The title compounds, (I) and (II), are 2-[(diaminopyrimidin-2-yl)sulfanyl]acetamides. The molecules have a folded conformation, with the pyrimidine ring being inclined to the benzene ring by 42.25 (14)° in (I), and by 59.70 (16) and 62.18 (15)° in the two independent molecules of compound (II).
Keywords: crystal structure, diaminopyrimidin(2-yl) derivatives, thioacteamide, hydrogen bonding, C—H⋯π interactions, offset π–π interactions
Abstract
The title compounds, C12H12ClN5OS, (I), and C12H12ClN5OS, (II), are 2-[(diaminopyrimidin-2-yl)sulfanyl]acetamides. Compound (II), crystallizes with two independent molecules (A and B) in the asymmetric unit. In each of the molecules, in both (I) and (II), an intramolecular N—H⋯N hydrogen bond forms an S(7) ring motif. The pyrimidine ring is inclined to the benzene ring by 42.25 (14)° in (I), and by 59.70 (16) and 62.18 (15)° in molecules A and B, respectively, of compound (II). In the crystal of (I), molecules are linked by pairs of N—H⋯N hydrogen bonds, forming inversion dimers with an R 2 2(8) ring motif. The dimers are linked via bifurcated N—H⋯O and C—H⋯O hydrogen bonds, forming corrugated layers parallel to the ac plane. In the crystal of (II), the A molecules are linked through N—H⋯O and N—H⋯Cl hydrogen bonds, forming layers parallel to (100). The B molecules are also linked by N—H⋯O and N—H⋯Cl hydrogen bonds, also forming layers parallel to (100). The parallel layers of A and B molecules are linked via N—H⋯N hydrogen bonds, forming a three-dimensional structure.
Chemical context
Diaminopyrimidine derivatives are reported to be therapeutic agents towards anti-cancer activity, selectively inhibiting c-Fms kinase of M-CSF-dependent myeloid leukemia cells (Xu et al., 2010 ▸). They have also shown antibacterial activity (Kandeel et al., 1994 ▸), are potential antimicrobial (Holla et al., 2006 ▸) and anti-AIDS agents (Nogueras et al., 1993 ▸) and antiviral agents (Hocková et al., 2003 ▸, 2004 ▸) and have shown promise as immunosuppressants (Blumenkopf et al., 2002 ▸). In this connection, the title 2-[(4,6-diaminopyrimidin-2-yl)sulfanyl] based analogues have been synthesized as antiviral agents against NS2B/NS3 Dengue protease. We report herein on the syntheses and crystal structures of the title compounds, (I) and (II).
Structural commentary
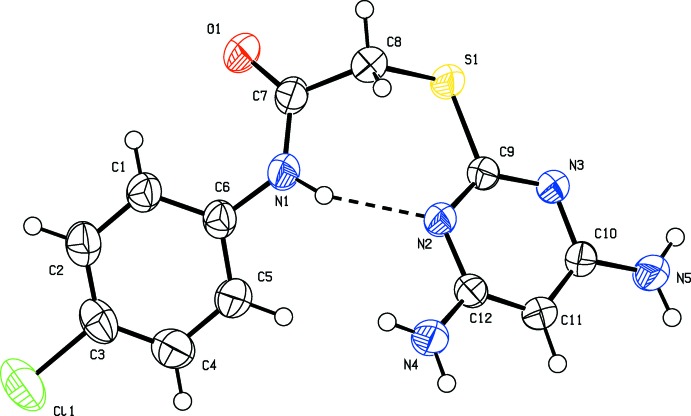
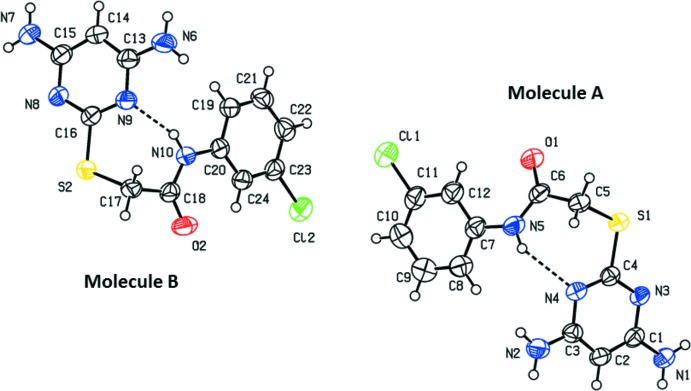
The molecular structures of compounds (I) and (II) are shown in Figs. 1 ▸ and 2 ▸, respectively. Compound (II) crystallizes with two independent molecules (A and B) in the asymmetric unit, which have similar conformations (Fig. 3 ▸). The molecules of both compounds are folded with the pyrimidine ring being inclined to the benzene ring by 42.25 (14)° in (I), and by 59.70 (16) and 62.18 (15)° in molecules A and B, respectively, of compound (II). In compound (I), the N1—C7—C8—S1 torsion angle is 77.2 (3)°, while in molecule A of compound (II), N5—C6—C5—S1 is 85.2 (3)°, and in molecule B N10—C18—C17—S2 is 68.4 (3)°.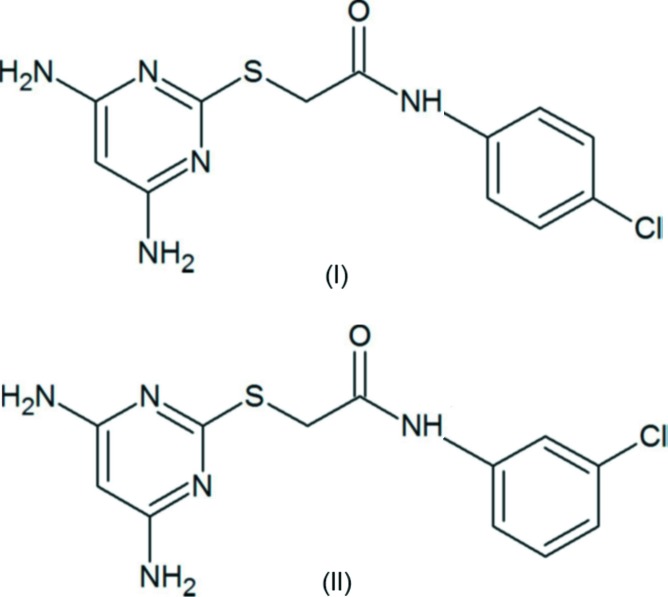
Figure 1.
The molecular structure of compound (I), with the atom labelling. Displacement ellipsoids are drawn at the 30% probability level. The intramolecular N—H⋯N hydrogen bond is shown as a dashed lines (see Table 1 ▸).
Figure 2.
The molecular structure of the two independent molecules (A and B) of compound (II), with the atom labelling. Displacement ellipsoids are drawn at the 30% probability level. The intramolecular N—H⋯N hydrogen bonds are shown as dashed lines (see Table 2 ▸).
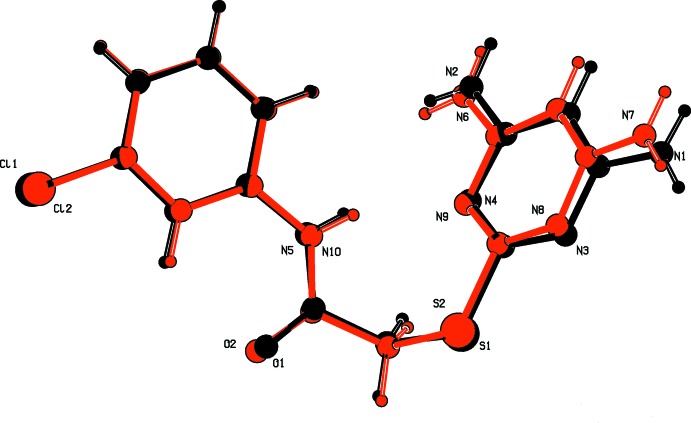
Figure 3.
An AutoMolFit (PLATON; Spek, 2009 ▸) view of molecule B (red) on molecule A (back) of (II).
In compound (I), the intramolecular N1—H1A⋯N2 hydrogen bond (Table 1 ▸) generates an S(7) ring motif, as shown in Fig. 1 ▸. Amine atoms N4 and N5 attached to the pyrimidine ring deviate by 0.018 (3) and 0.060 (3) Å, respectively. The chlorine atom Cl1 attached to the benzene ring deviates by 0.058 (1) Å.
Table 1. Hydrogen-bond geometry (Å, °) for (I) .
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1A⋯N2 | 0.86 | 2.06 | 2.856 (3) | 154 |
| N5—H5A⋯N3i | 0.86 | 2.16 | 2.990 (3) | 162 |
| N4—H4B⋯O1ii | 0.86 | 2.22 | 2.969 (3) | 146 |
| C11—H11⋯O1ii | 0.93 | 2.45 | 3.144 (3) | 132 |
Symmetry codes: (i) 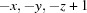 ; (ii)
; (ii) 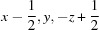 .
.
In compound (II), intramolecular N5—H5⋯N4 and N10—H10⋯N9 hydrogen bonds (Table 2 ▸ and Fig. 2 ▸) in molecules A and B, respectively, also generate S(7) ring motifs. In molecule A, the amine group atoms N1 and N2 attached to the pyrimidine ring deviate by 0.006 (3) and 0.004 (3) Å, respectively. The chlorine atom Cl1 attached to the benzene ring deviates by 0.013 (1) Å. In molecule B, the amine group atoms N6 and N7 attached to the pyrimidine ring deviate by −0.003 (3) and 0.050 (3) Å, respectively. Atom Cl2 attached to the benzene ring deviates by 0.074 (1) Å.
Table 2. Hydrogen-bond geometry (Å, °) for (II) .
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N5—H5⋯N4 | 0.86 | 2.25 | 2.962 (3) | 140 |
| N10—H10A⋯N9 | 0.86 | 2.02 | 2.826 (3) | 157 |
| N1—H1B⋯O1i | 0.86 | 2.19 | 2.931 (4) | 145 |
| N2—H2B⋯Cl1i | 0.86 | 2.76 | 3.405 (3) | 133 |
| N6—H6A⋯O2ii | 0.86 | 2.51 | 3.340 (4) | 162 |
| N6—H6B⋯Cl2iii | 0.86 | 2.70 | 3.556 (3) | 176 |
| N7—H7B⋯O2iii | 0.86 | 2.24 | 3.002 (4) | 148 |
| N1—H1A⋯N8iv | 0.86 | 2.21 | 3.070 (4) | 174 |
| N7—H7A⋯N3v | 0.86 | 2.19 | 3.046 (4) | 178 |
Symmetry codes: (i) 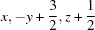 ; (ii)
; (ii) 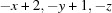 ; (iii)
; (iii) 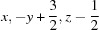 ; (iv)
; (iv) 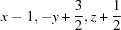 ; (v)
; (v) 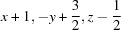 .
.
Supramolecular features
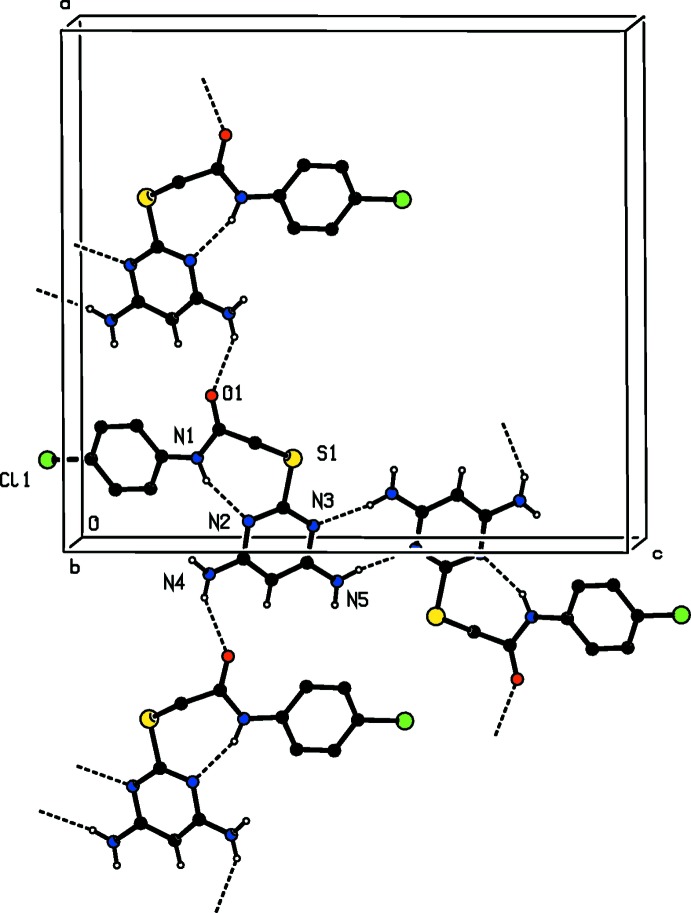
In the crystal of (I), molecules are linked by pairs of N—H⋯N hydrogen bonds, forming inversion dimers with an 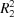 (8) ring motif (Table 1 ▸ and Fig. 4 ▸). The dimers are linked by via bifurcated N—H⋯O and C—H⋯O hydrogen bonds, forming corrugated layers parallel to the ac plane (Table 1 ▸ and Fig. 5 ▸).
(8) ring motif (Table 1 ▸ and Fig. 4 ▸). The dimers are linked by via bifurcated N—H⋯O and C—H⋯O hydrogen bonds, forming corrugated layers parallel to the ac plane (Table 1 ▸ and Fig. 5 ▸).
Figure 4.
The crystal packing of compound (I) viewed along the b axis. H atoms not involved in hydrogen bonding (see Table 1 ▸), have been excluded for clarity.
Figure 5.
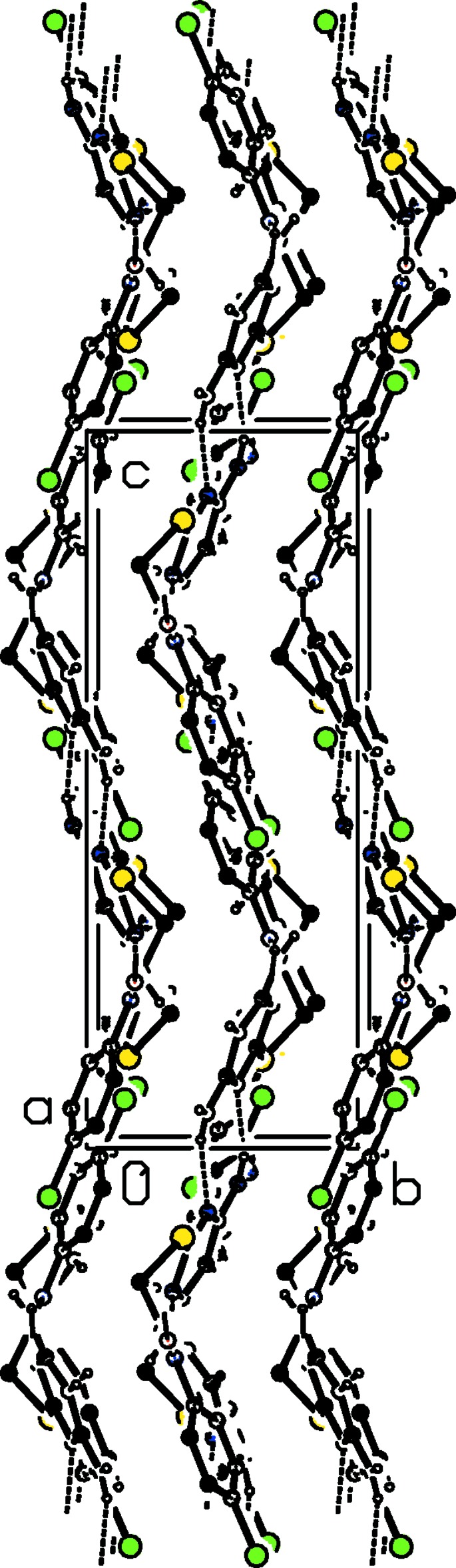
The crystal packing of compound (I) viewed along the a axis. H atoms not involved in hydrogen bonding (see Table 1 ▸), have been excluded for clarity.
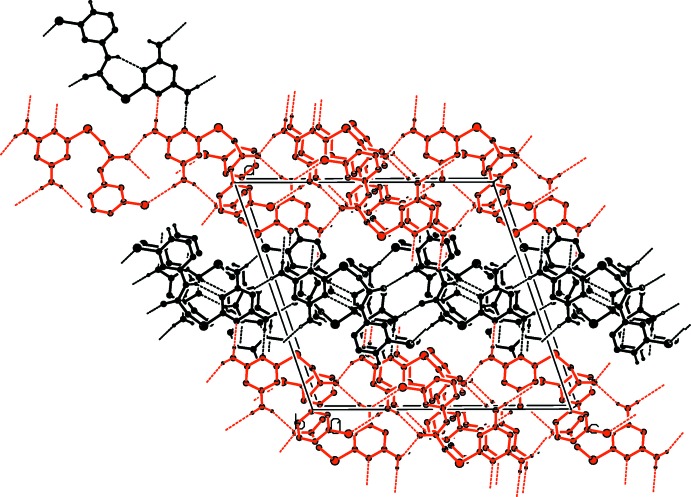
In the crystal of (II), the A molecules are linked through N—H⋯O and N—H⋯Cl hydrogen bonds, forming layers parallel to (100). Likewise the B molecules are also linked by N—H⋯O and N—H⋯Cl hydrogen bonds, forming layers parallel to (100). The parallel layers of A and layers of B molecules are linked via N—H⋯N hydrogen bonds, forming a three-dimensional structure (Table 2 ▸ and Fig. 6 ▸).
Figure 6.
The crystal packing of compound (II) viewed along the b axis (colour code: A molecules black, B molecules red). H atoms not involved in hydrogen bonding (see Table 2 ▸), have been excluded for clarity.
Database survey
A search of the Cambridge Structural Database (Version 5.37, update May 2016; Groom et al., 2016 ▸) for 2-[(pyrimidine-2-yl)sulfanyl]-N-phenylacetamide yielded five hits. Three of these involve (4,6-diaminopyrmidin-2-yl) groups. They include the 2-chlorophenyl analogue, N-(2-chlorophenyl)-2-[(4,6-diaminopyrimidin-2-yl)sulfanyl]acetamide (ARARUI; Subasri et al., 2016 ▸). Here the pyrimidine and benzene rings are inclined to one another by 67.84 (6)°, compared to 42.25 (14)° in (I), and 59.70 (16) and 62.18 (15)° in molecules A and B, respectively, of compound (II). As in the title compounds, there is also an intramolecular N—H⋯N hydrogen bond present, stabilizing the folded conformation of the molecule.
Synthesis and crystallization
Compound (I):
To a solution of 4,6-diamino-pyrimidine-2-sulfanyl (0.5 g; 3.52 mmol) in 25 ml of ethanol, was added potassium hydroxide (0.2g; 3.52 mmol) and the mixture was refluxed for 30 min, after which 3.52 mmol of 2-chloro-N-(4-chlorophenyl)acetamide derivative was added and refluxed for 4 h. At the end of the reaction (monitored by TLC), the ethanol was evaporated in vacuo and cold water was added; the precipitate formed was filtered and dried to give compound (I) as a crystalline powder (yield 97%). Colourless block-like crystals were obtained from a solution in methanol and ethyl acetate (1:1) by slow evaporation of the solvents at room temperature.
Compound (II):
To a solution of 4,6-diamino-pyrimidine-2-thiol (0.5 g; 3.52 mmol) in 25 ml of ethanol was added potassium hydroxide (0.2g; 3.52 mmol) and the mixture was refluxed for 30 min. Then 3.52 mmol of 2-chloro-N-(3-chlorophenyl)acetamide was added and refluxed for 3 h. At the end of the reaction (monitored by TLC), the ethanol was evaporated in vacuo and cold water was added and the precipitate formed was filtered and dried to give compound (II) as a crystalline powder (yield 92%). Colourless block-like crystals were obtained from a solution in methanol and ethyl acetate (2:1) by slow evaporation of the solvents at room temperature.
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 3 ▸. For both (I) and (II), hydrogen atoms were placed in calculated positions and refined as riding: C—H = 0.93–0.97 Å and N—H = 0.86 Å, with U iso(H) = 1.2U eq(N,C).
Table 3. Experimental details.
| (I) | (II) | |
|---|---|---|
| Crystal data | ||
| Chemical formula | C12H12ClN5OS | C12H12ClN5OS |
| M r | 309.78 | 309.78 |
| Crystal system, space group | Orthorhombic, P b c a | Monoclinic, P21/c |
| Temperature (K) | 293 | 293 |
| a, b, c (Å) | 18.2743 (12), 7.4835 (5), 19.8021 (12) | 18.220 (2), 8.1180 (12), 19.628 (2) |
| α, β, γ (°) | 90, 90, 90 | 90, 108.761 (8), 90 |
| V (Å3) | 2708.1 (3) | 2748.9 (6) |
| Z | 8 | 8 |
| Radiation type | Mo Kα | Mo Kα |
| μ (mm−1) | 0.44 | 0.43 |
| Crystal size (mm) | 0.28 × 0.25 × 0.18 | 0.31 × 0.22 × 0.16 |
| Data collection | ||
| Diffractometer | Bruker SMART APEXII area-detector | Bruker SMART APEXII area-detector |
| Absorption correction | Multi-scan (SADABS; Bruker, 2008 ▸) | Multi-scan (SADABS; Bruker, 2008 ▸) |
| T min, T max | 0.741, 0.863 | 0.742, 0.892 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 22685, 3372, 2007 | 25810, 6858, 3462 |
| R int | 0.060 | 0.075 |
| (sin θ/λ)max (Å−1) | 0.669 | 0.668 |
| Refinement | ||
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.050, 0.161, 1.08 | 0.050, 0.155, 0.96 |
| No. of reflections | 3372 | 6858 |
| No. of parameters | 181 | 361 |
| H-atom treatment | H-atom parameters constrained | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.42, −0.69 | 0.42, −0.42 |
Supplementary Material
Crystal structure: contains datablock(s) global, I, II. DOI: 10.1107/S2056989017003243/su5348sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989017003243/su5348Isup4.hkl
Structure factors: contains datablock(s) II. DOI: 10.1107/S2056989017003243/su5348IIsup3.hkl
Supporting information file. DOI: 10.1107/S2056989017003243/su5348Isup4.cml
Supporting information file. DOI: 10.1107/S2056989017003243/su5348IIsup5.cml
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
The authors thank the TBI X-ray facility, CAS in Crystallography and Biophysics, University of Madras, India, for the data collection. SS and DV thank the UGC (SAP–CAS) for the departmental facilities. SS also thanks UGC for the award of a meritorious fellowship.
supplementary crystallographic information
(I) N-(4-Chlorophenyl)-2-[(4,6-diaminopyrimidin-2-yl)sulfanyl]acetamide . Crystal data
| C12H12ClN5OS | Dx = 1.520 Mg m−3 |
| Mr = 309.78 | Mo Kα radiation, λ = 0.71073 Å |
| Orthorhombic, Pbca | Cell parameters from 3372 reflections |
| a = 18.2743 (12) Å | θ = 2.1–28.4° |
| b = 7.4835 (5) Å | µ = 0.44 mm−1 |
| c = 19.8021 (12) Å | T = 293 K |
| V = 2708.1 (3) Å3 | Block, colourless |
| Z = 8 | 0.28 × 0.25 × 0.18 mm |
| F(000) = 1280 |
(I) N-(4-Chlorophenyl)-2-[(4,6-diaminopyrimidin-2-yl)sulfanyl]acetamide . Data collection
| Bruker SMART APEXII area-detector diffractometer | 2007 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.060 |
| ω and φ scans | θmax = 28.4°, θmin = 2.1° |
| Absorption correction: multi-scan (SADABS; Bruker, 2008) | h = −23→24 |
| Tmin = 0.741, Tmax = 0.863 | k = −9→9 |
| 22685 measured reflections | l = −26→26 |
| 3372 independent reflections |
(I) N-(4-Chlorophenyl)-2-[(4,6-diaminopyrimidin-2-yl)sulfanyl]acetamide . Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.050 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.161 | H-atom parameters constrained |
| S = 1.08 | w = 1/[σ2(Fo2) + (0.0795P)2 + 0.3384P] where P = (Fo2 + 2Fc2)/3 |
| 3372 reflections | (Δ/σ)max < 0.001 |
| 181 parameters | Δρmax = 0.42 e Å−3 |
| 0 restraints | Δρmin = −0.69 e Å−3 |
(I) N-(4-Chlorophenyl)-2-[(4,6-diaminopyrimidin-2-yl)sulfanyl]acetamide . Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
(I) N-(4-Chlorophenyl)-2-[(4,6-diaminopyrimidin-2-yl)sulfanyl]acetamide . Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.21682 (15) | 0.1105 (4) | 0.10059 (13) | 0.0467 (7) | |
| H1 | 0.259655 | 0.165974 | 0.114873 | 0.056* | |
| C2 | 0.21206 (16) | 0.0419 (4) | 0.03543 (13) | 0.0524 (7) | |
| H2 | 0.251506 | 0.052014 | 0.006002 | 0.063* | |
| C3 | 0.14875 (17) | −0.0409 (4) | 0.01479 (13) | 0.0523 (7) | |
| C4 | 0.08942 (17) | −0.0537 (4) | 0.05714 (15) | 0.0563 (8) | |
| H4 | 0.046632 | −0.108550 | 0.042405 | 0.068* | |
| C5 | 0.09364 (15) | 0.0152 (4) | 0.12158 (14) | 0.0505 (7) | |
| H5 | 0.053355 | 0.007491 | 0.150122 | 0.061* | |
| C6 | 0.15764 (13) | 0.0963 (4) | 0.14435 (12) | 0.0411 (6) | |
| C7 | 0.21262 (13) | 0.2114 (4) | 0.25020 (12) | 0.0417 (6) | |
| C8 | 0.18937 (14) | 0.2955 (4) | 0.31653 (13) | 0.0466 (7) | |
| H8A | 0.151615 | 0.383361 | 0.307401 | 0.056* | |
| H8B | 0.230953 | 0.358263 | 0.335629 | 0.056* | |
| C9 | 0.06049 (13) | 0.1256 (4) | 0.36033 (12) | 0.0370 (6) | |
| C10 | −0.04825 (13) | 0.0187 (4) | 0.39846 (12) | 0.0388 (6) | |
| C11 | −0.07960 (13) | 0.0629 (4) | 0.33698 (12) | 0.0423 (6) | |
| H11 | −0.127989 | 0.033707 | 0.327503 | 0.051* | |
| C12 | −0.03712 (13) | 0.1511 (4) | 0.29042 (12) | 0.0384 (6) | |
| N1 | 0.15680 (11) | 0.1680 (3) | 0.20979 (10) | 0.0439 (6) | |
| H1A | 0.113996 | 0.186830 | 0.226380 | 0.053* | |
| N2 | 0.03568 (10) | 0.1801 (3) | 0.30081 (10) | 0.0393 (5) | |
| N3 | 0.02360 (10) | 0.0518 (3) | 0.41104 (10) | 0.0396 (5) | |
| N4 | −0.06459 (12) | 0.2133 (4) | 0.23153 (10) | 0.0513 (6) | |
| H4A | −0.036471 | 0.267317 | 0.203330 | 0.062* | |
| H4B | −0.110188 | 0.198752 | 0.222276 | 0.062* | |
| N5 | −0.08599 (12) | −0.0543 (4) | 0.44936 (11) | 0.0533 (7) | |
| H5A | −0.064552 | −0.077179 | 0.487041 | 0.064* | |
| H5B | −0.131697 | −0.078337 | 0.444396 | 0.064* | |
| O1 | 0.27697 (10) | 0.1893 (4) | 0.23652 (10) | 0.0662 (7) | |
| S1 | 0.15519 (3) | 0.13935 (11) | 0.37853 (3) | 0.0458 (2) | |
| CL1 | 0.14411 (6) | −0.13289 (15) | −0.06572 (4) | 0.0831 (4) |
(I) N-(4-Chlorophenyl)-2-[(4,6-diaminopyrimidin-2-yl)sulfanyl]acetamide . Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0401 (14) | 0.061 (2) | 0.0392 (13) | 0.0034 (13) | 0.0018 (11) | 0.0079 (13) |
| C2 | 0.0559 (17) | 0.060 (2) | 0.0414 (14) | 0.0041 (15) | 0.0078 (13) | 0.0057 (14) |
| C3 | 0.076 (2) | 0.0448 (17) | 0.0356 (13) | −0.0015 (15) | 0.0010 (13) | 0.0034 (12) |
| C4 | 0.0668 (19) | 0.054 (2) | 0.0478 (15) | −0.0159 (15) | −0.0061 (14) | 0.0062 (15) |
| C5 | 0.0508 (16) | 0.056 (2) | 0.0446 (15) | −0.0121 (14) | 0.0037 (12) | 0.0070 (14) |
| C6 | 0.0404 (14) | 0.0481 (17) | 0.0349 (12) | 0.0039 (12) | 0.0025 (10) | 0.0061 (12) |
| C7 | 0.0361 (13) | 0.0500 (17) | 0.0390 (13) | 0.0010 (12) | 0.0049 (11) | 0.0045 (12) |
| C8 | 0.0396 (14) | 0.0560 (18) | 0.0441 (14) | −0.0070 (12) | 0.0021 (11) | −0.0017 (13) |
| C9 | 0.0320 (12) | 0.0451 (16) | 0.0338 (11) | 0.0015 (11) | 0.0008 (9) | −0.0041 (11) |
| C10 | 0.0361 (13) | 0.0442 (16) | 0.0362 (12) | −0.0026 (11) | 0.0054 (10) | −0.0040 (11) |
| C11 | 0.0329 (13) | 0.0544 (18) | 0.0396 (13) | −0.0040 (11) | −0.0016 (11) | −0.0011 (12) |
| C12 | 0.0347 (13) | 0.0466 (17) | 0.0339 (12) | 0.0035 (11) | −0.0013 (10) | −0.0046 (11) |
| N1 | 0.0330 (11) | 0.0617 (16) | 0.0370 (11) | 0.0029 (10) | 0.0041 (9) | 0.0006 (10) |
| N2 | 0.0307 (10) | 0.0526 (14) | 0.0344 (10) | 0.0011 (9) | 0.0008 (9) | 0.0016 (10) |
| N3 | 0.0328 (11) | 0.0517 (15) | 0.0344 (10) | −0.0028 (9) | 0.0001 (8) | 0.0007 (10) |
| N4 | 0.0372 (12) | 0.0783 (19) | 0.0385 (11) | −0.0016 (12) | −0.0058 (10) | 0.0095 (12) |
| N5 | 0.0426 (13) | 0.0767 (19) | 0.0405 (11) | −0.0161 (12) | −0.0008 (10) | 0.0088 (12) |
| O1 | 0.0304 (10) | 0.114 (2) | 0.0538 (12) | 0.0020 (11) | 0.0062 (8) | −0.0101 (12) |
| S1 | 0.0312 (3) | 0.0698 (5) | 0.0363 (3) | −0.0015 (3) | −0.0016 (3) | 0.0060 (3) |
| CL1 | 0.1216 (9) | 0.0854 (7) | 0.0422 (4) | −0.0171 (6) | 0.0013 (4) | −0.0090 (4) |
(I) N-(4-Chlorophenyl)-2-[(4,6-diaminopyrimidin-2-yl)sulfanyl]acetamide . Geometric parameters (Å, º)
| C1—C6 | 1.390 (3) | C8—H8B | 0.9700 |
| C1—C2 | 1.391 (4) | C9—N2 | 1.327 (3) |
| C1—H1 | 0.9300 | C9—N3 | 1.329 (3) |
| C2—C3 | 1.375 (4) | C9—S1 | 1.771 (2) |
| C2—H2 | 0.9300 | C10—N5 | 1.338 (3) |
| C3—C4 | 1.374 (4) | C10—N3 | 1.359 (3) |
| C3—CL1 | 1.739 (3) | C10—C11 | 1.386 (3) |
| C4—C5 | 1.378 (4) | C11—C12 | 1.374 (4) |
| C4—H4 | 0.9300 | C11—H11 | 0.9300 |
| C5—C6 | 1.393 (4) | C12—N4 | 1.352 (3) |
| C5—H5 | 0.9300 | C12—N2 | 1.364 (3) |
| C6—N1 | 1.403 (3) | N1—H1A | 0.8600 |
| C7—O1 | 1.218 (3) | N4—H4A | 0.8600 |
| C7—N1 | 1.337 (3) | N4—H4B | 0.8600 |
| C7—C8 | 1.517 (4) | N5—H5A | 0.8600 |
| C8—S1 | 1.807 (3) | N5—H5B | 0.8600 |
| C8—H8A | 0.9700 | ||
| C6—C1—C2 | 120.1 (3) | H8A—C8—H8B | 107.6 |
| C6—C1—H1 | 120.0 | N2—C9—N3 | 128.7 (2) |
| C2—C1—H1 | 120.0 | N2—C9—S1 | 119.79 (18) |
| C3—C2—C1 | 119.7 (3) | N3—C9—S1 | 111.47 (17) |
| C3—C2—H2 | 120.2 | N5—C10—N3 | 115.8 (2) |
| C1—C2—H2 | 120.2 | N5—C10—C11 | 123.1 (2) |
| C4—C3—C2 | 120.9 (3) | N3—C10—C11 | 121.1 (2) |
| C4—C3—CL1 | 119.6 (2) | C12—C11—C10 | 118.1 (2) |
| C2—C3—CL1 | 119.5 (2) | C12—C11—H11 | 121.0 |
| C3—C4—C5 | 119.7 (3) | C10—C11—H11 | 121.0 |
| C3—C4—H4 | 120.2 | N4—C12—N2 | 116.0 (2) |
| C5—C4—H4 | 120.2 | N4—C12—C11 | 122.3 (2) |
| C4—C5—C6 | 120.7 (3) | N2—C12—C11 | 121.7 (2) |
| C4—C5—H5 | 119.7 | C7—N1—C6 | 129.6 (2) |
| C6—C5—H5 | 119.7 | C7—N1—H1A | 115.2 |
| C1—C6—C5 | 119.0 (2) | C6—N1—H1A | 115.2 |
| C1—C6—N1 | 123.7 (2) | C9—N2—C12 | 114.7 (2) |
| C5—C6—N1 | 117.2 (2) | C9—N3—C10 | 115.3 (2) |
| O1—C7—N1 | 124.8 (3) | C12—N4—H4A | 120.0 |
| O1—C7—C8 | 121.3 (2) | C12—N4—H4B | 120.0 |
| N1—C7—C8 | 113.9 (2) | H4A—N4—H4B | 120.0 |
| C7—C8—S1 | 114.6 (2) | C10—N5—H5A | 120.0 |
| C7—C8—H8A | 108.6 | C10—N5—H5B | 120.0 |
| S1—C8—H8A | 108.6 | H5A—N5—H5B | 120.0 |
| C7—C8—H8B | 108.6 | C9—S1—C8 | 103.73 (12) |
| S1—C8—H8B | 108.6 | ||
| C6—C1—C2—C3 | −0.4 (4) | O1—C7—N1—C6 | −2.5 (5) |
| C1—C2—C3—C4 | 1.4 (5) | C8—C7—N1—C6 | 176.8 (3) |
| C1—C2—C3—CL1 | −177.9 (2) | C1—C6—N1—C7 | −22.2 (5) |
| C2—C3—C4—C5 | −0.9 (5) | C5—C6—N1—C7 | 161.4 (3) |
| CL1—C3—C4—C5 | 178.4 (2) | N3—C9—N2—C12 | −2.0 (4) |
| C3—C4—C5—C6 | −0.5 (5) | S1—C9—N2—C12 | 175.36 (19) |
| C2—C1—C6—C5 | −1.0 (4) | N4—C12—N2—C9 | 176.9 (2) |
| C2—C1—C6—N1 | −177.3 (3) | C11—C12—N2—C9 | −3.5 (4) |
| C4—C5—C6—C1 | 1.5 (4) | N2—C9—N3—C10 | 4.2 (4) |
| C4—C5—C6—N1 | 178.1 (3) | S1—C9—N3—C10 | −173.28 (19) |
| O1—C7—C8—S1 | −103.4 (3) | N5—C10—N3—C9 | −179.5 (2) |
| N1—C7—C8—S1 | 77.2 (3) | C11—C10—N3—C9 | −1.0 (4) |
| N5—C10—C11—C12 | 174.5 (3) | N2—C9—S1—C8 | 17.9 (2) |
| N3—C10—C11—C12 | −3.8 (4) | N3—C9—S1—C8 | −164.3 (2) |
| C10—C11—C12—N4 | −174.2 (3) | C7—C8—S1—C9 | −88.3 (2) |
| C10—C11—C12—N2 | 6.3 (4) |
(I) N-(4-Chlorophenyl)-2-[(4,6-diaminopyrimidin-2-yl)sulfanyl]acetamide . Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1A···N2 | 0.86 | 2.06 | 2.856 (3) | 154 |
| N5—H5A···N3i | 0.86 | 2.16 | 2.990 (3) | 162 |
| N4—H4B···O1ii | 0.86 | 2.22 | 2.969 (3) | 146 |
| C11—H11···O1ii | 0.93 | 2.45 | 3.144 (3) | 132 |
Symmetry codes: (i) −x, −y, −z+1; (ii) x−1/2, y, −z+1/2.
(II) N-(3-Chlorophenyl)-2-[(4,6-diaminopyrimidin-2-yl)sulfanyl]acetamide . Crystal data
| C12H12ClN5OS | F(000) = 1280 |
| Mr = 309.78 | Dx = 1.497 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| a = 18.220 (2) Å | Cell parameters from 6858 reflections |
| b = 8.1180 (12) Å | θ = 1.2–28.4° |
| c = 19.628 (2) Å | µ = 0.43 mm−1 |
| β = 108.761 (8)° | T = 293 K |
| V = 2748.9 (6) Å3 | Block, colourless |
| Z = 8 | 0.31 × 0.22 × 0.16 mm |
(II) N-(3-Chlorophenyl)-2-[(4,6-diaminopyrimidin-2-yl)sulfanyl]acetamide . Data collection
| Bruker SMART APEXII area-detector diffractometer | 3462 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.075 |
| ω and φ scans | θmax = 28.4°, θmin = 1.2° |
| Absorption correction: multi-scan (SADABS; Bruker, 2008) | h = −24→24 |
| Tmin = 0.742, Tmax = 0.892 | k = −10→10 |
| 25810 measured reflections | l = −26→26 |
| 6858 independent reflections |
(II) N-(3-Chlorophenyl)-2-[(4,6-diaminopyrimidin-2-yl)sulfanyl]acetamide . Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.050 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.155 | H-atom parameters constrained |
| S = 0.96 | w = 1/[σ2(Fo2) + (0.0646P)2 + 0.6656P] where P = (Fo2 + 2Fc2)/3 |
| 6858 reflections | (Δ/σ)max = 0.001 |
| 361 parameters | Δρmax = 0.42 e Å−3 |
| 0 restraints | Δρmin = −0.42 e Å−3 |
(II) N-(3-Chlorophenyl)-2-[(4,6-diaminopyrimidin-2-yl)sulfanyl]acetamide . Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
(II) N-(3-Chlorophenyl)-2-[(4,6-diaminopyrimidin-2-yl)sulfanyl]acetamide . Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.43039 (17) | 0.6518 (4) | 0.39062 (15) | 0.0438 (7) | |
| C2 | 0.50489 (17) | 0.5880 (4) | 0.41809 (15) | 0.0461 (8) | |
| H2 | 0.533524 | 0.599968 | 0.466472 | 0.055* | |
| C3 | 0.53498 (16) | 0.5069 (4) | 0.37174 (15) | 0.0424 (7) | |
| C4 | 0.42495 (16) | 0.5606 (4) | 0.27980 (15) | 0.0400 (7) | |
| C5 | 0.42189 (17) | 0.4300 (4) | 0.14672 (16) | 0.0485 (8) | |
| H5A | 0.448033 | 0.345147 | 0.180668 | 0.058* | |
| H5B | 0.385949 | 0.375940 | 0.105242 | 0.058* | |
| C6 | 0.48119 (18) | 0.5233 (4) | 0.12307 (16) | 0.0473 (8) | |
| C7 | 0.61562 (17) | 0.6320 (4) | 0.17094 (16) | 0.0476 (8) | |
| C8 | 0.67034 (19) | 0.6788 (5) | 0.23523 (18) | 0.0592 (9) | |
| H8 | 0.663419 | 0.650619 | 0.278689 | 0.071* | |
| C9 | 0.7347 (2) | 0.7666 (5) | 0.2351 (2) | 0.0737 (11) | |
| H9 | 0.770444 | 0.800365 | 0.278367 | 0.088* | |
| C10 | 0.7465 (2) | 0.8045 (5) | 0.1718 (2) | 0.0663 (10) | |
| H10 | 0.790323 | 0.863055 | 0.171667 | 0.080* | |
| C11 | 0.69326 (19) | 0.7553 (4) | 0.10864 (18) | 0.0554 (9) | |
| C12 | 0.62693 (19) | 0.6684 (4) | 0.10619 (18) | 0.0538 (8) | |
| H12 | 0.591368 | 0.635782 | 0.062620 | 0.065* | |
| C13 | 1.07720 (18) | 0.5427 (4) | −0.18594 (16) | 0.0518 (8) | |
| C14 | 1.11114 (17) | 0.6066 (4) | −0.23360 (16) | 0.0545 (9) | |
| H14 | 1.085128 | 0.607744 | −0.282867 | 0.065* | |
| C15 | 1.18621 (18) | 0.6699 (4) | −0.20499 (16) | 0.0504 (8) | |
| C16 | 1.18492 (16) | 0.6063 (4) | −0.09333 (15) | 0.0422 (7) | |
| C17 | 1.17491 (16) | 0.5113 (4) | 0.04034 (15) | 0.0474 (8) | |
| H17A | 1.152219 | 0.418732 | 0.009504 | 0.057* | |
| H17B | 1.207528 | 0.467064 | 0.085940 | 0.057* | |
| C18 | 1.11047 (17) | 0.6094 (4) | 0.05351 (16) | 0.0457 (7) | |
| C19 | 0.98833 (16) | 0.7549 (4) | −0.01350 (15) | 0.0428 (7) | |
| C20 | 0.93861 (17) | 0.7880 (4) | −0.08218 (16) | 0.0488 (8) | |
| H20 | 0.948465 | 0.742561 | −0.121865 | 0.059* | |
| C21 | 0.87475 (18) | 0.8876 (4) | −0.09216 (17) | 0.0554 (9) | |
| H21 | 0.841809 | 0.907855 | −0.138552 | 0.066* | |
| C22 | 0.85896 (18) | 0.9577 (4) | −0.03444 (17) | 0.0533 (8) | |
| H22 | 0.815921 | 1.024970 | −0.041084 | 0.064* | |
| C23 | 0.90908 (17) | 0.9246 (4) | 0.03344 (16) | 0.0477 (8) | |
| C24 | 0.97354 (17) | 0.8249 (4) | 0.04526 (16) | 0.0460 (8) | |
| H24 | 1.006364 | 0.805075 | 0.091743 | 0.055* | |
| N1 | 0.39450 (15) | 0.7294 (4) | 0.43183 (13) | 0.0584 (8) | |
| H1A | 0.347948 | 0.765256 | 0.412896 | 0.070* | |
| H1B | 0.418159 | 0.742920 | 0.477051 | 0.070* | |
| N2 | 0.60725 (15) | 0.4414 (4) | 0.39269 (13) | 0.0573 (7) | |
| H2A | 0.624904 | 0.396367 | 0.361454 | 0.069* | |
| H2B | 0.635444 | 0.444816 | 0.437208 | 0.069* | |
| N3 | 0.38865 (13) | 0.6359 (3) | 0.31978 (12) | 0.0436 (6) | |
| N4 | 0.49560 (13) | 0.4945 (3) | 0.30007 (12) | 0.0412 (6) | |
| N5 | 0.55000 (14) | 0.5449 (3) | 0.17471 (13) | 0.0501 (7) | |
| H5 | 0.554587 | 0.499513 | 0.215468 | 0.060* | |
| N6 | 1.00456 (16) | 0.4803 (4) | −0.20701 (15) | 0.0811 (11) | |
| H6A | 0.984742 | 0.445292 | −0.175429 | 0.097* | |
| H6B | 0.978147 | 0.475742 | −0.252009 | 0.097* | |
| N7 | 1.22370 (15) | 0.7369 (4) | −0.24649 (14) | 0.0698 (9) | |
| H7A | 1.269792 | 0.775136 | −0.227375 | 0.084* | |
| H7B | 1.201739 | 0.741880 | −0.292327 | 0.084* | |
| N8 | 1.22428 (13) | 0.6664 (3) | −0.13364 (12) | 0.0461 (6) | |
| N9 | 1.11350 (14) | 0.5443 (3) | −0.11441 (12) | 0.0479 (6) | |
| N10 | 1.05376 (13) | 0.6562 (3) | −0.00640 (12) | 0.0448 (6) | |
| H10A | 1.058236 | 0.620508 | −0.046127 | 0.054* | |
| O1 | 0.46540 (13) | 0.5750 (4) | 0.06196 (11) | 0.0714 (7) | |
| O2 | 1.11150 (13) | 0.6397 (3) | 0.11485 (11) | 0.0643 (7) | |
| S1 | 0.36824 (4) | 0.56020 (11) | 0.18798 (4) | 0.0463 (2) | |
| S2 | 1.23535 (4) | 0.62488 (11) | −0.00038 (4) | 0.0492 (2) | |
| CL1 | 0.70803 (5) | 0.80248 (14) | 0.02776 (5) | 0.0738 (3) | |
| CL2 | 0.89352 (5) | 1.01784 (12) | 0.10723 (5) | 0.0649 (3) |
(II) N-(3-Chlorophenyl)-2-[(4,6-diaminopyrimidin-2-yl)sulfanyl]acetamide . Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0507 (17) | 0.045 (2) | 0.0406 (16) | −0.0074 (15) | 0.0221 (14) | −0.0013 (14) |
| C2 | 0.0525 (17) | 0.053 (2) | 0.0330 (15) | −0.0094 (16) | 0.0143 (13) | −0.0020 (15) |
| C3 | 0.0464 (16) | 0.0413 (19) | 0.0398 (16) | −0.0036 (14) | 0.0143 (13) | 0.0040 (14) |
| C4 | 0.0466 (16) | 0.0371 (18) | 0.0393 (15) | −0.0063 (14) | 0.0178 (13) | −0.0023 (13) |
| C5 | 0.0539 (17) | 0.052 (2) | 0.0403 (16) | −0.0006 (16) | 0.0166 (14) | −0.0116 (15) |
| C6 | 0.0552 (18) | 0.055 (2) | 0.0380 (16) | 0.0100 (16) | 0.0232 (15) | 0.0024 (15) |
| C7 | 0.0521 (17) | 0.046 (2) | 0.0499 (18) | 0.0132 (16) | 0.0232 (15) | 0.0032 (16) |
| C8 | 0.061 (2) | 0.069 (3) | 0.0482 (19) | 0.0039 (19) | 0.0180 (17) | 0.0030 (18) |
| C9 | 0.066 (2) | 0.089 (3) | 0.062 (2) | −0.001 (2) | 0.0159 (19) | −0.002 (2) |
| C10 | 0.054 (2) | 0.073 (3) | 0.069 (2) | −0.0075 (19) | 0.0165 (19) | −0.003 (2) |
| C11 | 0.059 (2) | 0.056 (2) | 0.058 (2) | 0.0111 (18) | 0.0301 (17) | 0.0080 (18) |
| C12 | 0.0582 (19) | 0.055 (2) | 0.0527 (19) | 0.0064 (17) | 0.0240 (16) | 0.0013 (17) |
| C13 | 0.0544 (18) | 0.059 (2) | 0.0405 (17) | −0.0068 (17) | 0.0129 (15) | −0.0037 (16) |
| C14 | 0.0539 (18) | 0.075 (3) | 0.0339 (16) | −0.0053 (18) | 0.0128 (14) | −0.0001 (16) |
| C15 | 0.0532 (18) | 0.059 (2) | 0.0424 (17) | 0.0088 (16) | 0.0201 (15) | 0.0042 (16) |
| C16 | 0.0478 (16) | 0.0424 (19) | 0.0394 (15) | 0.0039 (15) | 0.0182 (13) | 0.0018 (14) |
| C17 | 0.0470 (16) | 0.055 (2) | 0.0384 (16) | −0.0037 (15) | 0.0117 (13) | 0.0106 (15) |
| C18 | 0.0487 (17) | 0.050 (2) | 0.0405 (17) | −0.0135 (15) | 0.0176 (14) | −0.0007 (15) |
| C19 | 0.0411 (15) | 0.0448 (19) | 0.0436 (17) | −0.0150 (14) | 0.0151 (13) | −0.0049 (15) |
| C20 | 0.0448 (17) | 0.060 (2) | 0.0409 (17) | −0.0111 (16) | 0.0123 (14) | 0.0017 (16) |
| C21 | 0.0499 (18) | 0.064 (2) | 0.0470 (18) | −0.0147 (17) | 0.0079 (15) | 0.0024 (17) |
| C22 | 0.0452 (17) | 0.053 (2) | 0.060 (2) | −0.0072 (16) | 0.0147 (16) | 0.0037 (17) |
| C23 | 0.0510 (17) | 0.050 (2) | 0.0471 (18) | −0.0123 (16) | 0.0227 (15) | −0.0012 (15) |
| C24 | 0.0470 (17) | 0.050 (2) | 0.0417 (17) | −0.0109 (15) | 0.0149 (14) | 0.0024 (15) |
| N1 | 0.0530 (15) | 0.086 (2) | 0.0404 (14) | 0.0043 (15) | 0.0203 (12) | −0.0148 (14) |
| N2 | 0.0543 (15) | 0.071 (2) | 0.0427 (15) | 0.0115 (15) | 0.0108 (12) | 0.0000 (14) |
| N3 | 0.0461 (13) | 0.0513 (17) | 0.0367 (13) | −0.0030 (12) | 0.0180 (11) | −0.0083 (12) |
| N4 | 0.0487 (14) | 0.0416 (15) | 0.0345 (13) | 0.0011 (12) | 0.0153 (11) | −0.0003 (11) |
| N5 | 0.0552 (15) | 0.0582 (19) | 0.0387 (14) | 0.0041 (14) | 0.0178 (12) | 0.0082 (13) |
| N6 | 0.0650 (18) | 0.131 (3) | 0.0406 (16) | −0.044 (2) | 0.0086 (14) | −0.0034 (18) |
| N7 | 0.0558 (16) | 0.114 (3) | 0.0421 (15) | −0.0083 (17) | 0.0198 (13) | 0.0182 (16) |
| N8 | 0.0448 (13) | 0.0557 (18) | 0.0399 (14) | 0.0009 (12) | 0.0167 (11) | 0.0077 (12) |
| N9 | 0.0468 (14) | 0.0583 (18) | 0.0372 (14) | −0.0109 (13) | 0.0115 (11) | −0.0003 (13) |
| N10 | 0.0414 (13) | 0.0570 (18) | 0.0359 (13) | −0.0045 (12) | 0.0126 (11) | −0.0002 (12) |
| O1 | 0.0594 (14) | 0.113 (2) | 0.0437 (13) | 0.0075 (14) | 0.0188 (11) | 0.0222 (14) |
| O2 | 0.0714 (15) | 0.0839 (18) | 0.0362 (12) | 0.0075 (14) | 0.0152 (11) | −0.0032 (12) |
| S1 | 0.0464 (4) | 0.0585 (6) | 0.0351 (4) | 0.0021 (4) | 0.0146 (3) | −0.0024 (4) |
| S2 | 0.0415 (4) | 0.0653 (6) | 0.0391 (4) | −0.0092 (4) | 0.0107 (3) | 0.0047 (4) |
| CL1 | 0.0698 (6) | 0.0948 (8) | 0.0647 (6) | −0.0090 (5) | 0.0327 (5) | 0.0072 (5) |
| CL2 | 0.0665 (5) | 0.0756 (7) | 0.0602 (5) | 0.0000 (5) | 0.0308 (4) | −0.0049 (5) |
(II) N-(3-Chlorophenyl)-2-[(4,6-diaminopyrimidin-2-yl)sulfanyl]acetamide . Geometric parameters (Å, º)
| C1—N1 | 1.348 (4) | C15—N7 | 1.336 (4) |
| C1—N3 | 1.360 (3) | C15—N8 | 1.348 (4) |
| C1—C2 | 1.389 (4) | C16—N8 | 1.320 (3) |
| C2—C3 | 1.372 (4) | C16—N9 | 1.331 (4) |
| C2—H2 | 0.9300 | C16—S2 | 1.766 (3) |
| C3—N2 | 1.355 (4) | C17—C18 | 1.508 (4) |
| C3—N4 | 1.362 (3) | C17—S2 | 1.808 (3) |
| C4—N3 | 1.327 (3) | C17—H17A | 0.9700 |
| C4—N4 | 1.332 (4) | C17—H17B | 0.9700 |
| C4—S1 | 1.766 (3) | C18—O2 | 1.223 (3) |
| C5—C6 | 1.510 (4) | C18—N10 | 1.346 (4) |
| C5—S1 | 1.800 (3) | C19—C24 | 1.388 (4) |
| C5—H5A | 0.9700 | C19—C20 | 1.387 (4) |
| C5—H5B | 0.9700 | C19—N10 | 1.405 (4) |
| C6—O1 | 1.215 (3) | C20—C21 | 1.378 (4) |
| C6—N5 | 1.346 (4) | C20—H20 | 0.9300 |
| C7—C12 | 1.383 (4) | C21—C22 | 1.378 (4) |
| C7—C8 | 1.386 (4) | C21—H21 | 0.9300 |
| C7—N5 | 1.411 (4) | C22—C23 | 1.378 (4) |
| C8—C9 | 1.374 (5) | C22—H22 | 0.9300 |
| C8—H8 | 0.9300 | C23—C24 | 1.383 (4) |
| C9—C10 | 1.361 (5) | C23—CL2 | 1.736 (3) |
| C9—H9 | 0.9300 | C24—H24 | 0.9300 |
| C10—C11 | 1.366 (5) | N1—H1A | 0.8600 |
| C10—H10 | 0.9300 | N1—H1B | 0.8600 |
| C11—C12 | 1.387 (4) | N2—H2A | 0.8600 |
| C11—CL1 | 1.736 (3) | N2—H2B | 0.8600 |
| C12—H12 | 0.9300 | N5—H5 | 0.8600 |
| C13—N9 | 1.346 (4) | N6—H6A | 0.8600 |
| C13—N6 | 1.352 (4) | N6—H6B | 0.8600 |
| C13—C14 | 1.378 (4) | N7—H7A | 0.8600 |
| C14—C15 | 1.398 (4) | N7—H7B | 0.8600 |
| C14—H14 | 0.9300 | N10—H10A | 0.8600 |
| N1—C1—N3 | 116.0 (3) | C18—C17—S2 | 115.1 (2) |
| N1—C1—C2 | 122.9 (3) | C18—C17—H17A | 108.5 |
| N3—C1—C2 | 121.1 (3) | S2—C17—H17A | 108.5 |
| C3—C2—C1 | 118.1 (3) | C18—C17—H17B | 108.5 |
| C3—C2—H2 | 121.0 | S2—C17—H17B | 108.5 |
| C1—C2—H2 | 121.0 | H17A—C17—H17B | 107.5 |
| N2—C3—N4 | 115.0 (3) | O2—C18—N10 | 124.6 (3) |
| N2—C3—C2 | 122.9 (3) | O2—C18—C17 | 120.6 (3) |
| N4—C3—C2 | 122.0 (3) | N10—C18—C17 | 114.9 (2) |
| N3—C4—N4 | 128.8 (3) | C24—C19—C20 | 119.2 (3) |
| N3—C4—S1 | 111.4 (2) | C24—C19—N10 | 122.4 (3) |
| N4—C4—S1 | 119.8 (2) | C20—C19—N10 | 118.3 (3) |
| C6—C5—S1 | 112.9 (2) | C21—C20—C19 | 120.6 (3) |
| C6—C5—H5A | 109.0 | C21—C20—H20 | 119.7 |
| S1—C5—H5A | 109.0 | C19—C20—H20 | 119.7 |
| C6—C5—H5B | 109.0 | C22—C21—C20 | 121.0 (3) |
| S1—C5—H5B | 109.0 | C22—C21—H21 | 119.5 |
| H5A—C5—H5B | 107.8 | C20—C21—H21 | 119.5 |
| O1—C6—N5 | 124.5 (3) | C23—C22—C21 | 117.8 (3) |
| O1—C6—C5 | 120.8 (3) | C23—C22—H22 | 121.1 |
| N5—C6—C5 | 114.8 (3) | C21—C22—H22 | 121.1 |
| C12—C7—C8 | 120.1 (3) | C22—C23—C24 | 122.6 (3) |
| C12—C7—N5 | 122.3 (3) | C22—C23—CL2 | 119.1 (3) |
| C8—C7—N5 | 117.6 (3) | C24—C23—CL2 | 118.2 (2) |
| C9—C8—C7 | 120.3 (3) | C23—C24—C19 | 118.8 (3) |
| C9—C8—H8 | 119.8 | C23—C24—H24 | 120.6 |
| C7—C8—H8 | 119.8 | C19—C24—H24 | 120.6 |
| C10—C9—C8 | 120.3 (4) | C1—N1—H1A | 120.0 |
| C10—C9—H9 | 119.8 | C1—N1—H1B | 120.0 |
| C8—C9—H9 | 119.8 | H1A—N1—H1B | 120.0 |
| C9—C10—C11 | 119.2 (3) | C3—N2—H2A | 120.0 |
| C9—C10—H10 | 120.4 | C3—N2—H2B | 120.0 |
| C11—C10—H10 | 120.4 | H2A—N2—H2B | 120.0 |
| C10—C11—C12 | 122.5 (3) | C4—N3—C1 | 115.3 (2) |
| C10—C11—CL1 | 119.5 (3) | C4—N4—C3 | 114.6 (2) |
| C12—C11—CL1 | 118.0 (3) | C6—N5—C7 | 128.7 (3) |
| C7—C12—C11 | 117.5 (3) | C6—N5—H5 | 115.6 |
| C7—C12—H12 | 121.2 | C7—N5—H5 | 115.6 |
| C11—C12—H12 | 121.2 | C13—N6—H6A | 120.0 |
| N9—C13—N6 | 115.3 (3) | C13—N6—H6B | 120.0 |
| N9—C13—C14 | 121.8 (3) | H6A—N6—H6B | 120.0 |
| N6—C13—C14 | 122.8 (3) | C15—N7—H7A | 120.0 |
| C13—C14—C15 | 117.4 (3) | C15—N7—H7B | 120.0 |
| C13—C14—H14 | 121.3 | H7A—N7—H7B | 120.0 |
| C15—C14—H14 | 121.3 | C16—N8—C15 | 115.7 (3) |
| N7—C15—N8 | 116.7 (3) | C16—N9—C13 | 115.5 (2) |
| N7—C15—C14 | 122.0 (3) | C18—N10—C19 | 129.5 (3) |
| N8—C15—C14 | 121.3 (3) | C18—N10—H10A | 115.3 |
| N8—C16—N9 | 128.2 (3) | C19—N10—H10A | 115.3 |
| N8—C16—S2 | 112.7 (2) | C4—S1—C5 | 103.66 (14) |
| N9—C16—S2 | 119.0 (2) | C16—S2—C17 | 102.99 (14) |
| N1—C1—C2—C3 | 178.0 (3) | N10—C19—C24—C23 | 178.0 (3) |
| N3—C1—C2—C3 | −0.6 (4) | N4—C4—N3—C1 | 2.2 (5) |
| C1—C2—C3—N2 | 179.3 (3) | S1—C4—N3—C1 | −175.7 (2) |
| C1—C2—C3—N4 | 3.1 (4) | N1—C1—N3—C4 | 179.5 (3) |
| S1—C5—C6—O1 | −93.7 (3) | C2—C1—N3—C4 | −1.9 (4) |
| S1—C5—C6—N5 | 85.2 (3) | N3—C4—N4—C3 | 0.1 (4) |
| C12—C7—C8—C9 | 2.3 (5) | S1—C4—N4—C3 | 177.8 (2) |
| N5—C7—C8—C9 | −178.8 (3) | N2—C3—N4—C4 | −179.4 (3) |
| C7—C8—C9—C10 | −1.9 (6) | C2—C3—N4—C4 | −2.9 (4) |
| C8—C9—C10—C11 | 0.6 (6) | O1—C6—N5—C7 | 2.0 (5) |
| C9—C10—C11—C12 | 0.3 (6) | C5—C6—N5—C7 | −176.9 (3) |
| C9—C10—C11—CL1 | −179.8 (3) | C12—C7—N5—C6 | −20.0 (5) |
| C8—C7—C12—C11 | −1.4 (5) | C8—C7—N5—C6 | 161.1 (3) |
| N5—C7—C12—C11 | 179.7 (3) | N9—C16—N8—C15 | 2.3 (5) |
| C10—C11—C12—C7 | 0.1 (5) | S2—C16—N8—C15 | −174.2 (2) |
| CL1—C11—C12—C7 | −179.8 (2) | N7—C15—N8—C16 | 177.1 (3) |
| N9—C13—C14—C15 | 1.8 (5) | C14—C15—N8—C16 | −2.7 (5) |
| N6—C13—C14—C15 | 179.0 (3) | N8—C16—N9—C13 | 0.1 (5) |
| C13—C14—C15—N7 | −178.9 (3) | S2—C16—N9—C13 | 176.4 (2) |
| C13—C14—C15—N8 | 0.8 (5) | N6—C13—N9—C16 | −179.7 (3) |
| S2—C17—C18—O2 | −112.5 (3) | C14—C13—N9—C16 | −2.2 (5) |
| S2—C17—C18—N10 | 68.4 (3) | O2—C18—N10—C19 | 4.6 (5) |
| C24—C19—C20—C21 | −0.7 (4) | C17—C18—N10—C19 | −176.3 (3) |
| N10—C19—C20—C21 | −178.3 (3) | C24—C19—N10—C18 | 1.4 (5) |
| C19—C20—C21—C22 | 0.5 (5) | C20—C19—N10—C18 | 178.8 (3) |
| C20—C21—C22—C23 | −0.1 (5) | N3—C4—S1—C5 | −172.4 (2) |
| C21—C22—C23—C24 | −0.1 (5) | N4—C4—S1—C5 | 9.5 (3) |
| C21—C22—C23—CL2 | 177.2 (2) | C6—C5—S1—C4 | −88.7 (2) |
| C22—C23—C24—C19 | −0.1 (4) | N8—C16—S2—C17 | −172.9 (2) |
| CL2—C23—C24—C19 | −177.5 (2) | N9—C16—S2—C17 | 10.3 (3) |
| C20—C19—C24—C23 | 0.6 (4) | C18—C17—S2—C16 | −86.9 (2) |
(II) N-(3-Chlorophenyl)-2-[(4,6-diaminopyrimidin-2-yl)sulfanyl]acetamide . Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N5—H5···N4 | 0.86 | 2.25 | 2.962 (3) | 140 |
| N10—H10A···N9 | 0.86 | 2.02 | 2.826 (3) | 157 |
| N1—H1B···O1i | 0.86 | 2.19 | 2.931 (4) | 145 |
| N2—H2B···Cl1i | 0.86 | 2.76 | 3.405 (3) | 133 |
| N6—H6A···O2ii | 0.86 | 2.51 | 3.340 (4) | 162 |
| N6—H6B···Cl2iii | 0.86 | 2.70 | 3.556 (3) | 176 |
| N7—H7B···O2iii | 0.86 | 2.24 | 3.002 (4) | 148 |
| N1—H1A···N8iv | 0.86 | 2.21 | 3.070 (4) | 174 |
| N7—H7A···N3v | 0.86 | 2.19 | 3.046 (4) | 178 |
Symmetry codes: (i) x, −y+3/2, z+1/2; (ii) −x+2, −y+1, −z; (iii) x, −y+3/2, z−1/2; (iv) x−1, −y+3/2, z+1/2; (v) x+1, −y+3/2, z−1/2.
References
- Blumenkopf, T. A., Mueller, E. E. & Roskamp, E. J. (2002). Google Patents. WO2001040215 A1. PCT/IB2000/001628.
- Bruker (2008). SMART, SAINT and SADABS. Bruker AXS Inc., Madison, Wisconsin, USA.
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Hocková, D., Holý, A., Masojídková, M., Andrei, G., Snoeck, R., De Clercq, E. & Balzarini, J. (2003). J. Med. Chem. 46, 5064–5073. [DOI] [PubMed]
- Hocková, D., Holý, A. N., Masoj\?ídková, M., Andrei, G., Snoeck, R., De Clercq, E. & Balzarini, J. (2004). Bioorg. Med. Chem. 12, 3197–3202. [DOI] [PubMed]
- Holla, B. S., Mahalinga, M., Karthikeyan, M. S., Akberali, P. M. & Shetty, N. S. (2006). Bioorg. Med. Chem. 14, 2040–2047. [DOI] [PubMed]
- Kandeel, M., El-Meligie, S., Omar, R., Roshdy, S. & Youssef, K. (1994). J. Pharm. Sci 3, 197–205.
- Nogueras, M., Sánchez, A., Melguizo, M., Quijano, M. L. & Melgarejo, M. (1993). Bioorg. Med. Chem. Lett. 3, 601–606.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sheldrick, G. M. (2015). Acta Cryst. C71, 3–8.
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Subasri, S., Timiri, A. K., Barji, N. S., Jayaprakash, V., Vijayan, V. & Velmurugan, D. (2016). Acta Cryst. E72, 1171–1175. [DOI] [PMC free article] [PubMed]
- Xu, L. B., Sun, W., Liu, H. Y., Wang, L. L., Xiao, J. H., Yang, X. H. & Li, S. (2010). Chin. Chem. Lett. 21, 1318–1321.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I, II. DOI: 10.1107/S2056989017003243/su5348sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989017003243/su5348Isup4.hkl
Structure factors: contains datablock(s) II. DOI: 10.1107/S2056989017003243/su5348IIsup3.hkl
Supporting information file. DOI: 10.1107/S2056989017003243/su5348Isup4.cml
Supporting information file. DOI: 10.1107/S2056989017003243/su5348IIsup5.cml
Additional supporting information: crystallographic information; 3D view; checkCIF report