Abstract
Single nucleotide polymorphisms (SNPs) in the 3′-untranslated regions targeted by putative mircoRNA can change its binding strength, affecting the susceptibility and prognosis of cancer. We aimed to investigate the associations between SNPs within miR-148a binding sites and gastric cancer (GC) risk and prognosis. Using bioinformatics tools, we selected two SNPs (SCRN1 rs6976789 and PDYN rs2235749) located in miR-148a target sites. We genotyped the two SNPs in a case-control study comprising 753 GC patients and 949 cancer-free subjects. We found a significantly increased risk of GC associated with the SCRN1 rs6976789 C>T polymorphism [adjusted OR = 1.25, 95% confidence interval (CI) = 1.02–1.53; CT/TT vs. CC]. However, no significant association was found between the PDYN rs2235749 and GC risk in all genetic models. Furthermore, we evaluated whether SCRN1 rs6976789 affected the survival of GC patients. Results showed that individuals with SCRN1 rs6976789 TT genotype had poorer overall survival compared with those carried CC/CT genotypes in intestinal-type GC (adjusted HR = 2.47, 95% CI = 1.21–5.05). Luciferase report assay showed that the rs6976789 variant T allele influenced the binding ability of miR-148a. Our results suggested that the SCRN1 rs6976789 polymorphism may play an important role in the GC development and progression.
Gastric cancer continues to be a major problem accounting for about one million new cases and estimated 700,000 deaths per year worldwide1. Until now, it is also the second leading cause of global cancer mortality. Especially in Eastern Asia, predominantly in China, the incidence and mortality rates for gastric cancer remain highest2. Although accumulating evidence has indicated that living habits, environmental and genetic factors play important roles in the development and progression of gastric carcinoma, the precise mechanisms are still unknown. Even following the same exposure to environmental carcinogenic factors, only a fraction of the exposed individuals eventually develop cancer3. It suggests that genetic susceptibility may act substantially in the etiology of gastric cancer. Genetic susceptibility to some extent can be interpreted as single nucleotide polymorphisms (SNPs)4,5.
MicroRNAs (miRNAs) are a class of small, naturally occurring, noncoding and single-stranded RNA molecules (18~22 nucleotides). It plays a role in post-transcriptional regulatory by pairing to complementary sequences in the 3′-untranslated regions (UTRs) of messenger RNA (mRNA) of target gene, resulting in mRNA degradation and gene silencing6,7. If a genetic variant occurs in the miRNA binding site, the function of miRNA may be affected. Recently, several studies have demonstrated that SNPs located in miRNA binding sites can modify the miRNA regulatory function and affect tumor development8,9,10. Many studies have shown that miR-148a is related to various human malignancies, such as gastric cancer11,12, colorectal cancer13, liver cancer14, pancreatic cancer15, breast cancer16, renal cancer17, prostate cancer18 and so on. As for gastric cancer, miR-148a functions as a tumor metastasis suppressor, and down-regulation of miR-148a contributes to lymph node metastasis and poor progression11,12. In this study, we evaluated the effects of genetic polymorphisms within miR-148a binding sties on the genetic susceptibility and prognosis of gastric cancer.
Results
Characteristics and clinical features of subjects
Characteristics and clinical features of case-control and follow-up studies were summarized in Supplementary Table S1. The TNM stage classification was according to the 6th edition staging manual of the American Joint Committee on Cancer (AJCC) based on tumor size (T), lymph node metastasis (N), and distant metastasis (M). Lauren's criteria were used to classify the tumors into intestinal-type or diffuse-type gastric cancer. The cases and controls were matched on age (P = 0.501) and sex (P = 0.428). Among the 753 cases, there were 295 (39.2%) cardia gastric cancer patients and 458 (60.8%) noncardia gastric cancer patients; 437 (58.0%) patients with diffuse type of gastric cancer and 316 (42.0%) as intestinal type. Besides, 26.8%, 21.9%, 35.3%, and 15.9% of patients were identified to TNM stage I, II, III, and IV, respectively. There were 721 males (76.9%) and 216 females (23.1%) in the follow-up study. In a period of up to 119.0 months follow-up, 437 patients died of disease directly related to gastric cancer. The clinicopatholgical characteristics such as tumor size, histological types, depth of invasion, lymph node metastasis, distant metastasis and TNM stage were significantly associated with overall survival (P < 0.05, log-rank test).
Association of selected SNPs SCRN1 rs6976789 and PDYN rs2235749 with gastric cancer risk
Genotype distributions of SCRN1 rs6976789 and PDYN rs2235749 among the patients and controls were shown in Table 1. The genotype frequencies in controls were both conformed to the Hardy-Weinberg equilibrium (P = 0.173 for rs6976789 and P = 0.871 for rs2235749). We found that the genotype distributions of the rs6976789 and rs2235749 between the cases and controls was not statistically different (P = 0.100 for rs6976789 and P = 0.545 for rs22335749). For rs6976789, the combined CT/TT genotypes frequency were higher among cases than controls (36.1% vs. 31.2%, P = 0.032). In addition, we conducted logistic regression to evaluate the associations between genotypes and risk of gastric cancer. As shown in Table 1, when the rs6976789 CC genotype was used as the reference, the CT genotype had a significant increased gastric cancer risk (adjusted OR = 1.25, 95% CI = 1.02–1.54). The rs6976789 T carriers (CT/TT) had an adjusted OR (95% CI) of 1.25 (1.02–1.53), compared with the rs6976789 CC genotype. However, we didn't find any association between rs2235749 polymorphism and gastric cancer risk. The adjusted OR (95%CI) for AG, GG, and G carriers (AG/GG) were 0.89 (0.71–1.11), 1.01 (0.54–1.87), and 0.90 (0.72–1.12), respectively, compared with the AA homozygotes.
Table 1. Associations between the selected SNPs and gastric cancer risk.
| SNPs | Genetic model | Genotype | Cases | Controls | Pa | Crude OR (95% CI) | Adjusted OR (95% CI)b |
|---|---|---|---|---|---|---|---|
| rs6976789 | Codominant | CC | 481 | 653 | 0.100 | 1.00 (reference) | 1.00 (reference) |
| CT | 255 | 278 | 1.25 (1.01–1.53) | 1.25 (1.02–1.54) | |||
| TT | 17 | 18 | 1.28 (0.66–2.51) | 1.28 (0.65–2.51) | |||
| Dominant | CC | 481 | 653 | 0.032 | 1.00 (reference) | 1.00 (reference) | |
| CT+TT | 272 | 296 | 1.25 (1.02–1.53) | 1.25 (1.02–1.53) | |||
| Recessive | CC+CT | 736 | 931 | 0.602 | 1.00 (reference) | 1.00 (reference) | |
| TT | 17 | 18 | 1.20 (0.61–2.33) | 1.19 (0.61–2.32) | |||
| rs2235749 | Codominant | AA | 564 | 690 | 0.545 | 1.00 (reference) | 1.00 (reference) |
| AG | 170 | 236 | 0.88 (0.70–1.11) | 0.89 (0.71–1.11) | |||
| GG | 19 | 23 | 1.01 (0.55–1.87) | 1.01 (0.54–1.87) | |||
| Dominant | AA | 564 | 690 | 0.308 | 1.00 (reference) | 1.00 (reference) | |
| AG+GG | 189 | 259 | 0.89 (0.72–1.11) | 0.90 (0.72–1.12) | |||
| Recessive | AA+AG | 734 | 926 | 0.900 | 1.00 (reference) | 1.00 (reference) | |
| GG | 19 | 23 | 1.04 (0.56–1.93) | 1.04 (0.56–1.92) |
aTwo-sided χ2 test for either genotype distributions between cases and controls.
bAdjusted for age and sex in logistic regression model.
We then evaluated the effect of selected SNPs rs6976789 and rs2235749 with gastric cancer by different clinical variables. As shown in Table 2, when the rs6976789 CC genotype was used as the reference, the increased risk of gastric cancer for CT/TT genotypes were also observed among subgroup of age > 65 (adjusted OR = 1.40, 95% CI = 1.02–1.92), male (adjusted OR = 1.30, 95% CI = 1.01–1.68), noncardia (adjusted OR = 1.36, 95% CI = 1.07–1.71), N0 (adjusted OR = 1.44, 95% CI = 1.10–1.88), stage I/II (adjusted OR = 1.48, 95% CI = 1.05–2.08). No significant difference was observed for rs2235749 among subgroup analysis.
Table 2. Stratified analysis between the rs6976789 polymorphism and gastric cancer risk.
| CC, n | CT/TT, n | ||||||
|---|---|---|---|---|---|---|---|
| Varibales | Cases | Controls | Cases | Controls | Pa | Crude OR (95% CI) | Adjusted OR (95% CI)b |
| Overall | 481 | 653 | 272 | 296 | 0.032 | 1.25 (1.02–1.53) | 1.25 (1.02–1.53) |
| Age (years) | |||||||
| ≤65 | 283 | 360 | 149 | 169 | 0.405 | 1.12 (0.86–1.47) | 1.15 (0.87–1.51) |
| >65 | 198 | 293 | 123 | 127 | 0.021 | 1.43 (1.06–1.95) | 1.40 (1.02–1.92) |
| Sex | |||||||
| Male | 330 | 440 | 182 | 188 | 0.044 | 1.29 (1.00–1.66) | 1.30 (1.01–1.68) |
| Female | 151 | 213 | 90 | 108 | 0.364 | 1.18 (0.83–1.67) | 1.15 (0.80–1.65) |
| Tumor site | |||||||
| Cardia | 196 | 653 | 99 | 296 | 0.445 | 1.11 (0.84–1.47) | 1.11 (0.84–1.47) |
| Noncardia | 285 | 653 | 173 | 296 | 0.014 | 1.34 (1.06–1.69) | 1.36 (1.07–1.71) |
| Histological types | |||||||
| Intestinal | 201 | 653 | 115 | 296 | 0.087 | 1.26 (0.97–1.65) | 1.27 (0.97–1.67) |
| Diffuse | 280 | 653 | 157 | 296 | 0.081 | 1.23 (0.97–1.57) | 1.24 (0.98–1.57) |
| Lymph node metastasis | |||||||
| N0 | 180 | 653 | 117 | 296 | 0.009 | 1.43 (1.09–1.88) | 1.44 (1.10–1.88) |
| N1/N2/N3 | 301 | 653 | 155 | 296 | 0.293 | 1.14 (0.90–1.44) | 1.14 (0.90–1.45) |
| Clinical stage | |||||||
| I/II | 225 | 653 | 142 | 296 | 0.010 | 1.39 (1.08–1.79) | 1.39 (1.08–1.79) |
| III/IV | 256 | 653 | 130 | 296 | 0.377 | 1.12 (0.87–1.44) | 1.13 (0.88–1.45) |
aTwo-sided χ2 test for either genotype distributions between cases and controls.
bAdjusted for age and sex in logistic regression model.
SCRN1 rs6976789 polymorphism and gastric cancer survival
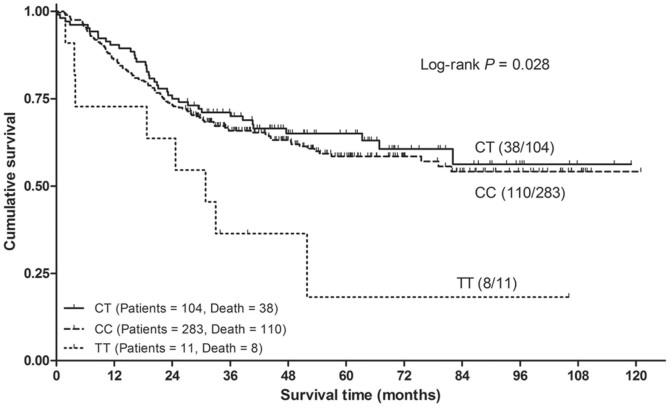
In this study, we further analyzed whether the SNP SCRN1 rs6976789 had an association with gastric cancer survival. There was no significant association between the rs6876789 polymorphism and survival in any genetic model. However, when stratified by histology with intestinal-type and diffuse-type gastric cancer, we observed significant difference in overall survival among intestinal-type gastric cancer (log-rank P = 0.028; Fig. 1). Cox regression analysis revealed that the intestinal-type gastric cancer patients who were TT genotype carriers had a lower survival than those carried the CC/CT genotypes (adjusted HR = 2.47, 95% CI = 1.21–5.05, Table 3).
Figure 1. Kaplan-Meier curves of overall survival for the SCRN1 rs6976789 codominant genotypes in intestinal-type gastric cancer patients.
Table 3. Stratified analysis of the rs6976789 genotypes associated with patients' survival.
| Variables | Genotypes (deaths/patients) | Crude HR (95% CI) | Adjusted HR (95% CI)a | |
|---|---|---|---|---|
| CC/CT | TT | |||
| Total | 422/908 | 15/29 | 1.18 (0.71–1.98) | 1.15 (0.69–1.93) |
| Age (years) | ||||
| ≤ 65 | 267/585 | 11/17 | 1.79 (0.98–3.28) | 1.78 (0.97–3.25) |
| >65 | 155/323 | 4/12 | 0.59 (0.22–1.60) | 0.59 (0.22–1.60) |
| Sex | ||||
| Male | 321/700 | 12/21 | 1.28 (0.72–2.28) | 1.25 (0.70–2.23) |
| Female | 101/208 | 3/8 | 0.90 (0.28–2.84) | 0.89 (0.28–2.81) |
| Histological types | ||||
| Intestinal | 148/387 | 8/11 | 2.49 (1.22–5.07) | 2.47 (1.21–5.05) |
| Diffuse | 274/521 | 7/18 | 0.69 (0.33–1.47) | 0.68 (0.32–1.43) |
| Lymph node metastasis | ||||
| N0 | 123/362 | 7/11 | 2.37 (1.10–5.08) | 2.31 (1.07–4.97) |
| N1/N2/N3 | 299/546 | 8/18 | 0.77 (0.38–1.55) | 0.75 (0.37–1.52) |
| Clinical stage | ||||
| I/II | 155/433 | 7/13 | 1.68 (0.79–3.58) | 1.64 (0.77–3.51) |
| III/IV | 267/475 | 8/16 | 0.88 (0.44–1.79) | 0.87 (0.43–1.76) |
aAdjusted for age and sex.
In addition, stepwise Cox proportional hazard analysis was performed to evaluate the correlation between selected demographic characteristics, clinical features and the different genetic models of the SCRN1 rs6976789 on intestinal-type gastric cancer survival. Two variables (rs6976789 and lymph node metastasis) were remained in the regression model. When age and sex were included in the final predictive model, the result indicated that the rs6976789 polymorphism was an independent risk factor for the survival of intestinal-type gastric cancer (TT vs. CC/CT, HR = 2.61, 95% CI = 1.28–5.33, Table 4).
Table 4. Stepwise Cox regression analysis on intestinal-type gastric cancer patients' survival.
| Entered variables | β | SE | HR | 95% CI | P value |
|---|---|---|---|---|---|
| Age | 0.01 | 0.01 | 1.01 | 0.99–1.03 | 0.191 |
| Sex | 0.29 | 0.18 | 1.34 | 0.93–1.94 | 0.120 |
| Lymph node metastasis (N1/N2/N3 vs. N0) | 0.52 | 0.16 | 1.68 | 1.22–2.03 | 0.001 |
| rs6976789 (TT vs. CC/CT) | 0.96 | 0.36 | 2.61 | 1.28–5.33 | 0.009 |
β, regression coefficient; SE, standard error; HR, hazard ratio; CI, confidence interval.
Effect of the SNP rs6976789 interaction between miR-148a and SCRN1 3′-UTR
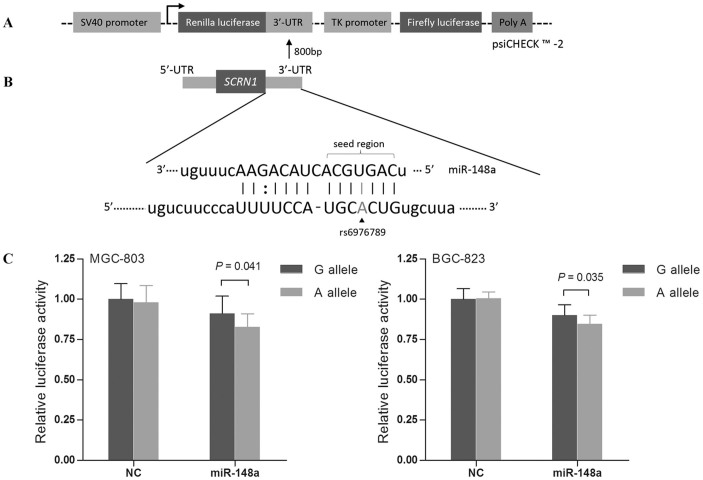
To investigate whether the SNP rs6976789 may influence SCRN1 regulation mediated by miR-148a, we constructed luciferase reporter vectors by using psiCHECK™-2 vector (Fig. 2A). Based on bioinformatics analysis, the SCRN1 rs6976789 was located on the target site of miR-148a (Fig. 2B). As shown in Fig. 2C, vectors within the rs6976789 A allele had a 0.91-fold decreased luciferase activities in MGC cells (P = 0.041) and 0.93-fold decreased in BGC cells (P = 0.035), compared with those of the G allele. In contrast, the negative control miRNA transient transfected with constructed vectors didn't affect luciferase expression. These findings indicated that miR-148a bound and negatively regulated the transcription of SCRN1 and that this regulation was more negatively influenced by the variant A allele in vitro.
Figure 2. Characterization and functional analyses of the 3′-UTR of SCRN1.
(A) Schematic representation of reporter plasmids containing the SCRN1 3′-UTR, which was inserted downstream of Renilla luciferase gene in the psiCHECK™ -2 vector. (B) Complementarity between miR-148 and the SCRN1 3′-UTR site targeted. The SNP rs6976789 was located within the ‘seed region' of the miR-148a binding site. (C) The effect of SNP rs6976789 on the interaction between the SCRN1 3′-UTR and miR-148a in MGC-803 and BGC-823 cells. The luciferase activity of each construct was normalized against the negative control miRNA (NC) transient transfected with constructed vectors with G allele.
Effect of the miR-148a and SNP rs6976789 on the expression of SCRN1 in gastric cancer tissues
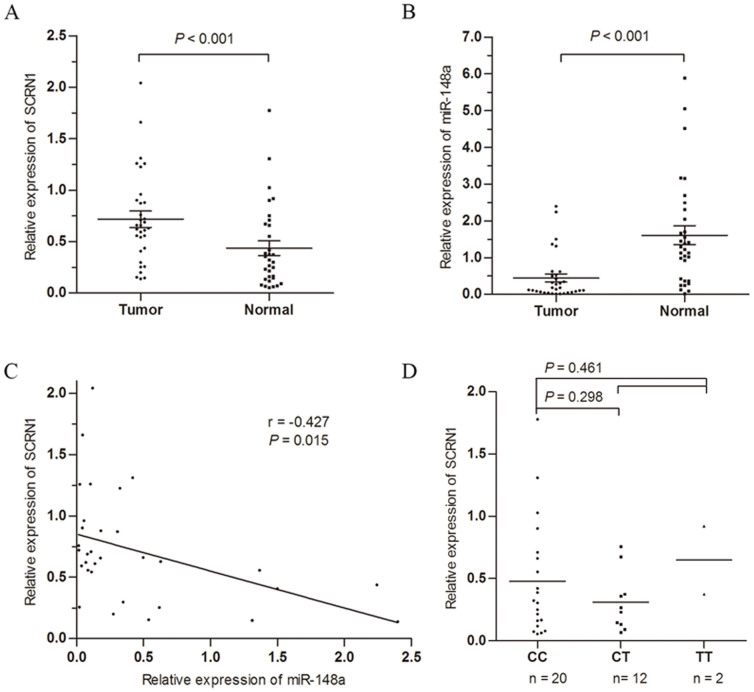
To determine the expression of SCRN1 and miR-148a in GC and correlate their expression to the different genotypes of the SNP rs6976789, we employed RT-PCR in 32 pairs of GC and non-GC sample. As shown in Fig. 3A, SCRN1 mRNA expression levels were significantly up-regulated in gastric carcinoma relative to adjacent normal mucosae. On the contrary, miR-148a was significantly down-regulated in GC tissues (Fig. 3B). A significant reverse correlation between miR-148a and SCRN1 mRNA expression was identified in 32 GC tissues (r = −0.427, P = 0.015, Fig. 3C). We further evaluated whether the rs6976789 polymorphism could influence SCRN1expression in GC. Our results showed that the C to T polymorphism did not change the expression level. The expression levels of SCRN1 in individuals with CT carriers (P = 0.298) or CT/TT carriers (P = 0.461) have no significant difference with those with CC carriers (Fig. 3D). Additionally, the expression levels of miR-148a were no obvious different in patients with or without variant allele (Supplementary Fig. S1).
Figure 3. Effect of the miR-148a and SNP rs6976789 on the expression of SCRN1 in gastric cancer.
Relative expression levels of (A) SCRN1 mRNA and (B) miR-148a were detected in 32 pairs of human gastric cancer tissues and adjacent normal tissues via qRT-PCR. Abundance of and SCRN1 mRNA and miRNA was normalized to GAPDH and U6 RNA, respectively. (C) Spearman's correlation analysis of SCRN1 mRNA expression levels to miR-148a in 32 gastric cancer tissues. (D) Association between rs6976789 polymorphism and SCRN1 mRNA levels in gastric cancer cases.
Discussion
In the present study, to explore SNPs within miR-148a binding sites and their associations with gastric cancer susceptibility and survival, we studied two SNPs SCRN1 rs6976789 (C>T) and PDYN rs2235749 (A>G). Results showed that the SCRN1 rs6976789 can influence the risk and prognosis to gastric cancer, and individuals with the rs6976789 variant genotypes (CT/TT) had a significantly increased gastric cancer risk compared with the CC genotypes. Moreover, the rs6976789 variant TT genotype had a significantly lower survival rate than the CC/CT genotypes in intestinal-type gastric cancer. To the best of our knowledge, this is the first study to evaluate the possible association between the SCRN1 rs6976789 polymorphism with gastric cancer susceptibility and survival.
Knowledge of the functional contribution of SCRN1 gene to the cancer risk was an important step. SCRN1, also called KIAA0193 or SES1, was a member of the secernin family, reportedly contained three secernin genes (SCRN1, 2 and 3). SCRN1 was localized on chromosome 7 (7p14.3–p14.1), and others located on chromosomes 17 (17q21.3) and 2 (2p14–q14.3), respectively. Secernin-1 is a 50-kDa cytosolic protein which expressed in normal organs such as the brain, prostate, thymus, and intestine and had a major role in the regulation of exocytosis in mast cells19,20,21. However, the mechanisms of its exocytosis were still not completely clarified. Secernin-1 might recruit secretory granules to the site of exocytosis in tumor cells, thus increased the granules swelling, core expulsion or breakdown20,21. Suda et al. found secernin-1 could serve as a tumor-associated antigen which could be recognized and killed by cytotoxic T lymphocyte (CTL)21. Previous studies had showed that secernin-1 was up-regulated in the tumor tissues of human cancer, especially in gastrointestinal tract cancer21,22. Increased expression of secernin-1 was closely associated with the poor prognosis of cancer patients22,23. Taken together, SCRN1 played an important role in the cancer development and progression.
Emerging evidences have demonstrated that mutations in the miRNA binding sites contributed to the process of various tumors, which may be caused by the changes of miRNA binding ability to the 3′-UTR, resulting in altered regulation of miRNA and possibly cellular miRNA level as well4,10,24. For example, Wang and colleagues found that the rs4143815 variant allele prevented miR-570 binding to the 3′-UTR of B7-H1 gene, thus enhanced the ability of B7-H1 to promote cancer cell growth and invasion, leading to increase gastric cancer risk10. We identified the SNP SCRN1 rs6976789 located in miR-148a binding sites. Recently, studies have showed that miR-148a was down-regulated in gastric cancer and significantly associated with TNM stage and lymph node metastasis11,12. Overexpression of miR-148a inhibited gastric cancer cell migration and invasion in vitro and lung metastasis formation in nude mice11. Besides, several miR-148a target genes had been discovered, including ACVR1, CAND1, ROCK1, and BCL211,25,26,27.
Our results from the luciferase assay indicated that miR-148a mediated SCRN1 translational regulation. Furthermore, the variant allele of rs6976789 might strengthen miR-148a bind to the SCRN1 3′-UTR, and decreased secernin-1 expression. This finding indicated that the SNP rs6976789 may weaken the ability of SCRN1 to confer the gastric cancer risk. Considering the low level of miR-148a in the gastric cancer contributing to cancer metastasis and progression, the mechanism that the variant allele generated a better binding site for miR-148a might not act as a critical mediator. Down-regulated miR-148a seemed to be more predominate during carcinogenesis. Intriguingly, in the stratification analysis, the increased risk of gastric cancer was more evident for patients carried the CT/TT genotypes with no lymph node metastasis. Whereas, Zheng et al. showed that gastric cancer patients with lymph node metastasis had much lower miR-148a expression than those without lymph node metastasis11. This finding indicated that the risk allele altered the dependence of the SCRN1 3′-UTR on miR-148a at the stage of tumor metastasis. Despite the exact mechanism about the rs6976789 polymorphism with gastric cancer risk remained to be elucidated, this difference could be partially owing to miR-148a.
To further explore the mechanism that patients who harbor the variant allele at high GC risk and poor prognosis, we investigated the effects of the SNP rs6976789 on expression of SCRN1. However, no significant association was found between SCRN1 mRNA expression and different genotypes. The reason for this result might be that we did not have sufficient samples to conduct PCR assay, especially for the TT genotype. We checked the SNP-gene association in expression quantitative trait loci (eQTL) studies using the Genevar software (http://www.sanger.ac.uk/resources/software/genevar/) based on HapMap3 database28. A significant association between the SNP rs6976789 and the expression of SCRN1 was found in LWK (Luhya in Webuye, Kenya) population, but not in CHB (Han Chinese in Beijing, China) population (Supplementary Fig. S2). As we know, it was plausible that some genetic variation was private to particular populations. The frequency of variant allele varied across the two populations (9.8% in CHB population, 43.3% in LWK population). Therefore, larger sample size studies with different populations are needed to reveal the possible correlation in the following research.
Although this was the first study to evaluate the SCRN1 rs6976789 and PDYN rs2235749 polymorphisms with gastric cancer development, some limitations of should be pointed out as follows. First, the case-control study was hospital-based, and the selection bias could not be excluded. Therefore we matched the controls to the cases by sex, age and other putative confounding factors to minimize the bias. Second, for the rs6876789, significant associations seemed to be found in particular strata subgroup (age > 65, male, noncardia, N0, stage I/II) with gastric cancer risk, and patients carried the TT genotype had poorer survival than those carried the CC/CT genotypes in intestinal-type gastric cancer. However, the sample size was relative smaller. We should treat the finding with caution. Third, as is known to all, Helicobacter pylori infection status, diet and alcohol consumption are well-known crucial factors in gastric carcinogenesis. Unfortunately, we did not have the detailed information of these risk factors, so we were not able to perform the gene-environment interactions analysis. Thus, studies with more detailed data on these risk factors were warranted to validate the results in present study.
In conclusion, our results indicated that the genetic variation SCRN1 rs6976789 within the miR-148a binding site was associated with the gastric cancer susceptibility and influenced the prognosis in a Chinese population. Further larger prospective studies are still needed to confirm our findings.
Methods
Study population
There were 753 histologically diagnosed gastric adenocarcinoma patients and 949 age- and sex- matched cancer-free controls in our case-control study, which were recruited from The Second Affiliated Hospital of Nanjing Medical University (Nanjing, China), Cancer Hospital of Nantong (Nantong, China) and Yixing Cancer Hospital (Yixing, China) from March 2006 to January 2010. Patients of follow-up study were recruited from the Yixing People's Hospital (Yixing, China), during January 1999 and December 2006, described in detail previously29. Briefly, a total of 940 patients within a maximum of 119.0 months (last follow-up in March 2009) follow-up time were entered into analyses. Date of death was obtained from inpatient and outpatient records or patients' relatives through follow-up telephone calls. Patients alive on the last follow-up date were considered as censored. Our study was approved by the Institutional Review Board of Nanjing Medical University, and signed informed consent was obtained from all participants or from patients' representatives if direct consent could not be obtained. All experiments were performed in accordance with relevant guidelines and regulations, and the Institutional Review Board of Nanjing Medical University had approved all experiments.
SNPs selection and genotyping
The online software miRNASNP (http://www.bioguo.org/miRNASNP/) was used to predict the possible miR-148a related SNPs. Minimum free energy (MFE) was computed using RNAhybrid30. We conducted comprehensive searches of predicted results based on 3 criteria: (1) SNPs located in the 3′-UTR of target mRNA, (2) targets gain by SNPs within the seed region of the miR-148a binding sites, and (3) minor allelic frequency (MAF) 10% or more in the Chinese population. Finally, 7 SNPs (i.e., PHC2 rs11061, YOD1 rs1044145, RAPGEFL1 rs3744806, MAP6D1 rs2255015, AFAP1 rs2269852, SCRN1 rs6976789, and PDYN rs2235749) matched those criteria. Among them, according to these genes related to genetic susceptibility of cancer and lower MFE for variant allele, we chose SCRN1 rs6976789 (C>T) and PDYN rs2235749 (A>G) for the further evaluation (Supplementary Table S2).
Genomic DNA of the case-control study was isolated from leucocytes of peripheral blood. Due to lacking blood samples, genomic DNA of follow-up study was obtained from paraffin sections of tumor tissues. Genotyping was performed with TaqMan MGB technology using ABI 7900HT Sequence Detection System version 2.4 (Applied Biosystems, Foster City, CA, USA). Each 384 well plate included four blank controls to ensure accuracy of the genotyping. However, one subject in case-control study and three patients in follow-up study were failure in genotyping because of DNA quality, which were excluded in final analysis. The genotyping assay was conducted by two persons independently in a blind fashion. In addition, more than 10% of the samples were randomly selected for confirmation and the results were in agreement with the results of the first assay. The structure of primers and probes were as follows: for rs6976789, forward primer: 5′-GCTCTGCAGTCCACCACACA-3′, reverse primer: 5′-CATCCTGGGCTCCTGAAGAA-3′, and probes: FAM-AAGCACAGTGCATGGA-MGB, HEX-AAGCACAGCGCATG-MGB; for rs2235749, forward primer: 5′-CCATGTTCTTGAGCAGCTGAAT-3′, reverse primer: 5′-TGGAGTCCCTTACCCAATGC-3′, and probes: FAM-CCCAACATATGCACTG-MGB, HEX-CCCAACATACGCACTG-MGB.
Construction of reporter plasmids and luciferase reporter assay
To construct luciferase reporter plasmids, 3′-UTR fragments (800-bp) of the SCRN1 (G allele or A allele for SNP rs6976789) were inserted at the XhoI/NotI site, downstream of the Renilla luciferase gene in the psiCHECK™-2 vector (Promega, Madison, WI, USA) by Generay Company (Shanghai, China). The nucleotide sequences of the constructed plasmids were confirmed by DNA sequencing method. MGC-803 and BGC-823 cells were seed on the 24-well dishes and 24 h later transfected with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA), respectively. Each transient transfected reaction contained 0.4 μg of constructed vectors, either with G allele or with A allele and 100 pmol/μl of chemically synthesized miR-148a or negative control miRNA. Forty-eight hours post-transfection, luciferase activity was measured with a Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA). Relative protein levels were expressed as Renilla luciferase normalized against Firefly luciferase signals. This assay was done in triplicate under the same conditions.
RNA extraction and quantitative real-time PCR analysis
Total RNA was extracted from 32 gastric cancer tissues, included in 753 cases, using Trizol Reagent (Invitrogen, CA, USA). Reverse transcription was done with Primescript RT Reagent Kit (TaKaRa, Osaka, Japan) or RNA PCR Kit (AMV) (TaKaRa, Dalian, China). Real-time PCR assay was performed on 7900 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) using SYBR Premix Ex Taq Kit (TaKaRa, Dalian, China). Relative quantification of SCRN1 mRNA was measured using 2-ΔΔCt method which normalized to GAPDH. To measure the expression level of miR-148a, U6 was used as an internal quantitative control. The primers used for amplification were F: 5′- GGATGGTCTGGTGGTATTTGG-3′ and R: 5′- CCTTGGAACTTGGTCGATTG -3′ for SCRN1, F: 5′-AAGGTGAAGGTCGGAGTCAAC-3′ and R: 5′-GGGGTCATTGATGGCAACAATA-3′ for GAPDH, F: 5′- ACACTCCAGCTGGGCAGCAGCACACTGTG -3′ and R: 5′- TGGTGTCGTGGAGTCG -3′ for miR-148a, F: 5′- CTCGCTTCGGCAGCACA -3′ and R: 5′- AACGCTTCACGAATTTGCGT -3′ for U6.
Statistical analysis
The Student's t-test (for continuous variables) and Pearson's χ2 test (for categorical variables) were used to examine differences in the distributions of demographic characteristics and selected variables between cases and controls. Associations between the genotypes and risk of gastric cancer were estimated by computing crude or adjusted odds ratios (ORs) and 95% confidence intervals (CIs) from unconditional logistic regression. Hardy-Weinberg equilibrium was conducted to evaluate the genotype frequencies among the controls. Kaplan-Meier method and log-rank test were employed to evaluate the associations between survival time and demographic characteristics, clinical features, and the SNP rs6876789. Mean survival time was provided when the median survival time (MST) could not be calculated. Univariate or multivariate Cox regression analysis was used to estimate crude or adjusted hazard ratios (HRs) and 95% CIs. The Cox stepwise regression analysis was used to determine predictive factors of gastric cancer survival, using P < 0.05 for entering and P > 0.10 for removal of the respective explanatory variables. Wilcoxon matched pairs test was used to analyze the results of miRNA or mRNA expression in GC and adjacent normal mucosae. All the statistical analyses were done with SAS software version 9.1 (SAS Institute, Inc, Cary, NC, USA) and two-sided P-value < 0.05 was considered statistically significant.
Supplementary Material
Supplementary data
Acknowledgments
This study was partly supported by National Natural Science Foundation of China (81230068, 81373091), Natural Science Foundation of Jiangsu Province (BK2011194, BK2012842), the Key Program for Basic Research of Jiangsu Provincial Department of Education (11KJB330002, 12KJA330002), Jiangsu Scientific and Technological Innovation Project and the Priority Academic Program Development of Jiangsu Higher Education Institutions (Public Health and Preventive Medicine).
Footnotes
The authors declare no competing financial interests.
Author Contributions Z.Z. and Q.Z. conceived and designed the experiments. P.S., H.Z. and D.Z. performed the experiments. P.S., H.C., D.W., M.K. and M.W.analyzed the data. H.C., M.W., W.G. and J.Z. contributed reagents/materials/analysis tools. P.S. wrote the paper.
References
- Blum M. A., Takashi T., Suzuki A. & Ajani J. A. Management of localized gastric cancer. J Surg Oncol 107, 265–270 (2013). [DOI] [PubMed] [Google Scholar]
- Jemal A. et al. Global cancer statistics. CA Cancer J Clin 61, 69–90 (2011). [DOI] [PubMed] [Google Scholar]
- Augenlicht L. H. et al. Environment-gene interactions in intestinal cancer. Eur J Cancer Prev 11 Suppl 2, S12–17 (2002). [PubMed] [Google Scholar]
- Shi Y. et al. A genome-wide association study identifies new susceptibility loci for non-cardia gastric cancer at 3q13.31 and 5p13.1. Nat Genet 43, 1215–1218 (2011). [DOI] [PubMed] [Google Scholar]
- Sakamoto H. et al. Genetic variation in PSCA is associated with susceptibility to diffuse-type gastric cancer. Nat Genet 40, 730–740 (2008). [DOI] [PubMed] [Google Scholar]
- Cullen B. R. MicroRNAs as mediators of viral evasion of the immune system. Nat Immunol 14, 205–210 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr M., Zampetaki A., Willeit P., Willeit J. & Kiechl S. MicroRNAs within the continuum of postgenomics biomarker discovery. Arterioscler Thromb Vasc Biol 33, 206–214 (2013). [DOI] [PubMed] [Google Scholar]
- Hu Z. et al. Genetic polymorphisms in the precursor MicroRNA flanking region and non-small cell lung cancer survival. Am J Respir Crit Care Med 183, 641–648 (2011). [DOI] [PubMed] [Google Scholar]
- Christensen B. C. et al. Mature microRNA sequence polymorphism in MIR196A2 is associated with risk and prognosis of head and neck cancer. Clin Cancer Res 16, 3713–3720 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W. et al. A miR-570 binding site polymorphism in the B7-H1 gene is associated with the risk of gastric adenocarcinoma. Hum Genet 132, 641–648 (2013). [DOI] [PubMed] [Google Scholar]
- Zheng B. et al. MicroRNA-148a suppresses tumor cell invasion and metastasis by downregulating ROCK1 in gastric cancer. Clin Cancer Res 17, 7574–7583 (2011). [DOI] [PubMed] [Google Scholar]
- Tchernitsa O. et al. Systematic evaluation of the miRNA-ome and its downstream effects on mRNA expression identifies gastric cancer progression. J Pathol 222, 310–319 (2010). [DOI] [PubMed] [Google Scholar]
- Cho W. C. Epigenetic alteration of microRNAs in feces of colorectal cancer and its clinical significance. Expert Rev Mol Diagn 11, 691–694 (2011). [DOI] [PubMed] [Google Scholar]
- Gailhouste L. et al. miR-148a plays a pivotal role in the liver by promoting the hepatospecific phenotype and suppressing the invasiveness of transformed cells. Hepatology 58, 1153–1165 (2013). [DOI] [PubMed] [Google Scholar]
- Schultz N. A. et al. Prognostic microRNAs in cancer tissue from patients operated for pancreatic cancer--five microRNAs in a prognostic index. World J Surg 36, 2699–2707 (2012). [DOI] [PubMed] [Google Scholar]
- Aydogdu E. et al. MicroRNA-regulated gene networks during mammary cell differentiation are associated with breast cancer. Carcinogenesis 33, 1502–1511 (2012). [DOI] [PubMed] [Google Scholar]
- Wotschofsky Z. et al. Identification of metastamirs as metastasis-associated microRNAs in clear cell renal cell carcinomas. Int J Biol Sci 8, 1363–1374 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y. et al. MiR-148a attenuates paclitaxel resistance of hormone-refractory, drug-resistant prostate cancer PC3 cells by regulating MSK1 expression. J Biol Chem 285, 19076–19084 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Way G., Morrice N., Smythe C. & O'Sullivan A. J. Purification and identification of secernin, a novel cytosolic protein that regulates exocytosis in mast cells. Mol Biol Cell 13, 3344–3354 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashida S. et al. Molecular features of the transition from prostatic intraepithelial neoplasia (PIN) to prostate cancer: genome-wide gene-expression profiles of prostate cancers and PINs. Cancer Res 64, 5963–5972 (2004). [DOI] [PubMed] [Google Scholar]
- Suda T. et al. Identification of secernin 1 as a novel immunotherapy target for gastric cancer using the expression profiles of cDNA microarray. Cancer Sci 97, 411–419 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi N. et al. SCRN1 is a novel marker for prognosis in colorectal cancer. J Surg Oncol 101, 156–159 (2010). [DOI] [PubMed] [Google Scholar]
- Suehara Y. et al. Secernin-1 as a novel prognostic biomarker candidate of synovial sarcoma revealed by proteomics. J Proteomics 74, 829–842 (2011). [DOI] [PubMed] [Google Scholar]
- Chin L. J. et al. A SNP in a let-7 microRNA complementary site in the KRAS 3′ untranslated region increases non-small cell lung cancer risk. Cancer Res 68, 8535–8540 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H. et al. ACVR1, a Therapeutic Target of Fibrodysplasia Ossificans Progressiva, Is Negatively Regulated by miR-148a. Int J Mol Sci 13, 2063–2077 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H. et al. MiR-148a promotes apoptosis by targeting Bcl-2 in colorectal cancer. Cell Death Differ 18, 1702–1710 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata T. et al. miR-148a is an androgen-responsive microRNA that promotes LNCaP prostate cell growth by repressing its target CAND1 expression. Prostate Cancer Prostatic Dis 13, 356–361 (2010). [DOI] [PubMed] [Google Scholar]
- Yang T. P. et al. Genevar: a database and Java application for the analysis and visualization of SNP-gene associations in eQTL studies. Bioinformatics 26, 2474–2476 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M. et al. Genetic variant in PSCA predicts survival of diffuse-type gastric cancer in a Chinese population. Int J Cancer 129, 1207–1213 (2011). [DOI] [PubMed] [Google Scholar]
- Kruger J. & Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res 34, W451–454 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data