Abstract
Phage display technology provides a powerful tool to screen a library for a binding molecule via an enrichment process. It has been adopted as a critical technology in the development of therapeutic antibodies. However, a major drawback of phage display technology is that because the degree of the enrichment cannot be controlled during the bio-panning process, it frequently results in a limited number of clones. In this study, we applied next-generation sequencing (NGS) to screen clones from a library and determine whether a greater number of clones can be identified using NGS than using conventional methods. Three chicken immune single-chain variable fragment (scFv) libraries were subjected to bio-panning on prostate-specific antigen (PSA). Phagemid DNA prepared from the original libraries as well as from the Escherichia coli pool after each round of bio-panning was analyzed using NGS, and the heavy chain complementarity-determining region 3 (HCDR3) sequences of the scFv clones were determined. Subsequently, through two-step linker PCR and cloning, the entire scFv gene was retrieved and analyzed for its reactivity to PSA in a phage enzyme immunoassay. After four rounds of bio-panning, the conventional colony screening method was performed for comparison. The scFv clones retrieved from NGS analysis included all clones identified by the conventional colony screening method as well as many additional clones. The enrichment of the HCDR3 sequence throughout the bio-panning process was a positive predictive factor for the selection of PSA-reactive scFv clones.
Introduction
One of the most important products on the therapeutic recombinant protein market is the monoclonal antibody. More than 54 therapeutic antibodies have been approved for various indications, including cancer and autoimmune diseases.1 Traditionally, therapeutic antibodies have been generated by mouse B-cell hybridoma technology followed by chimerization or humanization.2 In the past few decades, technologies such as transgenic mice encompassing human antibody gene repertoires, and phage display of antibody libraries, have become available, facilitating the rapid flourishing of therapeutic antibodies in the drug discovery field.3
Phage display technology frequently allows the creation of libraries containing up to 1011 different variants, which can be used to screen antibody clones by bio-panning.4 Despite the development of alternative display technologies such as bacterial display, yeast display and ribosome display, phage display remains the most widely used display technology due to the robustness of the filamentous bacteriophage M13.5 Several therapeutic antibodies that are currently either approved or in clinical trials have been developed by phage display technology.6, 7
Recently, next-generation sequencing (NGS) technology has allowed a massive increase in capacity to sequence genomes at relatively low cost and in a short time frame.8 It has revolutionized multiple aspects of biological research5 and is also actively being adopted into antibody phage display technology. Several NGS platforms are currently available, with average read lengths of 75–8500 bp and different error rates.9 The CDR3 sequence of the VH and VL genes has been effectively determined by the MiSeq system;10 a single-domain antibody gene was successfully determined by the MiSeq system using a 2 × 250 paired-end module;11 and the entire VH gene was successfully sequenced using the 454 pyrosequencing system.12 However, sequencing of the entire single-chain variable fragment (scFv) gene, which contains 750–800 bases, could not be achieved using any of these NGS platforms, to the extent of the authors' knowledge. In one study, to obtain the whole scFv gene sequence, HCDR3 sequences were first determined by the MiSeq system; the entire scFv gene was then generated by two-step linker PCR using primers based on the heavy chain complementarity-determining region 3 (HCDR3) sequences, and its sequence was determined by Sanger sequencing analysis.6 In another similar study, HCDR3/FR4 sequences were determined from Ion Torrent PGM sequence analysis using the 318 chip. Then, the entire scFv gene was retrieved by inverse PCR using primers based on the HCDR3/FR4 sequences.13
Following NGS analysis, the antibody gene is typically cloned and expressed. And the binding reactivity of the antibody to its target as well as its biological activity are tested. However, this may prove to be unproductive when the fraction of positive clones is not high following bio-panning. It has been extensively reported that positive clones tend to be enriched through bio-panning and negative clones show the opposite tendency. Therefore, NGS analysis of clones after each round of bio-panning could provide insights on which clones are more likely to be positive. Furthermore, it is unknown whether there is a difference between scFv clones identified by conventional colony screening methods14 and those obtained from NGS.
In this study, we have attempted to answer these questions. We performed four rounds of bio-panning using three scFv libraries constructed from prostate-specific antigen (PSA)-immunized chickens. We then performed NGS analysis of scFv clones focusing on HCDR3 in the initial scFv library and in four enriched scFv libraries obtained from subsequent rounds of bio-panning. scFv clones were obtained after the last round of bio-panning using the conventional colony screening method from the output titer plate, or from phagemid DNA prepared following a previously reported procedure.6 The reactivity of these scFv clones was measured using a phage enzyme immunoassay. Based on these experiments, the sequences obtained using NGS and the conventional colony screening method were compared. We also classified scFv clones obtained from NGS into 3–4 clusters based on their enrichment or impoverishment patterns, and analyzed these patterns for clues regarding the reactivity of scFv clones.
Materials and methods
Library construction and bio-panning
Three white leghorn chickens were immunized and boosted four times with recombinant human PSA (Fitzgerald, Acton, MA, USA). After the final booster injection, total RNA was extracted from the spleen, bone marrow, and bursa of Fabricius using the TRI Reagent (Invitrogen, Grand Island, NY, USA). First-strand cDNA was synthesized using SuperScript reverse transcriptase with oligo (dT) priming (Invitrogen). Using this cDNA, three phage-displayed libraries of chicken scFvs were constructed using the pComb3XSS phagemid vector, as described previously.14 Four rounds of bio-panning were performed to screen scFv clones from the library following a previously reported procedure.15 For each round of bio-panning, 5 × 106 magnetic beads (Dynabeads M-270 epoxy) (Invitrogen) coated with 1.5 μg recombinant PSA protein were used.
Phage enzyme immunoassay
The scFv-displaying phages were rescued from titer plates after transformation and subjected to phage enzyme immunoassay as described previously.14 The microtiter plates (Corning, NY, USA) were coated overnight at 4 °C with 20 μl recombinant human Fc-tagged PSA (5 μl ml−1) dissolved in phosphate-buffered saline (PBS). After blocking with 3% bovine serum albumin dissolved in PBS (w/v, PBS-B), the plates were then sequentially incubated with scFv-displaying phages in the culture supernatant, horseradish peroxidase (HRP)-conjugated mouse anti-M13 monoclonal antibody (GE Healthcare, Pittsburg, PA, USA) in PBS-B, and then finally with 2,2′-Azinobis [3-ethylbenzothiazoline-6-sulfonic acid]-diammonium salt (ABTS) substrate solutions (Amresco LLC, Solon, OH, USA), with intermittent washing using 0.05% Tween-20 in PBS (PBST). After incubating the plates at 37 °C for 10 min, the optical density was measured at 405 nm using a microtiter plate reader (Labsystems AiG SL, Barcelona, Spain).
Sanger sequencing analysis
Phagemid DNA from selected clones identified by phage enzyme immunoassays was prepared with a small-scale plasmid preparation kit (Qiagen, Hilden, Germany). The OmpSeq primer (5′-AAGACAGCTATCGCGATTGCAG-3′) and HRML-F primer (5′-GGTGGTTCCTCTAGATCTTCC-3′) were used to sequence the VH and VL chains of the antibody.14 Sequence analysis of positive clones (O.D.405nm>0.3) was performed by Macrogen (Seoul, Korea).
Next-generation sequencing analysis
NGS analysis was performed as described previously.16 A total of 15 sets of phagemid DNA including three initial chicken scFv libraries and three libraries obtained after each of four rounds of bio-panning were analyzed using a MiSeq system (Illumina Inc., San Diego, CA, USA). The MiSeq library for DNA sequencing was prepared using Illumina Nextera XT chemistry (Illumina) following the protocol provided by the manufacturer. The genes from the chicken library were amplified using the forward primer (pre-adaptor, 5′-TCGTCGGCAGCGTC-3′ sequencing primer, 5′-AGATGTGTATAAGAGACAG-3′ specific locus primer, 5′-TCAGCCTCGTCTGCAAGG-3′), and reverse primer (pre-adaptor, 5′-GTCTCGTGGGCTCGG-3′ sequencing primer, 5′-AGATGTGTATAAGAGACAG-3′ specific locus primer, 5′-AGTGGAGGAGACGATGACTTC-3′), respectively. The final libraries were normalized by quantification with LightCycler 480 II (Roche Applied Science, Indianapolis, IN, USA) and qualification with Bioanalyzer (Agilent, Palo Alto, CA, USA). The final loading concentration was adjusted to 11 pM following the MiSeq loading protocol. The MiSeq reagent kit v3 (Illumina) was used for long paired-end reads (2 × 300 bp) sequencing reactions. The sequencing data was processed by CLC Genomics Workbench version 5 (CLC Bio, Aarhus, Denmark) software. Low-quality sequencing data were first trimmed depending on quality scores using PHRED with the minimum quality score of 20 and reads with less than 150 bases in length were discarded.17 The cleaned-up sequencing data were processed by merging the paired-end sequence reads using fast length adjustment of short reads to obtain complete sequences of the chicken scFv libraries.18 Sequencing data were further cleaned up using PRINSEQ (San Diego State University, San Diego, CA, USA), setting the minimum quality score at 20 and read length at 150.17 EMBOSS Needle 6.5.0.0 (The European Bioinformatics Institute (EMBL-EBI), UK) was used to map sequence read in the HCDR3 region, with a threshold score of 300.19 Subsequently, a custom Perl script was used to summarize and count sequence reads in 15 sets of phagemid DNA. We merged the read counts across all the panning rounds, but for computational and statistical analysis, we only counted the reads existing in the phagemid DNA after the fourth bio-panning round.
Clustering analysis
An optimized number of clusters in the merged sequence read counts was estimated using the clValid algorithm, to facilitate pattern analysis of NGS data for population shifts in antibody clones throughout the bio-panning process.20 The clValid algorithm validated number of clusters by assessing intra-cluster homogeneity and inter-cluster separation, and the assessment for each and every clustering is represented in the Dunn index.20 A higher Dunn index indicates better clustering. The ‘Internal' cluster validation metrics were chosen, which consider only the data set and the clustering partition, and the intrinsic properties of the data were used to evaluate the quality of the clustering results in designated clustering algorithms such as hierarchical clustering and k-mean clustering.21 Unsupervised hierarchical clustering analysis was used to cluster HCDR3 sequences according to the number of clusters estimated by clValid. Ward's method was used to measure distances between sequence reads based on read counts throughout the bio-panning, and a heat map visualizing the sequence read changes in each cluster was generated using Gene Pattern v3.9.2 software.22 Line charts representing the pattern of sequence read changes in each cluster across all the bio-panning rounds were then generated as in a previous study.
Cloning to retrieve scFvs
To rebuild real scFv clones from the virtual HCDR3 sequences in the clusters, we performed two-step linker PCR. In the first PCR step, primers targeting both LFR1-HCDR3 (LFR1_F primer, 5′-GTGGCCCAGGCGGCCCTG-3′) and HCDR3-HFR4 fragments (HFR4_R primer, 5′-CTGGCCGGCCTGGCCACT-3′) were synthesized, based on HCDR3 sequences determined in NGS analysis and phagemid DNA obtained after the 4th round of bio-panning. The second PCR step linked these two gene fragments into a single scFv gene using primers annealing to LFR1 and HFR4 (LFR1_F primer, 5′-GTGGCCCAGGCGGCCCTG-3′ HFR4_R primer, 5′-CTGGCCGGCCTGGCCACT-3′). The scFv gene was ligated into the pComb3XSS phagemid vector and rescued as scFv-displaying phages, as described previously.14 To measure the binding reactivity of these scFv-displaying phages, we rescued more than 15 clones per HCDR3 sequence, and performed phage enzyme immunoassay as described earlier. We regarded the clone providing the highest optical density at 405 nm as the retrieved clone.
Statistical analysis
Statistical analysis was performed with GraphPad Prism 5 software. Specific P-values and statistical methods are provided in the figure legends.
Results
Generation of antibody library and screening for positive clones using the conventional colony screening method
Using mRNA prepared from spleen, bone marrow, and bursa of Fabricius from three PSA-immunized chickens, we generated scFv libraries with complexities of 6.09 × 1010, 3.64 × 1010 and 5.16 × 1010 clones, respectively, referred to as chicken libraries 1, 2 and 3. Next, we performed four rounds of bio-panning, rescued phage clones from the output titer plate of the fourth round, and performed a phage enzyme immunoassay to screen for positive clones. A total of 300 clones (100 clones in each library) exhibiting an optical density of >0.3 at 405 nm were considered to be positive, and their scFv gene sequence was determined by Sanger sequencing analysis. We finally obtained 22 clones with unique HCDR3 sequences (Table 1).
Table 1. HCDR3 amino-acid sequences selected using the conventional colony screening method, and binding reactivity measurement of the antibody clones.
| Library | Cluster label | Sequence of HCDR3 | Proportion of NGS (%) | Proportion of conventional method (%) | Binding reactivity (O.D.405 nm) |
|---|---|---|---|---|---|
| Library 1 | Cluster 1 | DFGSGVGEIDA | 3.81 | 1.04 | 1.010 |
| GIESDSDGYMTAEEIDA | 0.13 | 1.04 | 0.977 | ||
| Cluster 2 | AAHSTYIWGGYEAGSIDA | 6.49 | 4.17 | 0.669 | |
| SAVSSCSSGSCSASWIDA | 1.16 | 2.08 | 0.873 | ||
| TADDGFSCGGYGLCADRIDA | 0.39 | 1.04 | 0.723 | ||
| ESGNGGWITAARIDA | 0.08 | 1.04 | 0.767 | ||
| SSHSTYIWGAYEAGSIDA | 0.03 | 2.08 | 0.651 | ||
| Cluster 4 | APGTGSGYCGIWTYTTAGCIDA | 0.03 | 1.04 | 0.964 | |
| GRISYICADYDAGCIDA | 0.02 | 5.21 | 1.063 | ||
| SSHSTYIWGGYEAGSIDA | 0.01 | 2.08 | 0.916 | ||
| Library 2 | Cluster 2 | SSYSDGATVIYNIDA | 0.69 | 1.04 | 0.870 |
| Cluster 3 | GRISYICADYDAGCIDA | 0.04 | 6.25 | 1.063 | |
| AAGSWCAWGTGSCAGSIDA | 0.02 | 5.21 | 1.067 | ||
| AAGSWCAWGTGSCAGNIDA | 0.01 | 1.04 | 0.985 | ||
| TTGGDFYSGIDTAGYIDA | 0.01 | 5.21 | 0.938 | ||
| APGTGSGYCGIWTYTTAGCIDA | 0.01 | 3.13 | 0.964 | ||
| Library 3 | Cluster 2 | AAGSGYIYSGSAGWIDA | 1.07 | 3.13 | 0.941 |
| Cluster 3 | AAGSWCAWGTGSCAGSIDA | 0.03 | 4.17 | 0.918 | |
| GRISYICADYDAGCIDA | 0.02 | 8.33 | 1.063 | ||
| TTGGDFYSGIDTAGYIDA | 0.02 | 2.08 | 0.889 | ||
| AAGSWCAWGAGSCAGSIDA | 0.01 | 1.04 | 0.914 | ||
| AAGSGYVYSGSAGWIDA | 0.01 | 2.08 | 1.021 |
Abbreviations: HCDR3; heavy chain complementarity-determining region 3; NGS, next-generation sequencing; O.D., optical density.
Diversity analysis of antibody clones using next-generation sequencing
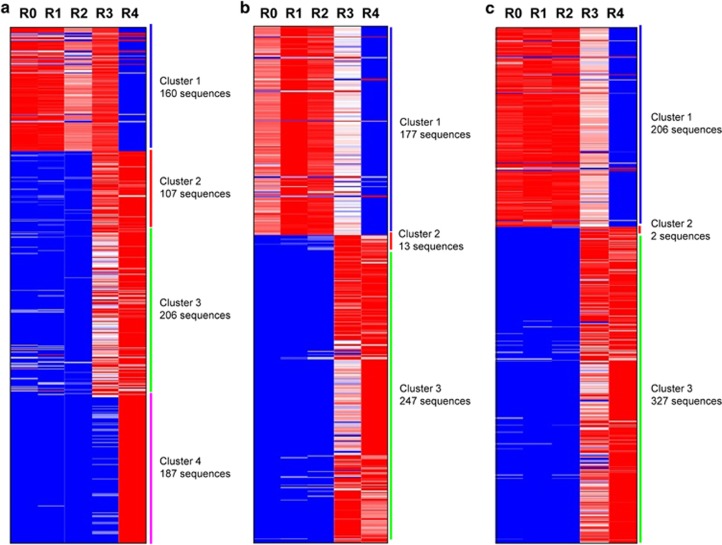
A total of 15 sets of phagemid DNA (three chicken libraries from bio-panning rounds 0, 1, 2, 3, and 4) were used for NGS analysis. After the NGS experiment, we obtained 60,000–180,000 VH sequences. Raw paired-end nucleotide sequences were merged, filtered, aligned and trimmed by uniformly applying pre-specified criteria to remove low-quality and meaningless short sequences. The numbers of nucleotide sequences remaining after each preprocess are summarized in Table 2; 44–53% of the original sequences were retained after aligning with OmpSeq primer sequence14 by Needle, and were used in subsequent analyses. From the NGS results, the total population of VH fragment nucleotides decreased as the bio-panning rounds proceeded. To analyze HCDR3 diversity and frequency, we used HCDR3 sequences existing only in the fourth bio-panning round. clValid predicted that 2–6 clusters would be the most dependable in the HCDR3 sequence count profile data (Table 3). The sequence reads in chicken library 1 showed the maximum Dunn index (0.1048) with 4–6 clusters, and chicken libraries 2 and 3 had maximum Dunn indices with 2–3 clusters. We clustered HCDR3 sequences into 2–6 clusters using hierarchical clustering, and generated heat maps for each cluster to examine the patterns of HCDR3 sequence enrichment and population shift throughout the bio-panning rounds. The pattern of HCDR3 sequence enrichment and population shift in chicken library 1 showed four clear clusters, and the patterns in chicken libraries 2 and 3 showed three clear clusters (Figure 1).
Table 2. Sequence read counts by preprocessing raw sequencing data.
| Trimmed by Prinseq | |||||||
|---|---|---|---|---|---|---|---|
| Panning round | Raw sequencing read count (paired-end FASTQ) | Read count after merging paired-end sequences by FLASH | Read ount of qualified sequences | Read count of disqualified sequences | Sequence read count aligned with HCDR3 region by NEEDLE (percentage in sequences merged by FLASH) | Unique nucleotide sequence count | |
| Library 1 | R0 | 664 955 | 393 749 | 393 624 | 125 | 310 589 (78.9) | 205 255 |
| R1 | 663 061 | 377 630 | 377 484 | 146 | 298 474 (79) | 198 150 | |
| R2 | 391 118 | 229 873 | 229 773 | 100 | 181 430 (78.9) | 128 513 | |
| R3 | 673 875 | 388 341 | 388 179 | 162 | 314 517 (81) | 148 787 | |
| R4 | 621 174 | 379 630 | 379 611 | 19 | 334 387 (88.1) | 27 141 | |
| Library 2 | R0 | 432 274 | 256 268 | 256 199 | 69 | 193 262 (75.4) | 148 862 |
| R1 | 661 248 | 417 426 | 417 323 | 103 | 316 150 (75.7) | 221 423 | |
| R2 | 608 850 | 363 553 | 363 460 | 93 | 274 100 (75.4) | 197 190 | |
| R3 | 547 353 | 342 189 | 342 123 | 66 | 289 287 (84.5) | 66 545 | |
| R4 | 455 119 | 290 741 | 290 722 | 19 | 274 635 (94.5) | 22 763 | |
| Library 3 | R0 | 616 410 | 360 830 | 360 783 | 47 | 279 996 (77.6) | 164 869 |
| R1 | 608 045 | 370 090 | 370 033 | 57 | 288 172 (77.9) | 167 249 | |
| R2 | 619 731 | 373 093 | 373 038 | 55 | 290 056 (77.7) | 168 084 | |
| R3 | 690 602 | 419 796 | 419 757 | 39 | 343 996 (81.9) | 74 611 | |
| R4 | 568 948 | 354 314 | 354 301 | 13 | 287 126 (81) | 21 884 | |
Abbreviations: FLASH, fast length adjustment of short reads; HCDR3, heavy chain complementarity-determining region 3.
Table 3. Dunn index on hierarchical clustering to estimate optimal number of clusters in scFv nucleotide sequence profile data.
| Number of clusters | |||||
|---|---|---|---|---|---|
| 2 | 3 | 4 | 5 | 6 | |
| Library 1 | 0.0863 | 0.0723 | 0.1048 | 0.1048 | 0.1048 |
| Library 2 | 0.2331 | 0.2331 | 0.0564 | 0.0564 | 0.0845 |
| Library 3 | 0.1508 | 0.1860 | 0.1544 | 0.0893 | 0.0893 |
Abbreviation: scFv, single-chain variable fragment. Bold numbers indicate the largest Dunn index in each library.
Figure 1.
Heat map representing the population of heavy chain complementarity-determining region 3 (HCDR3) sequences in each cluster through bio-panning rounds. Red and blue denote high and low proportions of the HCDR3 sequence, respectively. (a) scFv library 1, (b) scFv library 2 and (c) scFv library 3.
Population shift in HCDR3 sequences throughout bio-panning rounds
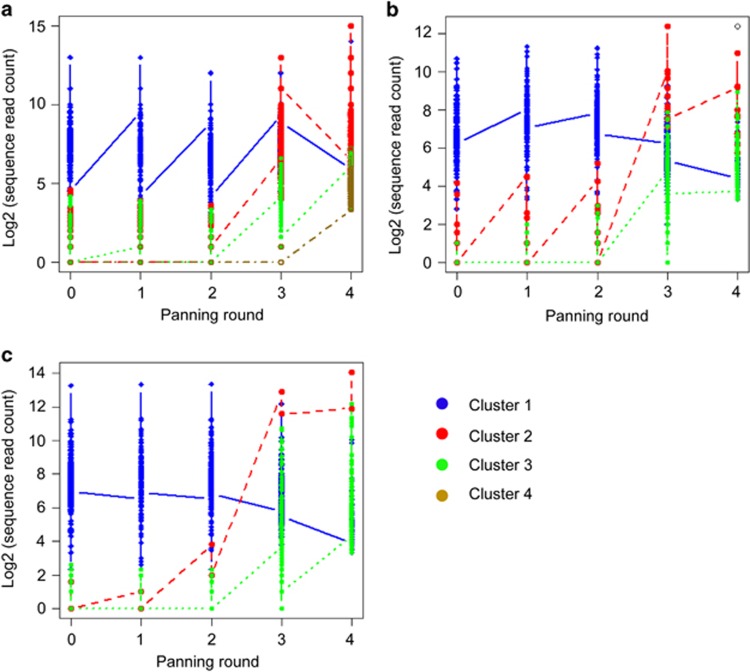
The diversity of the antibody clones is represented by the number of HCDR3 sequences that belong to each cluster (Figure 1). The abundance of the HCDR3 sequences in each cluster is represented by heat map color; high and low populations are indicated in red and blue, respectively. HCDR3 sequences in cluster 1 were highly abundant before bio-panning and up to the second bio-panning round. However, there was a sudden impoverishment in rounds 3 and 4 of bio-panning. In contrast, HCDR3 sequences that belonged to clusters 2 and 3 (including cluster 4 of library 1) showed the opposite pattern. Their populations were very low before bio-panning, remained low after the second round of bio-panning, and started to enrich from the third round of bio-panning. The increase continued in the fourth round of bio-panning. This population shift of HCDR3 sequences throughout bio-panning is represented in Figure 2. All 22 HCDR3 sequences in clones found via the conventional colony screening method existed among the HCDR3 sequences obtained from NGS analysis of phagemid DNA prepared after the fourth round of bio-panning (Table 1). Two out of the 22 unique HCDR3 sequences belonged to cluster 1, and the other 20 HCDR3 sequences belonged to clusters 2, 3 or 4.
Figure 2.
Line graph representing population shifts in HCDR3 sequences through bio-panning rounds. (a) scFv library 1, (b) scFv library 2 and (c) scFv library 3.
Reactivity of scFv clones identified in NGS analysis
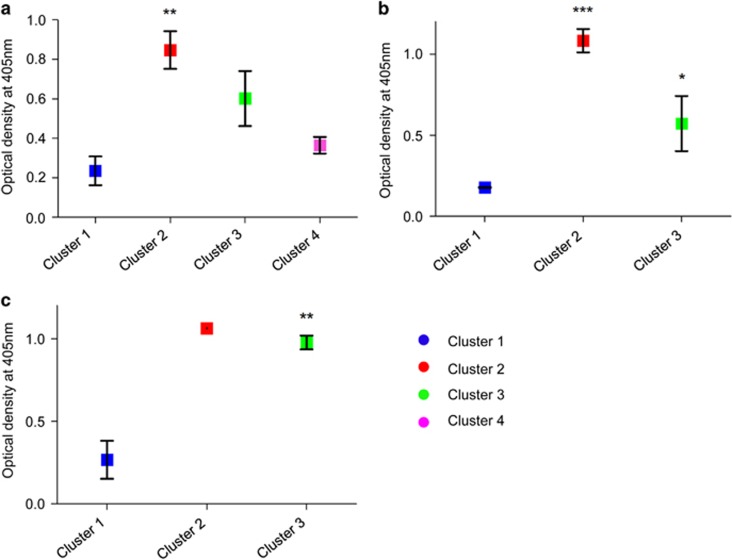
For each cluster, 1–5 HCDR3 sequences newly identified from the fourth round of bio-panning via NGS analysis were selected arbitrarily (Table 4). These selected sequences were used to synthesize the primers to retrieve the whole scFv gene from the phagemid DNA. The scFv gene was prepared in two-step linker PCR using the primers and cloned into a phagemid vector (Figure 3). After transformation of the phagemid vector-encoding scFv gene and rescue with helper phage, scFv-displaying phage was used to test their binding reactivity against PSA (Figure 4). In cluster 1, across the three libraries, 12 out of 14 antibody clones (85.7%) had negligible binding reactivity against PSA (O.D.450nm<0.2; Table 4, blue). In contrast, 21 out of 26 antibody clones (80.8%) in clusters 2~4 across the three libraries had significant binding reactivity (O.D.450nm>0.3; Table 4, red). These results imply that antibody clones with low reactivity tend to be impoverished throughout bio-panning (cluster 1), in contrast to the antibody clones with high reactivity, which showed enrichment throughout bio-panning (clusters 2~4).
Table 4. HCDR3 amino-acid sequences selected in each cluster from NGS and binding reactivity measurement of antibody clones.
| Library | Cluster label | HCDR3 Sequence | Proportion of the sequence in R4 |
Read count of identified sequences |
Binding reactivity (O.D.405 nm) | ||||
|---|---|---|---|---|---|---|---|---|---|
| R0 | R1 | R2 | R3 | R4 | |||||
| Library 1 | Cluster 1 | GVYSGSPDGYDIDA | 0.32% | 502 | 550 | 289 | 1133 | 1235 | 0.454 |
| TTCVGSSYCGGENIDA | 0.16% | 8061 | 8199 | 4786 | 6273 | 603 | 0.173 | ||
| GAYSDWGAGFIDA | 0.08% | 2016 | 2033 | 1237 | 1809 | 301 | 0.161 | ||
| DGDSGWGVYLNSAGNIDA | 0.03% | 39 | 25 | 19 | 76 | 133 | 0.153 | ||
| Cluster 2 | YAGSGWTYYSSDVGSIDA | 2.16% | 0 | 1 | 2 | 1498 | 8314 | 0.620 | |
| GVYSASGCCDSIDT | 1.93% | 0 | 0 | 2 | 1445 | 7443 | 1.032 | ||
| SAHSTYIWGGYEAGSIDA | 1.41% | 0 | 1 | 0 | 1049 | 5420 | 1.075 | ||
| GGGAGYGAPSIDT | 1.05% | 0 | 0 | 0 | 866 | 4034 | 0.871 | ||
| DVYSGLITANTIDA | 0.67% | 0 | 1 | 1 | 325 | 2607 | 0.639 | ||
| Cluster 3 | SSHSTYIWGAYEAGCIDA | 0.02% | 5 | 0 | 0 | 5 | 64 | 0.757 | |
| RAYGGGYCGCIEDIDA | 0.01% | 0 | 0 | 0 | 12 | 44 | 0.323 | ||
| AASTWSFYGSAEDIDA | 0.01% | 0 | 0 | 0 | 3 | 31 | 0.725 | ||
| Cluster 4 | APGTGSGYCGIWTYTTAGSIDA | 0.04% | 0 | 0 | 0 | 1 | 39 | 0.323 | |
| GRISYICADYEAGSIDA | 0.02% | 0 | 0 | 0 | 0 | 61 | 0.407 | ||
| Library 2 | Cluster 1 | GAYGHCDGWCAVDSIDT | 0.07% | 1673 | 2610 | 2430 | 823 | 196 | 0.175 |
| AAGSGYCGWGDCIAGSIDA | 0.07% | 108 | 159 | 139 | 184 | 193 | 0.167 | ||
| GIYGYSGGDYAAAEIDA | 0.06% | 1145 | 1815 | 1712 | 621 | 167 | 0.179 | ||
| GAGGSCDGGSWCSPGIIDA | 0.04% | 1423 | 2179 | 1964 | 595 | 121 | 0.187 | ||
| TRGGAGSGWYWYSGIAGIIDA | 0.03% | 782 | 1172 | 1118 | 399 | 96 | 0.180 | ||
| Cluster 2 | TAGCGPWSYITAGCIDA | 0.21% | 0 | 0 | 6 | 969 | 604 | 1.119 | |
| DAAYGYCGTWAGCAGRIDA | 0.21% | 12 | 22 | 37 | 5404 | 606 | 1.187 | ||
| CAYSGCTGGWSTSSIDA | 0.20% | 18 | 23 | 19 | 1046 | 592 | 1.007 | ||
| DVYGCNSYGCPYIGNTIDA | 0.09% | 0 | 2 | 3 | 190 | 259 | 1.254 | ||
| RAFSGCCDADSIDA | 0.07% | 4 | 5 | 3 | 275 | 195 | 0.845 | ||
| Cluster 3 | SSSGTTYYSSGVISAGGIDA | 0.17% | 0 | 0 | 0 | 62 | 488 | 0.167 | |
| GRISYICVDYDAGCIDA | 0.07% | 0 | 0 | 0 | 59 | 209 | 0.706 | ||
| NAYTSAYITDIDS | 0.06% | 0 | 1 | 1 | 103 | 188 | 0.944 | ||
| SAYSDSCCAEDIDA | 0.04% | 0 | 0 | 1 | 53 | 106 | 0.876 | ||
| SAFGGGACCYTAGTIDA | 0.03% | 0 | 4 | 0 | 15 | 103 | 0.165 | ||
| Library 3 | Cluster 1 | DGSGCGWSAAGCIDA | 0.35% | 9970 | 10385 | 10438 | 4639 | 924 | 0.160 |
| AATYSWLHSGIDA | 0.29% | 112 | 104 | 103 | 246 | 1045 | 0.728 | ||
| DGSDCGWSAAGCIDA | 0.06% | 2430 | 2498 | 2476 | 1164 | 222 | 0.146 | ||
| GTGSWCYSGADSIDT | 0.06% | 2206 | 2381 | 2367 | 1006 | 207 | 0.167 | ||
| SAAGYWYAGSIDA | 0.05% | 10 | 8 | 12 | 121 | 194 | 0.138 | ||
| Cluster 2 | TAGGDFYSGVDTAGYIDA | 4.79% | 1 | 1 | 4 | 3070 | 17187 | 1.064 | |
| Cluster 3 | GSGYSCWSYAGCIDA | 0.66% | 1 | 1 | 1 | 1034 | 2132 | 1.083 | |
| GRIYYICADYDAGCIDA | 0.53% | 0 | 0 | 1 | 429 | 1890 | 1.052 | ||
| TADSGFGCGGYGLCAAFIDA | 0.09% | 2 | 2 | 2 | 743 | 303 | 0.907 | ||
| TADIGYCFGGGIGCIDA | 0.08% | 0 | 0 | 0 | 86 | 289 | 0.984 | ||
| SAGGSYGYRYMDTAAAIDA | 0.07% | 2 | 1 | 1 | 195 | 269 | 0.861 | ||
Abbreviations: HCDR3; heavy chain complementarity-determining region 3; NGS, next-generation sequencing.
Figure 3.
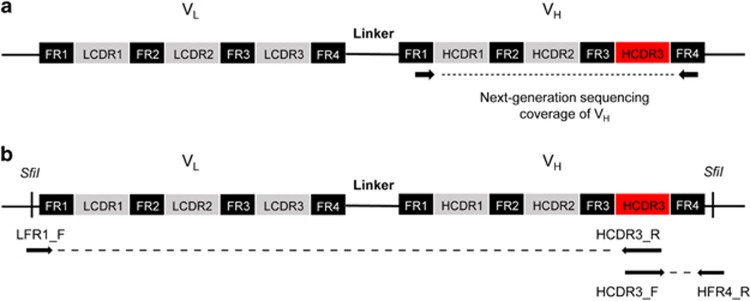
Schematic representation of next-generation sequencing and two-step linker PCR. The structure of scFv gene, CDRs and frameworks of variable regions are indicated by colored boxes. (a) For NGS analysis, most of VH region including HCDR3 was amplified and sequenced using specific primers as described in materials and methods. The sequencing coverage is indicated with dashed lines. (b) To retrieve scFv gene, two-step linker PCR was performed using primers annealing to HCDR3, LFR1 and HFR4. The first step of PCR was performed using LFR1_F and HCDR3_R primers and HCDR3_F and HFR4_R primers. The linker PCR was performed using LFR1_F and HFR4_R primers.
Figure 4.
Binding reactivity of scFv antibodies retrieved from selected HCDR3 amino-acid sequences in each cluster using NGS. (a) scFv library 1, (b) scFv library 2 and (c) scFv library 3. ANOVA with Turkey's multiple-comparison test was used to compare cluster 1 with other clusters. In library 3, the P-value was calculated using the Mann–Whitney U-test. *P-value <0.05; **P-value <0.01; ***P-value <0.001. ANOVA, analysis of variance.
Discussion
Phage display technology is commonly employed in the development of therapeutic antibodies.23 One of the major drawbacks of phage display technology lies in the fact that some clones become dominant through the bio-panning process, and frequently, only limited numbers of clones became available at the end of the screening.16 In this study, we showed that NGS analysis provided not only all of the HCDR3 sequences of clones identified using the conventional colony screening method but also new HCDR3 sequences. The proportion of HCDR3 sequences found by the conventional colony screening method varied from 0.01 to 6.49% of the HCDR3 sequences identified by NGS analysis (Table 1). There was no significant correlation between these two proportions.
After we successfully retrieved the entire scFv gene via two-step linker PCR using PCR primers designed based on the HCDR3 sequences from NGS, we measured the binding reactivity of these antibody clones. Of the 40 clones retrieved from the phagemid DNA pool prepared after the fourth round of bio-panning, positive binding reactivity was confirmed in 26 clones. Four clones in library 1 and one clone in library 3 exhibited a proportion of >1% among the HCDR3 sequences obtained after the fourth round. The proportion of two positive clones in library 1 and library 3 were 2.16 and 4.79%, respectively. All these clones successfully formed colonies after the retrieval process. Why these clones with such high proportions were not identified in the conventional colony screening method is not clear; however, it might be caused by either inherent toxicity with phage assembly or interference from bacterial growth.6, 24
Among the clones retrieved, thirteen clones with proportions less than 0.1% showed binding reactivity. Two of the positive clones were present at a proportion of 0.01%. Theoretically, screening for a clone with such low proportions via the conventional colony screening method requires either 1000 or 10000 positive colonies and Sanger sequencing analysis, which would require significant resources of time and cost.
Many scFv clones prove difficult to identify by the conventional colony screening method from the phage pool obtained after bio-panning, and a method for retrieving these scFv clones in a high-throughput way has not yet been developed. Using currently available NGS tools, it is not possible to sequence the entire scFv gene, which is about 750 bp in length, without error. Our study and the other previous studies6, 13 have proved that the scFv gene can be amplified by PCR primers designed based on HCDR3/FR4 sequences. However, there is always the possibility of cross-priming between clones during the PCR process. In this study, we also confirmed that clones retrieved via two-step linker PCR are typically a mixture of both negative and positive clones.
We also proved that when a certain HCDR3 sequence was enriched through the bio-panning process, the clone with the HCDR3 is more likely to be positive; in this study over 80% (21 out of 26 clones) (Table 4). Monitoring the enrichment or impoverishment pattern of HCDR3 during the bio-panning process might increase the efficiency of retrieving clones from NGS analysis. However, this two-step linker PCR and cloning process is very difficult to perform in a high-throughput manner and there is definitely a need for better way to retrieve whole scFv gene.
In summary, NGS analysis of the HCDR3 sequence, and two-step linker PCR using PCR primers based on this sequence, provide an effective way to retrieve antigen-specific scFv clones that are difficult to identify by the conventional colony screening method. Enrichment of the HCDR3 sequence over the bio-panning process is a positive predictive factor in the selection of scFv clones harboring binding reactivity.
Acknowledgments
This work was supported by a National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIP) (No 2012R1A5A2A44671346)
Footnotes
The authors declare no conflict of interest.
References
- Ecker DM, Jones SD, Levine HL. The therapeutic monoclonal antibody market. MAbs 2015; 7: 9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmen S, Jermutus L. Concepts in antibody phage display. Brief Funct Genomic Proteomic 2002; 1: 189–203. [DOI] [PubMed] [Google Scholar]
- Dantas-Barbosa C, de Macedo Brigido M, Maranhao AQ. Antibody phage display libraries: contributions to oncology. Int J Mol Sci 2012; 13: 5420–5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazan J, Calkosinski I, Gamian A. Phage display—a powerful technique for immunotherapy: 1. Introduction and potential of therapeutic applications. Hum Vaccin Immunother 2012; 8: 1817–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravn U, Gueneau F, Baerlocher L, Osteras M, Desmurs M, Malinge P et al. By-passing in vitro screening—next generation sequencing technologies applied to antibody display and in silico candidate selection. Nucleic Acids Res 2010; 38: e193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravn U, Didelot G, Venet S, Ng KT, Gueneau F, Rousseau F et al. Deep sequencing of phage display libraries to support antibody discovery. Methods 2013; 60: 99–110. [DOI] [PubMed] [Google Scholar]
- Shim H. Therapeutic antibodies antibodies by phage display. Curr Pharm Des 2016; 22: 6538–6559. [DOI] [PubMed] [Google Scholar]
- Luciani F, Bull RA, Lloyd AR. Next generation deep sequencing and vaccine design: today and tomorrow. Trends Biotechnol 2012; 30: 443–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodkinson BP, Grice EA. Next-generation sequencing: a review of technologies and tools for wound microbiome research. Adv Wound Care (New Rochelle) 2015; 4: 50–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Li R, Liu K, Li L, Zai X, Chi X et al. Identification of antigen-specific human monoclonal antibodies using high-throughput sequencing of the antibody repertoire. Biochem Biophys Res Commun 2016; 473: 23–28. [DOI] [PubMed] [Google Scholar]
- Turner KB, Naciri J, Liu JL, Anderson GP, Goldman ER, Zabetakis D. Next-generation sequencing of a single domain antibody repertoire reveals quality of phage display selected candidates. PLoS ONE 2016; 11: e0149393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Wang Z, Ren L, Zhang J, Feng G, Hong K et al. Study of antibody repertoires to the CD4 binding site of gp120 of a Chinese HIV-1-infected elite neutralizer, using 454 sequencing and single-cell sorting. Arch Virol 2016; 161: 789–799. [DOI] [PubMed] [Google Scholar]
- Spiliotopoulos A, Owen JP, Maddison BC, Dreveny I, Rees HC, Gough KC. Sensitive recovery of recombinant antibody clones after their in silico identification within NGS datasets. J Immunol Methods 2015; 420: 50–55. [DOI] [PubMed] [Google Scholar]
- Barbas CF, Burton DR, Scott JK, Silverman GJ. Phage Display: a Laboratory Manual. CSHL Press: NY, USA, 2001. [Google Scholar]
- Han J, Lee JH, Park S, Yoon S, Yoon A, Hwang DB et al. A phosphorylation pattern-recognizing antibody specifically reacts to RNA polymerase II bound to exons. Exp Mol Med 2016; 48: e271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki N, Kiyose N, Akazawa Y, Takashima M, Hagihara Y, Inoue N et al. Isolation and characterization of antigen-specific alpaca (Lama pacos) VHH antibodies by biopanning followed by high-throughput sequencing. J Biochem 2015; 158: 205–215. [DOI] [PubMed] [Google Scholar]
- Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011; 27: 863–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magoč T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011; 27: 2957–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Cowley A, Uludag M, Gur T, McWilliam H, Squizzato S et al. The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res 2015; 43: W580–W584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock G, Pihur V, Datta S, Datta S. clValid, an R package for cluster validation. J Stat Softw 2008; 25: 1–22. [Google Scholar]
- Hartigan JA, Wong MA. Algorithm AS 136: A k-means clustering algorithm. J R Stat Soc Ser C Appl Stat 1979; 28: 100–108. [Google Scholar]
- Kuehn H, Liberzon A, Reich M, Mesirov JP. Using GenePattern for gene expression analysis. Curr Protoc Bioinformatics 2008; Chapter 7: Unit 7.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CE, Lim AP, MacAry PA, Hanson BJ. The role of phage display in therapeutic antibody discovery. Int Immunol 2014; 26: 649–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammers CM, Stanley JR. Antibody phage display: technique and applications. J Invest Dermatol 2014; 134: e17. [DOI] [PMC free article] [PubMed] [Google Scholar]