Summary
Retinoic acid‐inducible gene I (RIG‐I) ‐like receptors (RLRs) are found conservatively present in teleost fish. All three members, RIG‐I, MDA5 and LGP2, together with the downstream molecules such as MITA, TRAF3 and TBK1, have been identified in a range of fish species. However, it is unexpected that RIG‐I has not been reported in fish of Acanthopterygii, and it would be important to clarify the presence and role of the RIG‐I gene in a broad range of taxa in Teleostei. RLRs in fish can be induced in vivo and in vitro by viral pathogens as well as synthetic dsRNA, poly(I:C), leading to the production of type I interferons (IFNs) and the expression of IFN‐stimulated genes (ISGs). Bacterial pathogens, such as Edwardsiella tarda, and their components, such as lipopolysaccharide are also found to induce the expression of RLRs, and whether such induction was mediated through the direct recognition by RLRs or through crosstalk with other pattern recognition receptors recognizing directly bacterial pathogen‐associated molecular patterns awaits to be investigated. On the other hand, RLR‐activated type I IFN production can be negatively regulated in fish by molecules, such as TBK‐1‐like protein and IRF10, which are found to negatively regulate RIG‐I and MAVS‐activated type I IFN production, and to block MITA or bind ISRE motifs, respectively. It is considered that the evolutionary occurrence of RLRs in fish, and their recognized ligands, especially those from their fish pathogens, as well as the mechanisms involved in the RLR signalling pathways, are of significant interest for further investigation.
Keywords: fish, LGP2, MDA5, retinoic acid‐inducible gene‐I, RIG‐I‐like receptor
Introduction
The innate immune system provides critical host defence against microorganism invasion through the recognition of conserved pathogen‐associated molecular patterns (PAMPs) by pattern recognition receptors (PRRs).1, 2 PAMPs can be nucleic acids from bacteria or viruses, and can also be other small molecular motifs or even molecules conserved in microbes, such as bacterial lipopolysaccharide (LPS), peptidoglycan, flagellin and lipoteichoic acid.3, 4, 5 To face the challenge of various and complicated PAMPs from microbes, hosts have evolved with different families of PRRs, currently including Toll‐like receptors (TLRs), C‐type lectin receptors, NOD‐like receptors (NLRs), retinoic acid‐inducible gene I (RIG‐I) ‐like receptors (RLRs) and cytosolic DNA sensors.6
Among all known PRR families, RLRs, which belong to DExD/H box RNA helicases, are the core cytosolic receptors in recognition of viral RNAs. In mammals, three members exist in the RLR family: retinoic acid‐inducible gene I (RIG‐I, or DEAD box polypeptide 58, DDX58), melanoma differentiation‐associated gene 5 (MDA5, or interferon induced with helicase C domain 1, IFIH1) and laboratory of genetics and physiology 2 (LGP2, or DExH box polypeptide 58, DHX58).7 Over the last few decades, significant progress has been achieved in the field of fish immunology, and orthologous genes of mammalian RIG‐I, MDA5 and LGP2 have been identified in teleost fish, and their function has been investigated in a range of fish species, including the model fish species, zebrafish (Danio rerio),8, 9 and some economically important fish species such as rainbow trout (Oncorhynchus mykiss) and Japanese flounder (Paralichthys olivaceus).10, 11, 12 It is considered that the knowledge of RLRs in teleost fish will improve our understanding of the immune system of fish, and also of the diversity and evolution of antiviral immunity in vertebrates. Hence, the recent discoveries in relation to RLRs in teleost fish are summarized in this review.
Findings of RLRs and their spliced variants in teleost fish
Obtained from available whole genome sequences, orthologues of MDA5 were first reported in 2008 from pufferfish using bioinformatic analysis.13 Later, RIG‐I, LGP2 and MDA5 were found in a few species of teleost fish using, again, a bioinformatics approach.14 Simultaneously, transcripts of RLRs in fish have been identified using different methods, such as the sequencing of reciprocal suppression subtractive hybridization cDNA libraries and rapid‐amplification of cDNA ends. RIG‐I, MDA5 and LGP2 genes were first cloned, respectively, in Atlantic salmon (Salmo salar) and a fish cell line, i.e. Epithelioma papulosum cyprini (EPC) cells,15 grass carp (Ctenopharyngodon idella) and Atlantic cod (Gadus morhua),16, 17 although the cod LGP2 gene was partially sequenced.
Currently, RIG‐I gene is found only in Cypriniformes, Siluriformes and Salmoniformes,8, 18, 19, 20, 21, 22 whereas both MDA5 and LGP2 genes are found in fish species belonging to Cypriniformes, Siluriformes, Salmoniformes and other fish in Acanthopterygii (Table 1).9, 10, 11, 12, 19, 21, 23, 24, 25, 26, 27, 28, 29, 30 It is uncertain whether RIG‐I genes have been lost in fish of Acanthopterygii, which requires certainly further research on this group of fish. Indeed, efforts to identify orthologues of RIG‐I have been unsuccessful in Japanese pufferfish (Takifugu rubripes), tetraodon (Tetraodon nigroviridis),13, 14 medaka (Oryzias latipes), three‐spined stickleback (Gasterosteus aculeatus),14 gilt‐head sea bream (Sparus aurata) and European sea bass (Dicentrarchus labrax).31 RIG‐I has not been found in mandarin fish, or so‐called Chinese perch (Siniperca chuatsi) (own unpublished data). Further research is required to clarify the occurrence of RIG‐I genes in a broad range of taxa in Teleostei.
Table 1.
Currently reported retinoic acid‐inducible gene I (RIG‐I) ‐like receptors (RLRs) in teleost fish
| Gene | Species/Cell line | GenBank accession number | Reference |
|---|---|---|---|
| RIG‐I | Crucian carp (Carassius auratus) | JF970225 | 19 |
| Grass carp (Ctenopharyngodon idella) | GQ478334 | 18 | |
| Common carp (Cyprinus carpio) | HQ850439 | 20 | |
| Zebrafish (Danio rerio) | JX462558 (RIG‐Ia) | 8 | |
| JX462559 (RIG‐Ib) | 8 | ||
| KM281808 | 22 | ||
| Channel catfish (Ictalurus punctatus) | JQ008940 | 21 | |
| Atlantic salmon (Salmo salar) | FN178459 | 15 | |
| EPC (Epithelioma papulosum cyprini) | FN394062 | 15 | |
| MDA5 | Crucian carp (Carassius auratus) | JF970226 | 19 |
| Grass carp (Ctenopharyngodon idella) | FJ542045 | 16 | |
| Zebrafish (Danio rerio) | JX462556 (MDA5a) | 9 | |
| JX462557 (MDA5b) | 9 | ||
| Orange spotted grouper (Epinephelus coioides) | AEX01716 | 24 | |
| Green chromide (Etroplus suratensis) | KM014661 | 25 | |
| Channel catfish (Ictalurus punctatus) | JQ008941 | 21 | |
| Large yellow croaker (Larimichthys crocea) | KU886064 | 23 | |
| Sea perch (Lateolabrax japonicas) | KU317137 | 26 | |
| Rainbow trout (Oncorhynchus mykiss) | FN396357 | 10 | |
| Japanese flounder (Paralichthys olivaceus) | HQ401014 | 11 | |
| LGP2 | Crucian carp (Carassius auratus) | JF970227 | 19 |
| Grass carp (Ctenopharyngodon idella) | ACI33640 | 27 | |
| Zebrafish (Danio rerio) | JF970230 | 19 | |
| Atlantic cod (Gadus morhua) | EU371924 | 17 | |
| Channel catfish (Ictalurus punctatus) | JQ008942 | 21 | |
| Large yellow croaker (Larimichthys crocea) | KU886062 | 23 | |
| Sea perch (Lateolabrax japonicas) | KRO62119 | 29 | |
| Black carp (Mylopharyngodon piceus) | KX344501 | 61 | |
| Rainbow trout (Oncorhynchus mykiss) | FN396358 | 10 | |
| Japanese flounder (Paralichthys olivaceus) | HM100666 | 12 | |
| Atlantic salmon (Salmo salar) | BT045378 | 28 |
Comparatively, RLRs in fish contain protein domains similar to those of mammalian RLRs. All RLR molecules comprise a DExD/H box helicase domain (DEXDc), a helicase C‐terminal domain (HELICc), a regulatory domain (RD), two caspase activation and recruitment domains (CARDs) at the N‐terminal region of RIG‐I and MDA5, but not LGP2, with CARD functioning critically in signalling transduction.8, 9, 10, 12, 26, 27
In fish, when being transcribed, RLRs may be spliced at RNA level, leading to sequence deletion or insertion in some functional domains (Table 2). Two spliced transcripts of MDA5, MDA5a and MDA5b, have been identified in zebrafish, which are derived from the same gene after searching the zebrafish genome database.9 MDA5a transcript contains all 16 exons, whereas a missing partial sequence in the 11th exon leads to the frameshift in MDA5b with the deletion of HELICc and RD domains from the predictive protein.9 RIG‐I gene in zebrafish also has two different transcripts, RIG‐Ia and RIG‐Ib, with insertion of a 114‐nucleotide sequence in the second CARD domain of RIG‐Ia, which shows no homology with those in other fish species or in mammals.8 In fact, RIG‐I genomic DNA sequence has not yet been completely assembled in the current zebrafish genome (GRCz10); therefore, it is still unclear whether the two isoforms are encoded by a single gene or by two duplicated genes. In addition, the LGP2 gene has a splicing variant in rainbow trout, with LGP2b being 54 amino acids shorter than LGP2a, resulting from premature termination due to an unspliced intron at the 3′ end region of the LGP2b open reading frame.10
Table 2.
Splicing variants of retinoic acid‐inducible gene I (RIG‐I) ‐like receptors (RLRs) in teleost
| Splicing variant | Type | Protein length (amino acids) | Fish species | Antiviral activity in vitro | Other functional description | Reference |
|---|---|---|---|---|---|---|
| RIG‐Ia | Insertion variant (114 nt sequence was inserted into the second CARD coding region) | 973 | Zebrafish (D. rerio) | No | Enhancing RIG‐Ib/MAVS‐mediated signalling pathway | 8 |
| RIG‐Ib | Wild‐type | 937 | Zebrafish | Yes | Inducing IFN and ISGs | 8 |
| MDA5a | Wild‐type | 997 | Zebrafish | Yes | Inducing type I IFN | 9 |
| MDA5b | Premature stop (deletion of partial exons 11 and 13 and entire exon 12) | 685 | Zebrafish | Yes | Inducing type I IFN, and enhancing MDA5a/MAVS‐mediated signalling pathway | 9 |
| LGP2a | Wild‐type | 678 | Rainbow trout (O. mykiss) | Yes | Inducing ISG | 10 |
| LGP2b | Premature stop (intron retention at the 3′‐end region of the open reading frame) | 624 | Rainbow trout | No | Regulating negatively LGP2a‐activated antiviral response | 10 |
In vivo and in vitro expression of RLRs in teleost fish
In mammals, the expression of RLRs can be induced by artificial synthetic RNAs, interferons (IFNs) and viruses.32 To date, the expression of RLRs has been examined in a variety of fish species, and it is well understood that RIG‐I, MDA5 and LGP2 can respond in vivo or in vitro to the stimulation of synthetic double‐stranded RNA (dsRNA) poly(I:C) and to viral infection. In zebrafish, the expression of RIG‐I was significantly up‐regulated in embryos at 24–36 hr post‐fertilization following the treatment of low‐molecular‐weight poly(I:C),22 which is the ligand for mammalian RIG‐I and is different from the ligand for MDA5 in length.33 Induction of RLR family members was observed in ZF4 cells infected with three kinds of single‐stranded RNA (ssRNA) viruses, i.e. snakehead fish vesiculovirus, nervous necrosis virus and spring viraemia of carp virus (SVCV), except that LGP2 has not been studied in SVCV‐infected cells.8, 9, 34, 35 In grass carp, RIG‐I, MDA5 and LGP2 were induced in vivo at mRNA level in several tissues or organs when healthy fish were challenged with a dsRNA viral pathogen, the grass carp reovirus.16, 18, 27 In rainbow trout, MDA5 and LGP2 were up‐regulated following the stimulation of poly(I:C) and two viruses, viral haemorrhagic septicaemia virus (VHSV) and salmon alphavirus, a negative and a positive ssRNA virus, respectively. On the other hand, RNA‐sequencing results revealed that RIG‐I, MDA5 and LGP2 in Atlantic salmon were all up‐regulated in a salmonid cell line (TO) cells infected with salmon alphavirus subtype 3.10, 36 RLRs have been reported in other species of fish in response to the infection of viruses. Infection of channel catfish virus enhanced the expression level of RIG‐I, MDA5 and LGP2 in channel catfish (Ictalurus punctatus).21 RLRs in perciform fish, such as MDA5 in orange spotted grouper (Epinephelus coioides) and green chromide (Etroplus suratensis), MDA5 and LGP2 in large yellow croaker (Larimichthys crocea) and sea perch (Lateolabrax japonicas), had increased transcripts following the treatment of poly(I:C) and infection of DNA or RNA viruses, such as VHSV, Singapore grouper iridovirus and nervous necrosis virus.23, 24, 25, 26, 37 In addition, the up‐regulation of MDA5 and LGP2 mRNA levels was detected in the kidney of VHSV‐infected Japanese flounder and in vitro in poly(I:C) stimulated whole kidney leucocytes.11, 12
It has been experimentally revealed that pathogenic bacteria or their components can significantly induce the expression of RLRs in teleost fish. The infection of an intracellular Gram‐negative bacterial pathogen, Edwardsiella tarda, caused the significant increase in RIG‐I and MDA5 mRNA contents in zebrafish ZF4 cells.8, 9 Edwardsiella ictaluri also induced the expression of RIG‐I, MDA5 and LGP2 in the liver of channel catfish.21 Similarly, induction of MDA5 and LGP2 was observed in vitro in LPS‐stimulated peripheral blood leucocytes and leucocytes from kidney of Japanese flounder,11, 12 In grass carp, RIG‐I, MDA5 and LGP2 were all up‐regulated in primary trunk kidney cells following LPS exposure.38 However, in sea perch, a decrease in the expression of MDA5 was observed in LPS‐stimulated brain and fry cells, and it was assumed that signal pathways mediated by TLRs or NLRs were likely to be inhibited following the expression of MDA5.26 To our knowledge, the mechanisms involved in the LPS recognition and signalling transduction remain unknown in fish, although TLR4 orthologues are reported in some teleost fish (not all teleosts) such as in zebrafish. In fact, TLR4 orthologues in fish are not involved in the recognition of LPS.39, 40 It is reported that fish can even tolerate higher levels of LPS than mammals,41 and TLR adaptor molecule 2 (TICAM2, also known as TRAM and TIRP), a critical adaptor for signal transduction activated by LPS in mammals,42 is absent in fish.43 Therefore, the mechanisms involved in the recognition of LPS and in expression of RLRs in response to LPS can be a valuable topic for further research. Moreover, it is implied that RLRs play possibly a role in antibacterial immunity in fish, along with their antiviral effect in fish.
Ligand recognition by RLRs in teleost fish
All three members in the RLR family have been confirmed to possess RNA‐binding activity in mammals.44 Chang et al.10 proved that recombinant proteins of MDA5 and LGP2 from rainbow trout can bind synthetic dsRNA poly(I:C) by pulldown assay in vitro. But, there has been no experimental evidence for the RNA‐binding activity of teleost RIG‐I. In mammals, differences exist in the ligands recognized by MDA5 and RIG‐I. MDA5 primarily recognizes poly(I:C) with high molecular weight,33 whereas the known ligands for RIG‐I include low‐molecular‐weight (around 300 bp) poly(I:C), short 5′ triphosphated dsRNA fragments, 5′ diphosphated dsRNA, 5′ triphosphated ssRNA with polyuridine signature.33, 45, 46, 47
There is an obvious gap in the knowledge of RLR‐recognized ligands in fish (Fig. 1), and it is necessary to analyse whether RLRs in different species of fish can recognize different RNA ligands from their unique or common viral pathogens. Furthermore, it is of great importance to determine whether RLRs in fish can directly recognize different ligands from bacteria or only have crosstalk with other PRRs involved in the recognition of bacterial PAMPs, as RLRs in fish can also be induced under bacterial infection.
Figure 1.
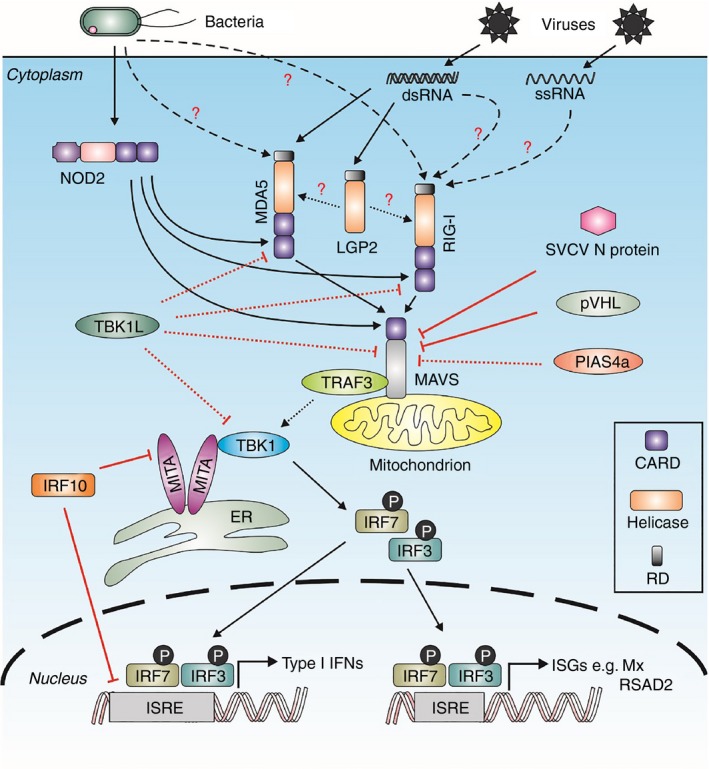
Fish retinoic acid‐inducible gene I (RIG‐I) ‐like receptor (RLR) ‐mediated signalling pathways in response to viral and bacterial infections. Upon the recognition of pathogen‐associated molecular patterns (PAMPs) from viruses or bacteria, MDA5/RIG‐I recruits mitochondrial antiviral signalling protein (MAVS), an adaptor protein located on mitochondria, and then become associated with TRAF3, MITA and TBK1, leading to the phosphorylation and activation of interferon (IFN) regulatory factor 3 (IRF3)/IRF7 for translocating into nucleus and then for binding IFN‐stimulated response element (ISRE) motif, and to the production of type I IFNs and IFN‐stimulated genes (ISGs). In addition, NOD2 can interact with MDA5, RIG‐I as well as MAVS in the signalling pathway, and IRF10 can inhibit type I IFN production through binding ISRE motif and interacting with MITA. TBK1‐like (TBK1L) and PIAS4a negatively regulate the RLRs–MAVS–TBK1‐mediated signalling pathway. PVHL and the N protein of spring viraemia of carp virus could induce the degradation of MAVS and block MAVS‐mediated type I IFN production. In the signalling schematics, the negative regulation cascades are marked with red lines, and broken lines indicate that the exact PAMPs recognized by fish RLRs have not been determined or the direct interaction or mechanism of the indicated molecules need to be confirmed.
Antiviral and antibacterial functions of RLRs in teleost fish
In fish, like their mammalian counterparts, RLRs possess capacities in the induction of IFNs, IFN‐stimulated genes (ISGs), inflammatory factors and antiviral state, although negative regulation and negative or positive regulation in antiviral immunity have been observed in in vitro overexpression of LGP2 in mammals and teleosts, respectively.
The overexpression of RIG‐I CARD domains in zebrafish embryos resulted in the activation of Mx and nuclear factor‐κB (NF‐κB) luciferase report plasmids and the significant up‐regulation of immune genes, such as IFN1, tumour necrosis factor‐α, interleukin‐8 (also known as CXCL8), ISG15 and radical S‐adenosyl methionine domain containing 2 (RSAD2, also known as viperin);22 but it is a surprise that such an effect was not observed with the overexpression of wild‐type RIG‐I.22 Simultaneously, Zou et al.8 reported two isoforms of RIG‐I, RIG‐Ia and RIG‐Ib, in zebrafish, and found that the overexpression of RIG‐Ib but not RIG‐Ia in EPC cells activated significantly zebrafish type I IFN promoter report plasmids, inducing the expression of Mx and interferon regulatory factor 7 (IRF7) and so protecting cells against SVCV infection. Zebrafish RIG‐Ia and RIG‐Ib, as discussed above, are spliced variant and full‐length transcript, respectively, and the unobserved effect with the overexpression of the zebrafish wild‐type RIG‐I gene in zebrafish embryos, as reported by Nie et al.22, may just reflect an undetected difference in the expression of one RIG‐I gene or two RIG‐I genes. Research from other species of fish supports that RIG‐I overexpression can enhance the antiviral response in vivo and in vitro. For example, crucian carp (Carassius auratus) RIG‐I strongly induced ISGs, such as RSAD2, and activated type I IFN promoters from crucian carp and zebrafish in CAB cells.19, 48 Similar results were observed in EPC cells, although zebrafish and EPC type I IFN luciferase report plasmids were promoted by N‐terminal region overexpression of zebrafish and EPC RIG‐I, respectively.49, 50 Transfection in EPC cells with N‐terminus of EPC RIG‐I resulted in the induction of antiviral genes, including IFN, RSAD2 and ISG15, together resisting VHSV attacks.15 In another study, zebrafish IFN1, IFN4 and myxovirus (influenza virus) resistance C (mxc) genes were significantly induced when full‐length open reading frame of zebrafish RIG‐I was overexpressed in embryos.51 These findings suggest that fish RIG‐I is capable of inducing IFNs and ISGs in antiviral functions, although there is discrepancy concerning the overexpression of zebrafish RIG‐I in its embryos as reported by Nie et al.22 and by Chen et al.51.
Like RIG‐I, MDA5 in teleost has been proved also to play an important role in antiviral immunity. Japanese flounder and rainbow trout are the first two species in which MDA5 was reported to possess antiviral function.10, 11 The overexpression of MDA5 in Japanese flounder provided protection for hirame natural embryo (HINAE) cells against the invasion of VHSV, hirame rhabdovirus and infectious pancreatic necrosis virus; and MDA5 induced IFN, Mx and ISG15 in the cells infected with VHSV.11 Similarly, rainbow trout MDA5 when overexpressed in RTG‐2 cells enhanced the expression of Mx gene and antiviral activity against VHSV.10 Subsequently, two splicing forms of MDA5, MDA5a and MDA5b, were identified in zebrafish, and activation of type I IFN promoters and immune defence against SVCV infection were found in EPC cells transfected with plasmids overexpressing the two isoforms.9 It is further confirmed in other studies that zebrafish MDA5 can increase the expression of type I IFNs in embryos and induce IFN promoter activities.49, 51, 52, 53 Moreover, the overexpression of zebrafish MDA5 in ZFL cells can provide resistance against snakehead rhabdovirus infection. The role of MDA5 is more clearly revealed through the establishment of dominant‐negative MDA5 gene (CARD deletion) transgenic zebrafish, in which higher mortality rate and virus titre were observed after snakehead rhabdovirus infection.53 In addition, a recent report showed that IFN and IFN‐stimulated response element (ISRE) promoters were activated in cells overexpressing grouper MDA5, leading to the induction of IRF3, IRF7 and tumour necrosis factor receptor‐associated factor 6 (TRAF6) and the expression of pro‐inflammatory cytokines, but the transcriptional level of viral genes was significantly reduced including MCP, Vp19 from Singapore grouper iridovirus, and CP and RdRp from red‐spotted grouper nervous necrosis virus.24
Mammal LGP2 overexpression in vitro, such as in L929 cells, negatively regulates RIG‐I and MDA5 signalling,54 but data from LGP2 knockout mice provides the opposite evidence that LGP2 is a positive regulator of RIG‐I and MDA5 in antiviral immune responses.55, 56 Recent studies have shown that mammal LGP2 is involved in MDA5 filament formation and MDA5‐mediated viral RNA recognition.57, 58, 59 Unexpectedly, some reports indicated that teleost LGP2 overexpression in vitro can protect cells against virus infection. For instance, HINAE cells transfected with Japanese flounder wild‐type LGP2, not mutant LGP2 (with RD region deletion), were resistant to infection by VHSV and hirame rhabdovirus, with the induction of antiviral related genes, including Mx, ISG15, ISG56 and IFN.12 Rainbow trout Mx gene was significantly up‐regulated in RTG‐2 cells overexpressing LGP2a but not LGP2b. LGP2a, which is the entire molecule also has antiviral function against VHSV, but LGP2b, a shorter variant, has no such role.10 In CIK cells, the overexpression of all grass carp LGP2 domains, including full‐length, RD region deletion and only RD region, provided protection against grass carp reovirus (097 strain) invasion.60 In addition, the overexpression of black carp (Mylopharyngodon piceus) LGP2 can significantly decrease the viral titre of SVCV and grass carp reovirus in infected EPC cells.61
However, it is a bit bewildering that some reports showed that fish LGP2 can be a negative regulator in antiviral immunity when overexpressed in vitro. Sun et al.19 reported that the activity of type I IFN promoters decreased with the overexpression of crucian carp LGP2 and LGP2‐RD (461–680 amino acids), not LGP2‐HD (1–507 amino acids), in CAB cells infected with poly(I:C), and that crucian carp LGP2 overexpression reduced the activity of IFN promoters mediated by RIG‐I and MDA5. Yu et al.62 observed the significant increase in the expression of viral genes, such as CP, RdRp in red‐spotted grouper nervous necrosis virus, and VP19, MCP, ICP‐18, VP49 in Singapore grouper iridovirus in GS cells infected with these viruses when transfected with grouper LGP2 plasmids, and the down‐regulation of antiviral immune genes, including IRF3, IRF7, ISG15, IFP35, MXI, MXII and MDA5 in cells overexpressing the grouper LGP2. It is therefore considered that the LGP2 overexpression has a negative effect on the induction of antiviral immune genes, but a positive effect on viral replications.62 It is therefore essential to investigate the regulatory effect of LGP2 during viral infection in future research.
Signalling pathways of teleost RLRs
After detecting intracellular non‐self RNA, such as viral RNA, RIG‐I and MDA5 in mammals can become associated with the adaptor molecule, the mitochondrial antiviral signalling protein (MAVS, also known as IPS‐1, VISA and Cardif), through interaction of common CARD domains to recruit TANK binding kinase 1 (TBK1) and to activate IRF3 and IRF7 (Fig. 1),32 which are the core transcription factors in regulation of type I IFNs and ISGs for immune protection of hosts against viral invasion.63
Unsurprisingly, MAVS orthologues also exist in teleost fish and have been cloned in several species, including zebrafish, crucian carp, grass carp, black carp, sea perch, large yellow croaker, rock bream (Oplegnathus fasciatus), Atlantic salmon, green spotted puffer and Japanese flounder.15, 23, 37, 51, 64, 65, 66, 67, 68, 69, 70, 71 Teleost MAVS, as an adaptor, seems to have conserved function on the RLR‐mediated signalling pathway, and contains similar protein domains as in mammals, with CARD domain and transmembrane (TM) region at the N‐ and C‐terminuses, respectively (Fig. 1). First, it is shown that fish MAVS is associated with both RIG‐I and MDA5, as supported by co‐immunoprecipitation assay,8, 9 and MAVS can enhance both RIG‐I‐ and MDA5‐induced type I IFN response and expression of ISGs, such as Mx, RSAD2 and IRF7.8, 9, 51 Indeed, MAVS‐ΔCARD (without CARD domain) and MAVS‐ΔTM (without TM domain) had a negative effect on RIG‐I‐mediated IFN promoter activity, which were CARD and TM region deletions of wild‐type MAVS, respectively.50, 65 Second, fish MAVS may depend on the TM region to locate at mitochondria as observed in mammal cells.15, 50, 66 Third, fish MAVS is able to induce type I IFNs and ISGs, and to provide resistance against infection by a variety of viruses. For example in EPC cells, the overexpression of MAVS from Atlantic salmon, zebrafish and EPC cells can protect the cells against VHSV infection, and Atlantic salmon MAVS also has antiviral function against infectious haematopoietic necrosis virus, SVCV, epizootic haematopoietic necrosis virus and infectious pancreatic necrosis virus.15, 66 Japanese flounder MAVS can induce ISGs, such as Mx, ISG15 and IRF3, and when overexpressed in HINAE cells, Japanese flounder MAVS up‐regulated further the expression of ISGs and type I IFN after VHSV infection, with the promotion of antiviral state.70
It has been also shown that fish MAVS is a positive regulator in antiviral immunity.50, 51, 64, 65, 67, 69, 71, 72 The MAVS is likely to activate IRF3 and IRF7 to trigger the IFN response through the recruitment of signalling molecules, such as TRAF3 and TBK1 (Fig. 1). It is revealed that fish TRAF3 can be associated with MAVS, whose biological function can be inhibited by the overexpression of mutant TBK1 (K38M), and that the overexpression of mutants IRF3 or IRF7 (IRF3‐DN and IRF7‐DN, N‐terminus deletion) diminishes MAVS‐induced IFN promoter activity and the expression level of IFN and ISGs, such as Mx and PKR.19, 65, 67 In fact, TBK1 possesses functions in inducing the expression of type I IFNs and is also associated with IRF3, and such function can be also inhibited by mutant IRF3‐DN or IRF7‐DN.19 Moreover, it was proved that fish IRF3 and IRF7 can activate the IFN promoter via ISRE sites.19, 49, 73 On the other hand, it was most recently shown that in grass carp, MAVS has its potential role in regulating type I IFN response dependent on IRF7 but not on IRF3, and in zebrafish MAVS variants suppress the induction of type I IFNs by targeting IRF7,64, 71 implying that fish RLR–MAVS‐mediated signalling pathways may have some functional variations, which are not as exactly observed as in mammals. Hence, it is considered that teleost fish are likely to have conserved RLR signalling pathways; but some detailed signalling regulatory mechanism remains unclear in fish, and IRF3 and IRF7, which function in RLR‐mediated immune responses, need to be further investigated.
Signalling crosstalk of RLRs in teleost fish
The RLR‐mediated pathway in antiviral immunity has not been clearly identified in fish, and can actually be influenced either directly or indirectly by several factors. Mediator of IRF3 activation (MITA), also known as STING, MPYS, ERIS and TMEM173, is a key molecule in the activation of IRF3 and NF‐κB in the course of type I IFN production (Fig. 1).74, 75 Because MITA is associated with MAVS and RIG‐I, but not MDA5, to enhance antiviral signalling, the molecule is involved in the RLR‐mediated immune response against RNA virus infections.76, 77 To date, MITA has been identified in many species of fish, and is similar to mammals in having an important role in innate immunity, with functions in endoplasmic reticulum localization, in TBK1 and IRF3 recruitment, as well as in antivirus and in type I IFN and ISG induction.19, 50, 78, 79 It has been also reported that fish MITA regulates RLR signalling. For example, RIG‐I‐induced and MDA5‐induced activity of type I IFN promoter was reduced by co‐overexpression of crucian carp MITA‐CT (mutational MITA with N‐terminus deletion) and Flag‐MITA (mutational MITA with flag tag addition at N‐terminus) in CAB cells, respectively.19 Overexpression of C‐terminal region of zebrafish MITA down‐regulated significantly the type I IFN promoter activity in EPC cells, which was induced by both RIG‐I and MAVS.50 However, it is still unclear whether fish MITA interacts directly with RLRs or MAVS, and how the RLR signal cascade is influenced by MITA.
In addition, the biological activity of IRF10 was recently identified in fish as a negative regulator in antiviral immunity;49 IRF10 belongs to the IRF family and exists at least in several vertebrate lineages from teleosts, to reptiles, birds and mammals with the exception of mouse and human.80 Overexpression of zebrafish IRF10 inhibited the type I IFN promoter response mediated by RIG‐I, MDA5, TBK1 and MITA, which probably resulted from the suppression of MITA and ISRE sites from IFNs (Fig. 1).49 In another report, fish RIG‐I and MDA5 were also found to crosstalk with NLRs, such as NOD2, the muramyl dipeptide sensor, to activate NF‐κB and antiviral function.81 Zebrafish NOD2 is associated with RIG‐I, MDA5 and MAVS (Fig. 1), and can increase MAVS‐mediated NF‐κB and IFN promoter activity.81 It is noteworthy that RIG‐I has no effect on the activation of NF‐κB promoter mediated by NOD2 in zebrafish,81 whereas studies in mammals revealed that RIG‐I negatively regulates NOD2‐induced NF‐κB signalling.82 The difference, however, cannot yet be explained.
Negative regulation of RLR signalling
To our knowledge, RLR signalling is strictly controlled by several molecules to avoid excess production of IFNs or inflammatory factors, and to maintain internal immune homeostasis. In fact, conserved mechanisms are also present in teleosts. For example, mammal PIAS4 (PIASy), which belongs to the protein inhibitor of activated signal transducer and activator of transcription (STAT) (PIAS) family, is involved in the repression of signalling mediated by some immune molecules, including STAT1, IRF3/7 and TRIF.83 Co‐overexpression of PIAS4a (PIAS4 homologous gene) and MAVS in zebrafish caused the reduction in expression of MAVS‐mediated type I IFN in the fish embryos (Fig. 1), and zebrafish embryos with the knockdown of PIAS4a had a higher transcriptional level of type I IFN than those with only overexpression of MAVS.84
In fact, PIAS4a is not the only negative regulator in teleosts. It is reported that zebrafish MAVS can be degraded by the overexpression of zebrafish pVHL in HEK293T cells (Fig. 1),85 which is encoded by the von Hippel–Lindau (VHL) tumour suppressor gene and is associated with genetic neoplasia syndromes.86 Indeed, homozygous zebrafish embryos for VHL knockout (vhl −/−) had higher protein level of MAVS than wild‐type embryos; and the expression of MAVS increased further after SVCV infection in vhl −/− embryos compared with wild‐type and embryos without viral infection.85
Furthermore, it has been reported that zebrafish TBK1‐like protein negatively regulates type I IFN promoter activity induced by RIG‐I and MAVS.87 On the other hand, RLR signalling is also inhibited by viruses, not for the prevention of autoimmune disease but for immune escape. In teleosts, a recent study showed that the aquatic virus SVCV suppresses the expression of type I IFN by targeting degradation of MAVS, which was mediated by the N protein of SVCV (Fig. 1).72 This study also indicated that N protein of SVCV caused the degradation of MAVS by K48‐linked ubiquitination.72 Confusingly, two groups obtained different or even opposite conclusions concerning the expression of MAVS in response to SVCV infection. Lu et al.72 reported SVCV‐induced degradation of zebrafish MAVS, while Du et al.85 detected a small increase in zebrafish MAVS after SVCV infection. These results were observed in vitro (EPC cells) and in vivo (zebrafish embryos), respectively,72, 85 and may indicate that the mechanisms in relation with SVCV‐triggered disruption of host innate immunity are possibly much more complicated than expected.
Conclusions and future perspectives
In teleost fish, RLRs, including RIG‐I, MDA5 and LGP2 and in spite of the absence of RIG‐I in some fish species, are essential PRRs responsible for the recognition of various viral PAMPs for inducing antiviral responses. Such antiviral responses of RLRs are provoked by recruiting the downstream adaptor MAVS located on mitochondria, then being associated with signalling molecules, MITA, TRAF3 and TBK1, which in turn facilitate IRF3 and IRF7 activation and phosphorylation, and their translocation into the nucleus, for the induction of type I IFNs as well as ISGs (Fig. 1). In the signalling pathway in teleost fish, some molecules can negatively regulate the production of type I IFNs and ISGs initiated by RLRs. TBK1‐like protein, as an example, can influence RLRs–MAVS–TBK1‐mediated type I IFN and ISG production. In addition, IRF10 in zebrafish inhibits type I IFN production by blocking MITA as well as binding to the ISRE motif (Fig. 1).
Although RLRs as well as the downstream molecules have currently been identified from various species of teleost fish, the origin and evolution of RLRs, the exact ligands recognized by different RLRs, and the signalling pathways of RLRs and the regulation mechanism involved in those pathways remain poorly understood. It is therefore of great importance for future studies to focus on the following interesting topics.
The origin and evolution of RLRs in fish is of significant interest for further research to understand the absence of RIG‐I in certain groups of fish species. It would be intriguing to clarify the antiviral response and the regulatory mechanisms mediated in RLR signalling in the presence as well as in the absence of RIG‐I in fish.
The exact ligands recognized by fish RLRs are poorly investigated, although they can be induced under the stimulation of ssRNA viruses, dsRNA viruses, and synthesized dsRNA poly(I:C). Further research should also be carried out to investigate if fish RLRs can sense bacterial components, or bacterial RNA or PAMPs, or even to interact with other bacterial PAMP‐sensing PRRs to explain the observed increase in the expression of RLRs in fish or fish cell lines following bacterial infection.
The regulatory mechanism involved in fish RLRs remains to be clearly illustrated. All possible molecules in the RLR‐mediated signalling pathway need to be identified functionally in future studies. In particular, molecules that are present possibly in the crosstalk with other PRRs should be functionally characterized. Additionally, it can be important to understand the function of different spliced variants of RLRs in signalling pathways. Pathogenic components which may interfere with fish RLRs or downstream molecules are of particular interests for understanding the pathogenesis of fish pathogens, which may shed light on the understanding of host–pathogen interactions, and may have significance in aquaculture.
In summary, the current knowledge on the composition and function of RLRs in teleost fish are reviewed in this paper. Future perspectives regarding the evolutionary presence of RLRs in teleost fish, the possible ligands from viral as well as from bacterial sources for fish RLRs, the regulatory mechanisms involved in fish RLRs‐mediated signalling pathways are proposed.
Disclosures
The authors declare that they have no competing interest.
Acknowledgements
This work was supported by grants (31320103913, 31402273) from the National Natural Science Foundation of China.
References
- 1. Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell 2006; 124:783–801. [DOI] [PubMed] [Google Scholar]
- 2. Janeway CA, Medzhitov R. Innate immune recognition. Annu Rev Immunol 2002; 20:197–216. [DOI] [PubMed] [Google Scholar]
- 3. Akira S, Hemmi H. Recognition of pathogen‐associated molecular patterns by TLR family. Immunol Lett 2003; 85:85–95. [DOI] [PubMed] [Google Scholar]
- 4. Keating SE, Baran M, Bowie AG. Cytosolic DNA sensors regulating type I interferon induction. Trends Immunol 2011; 32:574–81. [DOI] [PubMed] [Google Scholar]
- 5. Parvatiyar K, Zhang ZQ, Teles RM, Ouyang SY, Jiang Y, Iyer SS et al The helicase DDX41 recognizes the bacterial secondary messengers cyclic di‐GMP and cyclic di‐AMP to activate a type I interferon immune response. Nat Immunol 2012; 13:1155–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Broz P, Monack DM. Newly described pattern recognition receptors team up against intracellular pathogens. Nat Rev Immunol 2013; 13:551–65. [DOI] [PubMed] [Google Scholar]
- 7. Kumar H, Kawai T, Akira S. Pathogen recognition in the innate immune response. Biochem J 2009; 420:1–16. [DOI] [PubMed] [Google Scholar]
- 8. Zou PF, Chang MX, Li Y, Zhang SH, Fu JP, Chen SN et al Higher antiviral response of RIG‐I through enhancing RIG‐I/MAVS‐mediated signaling by its long insertion variant in zebrafish. Fish Shellfish Immunol 2015; 43:13–24. [DOI] [PubMed] [Google Scholar]
- 9. Zou PF, Chang MX, Xue NN, Liu XQ, Li JH, Fu JP et al Melanoma differentiation‐associated gene 5 in zebrafish provoking higher interferon‐promoter activity through signalling enhancing of its shorter splicing variant. Immunology 2014; 141:192–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chang M, Collet B, Nie P, Lester K, Campbell S, Secombes CJ et al Expression and functional characterization of the RIG‐I‐like receptors MDA5 and LGP2 in rainbow trout (Oncorhynchus mykiss). J Virol 2011; 85:8403–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ohtani M, Hikima J, Kondo H, Hirono I, Jung TS, Aoki T. Characterization and antiviral function of a cytosolic sensor gene, MDA5, in Japanese flounder, Paralichthys olivaceus . Dev Comp Immunol 2011; 35:554–62. [DOI] [PubMed] [Google Scholar]
- 12. Ohtani M, Hikima J, Kondo H, Hirono I, Jung TS, Aoki T. Evolutional conservation of molecular structure and antiviral function of a viral RNA receptor, LGP2, in Japanese flounder, Paralichthys olivaceus . J Immunol 2010; 185:7507–17. [DOI] [PubMed] [Google Scholar]
- 13. Sarkar D, Desalle R, Fisher PB. Evolution of MDA‐5/RIG‐I‐dependent innate immunity: independent evolution by domain grafting. Proc Natl Acad Sci U S A 2008; 105:17040–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zou J, Chang M, Nie P, Secombes CJ. Origin and evolution of the RIG‐I like RNA helicase gene family. BMC Evol Biol 2009; 9:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Biacchesi S, LeBerre M, Lamoureux A, Louise Y, Lauret E, Boudinot P et al Mitochondrial antiviral signaling protein plays a major role in induction of the fish innate immune response against RNA and DNA viruses. J Virol 2009; 83:7815–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Su J, Huang T, Dong J, Heng J, Zhang R, Peng L. Molecular cloning and immune responsive expression of MDA5 gene, a pivotal member of the RLR gene family from grass carp Ctenopharyngodon idella . Fish Shellfish Immunol 2010; 28:712–8. [DOI] [PubMed] [Google Scholar]
- 17. Rise ML, Hall J, Rise M, Hori T, Gamperl A, Kimball J et al Functional genomic analysis of the response of Atlantic cod (Gadus morhua) spleen to the viral mimic polyriboinosinic polyribocytidylic acid (pIC). Dev Comp Immunol 2008; 32:916–31. [DOI] [PubMed] [Google Scholar]
- 18. Yang C, Su J, Huang T, Zhang R, Peng L. Identification of a retinoic acid‐inducible gene I from grass carp (Ctenopharyngodon idella) and expression analysis in vivo and in vitro . Fish Shellfish Immunol 2011; 30:936–43. [DOI] [PubMed] [Google Scholar]
- 19. Sun F, Zhang YB, Liu TK, Shi J, Wang B, Gui JF. Fish MITA serves as a mediator for distinct fish IFN gene activation dependent on IRF3 or IRF7. J Immunol 2011; 187:2531–9. [DOI] [PubMed] [Google Scholar]
- 20. Feng H, Liu H, Kong R, Wang L, Wang Y, Hu W et al Expression profiles of carp IRF‐3/‐7 correlate with the up‐regulation of RIG‐I/MAVS/TRAF3/TBK1, four pivotal molecules in RIG‐I signaling pathway. Fish Shellfish Immunol 2011; 30:1159–69. [DOI] [PubMed] [Google Scholar]
- 21. Rajendran KV, Zhang J, Liu S, Peatman E, Kucuktas H, Wang X et al Pathogen recognition receptors in channel catfish: II. Identification, phylogeny and expression of retinoic acid‐inducible gene I (RIG‐I)‐like receptors (RLRs). Dev Comp Immunol 2012; 37:381–9. [DOI] [PubMed] [Google Scholar]
- 22. Nie L, Zhang YS, Dong WR, Xiang LX, Shao JZ. Involvement of zebrafish RIG‐I in NF‐κB and IFN signaling pathways: insights into functional conservation of RIG‐I in antiviral innate immunity. Dev Comp Immunol 2015; 48:95–101. [DOI] [PubMed] [Google Scholar]
- 23. Shen B, Hu Y, Zhang S, Zheng J, Zeng L, Zhang J et al Molecular characterization and expression analyses of three RIG‐I‐like receptor signaling pathway genes (MDA5, LGP2 and MAVS) in Larimichthys crocea . Fish Shellfish Immunol 2016; 55:535–49. [DOI] [PubMed] [Google Scholar]
- 24. Huang Y, Yu Y, Yang Y, Yang M, Zhou L, Huang X et al Antiviral function of grouper MDA5 against iridovirus and nodavirus. Fish Shellfish Immunol 2016; 54:188–96. [DOI] [PubMed] [Google Scholar]
- 25. Bhat A, Paria A, Deepika A, Sreedharan K, Makesh M, Bedekar MK et al Molecular cloning, characterisation and expression analysis of melanoma differentiation associated gene 5 (MDA5) of green chromide, Etroplus suratensis . Gene 2015; 557:172–81. [DOI] [PubMed] [Google Scholar]
- 26. Jia P, Jia K, Chen L, Le Y, Jin Y, Zhang J et al Identification and characterization of the melanoma differentiation‐associated gene 5 in sea perch, Lateolabrax japonicus . Dev Comp Immunol 2016; 61:161–8. [DOI] [PubMed] [Google Scholar]
- 27. Huang T, Su J, Heng J, Dong J, Zhang R, Zhu H. Identification and expression profiling analysis of grass carp Ctenopharyngodon idella LGP2 cDNA. Fish Shellfish Immunol 2010; 29:349–55. [DOI] [PubMed] [Google Scholar]
- 28. Leong JS, Jantzen SG, von Schalburg KR, Cooper GA, Messmer AM, Liao NY et al Salmo salar and Esox lucius full‐length cDNA sequences reveal changes in evolutionary pressures on a post‐tetraploidization genome. BMC Genom 2010; 11:279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jia P, Zhang J, Jin Y, Zeng L, Jia K, Yi M. Characterization and expression analysis of laboratory of genetics and physiology 2 gene in sea perch, Lateolabrax japonicus . Fish Shellfish Immunol 2015; 47:214–20. [DOI] [PubMed] [Google Scholar]
- 30. Cao XL, Chen JJ, Cao Y, Nie GX, Wan QY, Wang LF et al Identification and expression of the laboratory of genetics and physiology 2 gene in common carp Cyprinus carpio . J Fish Biol 2015; 86:74–91. [DOI] [PubMed] [Google Scholar]
- 31. Valero Y, Morcillo P, Meseguer J, Buonocore F, Esteban MA, Chaves‐Pozo E et al Characterization of the IFN pathway in the teleost fish gonad against vertically transmitted viral nervous necrosis virus. J Gen Virol 2015; 96:2176–87. [DOI] [PubMed] [Google Scholar]
- 32. Loo YM, Gale M Jr. Immune signaling by RIG‐I‐like receptors. Immunity 2011; 34:680–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kato H, Takeuchi O, Mikamo‐Satoh E, Hirai R, Kawai T, Matsushita K et al Length‐dependent recognition of double‐stranded ribonucleic acids by retinoic acid‐inducible gene‐I and melanoma differentiation‐associated gene 5. J Exp Med 2008; 205:1601–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen HY, Liu W, Wu SY, Chiou PP, Li YH, Chen YC et al RIG‐I specifically mediates group II type I IFN activation in nervous necrosis virus infected zebrafish cells. Fish Shellfish Immunol 2015; 43:427–35. [DOI] [PubMed] [Google Scholar]
- 35. Wang W, Asim M, Yi L, Hegazy AM, Hu X, Zhou Y et al Abortive infection of snakehead fish vesiculovirus in ZF4 cells was associated with the RLRs pathway activation by viral replicative intermediates. Int J Mol Sci 2015; 16:6235–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Xu C, Evensen O, Munang'andu HM. De Novo Transcriptome analysis shows that SAV‐3 infection upregulates pattern recognition receptors of the endosomal Toll‐like and RIG‐I‐like receptor signaling pathways in macrophage/dendritic like TO‐cells. Viruses 2016; 8:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jia P, Jin Y, Chen L, Zhang J, Jia K, Yi M. Molecular characterization and expression analysis of mitochondrial antiviral signaling protein gene in sea perch, Lateolabrax japonicus . Dev Comp Immunol 2016; 55:188–93. [DOI] [PubMed] [Google Scholar]
- 38. Chen L, Li Q, Su J, Yang C, Li Y, Rao Y. Trunk kidney of grass carp (Ctenopharyngodon idella) mediates immune responses against GCRV and viral/bacterial PAMPs in vivo and in vitro . Fish Shellfish Immunol 2013; 34:909–19. [DOI] [PubMed] [Google Scholar]
- 39. Sullivan C, Charette J, Catchen J, Lage CR, Giasson G, Postlethwait JH et al The gene history of zebrafish tlr4a and tlr4b is predictive of their divergent functions. J Immunol 2009; 183:5896–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sepulcre MP, Alcaraz‐Perez F, Lopez‐Munoz A, Roca FJ, Meseguer J, Cayuela ML et al Evolution of lipopolysaccharide (LPS) recognition and signaling: fish TLR4 does not recognize LPS and negatively regulates NF‐κB activation. J Immunol 2009; 182:1836–45. [DOI] [PubMed] [Google Scholar]
- 41. Berczi I, Bertok L, Bereznai T. Comparative studies on the toxicity of Escherichia coli lipopolysaccharide endotoxin in various animal species. Can J Microbiol 1966; 12:1070–1. [DOI] [PubMed] [Google Scholar]
- 42. Kawai T, Akira S. The role of pattern‐recognition receptors in innate immunity: update on Toll‐like receptors. Nat Immunol 2010; 11:373–84. [DOI] [PubMed] [Google Scholar]
- 43. Sullivan C, Postlethwait JH, Lage CR, Millard PJ, Kim CH. Evidence for evolving Toll‐IL‐1 receptor‐containing adaptor molecule function in vertebrates. J Immunol 2007; 178:4517–27. [DOI] [PubMed] [Google Scholar]
- 44. Wilkins C, Gale M Jr. Recognition of viruses by cytoplasmic sensors. Curr Opin Immunol 2010; 22:41–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schlee M, Roth A, Hornung V, Hagmann CA, Wimmenauer V, Barchet W et al Recognition of 5′ triphosphate by RIG‐I helicase requires short blunt double‐stranded RNA as contained in panhandle of negative‐strand virus. Immunity 2009; 31:25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Saito T, Owen DM, Jiang F, Marcotrigiano J, Gale M Jr. Innate immunity induced by composition‐dependent RIG‐I recognition of hepatitis C virus RNA. Nature 2008; 454:523–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Goubau D, Schlee M, Deddouche S, Pruijssers AJ, Zillinger T, Goldeck M et al Antiviral immunity via RIG‐I‐mediated recognition of RNA bearing 5'‐diphosphates. Nature 2014; 514:372–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang B, Zhang YB, Liu TK, Shi J, Sun F, Gui JF. Fish viperin exerts a conserved antiviral function through RLR‐triggered IFN signaling pathway. Dev Comp Immunol 2014; 47:140–9. [DOI] [PubMed] [Google Scholar]
- 49. Li S, Lu LF, Feng H, Wu N, Chen DD, Zhang YB et al IFN regulatory factor 10 is a negative regulator of the IFN responses in fish. J Immunol 2014; 193:1100–9. [DOI] [PubMed] [Google Scholar]
- 50. Biacchesi S, Merour E, Lamoureux A, Bernard J, Bremont M. Both STING and MAVS fish orthologs contribute to the induction of interferon mediated by RIG‐I. PLoS ONE 2012; 7:e47737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chen WQ, Hu YW, Zou PF, Ren SS, Nie P, Chang MX. MAVS splicing variants contribute to the induction of interferon and interferon‐stimulated genes mediated by RIG‐I‐like receptors. Dev Comp Immunol 2015; 49:19–30. [DOI] [PubMed] [Google Scholar]
- 52. Feng H, Zhang YB, Zhang QM, Li Z, Zhang QY, Gui JF. Zebrafish IRF1 regulates IFN antiviral response through binding to IFNφ1 and IFNκφ3 promoters downstream of MyD88 signaling. J Immunol 2015; 194:1225–38. [DOI] [PubMed] [Google Scholar]
- 53. Gabor KA, Charette JR, Pietraszewski MJ, Wingfield DJ, Shim JS, Millard PJ et al A DN‐mda5 transgenic zebrafish model demonstrates that Mda5 plays an important role in snakehead rhabdovirus resistance. Dev Comp Immunol 2015; 51:298–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yoneyama M, Kikuchi M, Matsumoto K, Imaizumi T, Miyagishi M, Taira K et al Shared and unique functions of the DExD/H‐box helicases RIG‐I, MDA5, and LGP2 in antiviral innate immunity. J Immunol 2005; 175:2851–8. [DOI] [PubMed] [Google Scholar]
- 55. Satoh T, Kato H, Kumagai Y, Yoneyama M, Sato S, Matsushita K et al LGP2 is a positive regulator of RIG‐I‐ and MDA5‐mediated antiviral responses. Proc Natl Acad Sci U S A 2010; 107:1512–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Venkataraman T, Valdes M, Elsby R, Kakuta S, Caceres G, Saijo S et al Loss of DExD/H box RNA helicase LGP2 manifests disparate antiviral responses. J Immunol 2007; 178:6444–55. [DOI] [PubMed] [Google Scholar]
- 57. Yoneyama M, Onomoto K, Jogi M, Akaboshi T, Fujita T. Viral RNA detection by RIG‐I‐like receptors. Curr Opin Immunol 2015; 32:48–53. [DOI] [PubMed] [Google Scholar]
- 58. Deddouche S, Goubau D, Rehwinkel J, Chakravarty P, Begum S, Maillard PV et al Identification of an LGP2‐associated MDA5 agonist in picornavirus‐infected cells. ELife 2014; 3:e01535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bruns AM, Leser GP, Lamb RA, Horvath CM. The innate immune sensor LGP2 activates antiviral signaling by regulating MDA5‐RNA interaction and filament assembly. Mol Cell 2014; 55:771–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chen X, Yang C, Su J, Rao Y, Gu T. LGP2 plays extensive roles in modulating innate immune responses in Ctenopharyngodon idella kidney (CIK) cells. Dev Comp Immunol 2015; 49:138–48. [DOI] [PubMed] [Google Scholar]
- 61. Xiao J, Yan J, Chen H, Li J, Tian Y, Feng H. LGP2 of black carp plays an important role in the innate immune response against SVCV and GCRV. Fish Shellfish Immunol 2016; 57:127–35. [DOI] [PubMed] [Google Scholar]
- 62. Yu Y, Huang Y, Yang Y, Wang S, Yang M, Huang X et al Negative regulation of the antiviral response by grouper LGP2 against fish viruses. Fish Shellfish Immunol 2016; 56:358–66. [DOI] [PubMed] [Google Scholar]
- 63. Barnes B, Lubyova B, Pitha PM. On the role of IRF in host defense. J Interferon Cytokine Res 2002; 22:59–71. [DOI] [PubMed] [Google Scholar]
- 64. Lu LF, Li S, Lu XB, Zhang YA. Functions of the two zebrafish MAVS variants are opposite in the induction of IFN1 by targeting IRF7. Fish Shellfish Immunol 2015; 45:574–82. [DOI] [PubMed] [Google Scholar]
- 65. Zhang J, Zhang YB, Wu M, Wang B, Chen C, Gui JF. Fish MAVS is involved in RLR pathway‐mediated IFN response. Fish Shellfish Immunol 2014; 41:222–30. [DOI] [PubMed] [Google Scholar]
- 66. Lauksund S, Svingerud T, Bergan V, Robertsen B. Atlantic salmon IPS‐1 mediates induction of IFNa1 and activation of NF‐κB and localizes to mitochondria. Dev Comp Immunol 2009; 33:1196–204. [DOI] [PubMed] [Google Scholar]
- 67. Xiang Z, Qi L, Chen W, Dong C, Liu Z, Liu D et al Characterization of a TnMAVS protein from Tetraodon nigroviridis . Dev Comp Immunol 2011; 35:1103–15. [DOI] [PubMed] [Google Scholar]
- 68. Zhou W, Zhou J, Lv Y, Qu Y, Chi M, Li J et al Identification and characterization of MAVS from black carp Mylopharyngodon piceus . Fish Shellfish Immunol 2015; 43:460–8. [DOI] [PubMed] [Google Scholar]
- 69. Kasthuri SR, Wan Q, Whang I, Lim BS, Yeo SY, Choi CY et al Functional characterization of the evolutionarily preserved mitochondrial antiviral signaling protein (MAVS) from rock bream, Oplegnathus fasciatus . Fish Shellfish Immunol 2014; 40:399–406. [DOI] [PubMed] [Google Scholar]
- 70. Simora RM, Ohtani M, Hikima J, Kondo H, Hirono I, Jung TS et al Molecular cloning and antiviral activity of IFN‐β promoter stimulator‐1 (IPS‐1) gene in Japanese flounder, Paralichthys olivaceus . Fish Shellfish Immunol 2010; 29:979–86. [DOI] [PubMed] [Google Scholar]
- 71. Feng X, Zhang Y, Yang C, Liao L, Wang Y, Su J. Functional characterizations of IPS‐1 in CIK cells: potential roles in regulating IFN‐I response dependent on IRF7 but not IRF3. Dev Comp Immunol 2015; 53:23–32. [DOI] [PubMed] [Google Scholar]
- 72. Lu LF, Li S, Lu XB, LaPatra SE, Zhang N, Zhang XJ et al Spring viremia of carp virus N protein suppresses fish IFNφ1 production by targeting the mitochondrial antiviral signaling protein. J Immunol 2016; 196:3744–53. [DOI] [PubMed] [Google Scholar]
- 73. Sun F, Zhang YB, Liu TK, Gan L, Yu FF, Liu Y et al Characterization of fish IRF3 as an IFN‐inducible protein reveals evolving regulation of IFN response in vertebrates. J Immunol 2010; 185:7573–82. [DOI] [PubMed] [Google Scholar]
- 74. Barber GN. Innate immune DNA sensing pathways: STING, AIMII and the regulation of interferon production and inflammatory responses. Curr Opin Immunol 2011; 23:10–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Cai X, Chiu YH, Chen ZJ. The cGAS‐cGAMP‐STING pathway of cytosolic DNA sensing and signaling. Mol Cell 2014; 54:289–96. [DOI] [PubMed] [Google Scholar]
- 76. Zhong B, Yang Y, Li S, Wang YY, Li Y, Diao F et al The adaptor protein MITA links virus‐sensing receptors to IRF3 transcription factor activation. Immunity 2008; 29:538–50. [DOI] [PubMed] [Google Scholar]
- 77. Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 2008; 455:674–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Huang Y, Ouyang Z, Wang W, Yu Y, Li P, Zhou S et al Antiviral role of grouper STING against iridovirus infection. Fish Shellfish Immunol 2015; 47:157–67. [DOI] [PubMed] [Google Scholar]
- 79. Ge R, Zhou Y, Peng R, Wang R, Li M, Zhang YB et al Conservation of the STING‐mediated cytosolic DNA sensing pathway in zebrafish. J Virol 2015; 89:7696–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Huang B, Qi ZT, Xu Z, Nie P. Global characterization of interferon regulatory factor (IRF) genes in vertebrates: glimpse of the diversification in evolution. BMC Immunol 2010; 11:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Zou PF, Chang MX, Li Y, Xue NN, Li JH, Chen SN et al NOD2 in zebrafish functions in antibacterial and also antiviral responses via NF‐κB, and also MDA5, RIG‐I and MAVS. Fish Shellfish Immunol 2016; 55:173–85. [DOI] [PubMed] [Google Scholar]
- 82. Morosky SA, Zhu J, Mukherjee A, Sarkar SN, Coyne CB. Retinoic acid‐induced gene‐I (RIG‐I) associates with nucleotide‐binding oligomerization domain‐2 (NOD2) to negatively regulate inflammatory signaling. J Biol Chem 2011; 286:28574–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Shuai K, Liu B. Regulation of gene‐activation pathways by PIAS proteins in the immune system. Nat Rev Immunol 2005; 5:593–605. [DOI] [PubMed] [Google Scholar]
- 84. Xiong R, Nie L, Xiang LX, Shao JZ. Characterization of a PIAS4 homologue from zebrafish: insights into its conserved negative regulatory mechanism in the TRIF, MAVS, and IFN signaling pathways during vertebrate evolution. J Immunol 2012; 188:2653–68. [DOI] [PubMed] [Google Scholar]
- 85. Du J, Zhang D, Zhang W, Ouyang G, Wang J, Liu X et al PVHL negatively regulates antiviral signaling by targeting MAVS for proteasomal degradation. J Immunol 2015; 195:1782–90. [DOI] [PubMed] [Google Scholar]
- 86. Nielsen SM, Rhodes L, Blanco I, Chung WK, Eng C, Maher ER et al Von Hippel–Lindau disease: genetics and role of genetic counseling in a multiple neoplasia syndrome. J Clin Oncol 2016; 34:2172–81. [DOI] [PubMed] [Google Scholar]
- 87. Zhang L, Chen WQ, Hu YW, Wu XM, Nie P, Chang MX. TBK1‐like transcript negatively regulates the production of IFN and IFN‐stimulated genes through RLRs‐MAVS‐TBK1 pathway. Fish Shellfish Immunol 2016; 54:135–43. [DOI] [PubMed] [Google Scholar]


