Abstract
Background
Arabian horses are believed to be one of the oldest and most influential horse breeds in the world. Blood is the main tissue involved in maintaining body homeostasis, and it is considered a marker of the processes taking place in the other tissues. Thus, the aim of our study was to identify the genetic basis of changes occurring in the blood of Arabian horses subjected to a training regimen and to compare the global gene expression profiles between different training periods (T1: after a slow canter phase that is considered a conditioning phase, T2: after an intense gallop phase, and T3: at the end of the racing season) and between trained and untrained horses (T0). RNA sequencing was performed on 37 samples with a 75-bp single-end run on a HiScanSQ platform (Illumina), and differentially expressed genes (DEGs) were identified based on DESeq2 (v1.11.25) software.
Results
An increase in the number of DEGs between subsequent training periods was observed, and the highest amount of DEGs (440) was detected between untrained horses (T0) and horses at the end of the racing season (T3). The comparisons of the T2 vs. T3 transcriptomes and the T0 vs. T3 transcriptomes showed a significant gain of up-regulated genes during long-term exercise (up-regulation of 266 and 389 DEGs in the T3 period compared to T2 and T0, respectively). Forty differentially expressed genes were detected between the T1 and T2 periods, and 296 between T2 and T3. Functional annotation showed that the most abundant genes up-regulated in exercise were involved in pathways regulating cell cycle (PI3K-Akt signalling pathway), cell communication (cAMP-dependent pathway), proliferation, differentiation and apoptosis, as well as immunity processes (Jak-STAT signalling pathway).
Conclusions
We investigated whether training causes permanent transcriptome changes in horse blood as a reflection of adaptation to conditioning and the maintenance of fitness to compete in flat races. The present study identified the overrepresented molecular pathways and genes that are essential for maintaining body homeostasis during long-term exercise in Arabian horses. Selected DEGs should be further investigated as markers that are potentially associated with racing performance in Arabian horses.
Electronic supplementary material
The online version of this article (doi:10.1186/s12863-017-0499-1) contains supplementary material, which is available to authorized users.
Keywords: Arabian horse, Training, Blood transcriptome, RNA-seq, Exercise adaptation
Background
It is well established that intensive training initiates long-term adaptation processes that are involved in the establishment of a new body homeostasis. During exercise, the oxygen demand significantly increases, and thus, one of the most important adaptations to training is optimization of O2 transport in the entire organism by both improvements in vascular endothelial function and increases in mitochondrial function [1, 2]. On the other hand, an imbalance between exercise and the recovery process can result in negative consequences, such as underperformance and a progressive fatigue called overtraining syndrome [3, 4]. It seems to be important to distinguish the signs of overtraining from the effects of balanced exercise on the molecular level in order to design molecular tools for training optimization. To date, only the protein levels of alpha-1 antitrypsin has been considered as a marker of overtraining in horses based on protein analysis during normal and intensive training [5].
In humans, several studies emphasized the need for identifying a possible genetic predisposition to athletic performance and for detecting the molecular pathways involved in overtraining syndrome [4]. The horse is considered as an excellent model for analysing the changes that occur in response to exercise-induced stress. In recent decades, huge improvements in molecular genetics methods have enabled genome-wide searches for genes associated with important phenotypic features. Due to the high profitability of sport and racing horses, a major area of research is to identify the genetic basis of athletic performance in Arabian horses.
The first analysis of changes in muscle gene expression profiles during exercise adaptation was performed in Thoroughbreds using cDNA microarrays [6]. In 2010, the research performed by McGivney et al. [7] using RNA sequencing (Next-Generation technology) confirmed the differential expression of 92 transcripts in equine muscle tissues before and after long-term exercise. Among the genes whose expression decreased after exercise, the authors identified the myostatin gene (MSTN). The strong association of MSTN with the best race distances and athletic performance in Thoroughbreds was later confirmed by other studies [8, 9]. The comparative transcriptome analysis of blood and muscle cells in six Thoroughbred horses showed the common expression of over 11,300 genes in both tissues, which constituted over 60% of all genes transcribed in muscle [10]. Furthermore, the authors confirmed the significant effects of intense exercise on the transcriptome profiles of both tissues [10].
The racing performance traits of Thoroughbred horses (TB) have been widely described. On the other hand, the exact mechanism that occurs in the blood and muscles of Arabian horses during exercise in relation to stamina and performance is still not well understood. In purebred Arabians, a main selection criteria is racetrack performance. Young (3- to 4-year-old) horses compete in races, and this age has a significant effect on their development. After their career on the race track is finished, the predisposed horses are prepared for endurance rides.
Capomaccio et al. [11], who analysed whole transcriptome profiles of blood in Arabian horses, pinpointed the biological processes that are potentially related to exercise-induced stress and identified groups of genes that are related to inflammation, immune interaction or cell signalling. Such research is intended to broaden our knowledge of the molecular mechanisms associated with adaptation to exercise in order to select the most suitable training schedules for obtaining better performance as well as maintaining animal health [12].
It has been established that repeated rounds of exercise lead to new basal levels of gene expression in resting muscles [13]. Blood is the main tissue involved in maintaining body homeostasis, and it is considered a marker of the processes taking place in the other tissues. Thus, the aim of our study was to identify the genetic basis of changes occurring in the blood of Arabian horses during their training regimen. We investigated whether the training period causes permanent transcriptome changes that are involved in the horse’s adaptation to conditioning and maintenance of its fitness to compete in flat races.
Methods
Animals
The profiling of blood transcriptomes was performed for 12 Arabian horses (3 years old) that were introduced to the training centre (to prepare for flat racing). Blood samples were collected at 3 different time points during the training procedure: after the slow canter phase, which is considered a conditioning phase (March; T1); after an intense gallop phase (May, trained horses before the racing season; T2); and at the end of the racing season (October; T3). The blood transcriptomes of 6 untrained Arabian horses (2.5 years old; T0) were also analysed. It was not possible to collect blood samples from all of the same horses in the control group as a training group (some horses sampled as untrained were not introduced to the training centre). The analysed group of animals included the same number of mares and stallions in order to avoid a gender effect.
To identify genes with the greatest effect on adaptation to long-term exercise, we compared transcriptomes starting from period T1 (conditioning phase) to T3 (end of training after the racing season): T1 → T2 → T3 (training comprised one year). Additionally, we compared transcriptome profiles between the T3 (trained horses) and T0 (untrained horses, control group) periods in order to detect potential training-induced modifications of gene expression: T1 → T2 → T3 → T0.
All animals were healthy (they were under veterinary supervision) and successfully performed on the racetrack during the racing season. In each period, samples were collected at the same time of the day (2 h after morning feeding and before training to avoid disruption of the training routine) and according to the same procedure for each horse. The exact training schedule and the time points of sample collection are precisely described in the Additional file 1: Figure S1. All horses were maintained in one stud, which was owned by one breeder. In addition, they were introduced in the Polish Arabian Stud Book (PASB) while also belonging to the pure Polish line of known origin since the late 1700s. They were also maintained under the same environmental and feeding conditions (fed a hay- and oat-based diet that was supplemented with a commercial feed mixture according to the stage of the training cycle).
The protocol was approved by the Animal Care and Use Committee of the Institute of Pharmacology, Polish Academy of Sciences in Kraków (no. 1173/2015). Owners consent for their horses’ inclusion in the study.
Transcriptome sequencing
In total, 42 blood samples were collected from the jugular vein into Tempus™ Blood RNA Tubes (Ambion, Life Technologies) and stored at −20 °C. Total RNA was isolated using the MagMAX™-96 Total RNA Isolation Kit (Ambion, Life Technologies) according to the manufacturer’s protocol. The quality and quantity of obtained RNA were examined using NanoDrop 2000 (Thermo Scientific; Wilmington, USA) and TapeStation 2200 instruments with Agilent RNA ScreenTape (Agilent, Perlan Technologies). The RNA samples with RIN (RNA integrity number) values above 8.5 were used for further analysis. Five samples were removed from RNA-seq analysis due to the exclusion of horses from the training cycle for various reasons (lameness or poor metabolic performance).
The cDNA libraries were prepared from 400 ng of total RNA with TruSeq RNA Sample Prep Kit v2 kit (Illumina) according to the protocol. The quality and quantity of the obtained libraries were established using Qubit 2.0 (Invitrogen, Life Technologies) and TapeStation 2200 (D1000 ScreenTape; Agilent). RNA sequencing was performed with a 75-bp single-end run on a HiScanSQ platform (Illumina) using TruSeq SR Cluster Kit v3- CBOT-HS and TruSeq SBS Kit v 3 - HS chemistry (Illumina). Indexed samples were pooled into two groups and loaded into 3–4 lanes on two flow-cells, which allowed for obtaining at least 3 technical replications per library.
Data analysis
Quality control (QC) of the raw sequence data was conducted using FastQC software (v0.11.4). The obtained QC results were used as an indicator to remove adapter sequences, reads shorter than 36 bp and/or reads with a quality score lower than 20 (Flexbar software v2.5) [14]. The quantification of transcript abundance was conducted using RSEM software (v1.2.22) [15] supported by STAR aligner software (STAR_2.4.2a) [16]. The raw reads were aligned to the Equus caballus genome (assembly Equus_caballus.EquCab2.83) as a reference, while the whole procedure was followed by alignment parameter evaluation using ENCODE3's STAR-RSEM pipeline. To detect the differentially expressed genes (DEGs), Bioconductor/R-project package (R version 3.3.1) was used with DESeq2 (1.12.4) (SummarizedExperiment_1.2.3; Biobase_2.32.0; GenomicRanges_1.24.2) (adjusted p-value was calculated using the Benjamini/Hochberg method) [17].
Gene Ontology (GO) annotation was performed for DEGs (fold change ≥ |1.3|; adjusted p-value < 0.05) using Panther software [18]. Because variability in the fold change (FC) of a gene is inversely related to its expression level [19], in the present study, DEGs with FC > 1.3 (p-value < 0.05) were considered significant in order to avoid overlooking the exercise–induced modifications of highly expressed genes. GO annotations were performed using the latest version of the Equus caballus reference, which includes 20,384 annotated genes. The p-value in enrichment tests was determined based on the Mann–Whitney U Test (Wilcoxon Rank-Sum Test) with a Bonferroni correction. The online DAVID software and the Kyoto Encyclopedia of Genes and Genomes (KEGG) with KEGG mapper – search pathway tools was applied for pathway detection and analysis. The significantly enriched pathways were identified based on the p-values obtained from a Fisher Exact test.
qPCR validation
The evaluation of mRNA levels of eight selected DEGs was performed by a real-time qPCR method. Genes selected for validation were identified as significantly differentially expressed in at least two comparisons between different training periods.
cDNA was synthesized for all analysed samples from 500 ng of total RNA with the use of a TranscriptME RNA kit (Blirt, Poland), according to manufacturer’s protocol. The determination of gene expression was performed on a 7500 Real-Time PCR System using AmpliQ 5x HOT EvaGreen® qPCR Mix Plus (ROX) (Novazym, Poland) in three technical replicates for each sample. Two different genes were used as endogenous controls: GAPDH (glyceraldehyde 3-phosphate dehydrogenase) and SDHA (succinate dehydrogenase complex subunit A) [20]. Primers for the target and control genes were designed using version 4.0.0 of Primer3 software based on the reference sequences annotated in the Ensembl database (Additional file 2: Table S1). For all genes except P2RY14, PCR products spanned two exons. The efficiency of the qPCR reactions was defined using the standard curve method, and mRNA abundance was calculated according to the ∆∆Ct method.
Pearson’s correlation was used to compare the normalized read counts and qPCR data. A one-way ANOVA with Duncan’s post hoc test was used to estimate the significance of differences in gene expression between training periods.
Results
Sequencing data
In total, 914.7 million raw reads were generated for all samples, ranging from 16.4 to 27.7 M per sample. On average, individual samples yielded 24.7 M (±3.8) reads. After quality trimming and filtration, 898 M high-quality reads (98.2% of all reads) were used for mapping. According to the STAR-RSEM pipeline, there were 7.8 – 28.2 M (19.7 M or 81%, on average) uniquely aligned reads per sample. Detailed statistics on the number of analysed reads and achieved mapping efficiency are presented in Additional file 3: Table S2. Approximately 81% of raw reads were mapped to the EquCab2.0 assembly, 51% of which matched annotated exonic regions, 16% matched intronic regions, and 33% matched intergenic regions. The analysis was submitted to the NCBI Gene Expression Omnibus (GEO) functional genomics data repository and assigned GEO accession number GSE83404 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE83404).
Exercise-regulated differentially expressed genes
In the present research, the blood transcriptome profiles between horses in successive training stages and a control group were compared as follows: T1:T2, T2:T3, and T0:T3. (T1 → T2 → T3 → T0). An increase in the number of DEGs (fold change at of least ±1.3; adjusted p-value < 0.05) between subsequent training periods was observed, and the highest amount of DEGs (440) was detected between untrained horses (T0) and horses at the end of the racing season (T3) (Table 1). The comparisons of the transcriptomes T2 vs. T3 and T0 vs. T3 showed a significant gain in up-regulated genes during long-term exercise (up-regulation of 266 and 389 DEGs in T3 period compared to T2 and T0, respectively). Forty differentially expressed genes were detected between the T1 and T2 periods, and 296 between T2 and T3.
Table 1.
The number of identified differentially expressed genes (DEGs) between different training periods
| Comparison | T1:T2 | T2:T3 | T0:T3 | |||
|---|---|---|---|---|---|---|
| Down | Up | Down | Up | Down | Up | |
| Annotated gene | 34 | 269 | 389 | |||
| 15 | 19 | 19 | 250 | 28 | 361 | |
| Novel gene | 6 | 27 | 51 | |||
| – | 6 | 11 | 16 | 23 | 28 | |
| Total DEGs | 40 | 296 | 440 | |||
Furthermore, when comparing the transcriptomes in the T0 vs. T3 exercise stages, 51 novel exercise-regulated genes (annotated as novel genes according to the Ensemble database) were detected: 23 were up-regulated and 28 were down-regulated, which accounted for approximately 11% of all identified DEGs. Due to the high number of uncharacterized DEGs, the functional classification of orthologues was performed using Panther software. The analysis showed that some of the gene orthologues classified as novel corresponded to protein families and/or protein classes (Additional file 4: Table S3). The most overrepresented functional group included a panel of orthologous genes related to binding activity (molecular function, GO:0005488; e.g., TBL1X, TAF1, SRCAP, AKAP11, POM121C, RBM27) and biological regulation (biological process, GO:0065007; TBL1X, TAF1, SRCAP, AKAP11 and LOC100062658 orthologs). Some of the genes identified as novel were differentially expressed in two out of three comparisons (T2:T3 and T0:T3), which suggested that selected genes can play an important role during the adaptation process to exercise (orthologues of TBL1X, RBM27, GOPC, KMT2C, LY5, PRR14L, TRAPPC10, LOC100062658) (Additional file 4: Table S3).
We also compared the sets of significant DEGs for T1 vs. T2, T2 vs. T3 and T0 vs. T3, and we detected several genes whose transcription was down- or up-regulated during at least two training periods (Table 2). According to their GO annotations, the identified genes play a role in the regulation of biological processes (GO:0050789; NCOA2, P2RY14, MBTD1, ZNF618, NFAT5), homeostatic processes (GO:0042592; ATP13A3) and metabolic processes (nucleobase-containing compound metabolic process, GO:0006139; NCOA2, P2RY14, MBTD1, ZNF618, NFAT5, WDR7).
Table 2.
List of exercise regulated (down or up-regulated) genes during at least two training periods
| Training periods | Gene name | Ensembl Accession number | T1:T2 | T2:T3 | T0:T3 | |||
|---|---|---|---|---|---|---|---|---|
| FC | adj p | FC | adj p | FC | adj p | |||
| Genes up-regulated | ||||||||
| 1 | ACVR2A | ENSECAG00000000138 | 1.56 | 0.01 | 1.55 | 0,003 | ns | |
| 2 | ZAB2 | ENSECAG00000019355 | 1.41 | 0.05 | 1.42 | 0.017 | ns | |
| 3 | P2RY14 | ENSECAG00000001554 | 1.54 | 0.05 | 1.69 | 0.009 | 1.59 | 0.05 |
| 4 | WDR7 | ENSECAG00000007050 | 1.37 | 0.006 | 1.60 | 0.01 | 1.60 | 0.005 |
| 5 | AGPAT1 | ENSECAG00000016106 | 1.55 | 0.011 | 1.60 | 0.02 | 1.52 | 0.02 |
| 6 | FOXN2 | ENSECAG00000008390 | ns | 1.80 | 0.007 | 2.01 | 8.78E-06 | |
| 7 | LARP4 | ENSECAG00000012479 | ns | 1.54 | 0.013 | 1.65 | 0.005 | |
| 8 | LPGAT1 | ENSECAG00000013187 | ns | 1.39 | 0.01 | 1.46 | 0.02 | |
| 9 | RICTOR | ENSECAG00000000308 | ns | 1.74 | 0.002 | 1.91 | 0.0002 | |
| 10 | ATP13A3 | ENSECAG00000006749 | ns | 1.65 | 0.0009 | 1.89 | 0.0005 | |
| 11 | HIPK1 | ENSECAG00000015193 | ns | 1.61 | 0.013 | 1.92 | 0.0001 | |
| 12 | NFAT5 | ENSECAG00000010266 | ns | 1.66 | 0.016 | 1.97 | 0.0008 | |
| 13 | ZNF618 | ENSECAG00000020621 | ns | 1.66 | 0.003 | 1.87 | 0.0006 | |
| 14 | GALNT7 | ENSECAG00000010243 | ns | 1.61 | 0.009 | 1.57 | 0.011 | |
| 15 | TMED5 | ENSECAG00000016426 | ns | 1.51 | 0.01 | 1.53 | 0.003 | |
| 16 | MBTD1 | ENSECAG00000022700 | ns | 1.65 | 0.009 | 1.61 | 0.005 | |
| 17 | NCOA2 | ENSECAG00000008394 | ns | 1.41 | 0.05 | 1.61 | 0.0003 | |
| Genes down-regulated | ||||||||
| 1 | GTSF1 | ENSECAG00000013618 | ns | −1.55 | 0.0005 | −1.48 | 0.034 | |
| 2 | novel | ENSECAG00000015782 | ns | −1.83 | 0.001 | −1.77 | 0.012 | |
| 3 | novel | ENSECAG00000017143 | ns | −1.49 | 0.03 | −1.66 | 0.02 | |
| 4 | ANKZF1 | ENSECAG00000016473 | 0.5 | 0.05 | −1.37 | 0.003 | ns | |
| 5 | C7orf50 | ENSECAG00000022930 | −1.39 | 0.007 | −1.39 | 0.04 | ns | |
| 6 | GZMH | ENSECAG00000008322 | −1.57 | 0.05 | −1.46 | 0.04 | ns | |
| 7 | SYCE1 | ENSECAG00000012421 | −1.43 | 0.01 | −1.83 | 0.001 | ns | |
| 8 | CRYGS | ENSECAG00000002645 | −1.39 | 0.007 | −1.43 | 0.004 | ns | |
| 9 | C12orf43 | ENSECAG00000014885 | −1.42 | 0.05 | ns | −1.36 | 0.04 | |
FC fold change, ns not significant
Functional annotation and identified pathways
Our results showed that the highest number of identified genes encoded transcription factors, nucleic acid binding proteins and G-protein modulators, which were mainly transcriptionally activated at the last training phase (T3) (Table 3). Moreover, in the T3 period, the identified DEGs represented genes encoding cytoskeletal proteins, including actin cytoskeletal proteins and kinases. In turn, at the beginning of the training cycle (T1), the differential expression of genes related to immune response (defence proteins, cytokine receptors) and genes encoding hydrolase were detected (Table 3).
Table 3.
The mostly affected genes encoding the most overrepresented protein classes during training regime in Arabian horses
| N | P | Gene | N | P | Gene | N | P | Gene | |
|---|---|---|---|---|---|---|---|---|---|
| Training periods | T1 vs T2 | T2 vs T3 | T0 vs T3 | ||||||
| transcription factors (PC00218) | 6 | ns | QKI; PPP1R37; PHTF2; MBTD1; FOXS1; ZEB2; | 34 | 0.0001 | AHR; GNPTAB; IKZF1; NCOA1; NFKBID; MBTD1; NFAT5; PDLIM5; RBBP8; RREB1 | 46 | 0.006 | NFATC1;NCOR2; FOXJ2;NFAT5;SATB1;NCOA1;SKI;QKI;AFF4 |
| nucleic acid binding (PC00171) | 13 | ns | LARP4; MBTD1; WDR7; RNASE1; OAS1; FOXS1 | 42 | 0.003 | MDM4; ZBTB38; ACAP2; TOP3B; BDP1; WDR7; ATM; LARP4; SATB1 | 74 | 0.0001 | CHD2; RREB1; TRA2A; MDM4; ZBTB38; SETX; ACAP2; NCOR2; WDR7; PRRC2C; |
| G-protein modulator (PC00022) | 2 | ns | P2RY14; CYSLTR1 | 10 | 0.004 | CCRL2; P2RY14; EVI5; GNB4; | 16 | 0.02 | S1PR4;P2RY14;ACAP2;APPL1 |
| cytoskeletal protein (PC00085) | 28 | 0.004 | CAMK4; MYO5A; DYNC1LI2; ACTR2; LMNB2; MSN; ASPM; SEPT11; MYH9; MYO9A; SYNE3 | ||||||
| actin family cytoskeletal protein (PC00041) | 15 | 0.02 | MYO5A; ACTR2; KLHL6; MSN; SPTBN1; MYO9A; ASPM; KLHL11; WASF2; SYNE3; MYH9 | ||||||
| kinase (PC00137) | 3 | 0.003 | ACVR2A, MAPK3, CASK | 17 | 0.0028 | PIK3CG; EEF2K; ACVR1B; ATM; MAP3K7; ACVR2A; WNK1; HIPK3; STK17B; LRRK2; HIPK1 | 19 | 0.002 | PIK3CG; CAMK4; PANK3; MAP3K7; EEF2K; DYRK2; ATM; AURKC; HIPK3; HIPK1 |
| TGF-beta receptor (PC00035) | 1 | 0.008 | ACVR2A | 1 | 0.0008 | ACVR1B; ACVR2A; | |||
| defense/immunity protein (PC00090) | 5 | 0.001 | CD1C; C2; OAS1; ENSECAG00000012920; ENSECAG00000000325 | 3 | 0.001 | TXLNG; IL6ST; CCRL2; | 4 | 0.003 | TXLNA;TXLNG; NOTCH1;NOTCH2; |
| cytokine receptor (PC00084) | 5 | 0.004 | ACVR2A; CD1C; ENSECAG00000012920; ENSECAG00000000325 | 4 | 0.01 | CCRL2; ACVR2A; ACVR1B; IL6ST | |||
| hydrolase (PC00121) | 5 | 0.002 | RNASE1; C2; WDR7; MX1; CHI3L1; | 8 | 0.01 | EVI5; ATP11C; NUDT17; ATP13A3; TPP1; GLS; PADI6; GNB4; | |||
N number of genes detected
Pathway analysis was performed with DAVID and KEGG software using as an input a set of differentially expressed genes (fold change of at least ± 1.3; adjusted p-value < 0.05) between subsequent training periods: T1 vs. T2; T2 vs. T3 and T0 vs. T3. The most abundant genes up-regulated in exercise were involved in pathways that are important for the regulation of cell cycle (PI3K-Akt signalling pathway), cell communication (cAMP-dependent pathway), proliferation, differentiation and apoptosis, as well as immunity processes (Jak-STAT signalling pathway). We also observed the exercise-induced expression of genes related to regulation of the actin cytoskeleton, gluconeogenesis (FoxO signalling pathway, insulin signalling pathway), glycerophospholipid metabolism and calcium signalling. The identified genes belonged to the two most overrepresented pathways, the FoxO and PI3K-Akt signalling pathways, which are highlighted in Figs. 1 and 2. Moreover, an increase in the transcriptional activity of genes belonging to the Notch signalling pathway (KAT2B, NCOR2, NOTCH1, NOTCH2 and RBPJ) was identified (Table 4). Due to the relatively low number of detected down-regulated genes, only one pathway was found to be significant, and it contained genes overexpressed at the start of the training schedule (T1): hypertrophic cardiomyopathy (HCM) pathway (CACNB1, TPM1, TPM2, TTN).
Fig. 1.
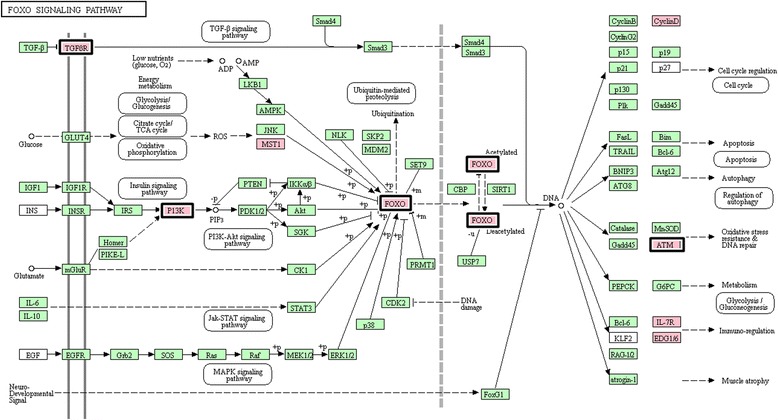
Significantly up-regulated genes during analysed training periods in Arabian horse blood that belong to the FoxO signalling pathway (KEGG ecb04068). Pathway presents the genes identified as differentially expressed (adjusted p-value < 0.05) between the T2 vs. T3 periods (bold highlight) and between T0 vs. T3 (pink highlight)
Fig. 2.
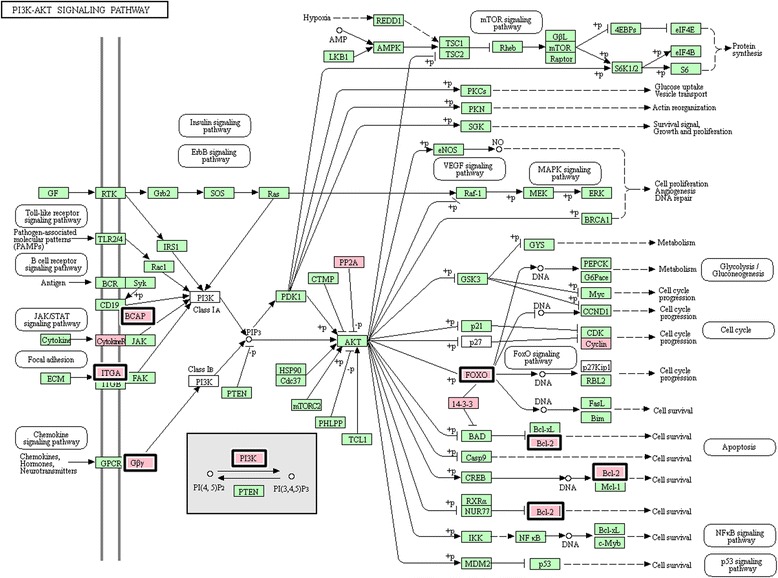
Significantly up-regulated genes during analysed training periods in Arabian horse blood that belong to the PI3K-Akt signalling pathway (KEGG ecb04151). Pathway presents the genes identified as differentially expressed (adjusted p-value < 0.05) between the T2 vs. T3 periods (bold highlight) and between T0 vs. T3 (pink highlight)
Table 4.
The significant exercise-regulated molecular pathways identified fallowing training in Arabian horses
| Pathway | Kegg number | Gene N | Gene symbol | p | Gene N |
Gene symbol | p | Gene N |
Gene symbol | p |
|---|---|---|---|---|---|---|---|---|---|---|
| Comparison | T1 vs T2 | T2 vs T3 | T0 vs T3 | |||||||
| Jak-STAT signaling pathway | ecb04630 | 4 | BCL2, IL6ST, PIK3CG, PTPN11, | 0.03 | 6 | BCL2, CCND2, IL6ST, IL7R, PIK3CG, PTPN11, | 0.01 | |||
| PI3K-Akt signaling pathway | ecb04151 | 6 | BCL2, FOXO3, GNBa, ITGA4, PIK3AP1, PIK3CG, | 0.01 | 11 | BCL2, CCND2, FOXO3, GNB4, IL7R, ITGA4, PIK3AP1, PIK3CG, PPP2R5E, YWHAG, GNGT2 | 0.001 | |||
| cAMP signaling pathway | ecb04024 | 2 | PDE3B, PIK3CG | 0.04 | 7 | ATP2B, CAMK4, NFATC1, PDE3B, PDE4A, PIK3CG, PPP1R12A | 0.003 | |||
| FoxO signaling pathway | ecb04068 | 4 | ATM, FOXO3, PIK3CG, TGFBR1, | 0.03 | 8 | ATM, CCND2, FOXO3, IL7R, STK4, PIK3CG, S1PR4, TGFBR1 | 0.001 | |||
| Glycerophospholipid metabolism | ecb00564 | 4 | AGPAT5, LGPAT1, GPD1, PLA2G4F | 0.001 | 4 | AGPAT3, AGPAT5, GPD2, LPGAT1, | 0.01 | 5 | AGPAT3, AGPAT5, GPD2, LPGAT1, PCYT1A | 0.002 |
| Insulin signaling pathway | ecb04910 | 4 | CBL, CBLB, PDE3B, PIK3CG | 0.02 | 5 | CBL, FASN, CBLB, PDE3B, PIK3CG | 0.02 | |||
| Calcium signaling pathway | ecb04020 | 2 | CYSLTR1, SLC25A4 | 0.01 | 3 | ITPR1, ITPR2, CYSLTR1 | 0.03 | 5 | ATP2A2, ATP2B4, CAMK4, ITPR1, ITPR2, | 0.03 |
| Regulation of actin cytoskeleton | ecb04810 | 1 | MYLPF | ns | 6 | ARHGEF6, IQGAP2, ITGA4, PAK2, PIK3CG, SSH2, | 0.01 | 8 | ARHGEF6, IQGAP2, ITGA4, MSN, PAK2, PIK3CG, SSH2, PPP1R12A | 0.02 |
| Notch signaling pathway | ecb04330 | 2 | KAT2B, RBPJ | ns | 5 | KAT2B, NCOR2, NOTCH1, NOTCH2, RBPJ | 0.001 | |||
| Hypertrophic cardiomyopathy (HCM) | ecb05410 | 4 | CACNB1, TPM1, TPM2, TTN, | 0.03 | 1 | ITGA4 | ns | 1 | ITGA4 | ns |
N number of identified genes, ns not significant; gene symbol indicated in bold were identified as down-regulated during training periods
Validation of results by qPCR
The validation of RNA-seq differential expression data was performed using a real-time PCR method for eight exercise-regulated genes. The significant positive correlation coefficients between normalized read counts (RNA-seq) and relative quantity results (real-time PCR) were observed for most of the analysed genes (Fig. 3). For six genes, significant differences in transcript levels were detected between training periods (CRYGS; LARP4; MBTD1; AGPAT5; LPGAT1; P2RY14). qPCR confirmed the increased expression of the P2RY14 and LPGAT1 genes during the training schedule when compared to untrained horses. For LARP4 and MBTD1, an increase in transcript levels during training was detected (from T1 to T3 periods), but the highest expression was identified in untrained horses. On the other hand, the expression level of the CRYGS gene decreased from the T1 to T3 training period, which was consistent with the RNA-seq results. The differences were not identified for the FOXN2 and ACVR2A genes; however, a trend similar to that of the RNA-seq data was observed (fold change values for both methods are presented in Additional file 5: Table S4). The scarce dissimilarities observed between the RNA-seq and qPCR data probably result from the incomplete annotation of the horse genome (for example, the presence of other, unknown gene splice variants).
Fig. 3.
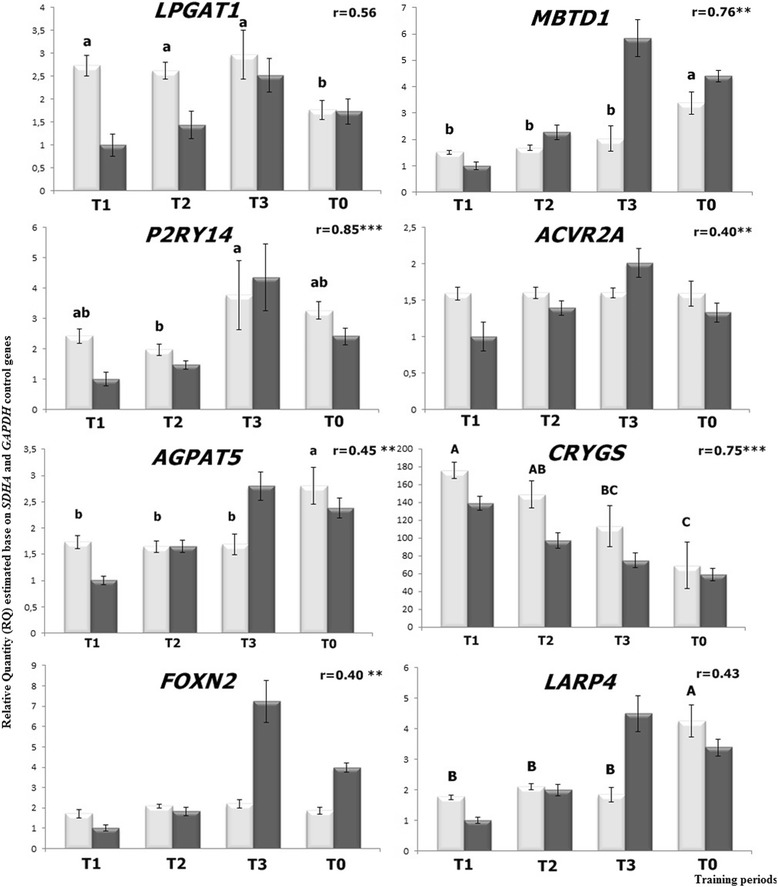
The exact gene expression levels estimated by real-time PCR (white column) and normalized counts detected by RNA-seq (grey column) for selected genes. The data are presented as mean ± standard error (training periods are T1: after the slow canter phase, T2: after the intense gallop phase, T3: at the end of the racing season; and T0 are untrained horses; the mean of Relative Quantity with different letters vary in significance with a,b: p < 0.05 and A,B: p < 0.01; r: correlation coefficient; * p < 0.05; ** p < 0.01; ***p < 0.001)
Discussion
Arabian horses are believed to be one of the oldest and most influential horse breeds in the world. The Arabian lineage was used to improve many breeds, such as the Thoroughbred and the Lipizzaner, and they are still contributing to the refinement of other breeds [21]. Compared to other breeds, Arabian horses are characterized by remarkable stamina and courage; thus, they dominate in endurance riding and racing. The racing performance traits in Thoroughbred horses have been widely described. On the other hand, the exact mechanisms that occur in the blood and muscles of the Arabian horses during exercise and how they are related to their skeletal muscle workload are still not well understood.
Arabians, when compared to TB, have a lower maximal rate of oxygen consumption (VO2max), lower running speed at VO2max, lower respiratory exchange ratio and higher free fatty acid consumption in substrate selection during low-intensity exercise [22]. Moreover, Arabian horses have a higher proportion of type I and IIa (oxidative) muscle fibres in comparison to Thoroughbreds, which tend to have more fibres that support ATP production via anaerobic metabolism [23–25]. These differences support the thesis that the performance traits of Thoroughbred and Arabian horses are conditioned in different ways [23, 25], but the genetic aspects of racing performance have not been established for Arabian horses.
In the present research, blood transcriptomes obtained from 18 Arabian horses were analysed using an NGS-based RNA-Seq approach. To identify the genetic basis of adaptive response to training, we compared global gene expression profiles between subsequent training periods and between two extreme points: horses in the last training phase and untrained horses. The analysis of overrepresented functional ontologies showed that the most abundant exercise-regulated transcripts formed a group of genes encoding transcription factors (34 DEGs for the T2 vs. T3 comparison and 43 DEGs for T0 vs. T3). This result was consistent with previous research performed on Thoroughbred horses before and after exercise [10]. The authors identified the differential expression of 91 transcription factors in equine blood and muscle tissues, which suggested that the identified genes are responsible for the regulation of exercise-triggered signalling pathways. The present study showed that a large number of DEGs encode proteins that are involved in nucleic acid binding, act as G-protein modulators or have kinase activity in Arabian horse blood during exercise. This result was probably an exercise-related response to restore body homeostasis through the activation and/or repression of many signalling pathways and adaptation to exercise.
During the training schedule, our results indicated a significant up-regulation of several pathways involved in cell cycle regulation, proliferation, differentiation and apoptosis (PI3K-Akt signalling and Jak-STAT signalling pathways), as well as cell-cell communication (cAMP-dependent and Notch signalling pathways) and cellular signalling (calcium signalling pathway). It has been established that exercise increases protein synthesis via activation of the PI3K-Akt signalling pathway, which regulates inter alia skeletal muscle hypertrophy [26, 27]. The PI3K-Akt acts by increasing protein synthesis though the activation of mTOR pathways and by inhibiting protein degradation through the inactivity of FOXO transcription factors [27, 28]. The present research also confirmed the significant impact of Arabian horse training on the overrepresentation of both the PI3K-Akt and FoxO signalling pathways, as represented inter alia by the differential expression of FOXO3 (Forkhead box protein O3), PIK3CG (Phosphoinositide-3-kinase PI3K), TGFBR1 (TGF-beta receptor type-1), BCL2 (Apoptosis regulator Bcl-2) and ATM (Serine-protein kinase ATM).
Comparisons of the T2 vs. T3 and T0 vs. T3 training periods showed exercise-induced expression of genes belonging to the Jak-STAT and cAMP signalling pathways; this expression was especially evident between the horses at the end of the training stage and the untrained horses (T0 vs. T3). In humans, Trenerry et al. [29] indicated that intensive exercise increased the levels of proteins involved in the JAK/STAT signalling pathway (interleukin-6, platelet-derived growth factor-BB), and the authors established that the investigated proteins are essential in adaptive responses to intensive training. Furthermore, it has been proven that cAMP signalling regulates the hypertrophic response in skeletal myofibres by increasing myofibre size and initiating fibre-type conversion [30–32]. In turn, in endurance horses, training induces the activity of oxidative fibres and decreases anaerobic metabolism, which is associated with an increase in the percentage of Type I fibres in muscle tissues [33]. During subsequent training stages, we observed an increase in the transcriptional activity of the PIK3CG gene (belonging to phosphoinositide-3-kinases, PI3Ks), which plays a critical role in most of the identified exercise-regulated pathways. In rats, Cheng et al. [34] showed that exercise enhances the protein levels of cardiac IGFI-R/PI3K/Akt pathways as well as the Bcl-2 family, including the BCL2 gene, which was also identified in our research. The significant function of PIK3 kinase during skeletal muscle development was confirmed in a mouse animal model [35]. According to the authors, PI3K promotes the role of myostatin (MSTN) in the modulation of cell survival. Based on the identified exercise-regulated pathways, it can be assumed that our results enabled detection of the molecular modifications that occur in horses and are the basis for metabolic changes in an organism’s adaptation to long-term training.
Moreover, the differential expression of genes related to metabolic processes that are critical to maintaining body homeostasis seems interesting. The results showed that the exercise-regulated genes belong to the FoxO and Insulin signalling pathways, which are essential for the glucose and glycerophospholipid metabolism pathways. In our study, eight genes from the FoxO signalling pathway were up-regulated in response to an increase in physical activity, including the FOXO3 (Forkhead Box O3) and TGFBR1 genes (Transforming Growth Factor, Beta Receptor 1). It has been proven that FoxO transcription factors play a key role in the regulation of energy homeostasis in muscle tissue during catabolic conditions [36]. Furthermore, FoxO3 can increase mitochondrial respiration by binding to the mitochondrial DNA-regulatory regions, and both FoxO1 and FoxO3 can regulate the two main proteolytic pathways (ubiquitin–proteasome and autophagy–lysosome pathways) [37]. The recent study performed by Sanchez [38] confirmed the significant function of FoxO factors in response to endurance training through the regulation of angiogenetic processes in muscle. The increased expression of the Forkhead family of transcription factors (FOXOs) that was observed in our research suggests that these genes also play an important role in adaptation to exercise in horses.
One of the most important groups of DEGs associated with training in Arabian horses are involved in metabolic processes, particularly in energy production (glycerophospholipid metabolism and insulin signalling pathways). Homeostasis between the utilization of glucose and lipids during intensive exercise is critical for the optimization of training efficiency. It has been shown that, in comparison to Thoroughbreds, the muscle tissue of Arabian horses is characterized by a higher proportion of slow twitch (Type I) muscle fibres [24; 25], which are characterized by a low glycogen content and a high reserve of triglycerides. The increased request of lipids during training can be confirmed by the observed up-regulation of genes belonging to the glycerophospholipid metabolism pathway in our results. The improved efficiency of fatty acid utilization was particularly evident between horses in the last training period and untrained horses, where five significant genes were related to lipids metabolism: AGPAT3, AGPAT5, GPD2, LPGAT1, and PCYT1A. Two of these genes belonged to the AGPAT family (acylglycerolphosphate acyltransferase), which catalyses an important step in triacylglycerol (TAG) synthesis and storage [39]. The glycerol-3-phosphate dehydrogenase 2 gene (GDP2) encodes the mitochondrial protein responsible for glycerone phosphate synthesis and is an element of glycerol degradation via the glycerol kinase pathway [40]. In turn, Choline-phosphate cytidylyltransferase A (PCYT1A) controls phosphatidylcholine biosynthesis, while lysophosphatidylglycerol acyltransferase 1 (LPGAT1) takes a part in the glycerophospholipid biosynthetic process [41].
Furthermore, our results pinpointed few transcripts that were affected by exercise and classified as part of the insulin signalling pathway (CBL, FASN, CBLB, PDE3B, PIK3CG – PI3Ks). Some of these genes control adipogenesis (FASN; fatty acid synthase), while others are considered negative regulators of many pathways, including the insulin signalling pathway (E3 ubiquitin-protein ligases: CBL and CBLB) [42]. Kim et al. [43] confirmed that endurance training increased the expression levels of several genes belonging to the insulin pathway (also PI3-kinase; Phosphoinositide 3-kinases) in rat skeletal muscles and suggested that this kind of exercise can be associated with alterations in insulin sensitivity. In 2002, the research performed on mice established that the increase in PI3-kinase activity after exercise resulted in the enhancement of insulin activity in skeletal muscles [44]. To date, the significant role of PI3Ks in insulin-mediated glucose uptake following training has been confirmed in humans and other species [45]. In Thoroughbred horses, McGivenly et al. [6] detected the up-regulation of insulin and Type II diabetes mellitus pathways after short-term exercise and proposed that these changes were transcriptional adaptations to training. Our results also highlighted the significance of molecular mechanisms for modulating glucose uptake and lipid metabolism. These mechanisms are essential for maintaining body homeostasis during long-term exercise in Arabian horses.
Regular training is associated with the increased expression of antioxidant enzymes and the activation of inflammatory cytokines [46], which are generally recognized by the organism as stress factors [4]. One of the most important adaptive responses to exercise is changes in gene expression that are responsible for metabolic modification and cell cycle progression, which ultimately result in a return to homeostasis. In Arabian endurance horses, Cappaelli et al. [20] identified four genes that had high sequence similarity to genes involved in a training-induced stress response: IL8 (interleukin 8), EIF4G3 (eukaryotic translation factor 4 gamma, 3), RBBP6 (retinoblastoma binding protein 6) and HSP90AA1 (heat shock protein). In 2010, Capomaccio et al. [12] showed the differential expression of several genes involved in inflammation in horses participating in endurance races. Global gene expression profiles of the blood were analysed in three periods (pre-, during and post-race). Among the 20 identified genes with the highest fold change, the authors detected three interleukins (IL18; IL8; IL1R2), three chemokines (CXCL2; CCR2; CCL5) and an interferon receptor 1 (IFNAR1). Similarly, Capomaccio et al. [11] compared the blood transcriptomes at two time points, at rest and immediately at the end of the race, and showed exercise-induced deregulation of pro-inflammatory molecules, such as IL8, IL18, CCR2, TLR1, IL1R1, CXCL1, CCL5 and integrins (ITGAL, ITGAM). In the present study, we attempted to estimate the influence of long-term exercise periods on gene expression patterns in the blood of Arabian horses. A significant up-regulation of interleukins (IL6ST; IL6R; IL7R) and integrin (ITGA4) was observed. The increased transcript abundance of acute-phase response molecules confirmed that an organism recognizes long-term training as a stress factor and that these inflammatory responses play an important function in triggering downstream signalling pathways, which are critical to the maintenance of homeostasis. Thus, understanding the molecular basis of exercise-induced stress can be helpful for identifying performance markers in horses.
Application of an RNA-seq method based on next-generation sequencing allowed us to detect a panel of uncharacterized DEGs (51 genes) with a potentially important function in exercise adaptation. Some of these genes were differentially expressed in more than two training periods, and orthologue analysis showed that these genes mainly encoded transcription factors, defence/immunity proteins, and proteins that bind nucleic acids and receptors. Similar results were presented by Park et al. [10], who analysed exercise-induced changes in the blood transcriptome of TB horses and detected 56 uncharacterized genes that code for transcription factors. Our approach provided a panel of candidate genes that may be responsible for adaptive training responses in horses; these genes should be investigated in the future.
Conclusions
Our results allowed for the identification of changes in gene expression profiles during a training schedule in Arabian horses. Based on a comparative analysis of blood transcriptomes, several exercise-regulated pathways and genes affected by exercise were detected. We pinpointed the overrepresented molecular pathways and genes that are essential for an exercise-related adaptive response for maintaining body homeostasis. The observed transcriptional activation of such genes as LPGAT1, AGPAT5, PIK3CG, GPD2, FOXN2, FOXO3, ACVR1B and ACVR2A can form the basis for further research to identify the genes that are potentially associated with racing performance in Arabian horses. Such markers may be essential for choosing a training regimen, and they can reveal what causes the differences in racing performance that are specific to various breeds.
Additional files
The training procedure and points of sample collection. (DOC 212 kb)
Primer sequences used in real-time qPCR validation. (DOC 40 kb)
The basic NGS data statistics for the analysed samples. (DOC 71 kb)
Identified gene orthologues with differential expression between analysed training periods. (DOC 112 kb)
The fold-change values obtained from comparisons between subsequent training periods using RNA-seq and qPCR methods. (DOC 38 kb)
Acknowledgments
Funding
The study was supported by the Polish Ministry of Science and Higher Education (project no. 2014/15/D/NZ9/05256).
Availability of data and material
All the raw data are available in GEO database (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE83404).
Authors’ contributions
KRM contributed to all steps, designed the study with MSS and KP, analyzed the data, and drafted the manuscript. MSS contributed to study design, analysis of the data, and writing and critical revision of the manuscript. KŻ contributed to data analysis and critical review of the manuscript. KP contributed to data collection and validation as well as critical review of the manuscript. AG contributed to NGS analysis, data collection and review of the manuscript. MBP contributed to data analysis and validation, and critical review of the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval
The protocol was approved by the Animal Care and Use Committee of the Institute of Pharmacology, Polish Academy of Sciences in Cracow (no. 1173/2015). The horses’ owners gave consent to be part of this study.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- DEGs
Differentially expressed genes
- TB
Thoroughbred horses
- PASB
Polish Arabian Stud Book
- RIN
RNA integrity number
- QC
Quality control
- GO
Gene Ontology
- VO2max
Maximal rate of oxygen consumption
- KEGG
Kyoto Encyclopedia of Genes and Genomes
Contributor Information
Katarzyna Ropka-Molik, Phone: +48 666 081 208, Email: katarzyna.ropka@izoo.krakow.pl.
Monika Stefaniuk-Szmukier, Email: m.k.stefaniuk@gmail.com.
Kacper Żukowski, Email: kacper.zukowski@izoo.krakow.pl.
Katarzyna Piórkowska, Email: katarzyna.piorkowska@izoo.krakow.pl.
Artur Gurgul, Email: artur.gurgul@izoo.krakow.pl.
Monika Bugno-Poniewierska, Email: monika.bugno@izoo.krakow.pl.
References
- 1.Holloszy JO, Coyle EF. Adaptations of skeletal muscle to endurance exercise and their metabolic consequences. J Appl Physiol. 1984;56:831–38. doi: 10.1152/jappl.1984.56.4.831. [DOI] [PubMed] [Google Scholar]
- 2.Jacobs RA, Rasmussen P, Siebenmann C, Diaz V, Gassmann M, Pesta D, Gnaiger E, Nordsborg NB, Robach P, Lundby C. Determinants of time trial performance and maximal incremental exercise in highly trained endurance athletes. J Appl Physiol. 2011;111:1422–30. doi: 10.1152/japplphysiol.00625.2011. [DOI] [PubMed] [Google Scholar]
- 3.Budgett R. Fatigue and underperformance in athletes: the overtraining syndrome. Br J Sports Med. 1998;32:107–10. doi: 10.1136/bjsm.32.2.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Purvis D, Gonsalves S, Deuster PA. Physiological and psychological fatigue in extreme conditions: overtraining and elite athletes. PM&R. 2010;2(5):442–450. doi: 10.1016/j.pmrj.2010.03.025. [DOI] [PubMed] [Google Scholar]
- 5.Bouwman FG, van Ginneken MM, Noben JP, Royackers E, de Graaf-Roelfsema E, Wijnberg ID, van der Kolk JH, Mariman EC, van Breda E. Differential expression of equine muscle biopsy proteins during normal training and intensified training in young standard bred horses using proteomics technology. Comp Biochem Physiol Part D Genomics Proteomics. 2010;5(1):55–64. doi: 10.1016/j.cbd.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 6.McGivney BA, Eivers SE, McHugh DE, MacLeod JN, O’Gorman GM, Park SDE, Katz LM, Hill EM. Transcriptomal adaptations following exercise in Thoroughbred horse skeletal muscle highlights molecular mechanisms that led to muscle hypertrophy. BMC Genomics. 2009;10:638. doi: 10.1186/1471-2164-10-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McGivney BA, McGettigan PA, Browne JA, Evans AC, Fonseca RG, Loftus BJ, Lohan A, MacHugh DE, Murphy BA, Katz LM, Hill EW. Characterization of the equine skeletal muscle transcriptome identifies novel functional responses to exercise training. BMC Genomics. 2010;11:398. doi: 10.1186/1471-2164-11-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hill EW, Gu J, Eivers SS, Fonseca RG, McGivney BA, Govindarajan P, Orr N, Katz LM, MacHugh DE. A sequence polymorphism in MSTN predicts sprinting ability and racing stamina in thoroughbred horses. PLoS One. 2010a; 5(1): e8645. doi: 10.1371/journal.pone.0008645 PMID: 20098749 [DOI] [PMC free article] [PubMed]
- 9.Hill EW, McGivney BA, Gu J, Whiston R, Machugh DE. A genome-wide SNP-association confirms a sequence variants (g.66493737C > T) in the equine myostatin (MSTN) gene at the most powerful predictor of optimum racing distance for Thoroughbred racehorses. BMC Genomics. 2010b;11,552. doi: 10.1186/1471-2164-11-552 PMID: 20932346 [DOI] [PMC free article] [PubMed]
- 10.Park KD, Park J, Ko J, Kim BC, Kim HS, Ahn K, Do KT, Choi H, Kim HM, Song S, Lee S, Jho S, Kong HS, Yang YM, Jhun BH, Kim C, Kim TH, Hwang S, Bhak J, Lee HK, Cho BW. Whole transcriptome analyses of six thoroughbred horses before and after exercise using RNA-seq. BMC Genomics. 2012;13:473. doi: 10.1186/1471-2164-13-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Capomaccio S, Vitulo N, Verini-Supplizi A, Barcaccia G, Albiero A, D’Angelo M, Campagna D, Valle G, Felicetti M, Silvestrelli M, Cappelli K. RNA sequencing of the exercise transcriptome in equine athletes. PLoS One. 2013;8(12):e83504. doi: 10.1371/journal.pone.0083504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Capomaccio S, Cappelli K, Barrey E, Felicetti M, Silvestrelli M, Verini-Supplizi A. Microarray analysis after strenuous exercise in peripheral blood mononuclear cells of endurance horses. Anim Genet. 2010;41(2):166–75. doi: 10.1111/j.1365-2052.2010.02129.x. [DOI] [PubMed] [Google Scholar]
- 13.Bickel CS, Slade J, Mahoney E, Haddad F, Dudley GA, Adams GR. Time course of molecular responses of human skeletal muscle to acute bouts of resistance exercise. J Appl Physiol. 2005;98(2):482–88. doi: 10.1152/japplphysiol.00895.2004. [DOI] [PubMed] [Google Scholar]
- 14.Dodt M, Roehr JT, Ahmed R, Dieterich C. FLEXBAR—Flexible Barcode and Adapter Processing for Next-Generation Sequencing Platforms. Biology. 2012;1(3):895–905. doi: 10.3390/biology1030895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Love M, Anders S, Huber W. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mi H, Poudel S, Muruganujan A, Casagrande JT, Thomas PD. PANTHER version 10: expanded protein families and functions, and analysis tools. Nucleic Acids Res. 2016;44:336–42. doi: 10.1093/nar/gkv1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mutch DM, Berger A, Mansourian R, Rytz A, Roberts MA. The limit fold change model: a practical approach for selecting differentially expressed genes from microarray data. BMC Bioinformatics. 2002;3:17. doi: 10.1186/1471-2105-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cappelli K, Felicetti M, Capomaccio S, Spinsanti G, Silvestrelli M, Supplizi AV. Exercise induced stress in horses: selection of the most stable reference genes for quantitative RT-PCR normalization. BMC Mol Biol. 2008;9:49. doi: 10.1186/1471-2199-9-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zechner P, Sölknera J, Bodob I, Drumla T, Baumunga R, Achmannc R, Martid E, Habee F, Bremc G. Analysis of diversity and population structure in the Lipizzan horse breed based on pedigree information. Livest Prod Sci. 2002;77(2–3):137–46. doi: 10.1016/S0301-6226(02)00079-9. [DOI] [Google Scholar]
- 22.Prince A, Geor R, Harris P, Hoekstra K, Gardner S, Hudson C, Pagan J. Comparison of the metabolic responses of trained Arabians and Thoroughbreds during high- and low intensity exercise. Equine Vet J Suppl. 2002;34:95–9. doi: 10.1111/j.2042-3306.2002.tb05398.x. [DOI] [PubMed] [Google Scholar]
- 23.JL L–R, Agüera E, Monterde JG, Rodríguez-Barbudo MV, Miró F. Comparative study of muscle fiber type compositions in the middle gluteal muscle of andalusioan, Thoroughbred and Arabian horses. JEquine VetSci. 1989;9:337–340. [Google Scholar]
- 24.Snow DH, Valberg SJ. Muscle anatomy: Adaptations to exercise and training. In: The Athletic Horse: Principles and Practice of Equine Sports Medicine, Sanders, 1994; New York. Rose et al. (eds.) pp 145–179.
- 25.Lopez-Rivero L, Letelier A. Skeletal muscle profile of show jumpers: Physiological and pathological considerations in the elite show jumper. In: The Elite Showjumper: Proceedings of the Conference on Equine Sports Medicine and Science, 2000; Ed: A. Lindner. pp 57–76.
- 26.Bodine SC, Stitt TN, Gonzalez M, Kline WO, Stover GL, Bauerlein R, Zlotchenko E, Scrimgeour A, Lawrence JC, Glass DJ, Yancopoulos GD. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat Cell Biol. 2001;3:1014–19. doi: 10.1038/ncb1101-1014. [DOI] [PubMed] [Google Scholar]
- 27.Camera DM, Edge J, Short MJ, Hawley JA, Coffey VG. Early time course of Akt phosphorylation after endurance and resistance exercise. Med Sci Sports Exerc. 2010;42:1843–52. doi: 10.1249/MSS.0b013e3181d964e4. [DOI] [PubMed] [Google Scholar]
- 28.Sandri M, Sandri C, Gilbert A, Skurk C, Calabria E, Picard A, Walsh K, Schiaffino S, Lecker SH, Goldberg AL. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 2004;117(3):399–12. doi: 10.1016/S0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trenerry MK, Della Gatta PA, Larsen AE, Garnham AP, Cameron-Smith D. Impact of resistance exercise training on interleukin-6 and JAK/STAT in young men. Muscle Nerve. 2011;43(3):385–92. doi: 10.1002/mus.21875. [DOI] [PubMed] [Google Scholar]
- 30.Chen M, Feng HZ, Gupta D, Kelleher J, Dickerson KE, Wang J, Hunt D, Jou W, Gavrilova O, Jin JP, Weinstein LS. G(s)alpha deficiency in skeletal muscle leads to reduced muscle mass, fiber-type switching, and glucose intolerance without insulin resistance or deficiency. Am J Physiol Cell Physiol. 2009;296(4):C930–40. doi: 10.1152/ajpcell.00443.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Minetti GC, Feige JN, Rosenstiel A, Bombard F, Meier V, Werner A, Bassilana F, Sailer AW, Kahle P, Lambert C, Glass DJ, Fornaro M. Gαi2 signaling promotes skeletal muscle hypertrophy, myoblast differentiation, and muscle regeneration. Sci Signal. 2011;4(201):ra80. doi: 10.1126/scisignal.2002038. [DOI] [PubMed] [Google Scholar]
- 32.Berdeaux R, Stewart R. cAMP signaling in skeletal muscle adaptation: hypertrophy, metabolism, and regeneration. Am J Physiol Endocrinol Metab. 2012;03(1):E1–E17. doi: 10.1152/ajpendo.00555.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lopez-Rivero JL, Morales-Lopez JL, Galisteo AM, Aguera E. Muscle fibre type composition in untrained and endurance-trained Andalusian and Arab horses. Equine Vet J. 1991;23(2):91–3. doi: 10.1111/j.2042-3306.1991.tb02727.x. [DOI] [PubMed] [Google Scholar]
- 34.Cheng SM, Ho TJ, Yang AL, Chen IJ, Kao CL, Wu FN, Lin JA, Kuo CH, Ou HC, Huang CY, Lee SD. Exercise training enhances cardiac IGFI-R/PI3K/Akt and Bcl-2 family associated pro-survival pathways in streptozotocin-induced diabetic rats. Int J Cardiol. 2013;167(2):478–85. doi: 10.1016/j.ijcard.2012.01.031. [DOI] [PubMed] [Google Scholar]
- 35.Chelh I, Meunier B, Picard B, Reecy MJ, Chevalier C, Hocquette JF, Cassar-Malek I. Molecular profiles of Quadriceps muscle in myostatin-null mice reveal PI3K and apoptotic pathways as myostatin targets. BMC Genomics. 2009;10:196. doi: 10.1186/1471-2164-10-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanchez AM, Candau RB, Bernardi H. FoxO transcription factors: their roles in the maintenance of skeletal muscle homeostasis. Cell Mol Life Sci. 2014a;71:1657–71. doi:10.1007/s00018-013-1513-z. PMID: 24232446. [DOI] [PMC free article] [PubMed]
- 37.Sanchez AM, Bernardi H, Py G, Candau R. Autophagy is essential to support skeletal muscle plasticity in response to endurance exercise. Am J Physiol Regul Integr Comp Physiol. 2014b;307:R956–69. doi:10.1152/ajpregu.00187.2014. PMID: 25121614. [DOI] [PubMed]
- 38.Sanchez AM. FoxO transcription factors and endurance training: a role for FoxO1 and FoxO3 in exercise-induced angiogenesis. J Physiol. 2015;593:363–364. doi: 10.1113/jphysiol.2014.285999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takeuchi K, Reue K. Biochemistry, physiology, and genetics of GPAT, AGPAT, and lipin enzymes in triglyceride synthesis. Am J Physiol Endocrinol Metab. 2009;296(6):E1195–E1209. doi: 10.1152/ajpendo.90958.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Geertman JM, van Maris AJ, van Dijken JP, Pronk JT. Physiological and genetic engineering of cytosolic redox metabolism in Saccharomyces cerevisiae for improved glycerol production. Metab Eng. 2006;8(6):532–42. doi: 10.1016/j.ymben.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 41.Yang Y, Cao J, Shi Y. Identification and characterization of a gene encoding human LPGAT1, an endoplasmic reticulum associated lysophosphatidylglycerol acyltransferase. J Biol Chem. 2004;279(53):55866–74. doi: 10.1074/jbc.M406710200. [DOI] [PubMed] [Google Scholar]
- 42.Nakao R, Hirasaka K, Goto J, Ishidoh K, Yamada C, Ohno A, Okumura Y, Nonaka I, Yasutomo K, Baldwin KM, Kominami E, Higashibata A, Nagano K, Tanaka K, Yasui N, Mills EM, Takeda S, Nikawa T. Ubiquitin ligase Cbl-b is a negative regulator for insulin-like growth factor 1 signaling during muscle atrophy caused by unloading. Mol Cell Biol. 2009;29(17):4798–811. doi: 10.1128/MCB.01347-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim YB, Inoue T, Nakajima R, Shirai-Morishita Y, Tokuyama K, Suzuki M. Effect of long-term exercise on gene expression of insulin signaling pathway intermediates in skeletal muscle. Biochem Biophys Res Commun. 1999;254(3):720–27. doi: 10.1006/bbrc.1998.9940. [DOI] [PubMed] [Google Scholar]
- 44.Howlett KF, Sakamoto K, Hirshman MF, Aschenbach WG, Dow M, White MF, Goodyear LJ. Insulin signaling after exercise in insulin receptor substrate-2-deficient mice. Diabetes. 2002;51(2):479–83. doi: 10.2337/diabetes.51.2.479. [DOI] [PubMed] [Google Scholar]
- 45.Hawley JA, Lessard SJ. Exercise training-induced improvements in insulin action. Acta Physiol. 2008;192:127–35. doi: 10.1111/j.1748-1716.2007.01783.x. [DOI] [PubMed] [Google Scholar]
- 46.Ennezat PV, Malendowicz SL, Testa M, Colombo PC, Cohen-Solal A, Evans T, LeJemtel TH. Physical training in patients with chronic heart failure enhances the expression of genes encoding antioxidative enzymes. J Am Coll Cardiol. 2001;38(1):194–98. doi: 10.1016/S0735-1097(01)01321-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The training procedure and points of sample collection. (DOC 212 kb)
Primer sequences used in real-time qPCR validation. (DOC 40 kb)
The basic NGS data statistics for the analysed samples. (DOC 71 kb)
Identified gene orthologues with differential expression between analysed training periods. (DOC 112 kb)
The fold-change values obtained from comparisons between subsequent training periods using RNA-seq and qPCR methods. (DOC 38 kb)


