoheme (about 35-40% related to the wild-type level). The Δnos mutant was unable to produce NO under aerobic conditions. The loss of NOS activity in S. aureus UAMS-1 mutant led to NO production of 65% as evaluated using fluorescent stain DAF-FM diacetate after growth on agar plates (Sapp et al., 2014), while the deletion of nos in S. aureus UAMS1182 completely abolished NO production after growth under aerobic conditions (van Sorge et al., 2013). These results revealed that some Staphylococcus strains are able to produce NO by other pathways under limited oxygenation. NO production can be achieved via molybdoenzymes, which are recognized for their ability to catalyze reduction of nitrite to NO radical (Maia and Moura, 2015). The molybdenum-containing respiratory nitrate reductase is believed to be an important source of NO (Maia and Moura, 2015). S. xylosus C2a has a nitrate reductase that was previously shown to be expressed and active under static conditions (Talon et al., 1999). The involvement of NOS under aerobic conditions seems relevant to the oxygen requirement of NOS activity (Crane et al., 2010). The potential for NOS activity has been investigated in 86 strains belonging to 17 CNS species (Sánchez-Mainar et al., 2014). Only one strain of S. haemolyticus showed NOS activity, based on its ability to produce L-citrulline in a meat-simulating medium supplemented with arginine under aerobic conditions (Sánchez-Mainar et al., 2014).
The deletion of nos in S. xylosus C2a slightly affected growth in complex medium under aerobic but not under microaerobic conditions. This nos deletion also increased H2O2 susceptibility. Such results are in accordance with previous studies showing that NO production through NOS activity protects Bacillus and S. aureus against oxidative stress generated by H2O2 exposure (Gusarov and Nudler, 2005; Shatalin et al., 2008; van Sorge et al., 2013). In B. subtilis, exogenous or endogenous NO protected cells from oxidative stress by boosting the activity of KatA, the major catalase (Gusarov and Nudler, 2005). Also, while sodA expression increased sharply in wild-type cells of B. subtilis during exponential growth, it was abolished in the Δnos mutant (Gusarov et al., 2009). In S. xylosus C2a Δnos mutant, the loss of NOS activity resulted in modulation of the expression of genes encoding catalases with upregulation of katA and downregulation of katB and katC. We showed in a previous study that S. xylosus responded to nitrosative stress generated by nitrite in a meat model, notably by the upregulation of katB and katC under the probable control of the repressor PerR and the downregulation of katA, which was not under PerR control (Vermassen et al., 2014). In this study, the limited amount of endogenous NO produced by the Δnos mutant may not derepress PerR, contributing to the downregulation of katB and katC. In contrast to B. subtilis, expression of the gene sodA was not modified in the S. xylosusΔnos mutant (sod was used as housekeeping gene in our study).
The S. xylosus C2a Δnos mutant displayed higher colony pigmentation than the wild-type strain after prolonged cultivation on agar medium, as observed for the S. aureusΔnos strain UAMS-1 (Sapp et al., 2014). S. aureus produced the intermediary yellow carotenoid 4,4′-diaponeurosporene, which, after prolonged cultivation, is converted to the yellow-orange end-product staphyloxanthin (Wieland et al., 1994). The yellow pigment in S. xylosus is likely to be a carotenoid pigment with peak absorbance at 460 nm. The carotenoid pigment of S. aureus protects against reactive oxygen species, so a non-pigmented mutant is more susceptible to oxidative killing (Clauditz et al., 2006). The increased colony pigmentation of the S. xylosus C2a Δnos mutant could be a mechanism to cope with oxidative stress in conditions of extended incubation only on agar medium, as no pigmentation was observed in liquid medium.
The S. aureus nos gene is co-transcribed with the pdt gene encoding a prephenate dehydratase, which catalyzes the penultimate reaction in phenylalanine biosynthesis (Sapp et al., 2014). This genetic arrangement is present in all publically available staphylococcal genome sequences and seems to be unique to the Staphylococcus genus (Sapp et al., 2014). As anticipated, co-transcription of the nos-pdt cluster was also demonstrated in S. xylosus. However, the role of the NOS and PDT enzymes of S. xylosus C2a did not appear to be interlinked. PDT was essential for S. xylosus growth only in the absence of phenylalanine, while NOS was not required to sustain phenylalanine biosynthesis.
Conclusion
Our results demonstrate NOS-dependent NO production in a coagulase-negative Staphylococcus. This endogenous NO production contributed to growth under aerobic conditions and to the cytoprotective effect against oxidative stress. S. xylosus is a species usually used as a starter culture in meat products. Therefore, NOS-dependent NO production in S. xylosus needs to be further characterized in meat products and optimized as a potential alternative to nitrate and nitrite in these products.
Author Contributions
SL and RT conceived the study. GR, SL and RT designed the experiments and wrote the manuscript. GR performed the laboratory experiments. GR, VZ, PD, TS, SL and RT analyzed data. All the authors contributed to preparing the final version of the manuscript, read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
GR is a Ph.D. Research Fellow funded by ANRT, CIFRE 2013/1490. We would like to thank Philippe Gatellier, INRA, for helpful discussions about nitrosoheme pigment, and Jean-Paul Chacornac and Carine Andant for technical assistance. The authors are grateful to David Marsh for correcting our English.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00598/full#supplementary-material
Staphylococcus xylosus strains in the Bioscreen Assay. S. xylosus C2a, Δnos, and ΔnospRBnos in TSB. Data represent means ± SD from n = 3 independent biological replicates.
Genetic cluster nos-pdt of S. xylosus C2a. The cluster of the two genes nos-pdt is depicted with primers used for PCR co-transcription.
Impact of stresses on growth under microaerobic conditions of S. xylosus strains. (A) Impact of salt and (B) impact of acid stresses. Data represent means ± SD from n = 3 independent biological replicates.
References
- Alderton W. K., Cooper C. E., Knowles R. G. (2001). Nitric oxide synthases: structure, function and inhibition. Biochem. J. 357(Pt 3) 593–615. 10.1042/bj3570593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrière C., Brückner R., Centeno D., Talon R. (2002). Characterisation of the katA gene encoding a catalase and evidence for at least a second catalase activity in Staphylococcus xylosus, bacteria used in food fermentation. FEMS Microbiol. Lett. 216 277–283. 10.1016/S0378-1097(02)01030-3 [DOI] [PubMed] [Google Scholar]
- Brückner R. (1992). A series of shuttle vectors for Bacillus subtilis and Escherichia coli. Gene 122 187–192. 10.1016/0378-1119(92)90048-T [DOI] [PubMed] [Google Scholar]
- Brückner R. (1997). Gene replacement in Staphylococcus carnosus and Staphylococcus xylosus. FEMS Microbiol. Lett. 151 1–8. 10.1016/S0378-1097(97)00116-X [DOI] [PubMed] [Google Scholar]
- Clauditz A., Resch A., Wieland K. P., Peschel A., Götz F. (2006). Staphyloxanthin plays a role in the fitness of Staphylococcus aureus and its ability to cope with oxidative stress. Infect. Immun. 74 4950–4953. 10.1128/IAI.00204-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Code of Federal Regulations (2016). Title 21 – Food and Drugs, Chapter I – Food and Drug Admistration, Department of Health and Human Services, Subchapter B – Food for Human Consumption, Part 172 – Food Additives Permitted for Direct Addition to Food for Human Consumption, Subpart B – Food Preservatives. Silver Spring, MD: U.S. Food and Drug Administration. [Google Scholar]
- Commission Regulation [EU] (2011). No 1129/2011 of November 2011 amending Annex II to Regulation (EC) No 1333/2008 of the European Parliament and of the Council by establishing a Union list of food additives. Off. J. Eur. Union L295 1–177. [Google Scholar]
- Crane B. R., Sudhamsu J., Patel B. A. (2010). Bacterial nitric oxide synthases. Annu. Rev. Biochem. 79 445–470. 10.1146/annurev-biochem-062608-103436 [DOI] [PubMed] [Google Scholar]
- Fiegler H., Brückner R. (1997). Identification of the serine acetyltransferase gene of Staphylococcus xylosus. FEMS Microbiol. Lett. 148 181–187. 10.1111/j.1574-6968.1997.tb10286.x [DOI] [PubMed] [Google Scholar]
- Gøtterup J., Olsen K., Knöchel S., Tjener K., Stahnke L. H., Møller J. K. (2007). Relationship between nitrate/nitrite reductase activities in meat associated staphylococci and nitrosomyoglobin formation in a cured meat model system. Int. J. Food Microbiol. 120 303–310. 10.1016/j.ijfoodmicro.2007.08.034 [DOI] [PubMed] [Google Scholar]
- Gøtterup J., Olsen K., Knöchel S., Tjener K., Stahnke L. H., Møller J. K. (2008). Colour formation in fermented sausages by meat-associated staphylococci with different nitrite- and nitrate-reductase activities. Meat Sci. 78 492–501. 10.1016/j.meatsci.2007.07.023 [DOI] [PubMed] [Google Scholar]
- Götz F., Zabielski J., Philipson L., Lindberg M. (1983). DNA homology between the arsenate resistance plasmid pSX267 from Staphylococcus xylosus and the penicillinase plasmid pI258 from Staphylococcus aureus. Plasmid 9 126–137. 10.1016/0147-619X(83)90015-X [DOI] [PubMed] [Google Scholar]
- Gündoğdu A. K., Karahan A. G., Çakmakç M. L. (2006). Production of nitric oxide (NO) by lactic acid bacteria isolated from fermented products. Eur. Food Res. Technol. 223 35–38. 10.1007/s00217-005-0097-8 [DOI] [Google Scholar]
- Gusarov I., Nudler E. (2005). NO-mediated cytoprotection: instant adaptation to oxidative stress in bacteria. Proc. Natl. Acad. Sci. U.S.A. 102 13855–13860. 10.1073/pnas.0504307102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusarov I., Shatalin K., Starodubtseva M., Nudler E. (2009). Endogenous nitric oxide protects bacteria against a wide spectrum of antibiotics. Science 325 1380–1384. 10.1126/science.1175439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusarov I., Starodubtseva M., Wang Z. Q., McQuade L., Lippard S. J., Stuehr D. J., et al. (2008). Bacterial nitric-oxide synthases operate without a dedicated redox partner. J. Biol. Chem. 283 13140–13147. 10.1074/jbc.M710178200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honikel K. O. (2008). The use and control of nitrate and nitrite for the processing of meat products. Meat Sci. 78 68–76. 10.1016/j.meatsci.2007.05.030 [DOI] [PubMed] [Google Scholar]
- Kabir M., Sudhamsu J., Crane B. R., Yeh S. R., Rousseau D. L. (2008). Substrate-ligand interactions in Geobacillus stearothermophilus nitric oxide synthase. Biochemistry 47 12389–12397. 10.1021/bi801491e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kers J. A., Wach M. J., Krasnoff S. B., Widom J., Cameron K. D., Bukhalid R. A., et al. (2004). Nitration of a peptide phytotoxin by bacterial nitric oxide synthase. Nature 429 79–82. 10.1038/nature02504nature02504 [DOI] [PubMed] [Google Scholar]
- Li P., Kong B., Chen Q., Zheng D., Liu N. (2013). Formation and identification of nitrosomyoglobin by Staphylococcus xylosus in raw meat batters: a potential solution for nitrite substitution in meat products. Meat Sci. 93 67–72. 10.1016/j.meatsci.2012.08.003 [DOI] [PubMed] [Google Scholar]
- Li P. J., Luo H. T., Kong B. H., Liu Q., Chen C. G. (2016). Formation of red myoglobin derivatives and inhibition of spoilage bacteria in raw meat batters by lactic acid bacteria and Staphylococcus xylosus. LWT Food Sci. Technol. 68 251–257. 10.1016/j.lwt.2015.12.035 [DOI] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Maia L. B., Moura J. J. (2015). Nitrite reduction by molybdoenzymes: a new class of nitric oxide-forming nitrite reductases. J. Biol. Inorg. Chem. 20 403–433. 10.1007/s00775-014-1234-2 [DOI] [PubMed] [Google Scholar]
- Montgomery H. J., Dupont A. L., Leivo H. E., Guillemette J. G. (2010). Cloning, expression, and purification of a nitric oxide synthase-like protein from Bacillus cereus. Biochem. Res. Int. 2010:489892 10.1155/2010/489892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikawa K., Maruyama A., Inose Y., Higashide M., Hayashi H., Ohta T. (2001). Overexpression of sigma factor, sigma(B), urges Staphylococcus aureus to thicken the cell wall and to resist beta-lactams. Biochem. Biophys. Res. Commun. 288 385–389. 10.1006/bbrc.2001.5774 [DOI] [PubMed] [Google Scholar]
- Morita H., Sakata R., Nagata Y. (1998). Nitric oxide complex of iron(II) myoglobin converted from metmyoglobin by Staphylococcus xylosus. J. Food Sci. 63 352–355. 10.1111/j.1365-2621.1998.tb15740.x [DOI] [Google Scholar]
- Patel B. A., Moreau M., Widom J., Chen H., Yin L., Hua Y., et al. (2009). Endogenous nitric oxide regulates the recovery of the radiation-resistant bacterium Deinococcus radiodurans from exposure to UV light. Proc. Natl. Acad. Sci. U.S.A 106 18183–18188. 10.1073/pnas.0907262106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Mainar M., Weckx S., Leroy F. (2014). Coagulase-negative Staphylococci favor conversion of arginine into ornithine despite a widespread genetic potential for nitric oxide synthase activity. Appl. Environ. Microbiol. 80 7741–7751. 10.1128/AEM.02298-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapp A. M., Mogen A. B., Almand E. A., Rivera F. E., Shaw L. N., Richardson A. R., et al. (2014). Contribution of the nos-pdt operon to virulence phenotypes in methicillin-sensitive Staphylococcus aureus. PLoS ONE 9:e108868 10.1371/journal.pone.0108868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shatalin K., Gusarov I., Avetissova E., Shatalina Y., McQuade L. E., Lippard S. J., et al. (2008). Bacillus anthracis-derived nitric oxide is essential for pathogen virulence and survival in macrophages. Proc. Natl. Acad. Sci. U.S.A. 105 1009–1013. 10.1073/pnas.0710950105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talon R., Leroy S. (2006). “Chapter 16–Latest developments in meat bacterial starters,” in Advanced Technologies for Meat Processing eds Nollet L. M. L., Toldra F. (New York, NY: CRC Press, Taylor and Francis group; ) 401–418. [Google Scholar]
- Talon R., Walter D., Chartier S., Barrière C., Montel M. C. (1999). Effect of nitrate and incubation conditions on the production of catalase and nitrate reductase by staphylococci. Int. J. Food Microbiol. 52 47–56. 10.1016/S0168-1605(99)00127-0 [DOI] [PubMed] [Google Scholar]
- Vaish M., Singh V. K. (2013). Antioxidant functions of nitric oxide synthase in a methicillin sensitive Staphylococcus aureus. Int. J. Microbiol. 2013:312146 10.1155/2013/312146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Sorge N. M., Beasley F. C., Gusarov I., Gonzalez D. J., von Köckritz-Blickwede M., Anik S., et al. (2013). Methicillin-resistant Staphylococcus aureus bacterial nitric-oxide synthase affects antibiotic sensitivity and skin abscess development. J. Biol. Chem. 288 6417–6426. 10.1074/jbc.M112.448738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermassen A., de la Foye A., Loux V., Talon R., Leroy S. (2014). Transcriptomic analysis of Staphylococcus xylosus in the presence of nitrate and nitrite in meat reveals its response to nitrosative stress. Front. Microbiol. 5:691 10.3389/fmicb.2014.00691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieland B., Feil C., Gloria-Maercker E., Thumm G., Lechner M., Bravo J. M., et al. (1994). Genetic and biochemical analyses of the biosynthesis of the yellow carotenoid 4,4’-diaponeurosporene of Staphylococcus aureus. J. Bacteriol. 176 7719–7726. 10.1128/jb.176.24.7719-7726.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C., Jiao J., Ma L., Sun W. (2016). Effect of pH on the stability and molecular structure of nitrosyl hemochromogen. Food Chem. 196 503–508. 10.1016/j.foodchem.2015.09.067 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Staphylococcus xylosus strains in the Bioscreen Assay. S. xylosus C2a, Δnos, and ΔnospRBnos in TSB. Data represent means ± SD from n = 3 independent biological replicates.
Genetic cluster nos-pdt of S. xylosus C2a. The cluster of the two genes nos-pdt is depicted with primers used for PCR co-transcription.
Impact of stresses on growth under microaerobic conditions of S. xylosus strains. (A) Impact of salt and (B) impact of acid stresses. Data represent means ± SD from n = 3 independent biological replicates.


