Abstract
The assembly of respiratory complexes into macromolecular supercomplexes is currently a hot topic, especially in the context of newly available structural details. However, most work to date has been done with purified detergent-solubilized material and in situ confirmation is absent. We here set out to enable the recording of respiratory supercomplex formation in living cells. Fluorescent sensor proteins were placed at specific positions at cytochrome c oxidase suspected to either be at the surface of a CI1CIII2CIV1 supercomplex or buried within this supercomplex. In contrast to other loci, sensors at subunits CoxVIIIa and CoxVIIc reported a dense protein environment, as detected by significantly shortened fluorescence lifetimes. According to 3D modelling CoxVIIIa and CoxVIIc are buried in the CI1CIII2CIV1 supercomplex. Suppression of supercomplex scaffold proteins HIGD2A and CoxVIIa2l was accompanied by an increase in the lifetime of the CoxVIIIa-sensor in line with release of CIV from supercomplexes. Strikingly, our data provide strong evidence for defined stable supercomplex configuration in situ.
Under oxygenic conditions, mitochondria are pivotal for cellular ATP supply in oxidative phosphorylation (OXPHOS)1. The oxidative part mainly consists of three integral membrane protein complexes that are also proton pumps: Complex I (NADH dehydrogenase; CI), complex III (cytochrome bc1 complex; ubiquinol:cytochrome c reductase; CIII), and complex IV (cytochrome c oxidase; CIV). Assembly of these complexes into supercomplexes or respirasomes was already hypothesized in the 60′s of the last century based on electron microscopic images2 and later supported by biochemical analysis3,4. Structures were provided for CI1CIII25 and CI1CIII2CIV1 assemblies6,7,8,9,10,11,12, supporting isolation-persistent supercomplexes assemblies. The assembly is supported by scaffold proteins, such as HIGD2A from the hypoxia inducible gene 1 (HIG1) family member and CoxVIIa2l (Cox7RP; SCAFI)13,14,15. However, several observations argue against stable supercomplex formation. First, the plasticity model demands flexible association – dissociation16, and second, single molecule and FRAP studies have shown that OXPHOS complexes are in principle mobile17,18. Furthermore, the functional relevance of supercomplex formation is still under debate19,20. Thus, a non-invasive live cell compatible technique that enables monitoring of dynamic supercomplex assembly in situ under live cell conditions, in addition to biochemical and genetic analysis methods, would be desirable. Here, we have implemented fluorescence lifetime imaging microscopy (FLIM) as such a method. FLIM is a fluorescence based technique takes advantage of the fact that the fluorescence lifetime τ of a fluorophore (defined as the average time that a molecule remains in the excited state after absorption of light prior to returning to the ground state) depends on its molecular environment. Fluorescence lifetime determination is therefore a feasible means of monitoring the local environment of proteins, such as the association of proteins into complexes and the re-location of proteins between different cellular micro-compartments21,22,23,24,25.
Here, we set out to prove that FLIM might also be a suitable technique to monitor respiratory supercomplex assembly in live cells.
Results
To test this hypothesis, we designed several lifetime probes by attaching fluorescent proteins to specific respiratory subunits which are embedded in a supercomplex based on structural information. We anticipate that positioning a fluorescence probe in a crowded environment, dense with proteins, should correlate with a shortened lifetime due to multiple dipole-dipole interactions. As a FLIM sensor probe, we used the fluorescent protein superecliptic pHluorin (sEcGFP), a pH sensitive monomeric EGFP variant (F64L, S65T, S147D, N149Q, V163A, S175G, S202F, Q204T, A206T) referred to as sEcGFP26. The fluorescence lifetime τ was determined in the time-domain by time-correlated single photon counting (TCSPC) fluorescence lifetime imaging microscopy (FLIM). Soluble purified sEcGFP in PBS had a lifetime of τ = 2.23 ns analogous to that reported for GFP27. In aqueous solutions with increasing glycerol concentrations, which mimic increasing molecular crowding, the lifetime of sEcGFP decreased as expected (Supplementary Fig. S1)28.
Probes at CoxVIIIa and CoxVIIc report supercomplex formation
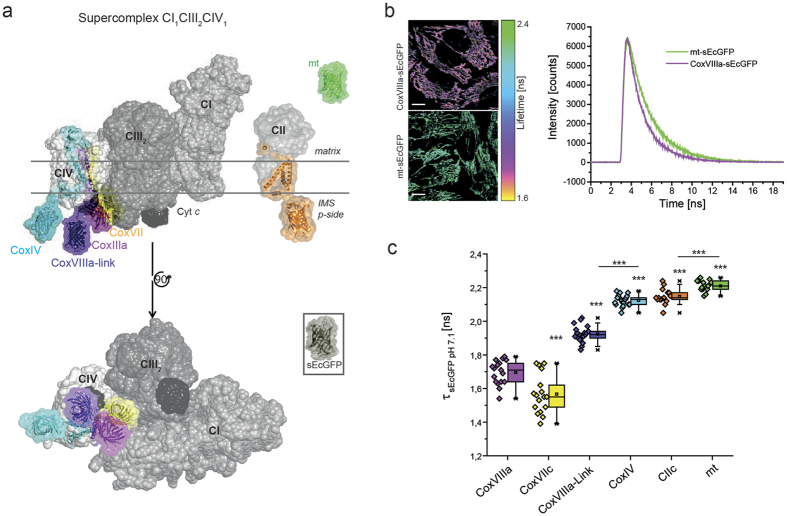
We then tested the appropriateness of FLIM to detect supercomplex formation, which is a specific form of molecular crowding, in situ. Therefore, sEcGFP was fused to the C-terminus of subunit CoxVIIIa of cytochrome c oxidase in the inner mitochondrial membrane (Fig. 1a, pink structure)29. According to previous and recent structures, CoxVIIIa is buried at the interface between complexes I, III and IV. As a control, a matrix-targeted mt-sEcGFP was generated (Fig. 1a, green structure). Lifetime intensity images from cells expressing either of the constructs showed a clear difference (Fig. 1b, left panel). The corresponding time constant τ of the sEcGFP fluorescence lifetime decay was determined by fitting the respective TCSPC diagram (Fig. 1b, right panel). For soluble mt-sEcGFP, a mono-exponential fit can be used30, while for the membrane bound form, a bi-exponential fit was more appropriate (Supplementary Fig. S2). The averaged lifetime for CoxVIIIa-sEcGFP τamp = 1.69 ns was significantly lower than for mt-sEcGFP with τamp = 2.21 ns (δamp = 0.52 ns). To test the possible influence of pH on the result (basic matrix pH and acidic intermembrane space pH), the lifetime of both constructs was recorded in cells at different pH values in the range of pH 6.1 to pH 8.5 as described before29. The lifetime differences due to altered pH were small in the physiological range (pH 7–8), and thus were not responsible for the lifetime differences between mt-sEcGFP and CoxVIIIa-sEcGFP (Supplementary Fig. S3).
Figure 1. Lifetime variations of sEcGFP attached to OXPHOS subunits CoxVIIIa and CoxVIIc report specific molecular crowding at the contact site of CI1CIII2CIV1.
(a) 3D maps of CII and the CI1CIII2CIV1 supercomplex7 showing the side view (top) and the view of the supercomplex from the p-side (complex I in grey, complex III in darker grey and complex IV in light grey, cytochrome c in black). sEcGFP was fused to different subunits of respiratory complexes: CoxVIIc (yellow), CoxVIIIa (pink), CoxVIIIa-Link (violet), CoxIV (cyan), CIIC/SDHC (orange) and to a short matrix targeting sequence mt (green). Selected fitting models are shown. (b) Intensity lifetime image and time correlated single photon counting (TCSCP) histograms of mt-sEcGFP and CoxVIIIa-sEcGFP showing clear differences in lifetime revealed from the decay of fluorescence lifetime. Scale bars: 10 μm. (c) Lifetime constants τ for sEcGFP at different subunits of cytochrome c oxidase (CoxVIIIa - pink, CoxVIIc - yellow, and CoxIV - cyan), at subunit C of SDH (orange) and of matrix-targeted soluble sEcGFP (green). Each data point in the box-and-whisker plot represents the average fluorescence lifetime of a cell ( = one mitochondrial network). The error bars denote s.d., the boxes represent the 25th to 75th percentiles. The vertical lines in the boxes represent the median values, whereas the square symbols in the boxes denote the respective mean values. The minimum and maximum values are denoted by x. All measurements were performed at pH 7.1 to exclude pH effects28. One data point per cell, error bars represent s.d. of ∼18 cells (n = 3 replicates). ***P < 0.001 (by one-way ANOVA).
The low fluorescence lifetime τamp of CoxVIIIa-sEcGFP was therefore relevant and independent of pH. When fitted into a 3D model of supercomplex, the CoxVIIIa-sEcGFP sensor is buried between CIV and CIII (Fig. 1a, pink barrel). As a consequence, multiple dipole-dipole interactions with adjacent proteins from complex I and III are possible that could account for the lifetime reduction of the excited state for sEcGFP at this specific CoxVIIIa position.
We next tested CoxVIIc as a further candidate to probe supercomplex formation since it is probably also exposed to a high protein density (Fig. 1a, yellow)10. sEcGFP at CoxIV was used as a control as it is located at the surface of the supercomplex where protein density probably is lower (Fig. 1a, light blue; Supplementary Fig. S4). To exclude interference with metabolic activity, pH, or redox potential, cells transfected with the respective sEcGFP-subunits were permeabilized, the OXPHOS activity was inhibited and the pH set to pH 7.1 during FLIM measurements. Biexponential fits of TSCPC decays were more appropriate than mono-exponential fits and thus further used for determination of lifetimes (Supplementary Fig. S2). Strikingly, the lifetime of the CoxVIIc sensor was very short (τ = 1.57 ± 0.12 ns, s.d.), while the lifetime of the sensor at CoxIV was significantly longer (τ = 2.12 ± 0.03 ns, s.d.) than that for CoxVIIIa and CoxVIIc. The calculated distance of sEcGFP to the last amino acid of the respective transmembrane domain is 1–2 Å in CoxVIIc-sEcGFP and 5 Å in CoxVIIIa-sEcGFP, which implies positions buried deep in the supercomplex. To elevate the sensor out of the supercomplex, we next inserted a linker composed of the C-terminal sequence of CoxVIIb between CoxVIIIa and sEcGFP (Table 1). The introduction of the linker resulted in a significantly longer lifetime τ = 1.92 ns ( ± 0.05 ns, s.d.) of the fusion protein CoxVIIIa-link-sEcGFP, which let us conclude that this sensor attached via a linker could escape from the crowded protein environment of the supercomplex (Fig. 1a, CoxVIIIa-link, violet).
Table 1. Linker length and distances of sEcGFP to the membrane surface.
| Construct | AA residues of flexible linker part | Calculated distance [Å] between sEcGFP and IMM surface in a supercomplex | Calculated distance [Å] between sEcGFP and IMM surface in a single complex |
|---|---|---|---|
| CoxVIIc-sEcGFP | 7 | 1–2 | 5 |
| CoxVIIIa-sEcGFP | 11 | 5 | 15 |
| CoxVIIIa-Link-sEcGFP | 27 | 50 | 50 |
| CoxIV-sEcGFP | 15 | 44 | 44 |
| CII-sEcGFP | 7 | — | 15 |
Number of amino acid (AA) residues belonging to the flexible linker part between sEcGFP and C-termini of different subunits (see also Supplementary Fig. 4a). Distances [Å] between N-terminus of sEcGFP and the AA residue directly at the inner membrane surface at the different subunits.
To verify that the decreased lifetime of sEcGFP at CoxVIIIa was an effect of defined supercomplex assembly but not caused by random molecular crowding or by the short distance of the probe to the membrane surface, we determined the lifetime of sEcGFP attached to subunit C of complex II. OXPHOS complex CII is a monomeric protein complex16 found in cristae as CI, CIII and CIV31. sEcGFP was fused to the C-terminus of the C-subunit of CII (SDHC) with a 7 amino acid residues linker that should result in a short distance between the probe and the membrane surface (Table 1). The resulting fluorescence lifetime τ = 2.15 ns ( ± 0.05 ns, s.d.) for CIIC-sEcGFP was similar to the lifetime of CoxIV- sEcGFP (τ = 2.12 ns ± 0.03 ns, s.d.) at the surface of CIV. It was about 0.47 ns higher than that for CoxVIIc-sEcGFP with the same linker length between the membrane domain and the sEcGFP (Table 1). Overall, the fluorescence lifetimes of CoxVIIc-sEcGFP and CoxVIIIa-sEcGFP were the shortest of all lifetimes measured by a significant margin, and this is consistent with a position in the center of a supercomplex (Fig. 2c). Similar results were obtained with mCitrine as sensor – except that the τ values were shifted to longer τ(Δ~0.59 ns) due to the generally longer lifetime of mCitrine (Supplementary Fig. S5)32.
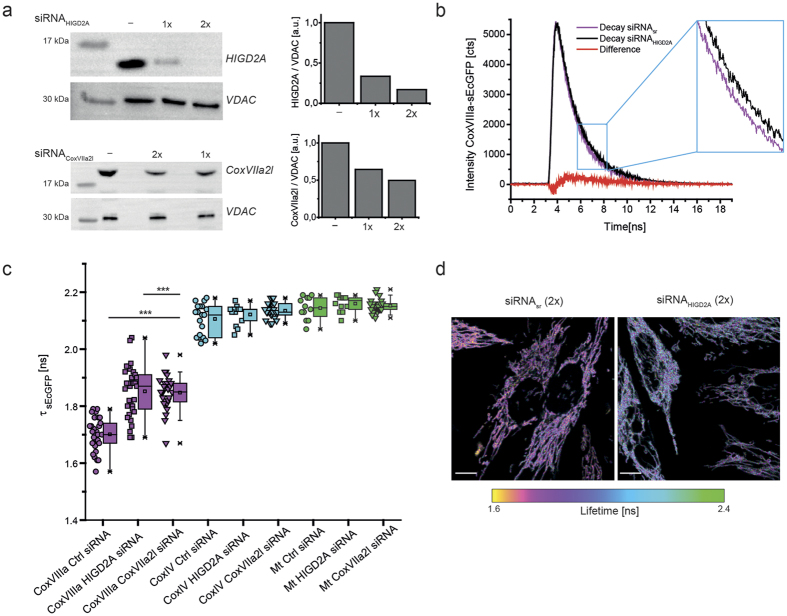
Figure 2. The lifetime of CoxVIIIa-sEcGFP increases in cells with decreased levels of scaffold proteins.
(a) Silencing of HIGD2A and CoxVIIa2l in stable CoxVIIIa-sEcGFP cells as shown by immuno-staining. Scrambled siRNA was used as a control, loading control was VDAC (middle panel). Right panel: ratio between HIGD2A respectively CoxVIIa2l and VDAC levels. (b) TCSPC diagram showing the change in fluorescence decay with decreased scaffold proteins. (c) Average fluorescence lifetimes τamp of stable HeLa cell lines expressing CoxVIIIa-sEcGFP, CoxIV-sEcGFP and mt-sEcGFP, respectively with downregulated HIGD2A or CoxVIIa2l. One data point per cell, error bars represent s.d. of ∼24 cells (n = 3 biological replicates). (d) Fluorescence intensity/lifetime images of CoxVIIIa-sEcGFP in cells with decreased scaffold protein HIGD2A or All-Stars non-targeting siRNA. Scale bars: 10 μm. Significance: ***P < 0.001 compared to CoxVIIIa-sEcGFP (ANOVA one-way).
Because mitochondria of cells expressing CoxVIIc-sEcGFP appeared stressed with few mitochondria in the cell periphery (data not shown) and slightly decreased basal and ATP-linked respiration (Supplementary Fig. S6), we decided to use CoxVIIIa-sEcGFP for further studies. In stable CoxVIIIa-sEcGFP cells no alterations of mitochondrial morphology, distribution or respiration rates were observed (Figure 1b, Supplementary Fig. S6). Successful assembly of a CoxVIIIa-GFP fusion construct into complex IV was shown earlier33.
Influence of supercomplex scaffold factors
We next decreased levels of supercomplex scaffold factors HIGD2A and CoxVIIa2l by siRNA treatment. Twofold siRNA treatment resulted in 80% decreased HIGD2A levels, while CoxVIIa2l levels were decreased to 50% (Fig. 2a). The TCSPC decays of CoxVIIIa-sEcGFP in cells with decreased scaffold factor HIGD2A showed a significant shift towards longer lifetimes (Fig. 2b). The HIG2DA suppression obviously did not influence the cellular or mitochondrial morphology (Fig. 2d, lower panel).
Overall, the suppression of the supercomplex assembly factors HIGD2A and CoxVIIa2l increased the mean lifetime of the CoxVIIIa probe but not of the CoxIV- or mt-sEcGFP probe (Fig. 2c). Moreover, according to the residuals (Supplementary Fig. S2) mono-exponential fits for CoxVIIIa-sEcGFP with suppressed HIGD2A and CoxVIIa2l became appropriate in most cases. This is what we had expected as an outcome of supercomplex disassembly and CoxVIIIa-sEcGFP release. While CoxVIIIa-sEcGFP in a supercomplex will be subject to a dense protein environment, CoxVIIIa-sEcGFP in monomeric cytochrome c oxidase or in cytochrome c oxidase homo-dimers will rather be exposed to bulk water, resulting in less dipole-dipole interactions and thus an increase in the corresponding fluorescence lifetime of CoxVIIIa-sEcGFP (Fig. 3). CoxVIIIa-sEcGFP is localized at the outward surface, since subunits VIa, VIb and VIIb are the subunits that form the major protein-protein contacts between the two monomeric units within the dimeric complex (Supplementary Fig. S7)34,35,36. We thus interpret the prolonged fluorescence lifetime in HIGD2A down-regulated cells as a shift towards higher amounts of free CIV and less CoxVIIIa-sEcGFP buried within a supercomplex. However, the lifetime of CoxVIIIa-link-sEcGFP was not changed in cells with downregulated HIGD2A (Supplementary Fig. S8). In general, the increase of τamp was not accompanied by an increase in fluorescence intensity (Supplementary Fig. S9) which is not surprising since fluorescence intensity is not necessarily a parameter that responds to the nano-environment28. There is also no possibility of using fluorescence intensity read out for quantification, since expression levels of the different fluorescent fusion-proteins were not comparable (Supplementary Fig. S9).
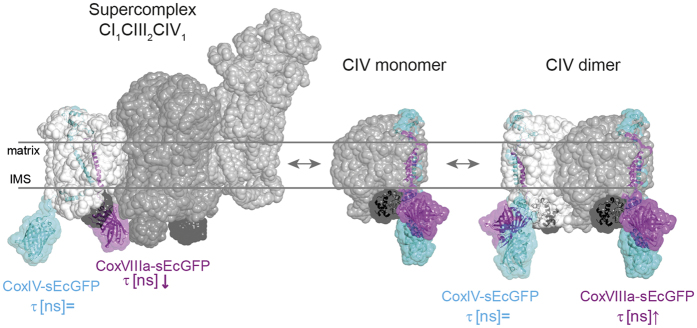
Figure 3. Positioning of CoxIV-sEcGFP and CoxVIIIa-sEcGFP in a supercomplex and monomeric and dimeric CIV.
Models illustrating conformations expected for CoxVIIIa-sEcGFP in a supercomplex and in monomeric and homo-dimeric CIV(2) with respective environments: CoxVIIIa-sEcGFP senses a dense molecular environment in a supercomplex and an aqueous environment in free CIV(2) and CIV, while CoxIV-sEcGFP is always exposed to an aqueous environment. Supercomplex sensor CoxVIIIa-sEcGFP in pink framed with a dotted line, control CoxIV-sEcGFP in cyan, cytochrome c framed with disrupted lines.
In summary, our live cell FLIM approach with probes at different Cox subunits clearly demonstrates the exceptional position of subunit CoxVIIIa (and CoxVIIc) and is consistent with localization of CIV inside a supercomplex but not by CIV monomers or dimers (Fig. 3).
FRET between CIII and CIV confirms supercomplex formation
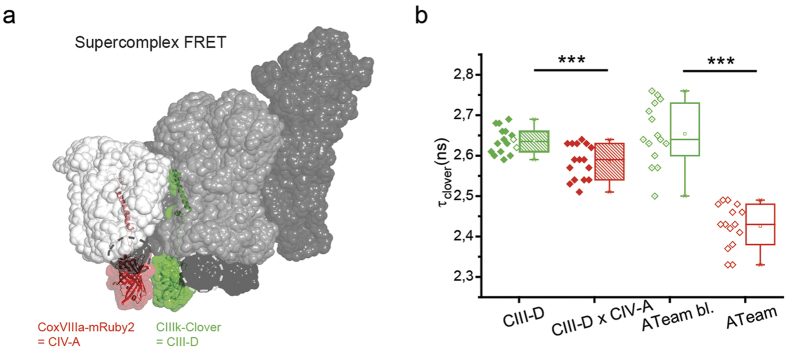
We next tested possible colocalization of CIII and CIV in a supercomplex by Förster Resonance Energy Transfer (FRET) using CIII subunit k fused to Clover as donor and CoxVIIIa-mRuby2 as acceptor. The C-terminus of CIII k is localized at the p-side according to the supercomplex models and the distance between the fused Clover and CoxVIIIa-mRuby2 should be short enough to allow FRET (Fig. 4a). Cells were transiently transfected with donor only (CIIIk-Clover, CIII-D) or both constructs (CIII-D × CIV-A) and fluorescence lifetime images were recorded. As a control, ATeam37 was used that was constructed with Clover as donor and mRuby2 as acceptor within the same ATeam molecule. The lifetime of Clover was obtained from bi-exponential fits from the TCSPC decays. In the presence of the acceptor mRuby2, the lifetime of the donor significantly decreased. This indicates proximity of CIII and CIV in a supercomplex. The decrease in fluorescence lifetime was stronger in an ATeam control construct, since here the expression and proximity of both fluorescent proteins was guaranteed (Fig. 4b, Supplementary Fig. S10). The FRET results thus are in line with the preceding FLIM data with one sensor only at CoxVIIIa and suggest the existence of a CICIII2CIV supercomplex in situ.
Figure 4. FRET measurements showing close proximity of CIV subunit CoxVIIIa and CIII-subunit k in a supercomplex.
(a) Positions of CoxVIIIa-mRuby (acceptor, CIV-A) and CIIIk-Clover (donor, CIII-D) within a CICIII2CIV supercomplex. Only one subunit k of one CIII is labeled. (b) Fluorescence lifetime of Clover as donor in the presence and absence of acceptor mRuby. Pair 1: CoxVIIIa-mRuby as acceptor and CIIIk-clover as donor. Pair 2: modified ATeam37 composed of clover as donor and mRuby as acceptor was used as FRET control. Significance: ***P < 0.001 compared to CIIIk-Clover (ANOVA one-way).
Discussion
The architecture of respiratory supercomplexes has been revealed and refined by several groups6,7,8,9,10,11,12. Purification and re-constitution involve some critical steps16 and freeze the supercomplex in a certain state38. Thus, dynamic supercomplex assembly is difficult to determine from the analysis of purified supercomplexes. Notably, approximately 80% of total CI, and 66% of total CIII2 was found to be supercomplexed, but 85% of total CIV – according to BN-PAGE data – was found in the free form in bovine heart mitochondria39. This probably makes CIV1 the more dynamically regulated complex. For that reason, we focused on CIV1 for in situ studies. Moreover, it was reported that interactions between CIV and CI/CIII were weaker than interactions between CI and CIII9. By FLIM using fluorescent sensor proteins at specific positions we provide a tool to study supercomplex assembly in live cells and monitor supercomplex plasticity. We focused on single probes since FRET is much more difficult and not straight forward for several reasons: (i) The successful labeling of two subunits within one supercomplex for successful FRET studies is more challenging than working with a single sensor, (ii) The recording time after transfection is critical, since little is known about the turnover and assembly of SU subunits into complexes and supercomplexes, (iii) Attachment of fluorescent proteins to two subunits might interfere with supercomplex assembly, (iv) CIII2 was always found as a dimeric complex and this might complicate the analysis. However, we were able to obtain positive FRET between labeled subunits CoxVIIIa of CIV and subunit k of CIII indicating proximity within the Förster radius. FRET was used before to show F1FO ATP synthase interaction40. Instead, we decided to go for a simpler assay: the lifetime of a single fluorescent probe already reports on the local molecular environment and becomes shorter in protein dense environments. We genetically sited fluorescent proteins to different subunits of cytochrome c oxidase (CIV) for FLIM analysis. Lifetime measurements with fluorescent proteins are feasible since interactions with proximal proteins are translated into the interior of the fluorescent protein barrel and decrease the lifetime41. CIV is part of several supercomplexes described to date with specific configurations3,4. Assembly of complexes into a supercomplex explains the finding that individual complex stability depends on the interaction with other complexes42,43,44 better than random collision in a crowded membrane. Also, the isolation of mammalian supercomplex CI1CIII2CIV1 indicates a rather stable supercomplex association with KD values in the ms to s time range39. The short sEcGFP fluorescence lifetimes that we measured at subunits CoxVIIIa and CoxVIIc of cytochrome c oxidase fit exactly into the models of the CI1CIII2CIV1 supercomplexes, indicating a dense protein environment at the interface between CIV, CI and CIII. From our results, we can rule out random collision as the general cause for the observed lifetime reduction, since the fluorescence lifetime of CIIC-sEcGFP in the same microcompartment was significantly higher than of CoxVIIIa/CoxVIIc-sEcGFP. Rather, the shortened lifetimes of the CoxVIIIa and CoxVIIc sensors is caused by higher protein density at their location due to a specific, defined protein environment in a supercomplex. This was supported by the finding that suppression of supercomplex scaffold factors specifically increased the lifetime of CoxVIIIa, the sensor embedded within the supercomplex, but not of sensors that were located only at the surface or a sensor attached with a long link to the CoxVIIIa subunit. This is in line with a shift to monomeric and dimeric forms of CIV45.
Our data show that FLIM with one sensor is a valuable alternative to the Förster Resonance Energy Transfer technique. Not only, FLIM is a tool to investigate supercomplex formation and dissociation non-invasively in a functional context but has the potential to be used to study other events involving complex formation such as G protein coupled signalling, or supercomplex formation in photosynthetic membranes.
Material and Methods
FLIM sensor construction
For eukaryotic expression of sEcGFP, the full-length protein-coding region of superecliptic pHluorin (F64L/S65T/S147D/N149Q/V163A/S175G/S202F/Q204T/A206T) (a gift from Prof. Jürgen Klingauf) was inserted by PCR amplification into a modified pSEMS-26 m vector from NEB Biosciences (formerly Covalys Biosciences). For primers see Rieger et al. 201429. For determination of the fluorescence lifetime in the matrix bulk, the mitochondrial targeting sequence of the mitochondrial processing peptidase MPP, consisting of the N-terminal 60 amino acid residues (MPP60, referred to as mt), was included behind the CMV promoter and at the 3′end of sEcGFP in this vector resulting in mt-sEcGFP. This targeting sequence is likely removed from the fusion construct by MPP, leaving sEcGFP in the matrix. For measurements in the IMS, CoxVIIc-, CoxVIIIa-, CoxIV-, and CIIC-sEcGFP were assembled behind the CMV promoter and at the 3′end of sEcGFP in the sEcGFP-vector. The respective subunits of OXPHOS complexes were fused with their C termini to the N terminus of sEcGFP. To increase sensor distance to the membrane by C-terminal extension of CoxVIIIa, we used part of the C-terminal sequence of CoxVIIb (TCCCCTGTTG GCAGAGTTAC CCCAAAGGAA TGGAGGAATC AG) to insert a linker between CoxVIIIa and sEcGFP, resulting in CoxVIIIa-Link-sEcGFP. For FLIM measurements in the presence of an acceptor, CoxVIIIa-mRuby2 was generated as an acceptor, while subunit k of CIII was C-terminally fused to Clover as donor.
Construction of pSems-Clover-ATeam-mRuby2
In CFP-ATeam-YFP (AT1.03; a gift from Hiromi Imamura)37 CFP was substituted by Clover and YFP was substituted by mRuby2. Therefore, primers for ATeam, Clover and mRuby2 were constructed: f-ATeam-BamHI GATTAGGGATCCATGAAAACTGTGAAA GTGAATATAAC, r-ATeam-XhoI GATTAGCTCGAGGTTTGCCTTCCCAGCCACGTCCAG, f-mRuby2-XhoI GAT TAGCTCGAGATGGTGTCTAAGGGCGAAGAGC, r-mRuby2-NotI GATTACGCGGCCGCCTTGTACAGCTCGTCC ATCC. ATeam and mRuby were amplified by PCR and inserted into a modified pSEMS-MCS vector from NEB Biosciences (formerly Covalys Biosciences). Therefore, the ATeam PCR construct was digested with BamHI and XhoI and the mRuby2 PCR construct with XhoI and NotI. Finally, Clover from pSems-FRB-Clover (cloning analogous to pSems-FRB-mCitrine8 was inserted in pSems-ATeam-mRuby2 after digestion with EcoRI and BamHI. PCR primers were purchased from Sigma Aldrich. Enzymes, restriction buffer and BSA were purchased from NEB Biosciences.
Cell culture of HeLa cells
For generation of stable cell lines, transfected HeLa cells were selected for stable neomycine resistance by growth in the presence of 0.8 mg/ml G418 (Calbiochem 345810). Untransfected HeLa cells and the stable HeLa cell lines (CoxVIIc-, CoxVIIIa-, CoxVIIIa-Link-, CoxIV- and mt-sEcGFP) were cultured in Minimal Essential Medium with Earle’s salts (MEM, PAA Lab GmbH, E15–888) with 5.6 mM glucose, 2 mM stable glutamine and sodium bicarbonate, supplemented with 10% (v/v) fetal bovine serum (FBS) superior (Biochrom AG), 1% MEM nonessential amino acids (NEAA, Biochrom AG) and 1% 4−(2−hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES, PAA Lab GmbH) at 37 °C with 5% CO2. Stable HeLa cell lines were kept in 0.8 mg/ml G418 in addition. Cells were split 2–3 times a week using Trypsin/EDTA (Biochrom), supplemented with HEPES (PAA), sodium bicarbonate (PAA), penicillin/streptomycin (Biochrom) and PBS (Biochrom). For nanoenvironment determination by fluorescence lifetime measurements, sEcGFP cell lines were seeded onto glass coverslips in 3.5 cm cell culture dishes. Imaging was performed 2 days after seeding.
siRNA mediated knockdown
HeLa cells were maintained in culture medium. Knockdowns were performed by treating cells with 5 nM total siRNA (Qiagen), using the Hiperfect transfection reagent, according to the manufacturer’s instructions for fast-forward transfection (Qiagen). The All-Stars non-targeting siRNA (1027295/SI03650318) was used as the control for siRNAs targeting HIGD2A (NM_ 138820, 1027419/SI04275957) and CoxVIIa2l (NM_004718, 1027419/SI00067445) – all with 3′ AlexaFluor647 modification. Cells were subjected to knockdown on day zero, again on day 1, and analysed on day 3.
SDS-PAGE analysis
For SDS-PAGE, cell lysates from samples of confluent T-25 flasks (1 flask would be sufficient for 10–15 gels) were heated at 95 °C for 5 min and separated on 12% Tricine-SDS-PAGEs and transferred to PVDF membranes. Membranes were blocked prior to detection using 10% skimmed milk powder in TBS-T (200 mM Tris, 1.37 M NaCl, + 0.1% Tween20). BN-PAGE was performed using a Tricine-Tris buffer system. SuperSignal West Pico from ThermoScientific (34080) was used as chemiluminescent substrate to visualize proteins. Protein ladder was purchased from ThermoScientific (ThermoScientific PageRuler Plus Prestained Protein Ladder #26619). Semi-Dry-Blotting was performed using a Tris-Glycine buffer with 13 mM SDS. Antibodies were purchased from CellSignaling (VDAC, D73D12 #4661), antikoerper-online.de (HIG1 Hypoxia Inducible Domain Family, Member 2 A (HIGD2A) (AA15–44) (N-Term) antibody, ABIN658252), Abcam (CoxVIIa2l, ab170696) or Dianova (Peroxidase-conjugated AffiniPure Goat Anti-Rabbit IgG (H + L), 111-035-045).
Time correlated single photon counting (TCSPC) fluorescence lifetime imaging microscopy (FLIM)
Time-resolved fluorescence measurements were recorded in situ at 37 °C and 5% CO2 using a confocal laser scanning microscope (FluoView FV1000, Olympus) equipped with a TCSPC extension module (PicoQuant GmbH). The excitation source was a pulsed LDH-D-C-485 laser (PicoQuant GmbH) operated at a repetition time rate of 40 MHz. The output pulses were coupled into an optical fiber. The output at the fiber end was reflected from a beam splitter onto the base of a multiwell plate (Ibidi®, 30 μL) or a glass cover slip via a 60 × oil-immersion objective (UPLSAPO oil, NA 1.35, N/0.17/FN26.5, Olympus) upgraded with an objective heater (BIOPTECHS). Emission was restricted to 525/50 nm with an emission filter and photons were detected with a single photon avalanche diode (SPAD). The acquisition was performed until at least 1000 photons in the brightest pixel were reached. For calibration, sEcGFP-GST was expressed in E.coli and purified via agarose beads. The purified sEcGFP was added to aqueous Dulbecco’s Phosphate Buffered Saline (PBS) solutions with increasing glycerol content (w/w%) at 37 °C. In parallel, the refractive index of the same glycerol/PBS mixtures was determined by an Abbe-refractometer heated to 37 °C. Data analysis was performed with SymphoTime® software (32 bit) and mono- respectively bi-exponential fitting of the fluorescence decay curves (substracting the IRF) from ROIs (approx. 50% of the mitochondrial network of a cell). From bi-exponential fits, the average lifetime was calculated as mean weighted average lifetime τamp = ((τ1.A1) + (τ2.A2))/(A1 + A2), with τ1 and τ2 as different lifetimes and A1 and A2 as their amplitudes. For measurement of sEcGFP proteins in HeLa cells, these were transiently transfected with the respective plasmid construct. FLIM measurements were performed 72 h after transfection. The resulting values for the fluorescence lifetime were displayed on false color scale.
Supercomplex modeling
For the modeling of respiratory supercomplex CI1CIII2CIV1 we mainly considered well-resolved experimental data, namely EM-53198, and previous models of respiratory supercomplex structures, including all experiments deposited in EMDB (see Supplementary information for details). The best fit was achieved with the following structures: complex I (PDB 4UQ8; EM 2676), cytochrome bc1 complex dimer (PDB 1PP9) and cytochrome oxidase (PDB 1V55). With electron density maps simulated from X-ray structures with a resolution of 10 Å and average map value 0.1669 the correlation with the experimental density was 0.8346. Fitting and analysis were performed with Chimera v.1.7. For each sequence of labelled COX subunit model structures were generated using structures of GFP (PDB 1GFL) and COX (PDB 1V55) with variable linker conformations. In each case 25 structures were generated with MODELLER v.9.25 and placed in the supercomplex model. All structures of labeled subunits with steric clashes between sEcGFP and membrane or supercomplex proteins were removed (see supplementary information). In each case, several structures were selected to represent available locations for sEcGFP label. Structural visualization and analysis were performed with Pymol v. 1.5.0.3.
Oxygen consumption measurements
Oxygen consumption of intact HeLa cells was recorded with the Seahorse XFe Extracellular Flux Analyzer (Seahorse Biosciences; North Billerica, MA, USA). ~30.000 cells were seeded into each well of a 96-well Seahorse plate 24 h before the experiment. 60 min before the experiment, cells were washed with XF base medium adjusted to pH 7.4 (Minimal DMEM, 0 mM Glucose, 102353–100 from Seahorse Biosciences), placed in fresh XF base medium pH 7.4 with supplements (1 mM pyruvate, 2 mM L-glutamine, and 5.6 mM D-glucose) and incubated at 37 °C before loading into the XFe Analyzer. Supplements were from Roth. After recording resting respiration in the analyzer, the following chemicals from Seahorse Biosciences were added sequentially to the cells: oligomycin (1 μm), to measure the nonphosphorylating OCR, FCCP (2 μm), to achieve maximal OCR, and antimycin A (0.5 μm) and rotenone (0.5 μm), for determination of the extramitochondrial OCR. For each experiment, three measurements were performed for the resting OCR, three after oligomycin addition, three after FCCP and three after antimycin A plus rotenone with a 2-min interval of recording followed by 2 min of mixing and 2 min of incubation for each measurement.
Additional Information
How to cite this article: Rieger, B. et al. Lifetime imaging of GFP at CoxVIIIa reports respiratory supercomplex assembly in live cells. Sci. Rep. 7, 46055; doi: 10.1038/srep46055 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This work supported by the CRC-944 and DFG grant Bu2288/1–2 (KB), the Osnabrueck University, the German Academic Exchange Service, and the grant 14-50-00029 from the Russian Science Foundation (DNS). We thank Iven Winckelmann for important preliminary studies and Guy Hanke for critically reading and improving the manuscript.
Footnotes
The authors declare no competing financial interests.
Author Contributions B.R., A.Y.M. and K.B.B. designed the experiments, A.C. contributed the calibration curve, W.K. and P.D. provided technical assistance with cloning and WESTERN, D.S. performed the modelling, B.R., and D.S. prepared figures and K.B.B. wrote the main manuscript text. All authors reviewed the manuscript.
References
- Mitchell P. Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism. Nature 191, 144–148 (1961). [DOI] [PubMed] [Google Scholar]
- Parsons D. F. Mitochondrial Structure: Two Types of Subunits on Negatively Stained Mitochondrial Membranes. Science 140, 985–987, doi: 10.1126/science.140.3570.985 (1963). [DOI] [PubMed] [Google Scholar]
- Wittig I., Carrozzo R., Santorelli F. M. & Schagger H. Supercomplexes and subcomplexes of mitochondrial oxidative phosphorylation. Biochim Biophys Acta 1757, 1066-1072, doi: 10.1016/j.bbabio.2006.05.006 (2006). [DOI] [PubMed] [Google Scholar]
- Cruciat C. M., Brunner S., Baumann F., Neupert W. & Stuart R. A. The cytochrome bc1 and cytochrome c oxidase complexes associate to form a single supracomplex in yeast mitochondria. J Biol Chem 275, 18093–18098, doi: 10.1074/jbc.M001901200 (2000). [DOI] [PubMed] [Google Scholar]
- Dudkina N. V., Eubel H., Keegstra W., Boekema E. J. & Braun H. P. Structure of a mitochondrial supercomplex formed by respiratory-chain complexes I and III. Proc Natl Acad Sci USA 102, 3225–3229 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer E., Dencher N. A., Vonck J. & Parcej D. N. Three-dimensional structure of the respiratory chain supercomplex I1III2IV1 from bovine heart mitochondria. Biochemistry 46, 12579–12585, doi: 10.1021/bi700983h (2007). [DOI] [PubMed] [Google Scholar]
- Althoff T., Mills D. J., Popot J. L. & Kuhlbrandt W. Arrangement of electron transport chain components in bovine mitochondrial supercomplex I1III2IV1. EMBO J 30, 4652–4664, doi: 10.1038/emboj.2011.324 [pii] (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudkina N. V., Kudryashev M., Stahlberg H. & Boekema E. J. Interaction of complexes I, III, and IV within the bovine respirasome by single particle cryoelectron tomography. Proceedings of the National Academy of Sciences 108, 15196–15200, doi: 10.1073/pnas.1107819108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J. et al. The architecture of the mammalian respirasome. Nature 537, 639–643, doi: 10.1038/nature19359 (2016). [DOI] [PubMed] [Google Scholar]
- Letts J. A., Fiedorczuk K. & Sazanov L. A. The architecture of respiratory supercomplexes. Nature 537, 644–648, doi: 10.1038/nature19774 (2016). [DOI] [PubMed] [Google Scholar]
- Sousa J. S., Mills D. J., Vonck J. & Kuhlbrandt W. Functional asymmetry and electron flow in the bovine respirasome. Elife 5, doi: 10.7554/eLife.21290 [pii] (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M., Gu J., Guo R., Huang Y. & Yang M. Structure of Mammalian Respiratory Supercomplex I1III2IV1. Cell 167, 1598–1609 e1510, doi: 10.1016/j.cell.2016.11.012 (2016). [DOI] [PubMed] [Google Scholar]
- Lapuente-Brun E. et al. Supercomplex assembly determines electron flux in the mitochondrial electron transport chain. Science 340, 1567–1570, doi: 10.1126/science.1230381 (2013). [DOI] [PubMed] [Google Scholar]
- Vukotic M. et al. Rcf1 mediates cytochrome oxidase assembly and respirasome formation, revealing heterogeneity of the enzyme complex. Cell Metab 15, 336–347, doi: 10.1016/j.cmet.2012.01.016 (2012). [DOI] [PubMed] [Google Scholar]
- Ikeda K., Shiba S., Horie-Inoue K., Shimokata K. & Inoue S. A stabilizing factor for mitochondrial respiratory supercomplex assembly regulates energy metabolism in muscle. Nature communications 4, 2147, doi: 10.1038/ncomms3147 (2013). [DOI] [PubMed] [Google Scholar]
- Acin-Perez R., Fernandez-Silva P., Peleato M. L., Perez-Martos A. & Enriquez J. A. Respiratory active mitochondrial supercomplexes. Mol Cell 32, 529–539, doi: 10.1016/j.molcel.2008.10.021 (2008). [DOI] [PubMed] [Google Scholar]
- Appelhans T. et al. Nanoscale organization of mitochondrial microcompartments revealed by combining tracking and localization microscopy. Nano Lett 12, 610–616, doi: 10.1021/nl203343a (2012). [DOI] [PubMed] [Google Scholar]
- Sukhorukov V. M. et al. Determination of protein mobility in mitochondrial membranes of living cells. Biochim Biophys Acta 1798, 2022–2032, doi: 10.1016/j.bbamem.2010.07.016 ( 2010). [DOI] [PubMed] [Google Scholar]
- Blaza J. N., Serreli R., Jones A. J., Mohammed K. & Hirst J. Kinetic evidence against partitioning of the ubiquinone pool and the catalytic relevance of respiratory-chain supercomplexes. Proc Natl Acad Sci USA 111, 15735–15740, doi: 10.1073/pnas.1413855111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trouillard M., Meunier B. & Rappaport F. Questioning the functional relevance of mitochondrial supercomplexes by time-resolved analysis of the respiratory chain. Proc Natl Acad Sci USA 108, E1027–1034, doi: 10.1073/pnas.1109510108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Manen H. J. et al. Refractive index sensing of green fluorescent proteins in living cells using fluorescence lifetime imaging microscopy. Biophys J 94, L67–L69, doi: 10.1529/biophysj.107.127837 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Festy F., Ameer-Beg S. M., Ng T. & Suhling K. Imaging proteins in vivo using fluorescence lifetime microscopy. Mol Biosyst 3, 381–391, doi: 10.1039/b617204k (2007). [DOI] [PubMed] [Google Scholar]
- Bastiaens P. I. & Squire A. Fluorescence lifetime imaging microscopy: spatial resolution of biochemical processes in the cell. Trends in cell biology 9, 48–52 (1999). [DOI] [PubMed] [Google Scholar]
- Nakabayashi T. et al. Stress-induced environmental changes in a single cell as revealed by fluorescence lifetime imaging. Photochem Photobiol Sci 7, 671–674, doi: 10.1039/b805032e (2008). [DOI] [PubMed] [Google Scholar]
- Sohnel A. C. et al. Probing of protein localization and shuttling in mitochondrial microcompartments by FLIM with sub-diffraction resolution. Biochimica et biophysica acta 1857, 1290–1299, doi: 10.1016/j.bbabio.2016.03.021 (2016). [DOI] [PubMed] [Google Scholar]
- Miesenbock G., De Angelis D. A. & Rothman J. E. Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature 394, 192–195, doi: 10.1038/28190 (1998). [DOI] [PubMed] [Google Scholar]
- Tregidgo C., Levitt J. A. & Suhling K. Effect of refractive index on the fluorescence lifetime of green fluorescent protein. J Biomed Opt 13, 031218, doi: 10.1117/1.2937212 (2008). [DOI] [PubMed] [Google Scholar]
- Suhling K. et al. Imaging the environment of green fluorescent protein. Biophys J 83, 3589–3595, doi: 10.1016/S0006-3495(02)75359-9 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieger B., Junge W. & Busch K. B. Lateral pH gradient between OXPHOS complex IV and F0F1 ATP-synthase in folded mitochondrial membranes. Nature communications 5, 3103, doi: 10.1038/ncomms4103 (2014). [DOI] [PubMed] [Google Scholar]
- Peter M. et al. Multiphoton-FLIM quantification of the EGFP-mRFP1 FRET pair for localization of membrane receptor-kinase interactions. Biophysical journal 88, 1224–1237, doi: 10.1529/biophysj.104.050153 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkens V., Kohl W. & Busch K. Restricted diffusion of OXPHOS complexes in dynamic mitochondria delays their exchange between cristae and engenders a transitory mosaic distribution. Journal of cell science 126, 103–116, doi: 10.1242/jcs.108852 (2013). [DOI] [PubMed] [Google Scholar]
- Griesbeck O., Baird G. S., Campbell R. E., Zacharias D. A. & Tsien R. Y. Reducing the environmental sensitivity of yellow fluorescent protein. Mechanism and applications. J Biol Chem 276, 29188–29194, doi: 10.1074/jbc.M102815200 (2001). [DOI] [PubMed] [Google Scholar]
- Muster B. et al. Respiratory chain complexes in dynamic mitochondria display a patchy distribution in life cells. PLoS One 5, e11910, doi: 10.1371/journal.pone.0011910 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedlak E. & Robinson N. C. Phospholipase A(2) digestion of cardiolipin bound to bovine cytochrome c oxidase alters both activity and quaternary structure. Biochemistry 38, 14966–14972 (1999). [DOI] [PubMed] [Google Scholar]
- Estey L. A. & Prochaska L. J. Detection of bovine heart mitochondrial cytochrome c oxidase dimers in Triton X-100 and phospholipid vesicles by chemical cross-linking. Biochemistry 32, 13270–13276 (1993). [DOI] [PubMed] [Google Scholar]
- Muramoto K. et al. A histidine residue acting as a controlling site for dioxygen reduction and proton pumping by cytochrome c oxidase. Proc Natl Acad Sci USA 104, 7881–7886, doi: 10.1073/pnas.0610031104 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura H. et al. Visualization of ATP levels inside single living cells with fluorescence resonance energy transfer-based genetically encoded indicators. P Natl Acad Sci USA 106, 15651–15656, doi: 10.1073/pnas.0904764106 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinzawa-Itoh K. et al. Purification of Active Respiratory Supercomplex from Bovine Heart Mitochondria Enables Functional Studies. J Biol Chem 291, 4178–4184, doi: 10.1074/jbc.M115.680553 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schagger H. & Pfeiffer K. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. Embo Journal 19, 1777–1783 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin P. D., Prescott M. & Devenish R. J. F1F0-ATP synthase complex interactions in vivo can occur in the absence of the dimer specific subunit e. J Bioenerg Biomembr 37, 55–66, doi: 10.1007/s10863-005-4128-8 (2005). [DOI] [PubMed] [Google Scholar]
- Berezin M. Y. & Achilefu S. Fluorescence lifetime measurements and biological imaging. Chem Rev 110, 2641–2684, doi: 10.1021/cr900343z (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acin-Perez R. et al. Respiratory complex III is required to maintain complex I in mammalian mitochondria. Mol Cell 13, 805–815 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz F., Fukui H., Garcia S. & Moraes C. T. Cytochrome c oxidase is required for the assembly/stability of respiratory complex I in mouse fibroblasts. Molecular and cellular biology 26, 4872–4881, doi: 10.1128/mcb.01767-05 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakely E. L. et al. A mitochondrial cytochrome b mutation causing severe respiratory chain enzyme deficiency in humans and yeast. The FEBS journal 272, 3583–3592, doi: 10.1111/j.1742-4658.2005.04779.x (2005). [DOI] [PubMed] [Google Scholar]
- Davoudi M., Kotarsky H., Hansson E., Kallijarvi J. & Fellman V. COX7A2L/SCAFI and Pre-Complex III Modify Respiratory Chain Supercomplex Formation in Different Mouse Strains with a Bcs1l Mutation. PLoS One 11, e0168774, doi: 10.1371/journal.pone.0168774 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.