Abstract
The default mode network (DMN) is a complex dynamic network that is critical for understanding cognitive function. However, whether dynamic topological reconfiguration of the DMN occurs across different brain states, and whether this potential reorganization is associated with prior learning or experience is unclear. To better understand the temporally changing topology of the DMN, we investigated both nodal and global dynamic DMN-topology metrics across different brain states. We found that DMN topology changes over time and those different patterns are associated with different brain states. Further, the nodal and global topological organization can be rebuilt by different brain states. These results indicate that the post-task, resting-state topology of the brain network is dynamically altered as a function of immediately prior cognitive experience, and that these modulated networks are assembled in the subsequent state. Together, these findings suggest that the changing topology of the DMN may play an important role in characterizing brain states.
Recent studies have revealed higher metabolic activity in a particular network of regions termed the default mode network (DMN)1,2,3. The DMN is a set of functionally and structurally connected brain regions that typically exhibit deactivation during the performance of an externally oriented attention-demanding task and high cerebral blood flow and oxygen consumption during the resting state4,5,6,7. DMN function has been largely linked to self-referential thought, mind-wandering, internal-oriented cognition, and autobiographical memory2,7. To date, a large body of studies use a stationary functional connectivity (FC) approach to characterize DMN organization via analyses of inter-regional temporal relationships during resting and task states8,9,10,11,12,13. The various neuroimaging techniques used for stationary FC analysis have revealed DMN spatial-temporal properties not only during attention-demanding cognitive task and at rest, but also during anaesthesia, in vegetative patients, and during different stages of sleep14,15,16,17. Moreover, accumulating evidence indicates that stationary FC within the DMN is linked to specialized cognitive functions and clinically relevant information8,12,18,19,20,21.
The brain is a complex system in which the dynamic adjustment of network organization over multiple time scales is crucial for mediating perception and cognition22,23,24,25,26,27,28,29,30,31,32. Characterizing the dynamics of brain FC or properties of network topology is thought to be important for gaining a better understanding of brain function and behavioural performance13,33,34,35. Further, growing evidence supports the idea that dynamic FC patterns are tightly linked to adaptive behaviour36.
The flexibility of functional network configurations during learning tasks has been shown to facilitate subsequent learning performance37,38,39. Additional evidence suggests that brain functional networks dynamically adjust the structure of their global and local network connectivity as a means to guide optimal behavioural performance, such as during an upcoming visual discrimination task40,41. These findings emphasize the importance of dynamic coordination within the brain network and the adaptability of its topological structure for supporting ongoing task performance42,43.
Until now, few studies have directly examined dynamic DMN during different brain states (i.e., pre-task resting state, task state, and post-task resting state). Answering how DMN temporal properties depend on different brain states, such as past cognitive learning experiences, could thus help provide a more complete understanding of DMN function. Studies have found that prior tasks can affect post-task resting-state brain activity44,45,46. Thus, modulation of learning-dependent spontaneous brain activity has been observed after people perform cognitive tasks involving working memory, emotion, visual perception, and motor training44,45,47,48,49,50. Such modulation of brain regions associated with a prior task can be understood as reflecting the gradual establishment, or reinforcement, of recent experiences in the offline resting state. A possible explanation for this is that recent cognitive processes require a functional interaction between multiple specialized local and remote brain regions as a result of a reorganized neuronal network structure. Whether these types of changes in DMN configuration result from the prior cognitive task remains unclear. Some studies have indicated that the spatial distribution of the DMN appears to persist unchanged across active and resting states, suggesting that the DMN reflects stable properties of brain-network architecture. However, this question cannot be fully answered without knowing how dynamic DMN connectivity changes across different brain states. Additionally, characterization of this dynamic DMN activity is critical for understanding brain functional stability and flexibility.
Indeed, characterizing brain-network topology plays an important role in understanding cognitive function51,52. Many researchers have explored the relationship between complex topological properties of functional brain networks and cognitive or behavioural measures27,41,43,47,53,54. Moreover, recent studies show that the community structure of the human brain network is associated with cognitive function47,55. Although many studies having investigated functional connectivity of the DMN during different tasks or at rest, none have similarly assessed DMN topological measures8,13,19,22,36. Furthermore, from the perspective of a brain network, the dynamic reorganization of DMN functional connectivity in response to an external cognitive task depends on topological properties.
Determining the properties of DMN topology is an important step toward understanding the mechanism of its functional reconfiguration. Here, we used complex network-analysis techniques to investigate the properties of global and nodal DMN topology across different brain states. Specifically, we answered the following questions: (1) Are changes in the topological structure of the DMN associated with different brain states? (2) If so, are these changes modulated by recent cognitive task performance?
Methods
Ethics statement and participants
All participants gave their written, informed consent before the experiment was conducted in compliance with an experimental protocol approved by the Ethics Committee of University of Trento, Italy. Experiments were carried out in accordance with the approved guidelines. Fourteen right-handed participants (mean age: 27.4 years, range: 23–35 years) were recruited for the experiment. All participants were healthy and without a history of neurological or psychiatric disorders.
Experimental design
To explore dynamic DMN functional connectivity across different brain states, we conducted three separate experiments. Each experiment consisted of a 10-min fMRI scan. First, participants underwent a 10-min resting-state scan before an attention task. They were instructed simply to keep their eyes closed and to not think about anything in particular. After the 10-min resting-state scan, participants performed a visual attention task that was administered in blocks. A detailed account of this task has been published elsewhere5. Briefly, participants were instructed to respond as quickly as possible to a lateralized visual target with an ipsilateral or a contralateral button press, according to non-verbal visual instructions (a square or diamond, respectively) that they saw at the beginning of each trial within a block. A masked instruction (smaller square or diamond) or neutral shape (star) subliminally preceded each visible instruction. After completing the attention task, participants sat through another 10-min resting-state scan. Figure 1 shows the experimental design.
Figure 1. Experimental design.
Task design order: pre-resting state, attention task, and post-resting state. For both resting-state conditions, participants were instructed to lie still, relax, and close their eyes. During the cognitive task condition, participants performed a visual attention task.
MRI data acquisition
Scanning was performed on a 4-T Bruker MedSpec Biospin MRI scanner with an eight-channel phase-array receiver head coil. To make sure the participants fit comfortably inside the scanner during the experiment, we fixed their heads with foam cushions to minimize head movement. The T1-weighted high-resolution anatomical images were acquired using a sagittal magnetization-prepared rapid gradient echo (MP-RAGE) three-dimensional T1-weighted sequence optimized for grey-white matter contrast (repetition time [TR] = 2700 ms, echo time [TE] = 4.18 ms, inversion time [TI] = 1020 ms, flip angle [FA] = 7°, slice number = 176, and matrix size = 224 × 256, 1 × 1 × 1 mm3, GRAPPA iPAT = 2). The functional images were acquired using an echo-planar (EPI) sequence corrected for distortion using the PSF method (TR = 2500 ms, TE = 33 ms, FA = 73°, 34 slices, 3 × 3 × 3 mm voxels, field of view [FOV] = 192).
Data Analysis
Functional connectivity preprocessing
The following preprocessing of fMRI data was performed using an analysis based on AFNI (http://afni.nimh.nih.gov/afni/) (Cox 1996) and FSL’s software Library (http://www.fmrib.ox.ac.uk/fsl/). The first four images were removed to avoid T1 equilibration effects. First, the fMRI data were motion-corrected using a linear registration algorithm tool. Then they were spatially smoothed with a Gaussian kernel with a FWHM of 6 mm. Next, we followed the following steps: (1) remove the linear trends; (2) temporally band pass filter (0.009 < f < 0.08) the data and (3) remove several sources of spurious variance from the data through linear regression that incorporated six rigid body motion correction parameters, the white matter signal, and the cerebral spinal fluid (CSF) signal. Recently, whole-brain signal regression has raised challenging interpretive issues. Relatively recent evidence suggests a neuronal origin for the global blood-oxygen-level dependent (BOLD) signal that is typically removed as a nuisance term in resting-state studies. The use of global signal regression is still under debate as a preprocessing step in resting-state fMRI analysis and is not universally recommended56,57. Here, we did not remove the global whole-brain signal. Each participant’s fMRI data were registered to the MNI152 standard template using FSL’s linear registration algorithm (FLIRT). Additionally, as participant motion can contribute to resting state functional connectivity, we further examined motion across different brain states. Framewise displacement (FD) and temporal derivative of the fMRI time series (DVARS) values were used to identify volumes in the fMRI time data to remove from data analysis. To minimize the motion effects, previous study suggests that each volume movement (FD) exceed the 0.5 mm should be censored58. To further compare head motion across pre-task, task, and post-task brain states, we calculated the FD and DVARS across the three brain states. One-way analyses of variance showed that neither FD nor DVARS differed significantly across three brain states (FD: p = 0.1075; DVARS: p = 0.911; Fig. S1).
Functional connectivity-mapping analysis
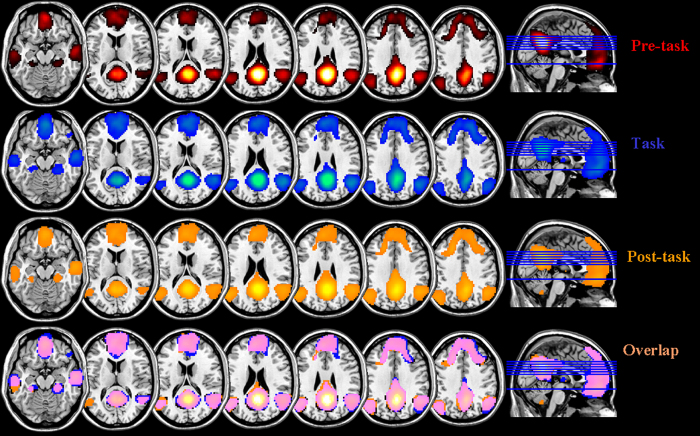
For each participant, seed-based functional connectivity (FC) analysis was conducted by extracting the time series from the PCC region. The seed was constructed using an 8-mm sphere located at (0, −53, 26) in Montreal Neurological Institute (MNI) space, in accordance with a previous study59. The correlation coefficients between the PCC-seed time series and those of the entire-brain voxels were calculated. FC maps of the DMN were further derived by Fisher’s r-to-z transformation. For the group-level analysis, a one-sample t-test was conducted to detect brain regions showing significant FC across participants. The group-level FC maps were further corrected for multiple comparisons using FWE-correction with a corrected threshold of p < 0.05.
DMN region of interest definition and dynamic correlation matrix construction
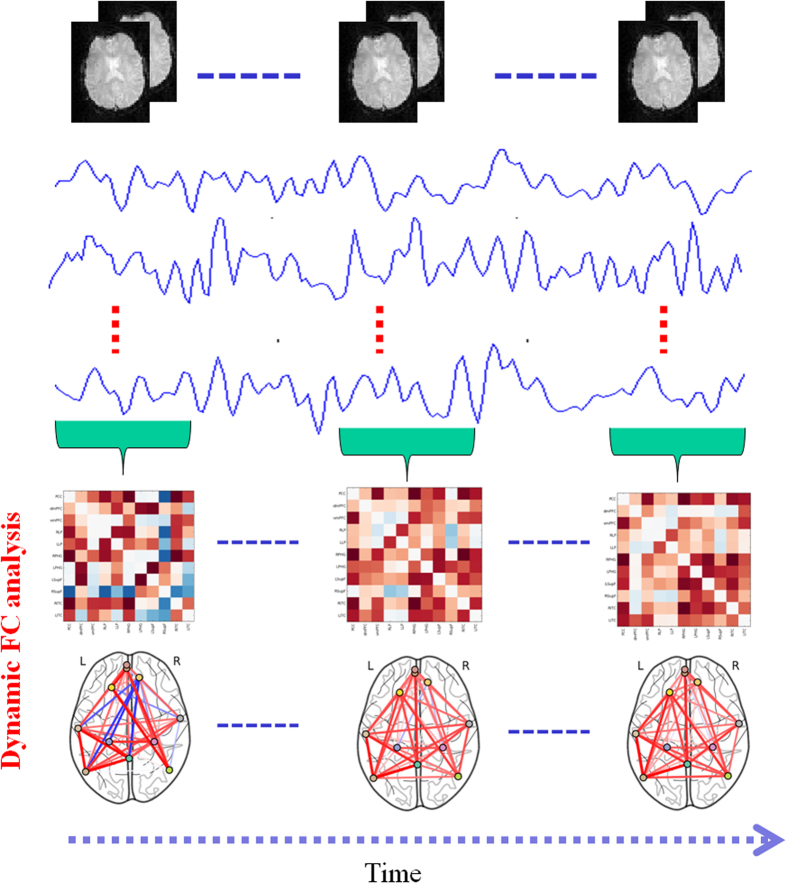
Here, group-based z-score maps of the PCC were corrected for multiple comparisons using FWE-correction for peak voxels. The corrected threshold was set at p < 0.05. The centres of each DMN region of interest (ROI) were determined by considering the coordinates of the z-value local maxima on the correlation map (see Fig. 2). For all DMN ROIs, an 8-mm sphere was created at the peak coordinates of the pre task resting-state FC mapping of the DMN. Thus, the following default network regions were defined in MNI space: posterior cingulate cortex (PCC; MNI coordinate: 0, −53, 26), dorsal medial prefrontal cortex (dmPFC; MNI coordinate: −3, 55, 22), ventral medial prefrontal cortex (vmPFC; MNI coordinate: −3 59, −7), left parahippocampal gyrus (LPHG; MNI coordinate: −24, 33, −27), right parahippocampal gyrus (RPHG; MNI coordinate: 30, −33, −27), left lateral parietal cortex (LLP; MNI coordinate: −52, 69, 26), right lateral parietal cortex (RLP; MNI coordinate: 48, −67, 36), left superior frontal cortex (LSupF; MNI coordinate: −21, 32, 47), right superior frontal cortex (RSupF; MNI coordinate: 12, 44, 48), left inferior temporal cortex (LITC; MNI coordinate: −61, −17, −30), and right inferior temporal cortex (RITC; MNI coordinate: 61, −5, −25). The mean time series for each ROI was then calculated by taking the mean of the voxel time series within each region. Dynamic Pearson’s correlation coefficients were computed between the time series for all pairs of brain regions for each participant based on different sliding windows (60 s, 75 s, and 90 s), and sliding within a step of one TR. Then, 11 × 11 dynamic correlation matrices were generated for all participants. For further statistical analysis, a Fisher’s r-to-z transformation was applied to improve the normality of the correlation coefficients (see Fig. 3).
Figure 2. Mapping of DMN functional connectivity in different brain states (corrected p < 0.05).
Figure 3. General scheme for the characterization of dynamic DMN, including the construction of dynamic correlation matrices and DMN topology analysis.
Topology metrics included nodal degree, clustering coefficient, local efficiency, and global efficiency.
Dynamic Network Analysis
Graph theory is a useful tool for brain-network analysis. To date, most graph measures have only been defined for the simplest case of an unweighted graph, such as by setting all edges with a weight above a certain threshold to binary. However, an unweighted graph approach has several disadvantages. For example, much of the information available in the weights is not used. To better understand the complex brain systems under study, information about the nature and strength of the underlying node interactions should be taken into account to build the weighted network. Thus, we characterized the dynamic changes in the coordinated pattern of DMN networks with a weighted-network analysis. The dynamic correlation matrix for each participant and each sliding window was then obtained based on this analysis. We set the negative functional connectivity to zero. We used this weighted-graph method to calculate the topological metrics of the DMN, and then characterized the dynamic temporal-topological metrics for each sliding window using a weighted complex network analysis that was based on the BCT Matlab toolbox (http://www.brain-connectivity-toolbox.net) (see Fig. 3). The network topology metrics used in this study included nodal degree, clustering coefficient, local efficiency, and global efficiency.
Statistical Analysis
We calculated the mean topology metrics (global and local efficiency, clustering coefficients, and degrees of topology metrics) across participants. To further assess the differences in brain-network topological properties across different brain states, the four metrics were statistically evaluated using the Wilcoxon rank sum test. Significant differences in their distributions across different brain states were calculated using the Kolmogorov–Smirnov test. In order to characterize the patterns of dynamic DMN topology across different brain states, we further used the k-means approach to analyse the topology metrics. The K-means approach is one of the simplest and fastest unsupervised learning techniques for clustering, and has been widely used to analyse numerous pattern-recognition problems. For more details regarding the k-means approach, see the supplemental Methods. To assess the relationships between whole-DMN topology metrics within different brain states, we used a robust regression approach that yields valid p-values while minimizing the influences of outliers.
Results
Functional connectivity mapping of the DMN across different mental states
Figure 2 shows the z-map for group functional connectivity of the posterior cingulate cortex (PCC) seed across different mental states (FWE-corrected; at a threshold of p < 0.05).
Dynamic DMN nodal network topology
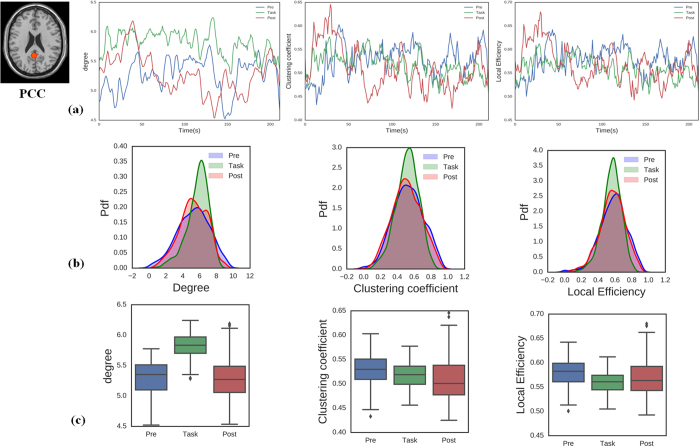
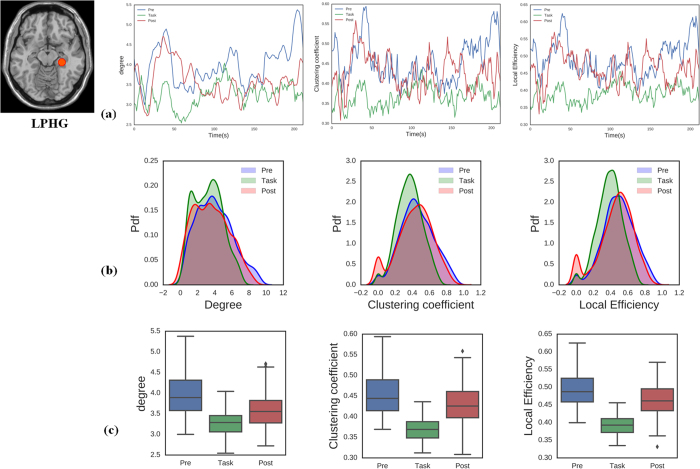
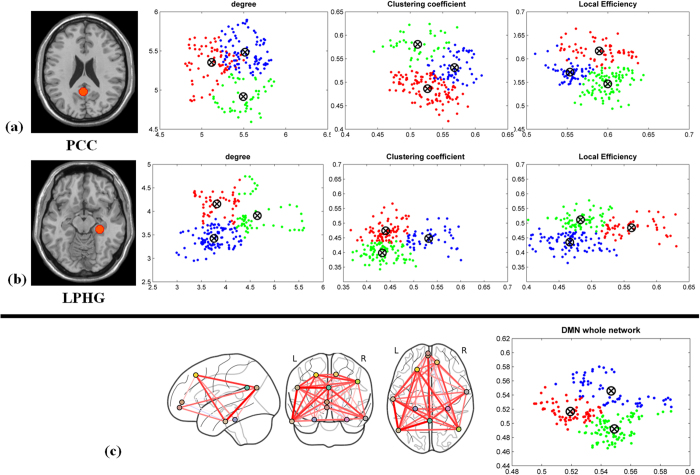
To investigate the properties of dynamic DMN nodal topology, we considered three metrics for data analysis (nodal degree, clustering coefficient, and local efficiency). The DMN can be viewed as being composed of sub-networks60, including the medial temporal sub-network associated with memory-related nodes (left and right parahippocampal gyri; LPHG and RPHG) and another sub-network associated with the PCC. Here, we primarily focused on analysing these two sub-networks during three different brain states (pre-task resting state network, task, post-task resting state network). Figures 4a and 5a show the average topology of each metric for the PCC and LPHG nodes across the three brain states. We calculated the statistical properties for the temporal evolution of the topology metrics. Figures 4b and 5b show that that the probability distribution functions for the PCC and LPHG topology metrics across all participants were significantly different from each other during the different brain states (p < 0.05, Kolmogorov–Smirnov test). Nodal degree, clustering coefficient, and local efficiency were all significantly altered across different brain states (Figs 4c and 5c, Wilcoxon rank sum test, p < 0.05). Other metrics of DMN nodal topology show similar patterns (see Supplementary Figs S2–S10). These results indicated that DMN nodal topology can become reorganized as brain states transition back and forth.
Figure 4. Temporal dynamic topology metrics of the PCC node across different brain states (pre-task resting state, task state, and post-task resting state).
(a) Properties of PCC topology metrics for nodal degree, clustering coefficient, and local efficiency. (b) The probability distribution functions for PCC nodal topology metrics differed significantly across different brain states (two-sample Kolmogorov–Smirnov test, p < 0.001). (c) Boxplots of the PCC topology metrics indicating that the metrics differ significantly across brain states (Wilcoxon rank sum test, p < 0.05).
Figure 5. Topology metrics over time for the LPHG node during the pre-task resting state, task state, and post-task resting state.
(a) Properties of LPHG topology metrics for nodal degree, clustering coefficient, and local efficiency. (b) The probability distribution functions for LPHG nodal topology metrics differed significantly across brain states (two-sample Kolmogorov–Smirnov test, p < 0.001). (c) Boxplots of the LPHG topology metrics indicating that the metrics differ significantly across brain states (Wilcoxon rank sum test, p < 0.05).
To further investigate whether the DMN topology metrics exhibit different patterns as a function of brain state, we analysed them using the k-mean clustering approach. Figure 6a and b show three clusters of topology metrics between different brain states, indicating that DMN nodal topology is dynamically linked to brain state. The other nodal topology metrics show similar patterns (see the Supplementary Fig. S11).
Figure 6. K-means clustering for DMN topology metrics analysis across different brain states.
(a–c) The DMN nodal and global topology metrics formed a separate cluster for each brain state.
Dynamic DMN global network topology
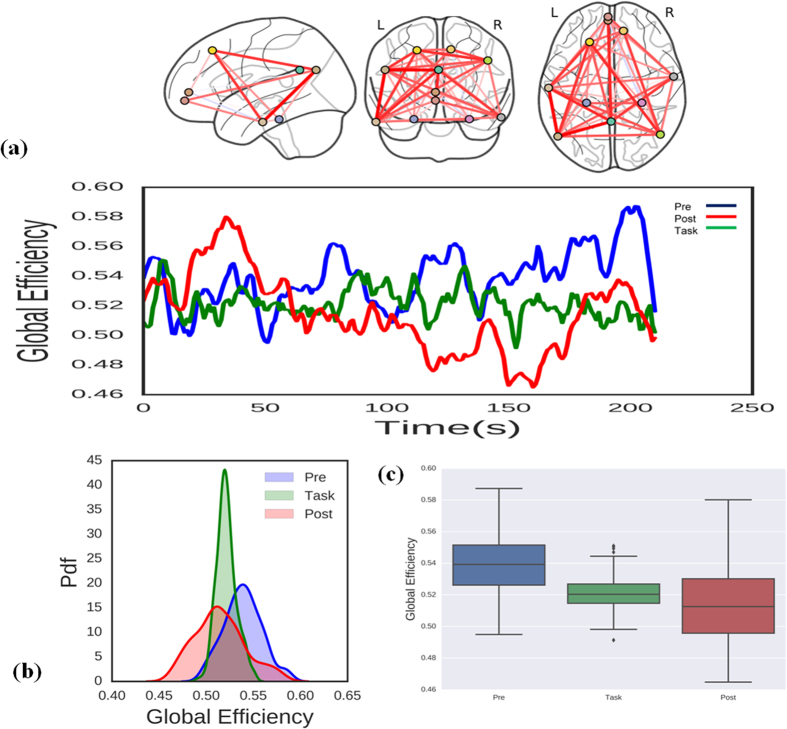
We also investigated whether global DMN topology metrics differed depending on brain state. Figure 7a shows the temporal properties of global DMN topology during the pre-task resting state, task state, and post-task resting state. Figure 7b shows that the probability distribution functions for the global topology metrics differed significantly across diverse brain states (p < 0.05, Kolmogorov–Smirnov test). Further analysis showed that the task altered DMN global topology (Fig. 7c, Wilcoxon rank sum test, p < 0.05), with the DMN global topology metrics for the three brain states forming three separate clusters. These results indicate that the DMN global network topology is also dynamically linked to brain state.
Figure 7. Changes in the DMN global efficiency during the pre-task resting state, task state, and post-task resting state.
(a) topological properties of DMN global efficiency. (b) probability distribution functions for global efficiency. The distribution functions differed significantly (two-sample Kolmogorov–Smirnov test, p < 0.001). (c) Boxplots of the DMN global efficiency indicating that it differed significantly across brain states (Wilcoxon rank sum test, p < 0.05).
The relationship between topology structures across different brain states
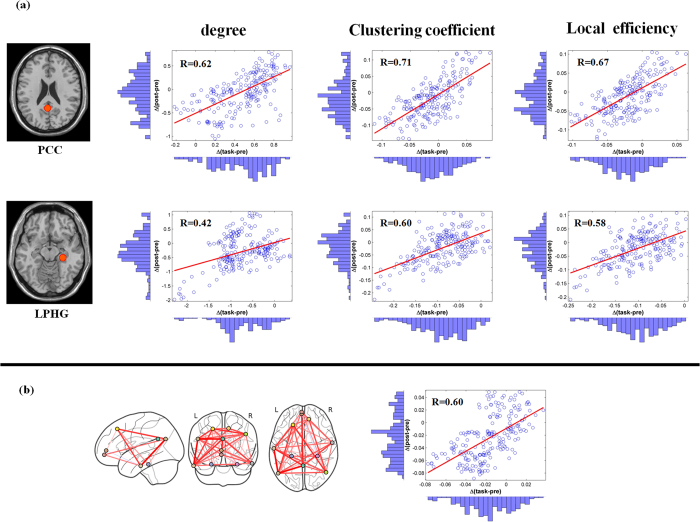
To explore the relationship between changes in topology metrics and different brain states, we used a robust regression approach. The change in network topology metrics Δ (task-pre) induced by task performance was associated with the change in network topology metrics Δ (post-pre). Our results show significant positive correlations between Δ (task-pre) and Δ (post-pre) in the PCC, LPHG nodal, and DMN global network topology metrics (see Fig. 8, p < 0.05). The other DMN nodal metrics showed similar results (Supplementary Fig. S12). Thus, the results indicate that task performance significantly modulates DMN topology structure.
Figure 8. Scatterplots of the association between Δ(task-pre) and Δ(post-pre) in the DMN.
(a) DMN nodal topology metrics show significant correlation between Δ(task-pre) and Δ(post-pre) in the PCC and LPHG across brain states (p < 0.0001). (b) DMN global topology metrics show significant correlation between Δ(task-pre) and Δ(post-pre) across brain states (p < 0.0001).
Dynamic DMN topology analysis with different sliding windows
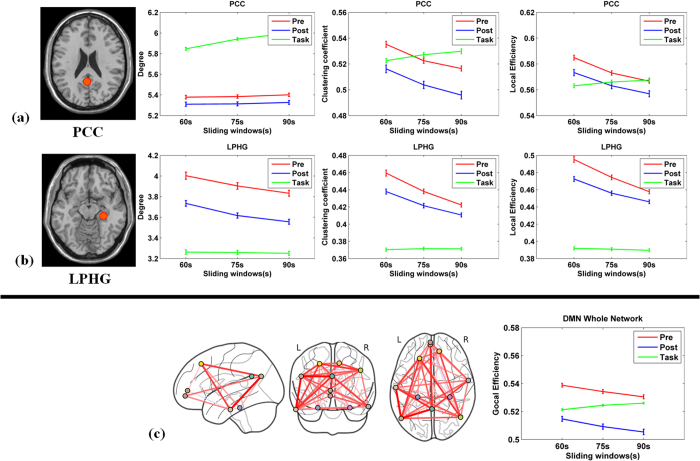
To investigate how the length of the sliding window affects the analysis of DMN topology, we further examined our data using different sliding-window lengths. Previous studies have suggested that longer sliding windows would not be able to adequately characterize dynamic FC properties. Thus, here we focused on shorter sliding-window lengths (60 s, 75 s and 90 s). We consistently observed that DMN nodal and global topology metrics differed significantly depending on task state, regardless of the window length (Fig. 9, Wilcoxon rank sum test, p < 0.05). We found similar results for the other DMN regions (Supplementary Fig. S13).
Figure 9. Dynamic DMN network topology metrics for different sliding windows.
(a–c) The PCC, LPHG, and DMN whole-network dynamic topology metrics for each sliding window length (60 s, 75 s, and 90 s). Error bars indicate SEM.
Discussion
We investigated the structure of DMN topology across different brain states. As we hypothesized, DMN topological structure dynamically changes over time during different brain states. Interestingly, this dynamic organization of nodal and global DMN topology structure can be rebuilt by different brain states. Differences between pre- and post-task DMN topology metrics significantly correlated with differences between pre-task and task DMN topology metrics, thus supporting our second hypothesis and indicating that the topologies of the post-task resting-state brain network are dynamically altered as a function of immediately preceding cognitive experiences. Most importantly, these findings suggest that dynamic changes in network topology within the DMN are related to establishing or reinforcing the most recently acquired novel information.
To date, most stationary FC studies of the DMN have indicated an interplay between the DMN and the dorsal attention network (DAN) during task performance5,24,61, which was associated with searching memory, attention, and spatial learning62,63. Stationary FC analysis however, may not fully capture the potential variation in correlation patterns over time, and as a result, some of the more subtle information related to network interactions may be lost33. Recently, several studies have suggested that the functional connectivity of dynamic brain networks may carry important cognitive and clinically relevant information33,36,47,64,65,66,67,68. Thus, to better understand the relationship between DMN functional connectivity and brain states over time, the functional connectivity must be investigated across different brain states. The functional connectivity of brain networks during resting and task states depends on time-varying coordination of the cerebral cortex that supports the self-conscious and adaptive processing of the ongoing task13,36. For example, functional magnetic resonance imaging (fMRI) and magnetoencephalography have revealed a time-varying relationship between the DMN and the DAN that supports human cognitive function69. Importantly, optimal behavioural performance is guided by the dynamic changes in the global and local network connectivity structure within the functional networks of the brain41.
Furthermore, the DMN topology metrics may index changes in neuronal activity patterns that underlie critical aspects of cognition and clinically relevant information13,33. For example, dynamic reconfiguration of DMN functional connectivity can track ongoing daydreaming, and changes DMN functional connectivity over time may carry important information associated with behaviour36. More importantly, the dynamic properties of network topology are critical for understanding mental disorders33. These observations raise the following critical open questions: (1) how can the dynamic-temporal properties of the DMN be characterized? and (2) how does the dynamic DMN configuration change across different brain states? Because the combination of graph analysis and fMRI offers a powerful tool for characterizing complex topological properties of functional brain networks and cognition52, determining the dynamic topological properties of the DMN is an important step towards understanding the relationship between DMN functional network organization and cognitive function.
In the present work, we combined a graph-theory approach that incorporated the sliding-windows method to investigate both global and local nodal DMN topology metrics across three brain states (pre-task resting state, task-state and post-task resting state). We observed that the topological structure of DMN local nodal topology metrics (degree, clustering coefficient, and local efficiency) and the DMN global network efficiency metric both dynamically change over time. These results are consistent with prior studies33. The fact that interaction within DMN regions was dominated by short intermittent events rather than being the result of a continuously sustained process has important implications for understanding how DMN organization adapts and reconfigures itself in response to an external task stimulus or to a change in cognitive state36. The dynamic topological structure of the DMN that we observed could represent a temporal signature of a brain microstate that supports internal and on-going different brain-state cognitive processes47,70. These results suggest that DMN dynamic properties could be linked to variations in brain-information processing across multiple time scales. Specifically, characterizing the dynamic and adaptive reconfiguration of the DMN may provide greater understanding of both the fundamental properties of normative brain cognition and the pathophysiology of mental illnesses.
The DMN is known to be most active during the resting state, with deactivation occurring during task performance2. The DMN shows a similar spatial architecture across rest and a variety of tasks71. However, whether its temporal topological pattern change as a function of brain states has been unclear. Here, our results show that metrics for DMN nodal and global network topology are dynamic and significantly differ across different brain states.
Previous studies have demonstrated that the dynamic interactions between DMN, central executive network (CEN) and salience network (SN) play an important role in the shift between resting and focusing attention, which involves the re-allocation of resources within the brain for supporting stimulus-related cognitive processing2,5,21,63,72,73,74. The organization of brain network recruitment is thought to occur in response to an external cognitive task. From the perspective of DMN deactivation, studies have shown a decrease in the activity of the DMN during performance of an external task. Additionally, research has found evidence of increased connectivity within the DMN during the resting state compared to the task state. Indeed, a number of studies have proved that the DMN exhibits a tendency to co-modulate during the resting state46. Based on this evidence, we expected that DMN topology during task performance would shift to support the ongoing task. In fact, our results show that PCC-, LPHG- and DMN-related nodal topology metrics are dynamic and change during the task state. At the level of global topology, we found that global efficiency decreased during the task state. Global efficiency has been shown to measure the speed of information transfer in the whole network75. One possible explanation is the suppression of DMN activity during task performance, which induces a decrease in global efficiency within the DMN. The dynamic configuration of local and global DMN topology metrics plays an important role in supporting cognitive function. A series of recent studies have suggested that dynamic changes in brain network topology are associated with task performance27,39,47,69. Our results support the idea that both local and global topology reorganization, in concert, assist in the shift between resting state and task state neuronal processing.
Our findings raise the issue of how a behavioural task can modulate subsequent post-task resting-state DMN topology. Here, we found that post-task resting-state DMN topology metrics exhibited significantly different topological patterns than the task state. The DMN nodal and global network topology metrics significantly changed during the post-task resting state. K-means clustering results indicate that the nodal and global network topology metrics exhibit different patterns during different brain states. Furthermore, switching from the task state to the resting state generated a topological pattern that was opposite what was generates when switching from the resting state to the task state. Previous studies have suggested that the DMN might recover to its pre-task state during the subsequent resting state76. Somewhat in contrast, we found that in addition to recovery of the pre-task state, significant differences in local and global DMN network topology metrics between the pre- and post-task resting states. A possible explanation for our finding is that the DMN has been modulated by the prior cognitive task, which would shape the DMN topology properties and lead to the development of a new DMN organization. Substantial change in DMN activity has been found to occur during the transitions from task states to the resting state68,77,78. Moreover, recent studies have shown that DMN activity could be differentially modulated even in the absence of an external task15,79,80. Taken together, our results demonstrate that the patterns of DMN topology vary across different brain states. The characterization of these dynamic DMN topological patterns might thus enable us to trace or distinguish different brain states.
Studies have shown that the post-task resting-state network might be shaped by the performance during a prior task or through training44,45,49. Here, we explored the relationship between post-task resting-state, pre-task resting-state, and task state nodal topology metrics. We found that post-task resting-state DMN nodal and global topology metrics were associated with task-state modulation. In particular, we found that differences between pre-task and post-task PCC and LPHG nodal topology and DMN whole-network metrics significantly correlated with differences between pre-task and task DMN topology metrics. These results suggest that the post-task resting-state DMN nodal and global topologies are related to prior network co-activation. A possible explanation is that prior learning or external task demand changes the DMN sub-network interactions within the DMN, which can lead to reshaping of the new DMN topology properties to fit the new cognitive behaviour. Recently, studies have suggested that post-task resting-state brain network reorganization is dependent on a prior cognitive task, which may be compatible with a long-term memory system81. Thus, learning, or experience-dependent alteration, has been observed in spontaneous brain activity following cognitive task performance, and brain activity related to the alterations are associated with the preceding task demands45,77. Interestingly, Peigneux et al. found that offline regional activity was modulated by a recent learning task82. The post-training activity significantly correlated with prior task performance81. Additionally, clinical studies show that reshaping of the DMN topology is also associated with neuropsychiatric diseases such as schizophrenia, Alzheimer’s disease and alcoholism18,53,83. Thus, temporally coherent dynamic change in the global brain network is dependent on the prior task type. Moreover, the whole-network topology changes smoothly over a range of temporal scales. Taken together with our findings, this shows that DMN global network topology metrics may be modulated under different brain states.
In conclusion, the topology of the DMN changes over time as individuals’ transition between resting and task states. The nodal and global topological structure is reshaped in different brain states and shows different patterns. Furthermore, the topology metrics are dynamically altered as a function of cognitive experiences, and the modulated networks are assembled in the subsequent resting state. These findings suggest that understanding the dynamic topology of the DMN can provide a new way to better characterize DMN function.
Additional Information
How to cite this article: Lin, P. et al. Dynamic Default Mode Network across Different Brain States. Sci. Rep. 7, 46088; doi: 10.1038/srep46088 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
Government of the Provincia Autonoma di Trento, Italy, Project PAT Post-doc 2006; Fondazione Cassa di Risparmio di Trento e Rovereto; and University of Trento, Italy. This work was supported by the National Natural Science Foundation of China (61473221, 61262034, 81271659), by Nature Science Foundation of Shaan Xi Province of China (2015JM3105), by the National “973 project” (2015CB351704), by the Fundamental Research Funds for the Central Universities of China (1191320118).
Footnotes
The authors declare no competing financial interests.
Author Contributions P.L. and N.D.P. designed this study. P.L. and N.D.P. performed the study. P.L., Y.Y., S.G., J.F.G., X.W., C.N. analyzed the data. P.L., Y.Y., X.W., C.N., C.S.Z. and J.J.L. wrote the main manuscript text. P.L. and C.N. prepared the figures and references. All authors contributed to and have approved the final manuscript.
References
- Raichle M. E. et al. A default mode of brain function. Proceedings of the National Academy of Sciences of the United States of America 98, 676–682 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckner R. L., Andrews-Hanna J. R. & Schacter D. L. The brain’s default network: anatomy, function, and relevance to disease. Ann N Y Acad Sci 1124, 1–38, doi: 10.1196/annals.1440.011 (2008). [DOI] [PubMed] [Google Scholar]
- Fransson P. Spontaneous low-frequency BOLD signal fluctuations: an fMRI investigation of the resting-state default mode of brain function hypothesis. Human brain mapping 26, 15–29, doi: 10.1002/hbm.20113 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin P., Hasson U., Jovicich J. & Robinson S. A neuronal basis for task-negative responses in the human brain. Cerebral cortex (New York, N Y: 1991) 21, 821–830 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Pisapia N., Turatto M., Lin P., Jovicich J. & Caramazza A. Unconscious priming instructions modulate activity in default and executive networks of the human brain. Cereb Cortex 22, 639–649, doi: 10.1093/cercor/bhr146 (2012). [DOI] [PubMed] [Google Scholar]
- Laurienti P. J. Deactivations, global signal, and the default mode of brain function. J Cogn Neurosci 16, 1481–1483, doi: 10.1162/0898929042568596 (2004). [DOI] [PubMed] [Google Scholar]
- Mantini D. & Vanduffel W. Emerging roles of the brain’s default network. Neuroscientist 19, 76–87, doi: 10.1177/1073858412446202 (2013). [DOI] [PubMed] [Google Scholar]
- Bluhm R. L. et al. Default Network Connectivity During a Working Memory Task. Human brain mapping 32, 1029–1035, doi: 10.1002/Hbm.21090 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnelle V. et al. Default mode network connectivity predicts sustained attention deficits after traumatic brain injury. Journal of Neuroscience 31, 13442–13451, doi: 10.1523/JNEUROSCI.1163-11.2011 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fair D. A. et al. The maturing architecture of the brain’s default network. Proceedings of the National Academy of Sciences of the United States of America 105, 4028–4032 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guldenmund P., Vanhaudenhuyse A., Boly M., Laureys S. & Soddu A. A default mode of brain function in altered states of consciousness. Arch Ital Biol 150, 107–121, doi: 10.4449/aib.v150i2.1373 (2012). [DOI] [PubMed] [Google Scholar]
- Sharp D. J. et al. Default mode network functional and structural connectivity after traumatic brain injury. Brain: a journal of neurology 134, 2233–2247, doi: 10.1093/brain/awr175 (2011). [DOI] [PubMed] [Google Scholar]
- Kucyi A., Salomons T. V. & Davis K. D. Mind wandering away from pain dynamically engages antinociceptive and default mode brain networks. Proceedings of the National Academy of Sciences of the United States of America 110, 18692–18697, doi: 10.1073/pnas.1312902110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power J. D. et al. Functional network organization of the human brain. Neuron 72, 665–678, doi: 10.1016/j.neuron.2011.09.006 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T. et al. Network-dependent modulation of brain activity during sleep. NeuroImage 98, 1–10, doi: 10.1016/j.neuroimage.2014.04.079 (2014). [DOI] [PubMed] [Google Scholar]
- Fernandez-Espejo D. et al. A role for the default mode network in the bases of disorders of consciousness. Ann Neurol 72, 335–343, doi: 10.1002/ana.23635 (2012). [DOI] [PubMed] [Google Scholar]
- Nallasamy N. & Tsao D. Y. Functional connectivity in the brain: effects of anesthesia. Neuroscientist 17, 94–106, doi: 10.1177/1073858410374126 (2011). [DOI] [PubMed] [Google Scholar]
- Chanraud S., Pitel A. L., Pfefferbaum A. & Sullivan E. V. Disruption of Functional Connectivity of the Default-Mode Network in Alcoholism. Cerebral cortex (New York, N Y: 1991), doi: 10.1093/cercor/bhq297 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucyi A. et al. Enhanced Medial Prefrontal-Default Mode Network Functional Connectivity in Chronic Pain and Its Association with Pain Rumination. Journal of Neuroscience 34, 3969–3975, doi: 10.1523/Jneurosci.5055-13.2014 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y. X. et al. Default-Mode Network Disruption in Mild Traumatic Brain Injury. Radiology 265, 882–892, doi: 10.1148/radiol.12120748 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callard F. & Margulies D. S. What we talk about when we talk about the default mode network. Frontiers in Human Neuroscience 8, doi: 10.3389/Fnhum.2014.00619 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer A. et al. Dynamic network participation of functional connectivity hubs assessed by resting-state fMRI. Frontiers in Human Neuroscience 8, 195, doi: 10.3389/fnhum.2014.00195 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin G. & van den Heuvel M. P. The Ontogeny of the Human Connectome: Development and Dynamic Changes of Brain Connectivity Across the Life Span. Neuroscientist 19, 616–628, doi: 10.1177/1073858413503712 (2013). [DOI] [PubMed] [Google Scholar]
- Hellyer P. J. et al. The control of global brain dynamics: opposing actions of frontoparietal control and default mode networks on attention. Journal of Neuroscience 34, 451–461, doi: 10.1523/JNEUROSCI.1853-13.2014 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D. T. et al. Non-Stationarity in the “Resting Brain’s” Modular Architecture. PloS one 7, e39731, doi: 10.1371/journal.pone.0039731 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala-Llonch R. et al. Dynamic functional reorganizations and relationship with working memory performance in healthy aging. Frontiers in Human Neuroscience 6, doi: 10.3389/Fnhum.2012.00152 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breckel T. P. et al. Long-term effects of attentional performance on functional brain network topology. PloS one 8, e74125, doi: 10.1371/journal.pone.0074125 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin P. et al. Static and dynamic posterior cingulate cortex nodal topology of default mode network predicts attention task performance. Brain imaging and behavior 10, 212–225, doi: 10.1007/s11682-015-9384-6 (2016). [DOI] [PubMed] [Google Scholar]
- Gao J. et al. Exploring time- and frequency- dependent functional connectivity and brain networks during deception with single-trial event-related potentials. Scientific reports 6, doi: 10.1038/srep37065 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R. et al. Random matrix theory for analyzing the brain functional network in attention deficit hyperactivity disorder. Physical Review E 94, 052411 (2016). [DOI] [PubMed] [Google Scholar]
- Wang R. et al. Spectral properties of the temporal evolution of brain network structure. Chaos An Interdisciplinary Journal of Nonlinear Science 25, 7641–7646 (2015). [DOI] [PubMed] [Google Scholar]
- Zhao C. Z. M., Yang Y., Gao J. F., Rao N. N. & Lin P. The Reorganization of Human Brain Networks Modulated by Driving Mental Fatigue. IEEE Journal of Biomedical and Health Informatics PP, 1, doi: 10.1109/JBHI.2016.2544061 (2016). [DOI] [PubMed] [Google Scholar]
- Hutchison R. M. et al. Dynamic functional connectivity: Promise, issues, and interpretations. NeuroImage 80, 360–378, doi: 10.1016/j.neuroimage.2013.05.079 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barttfeld P. et al. Signature of consciousness in the dynamics of resting-state brain activity. Proc Natl Acad Sci USA 112, 887–892, doi: 10.1073/pnas.1418031112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatansever D., Menon D. K., Manktelow A. E., Sahakian B. J. & Stamatakis E. A. Default mode network connectivity during task execution. NeuroImage 122, 96–104, doi: 10.1016/j.neuroimage.2015.07.053 (2015). [DOI] [PubMed] [Google Scholar]
- Kucyi A. & Davis K. D. Dynamic functional connectivity of the default mode network tracks daydreaming. NeuroImage 100, 471–480, doi: 10.1016/j.neuroimage.2014.06.044 (2014). [DOI] [PubMed] [Google Scholar]
- Sauvage C. et al. Reorganization of large-scale cognitive networks during automation of imagination of a complex sequential movement. J Neuroradiol 42, 115–125, doi: 10.1016/j.neurad.2014.04.001 (2014). [DOI] [PubMed] [Google Scholar]
- Hattori S., Yoon T., Disterhoft J. F. & Weiss C. Functional reorganization of a prefrontal cortical network mediating consolidation of trace eyeblink conditioning. Journal of Neuroscience 34, 1432–1445, doi: 10.1523/JNEUROSCI.4428-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldassarre A. et al. Individual variability in functional connectivity predicts performance of a perceptual task. Proc Natl Acad Sci USA 109, 3516–3521, doi: 10.1073/pnas.1113148109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidalgo C., Conejo N. M., Gonzalez-Pardo H. & Arias J. L. Dynamic functional brain networks involved in simple visual discrimination learning. Neurobiol Learn Mem 114, 165–170, doi: 10.1016/j.nlm.2014.06.001 (2014). [DOI] [PubMed] [Google Scholar]
- Ekman M., Derrfuss J., Tittgemeyer M. & Fiebach C. J. Predicting errors from reconfiguration patterns in human brain networks. Proc Natl Acad Sci USA 109, 16714–16719, doi: 10.1073/pnas.1207523109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitzbichler M. G., Henson R. N., Smith M. L., Nathan P. J. & Bullmore E. T. Cognitive effort drives workspace configuration of human brain functional networks. Journal of Neuroscience 31, 8259–8270, doi: 10.1523/JNEUROSCI.0440-11.2011 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doron K. W., Bassett D. S. & Gazzaniga M. S. Dynamic network structure of interhemispheric coordination. Proc Natl Acad Sci USA 109, 18661–18668, doi: 10.1073/pnas.1216402109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert N. B., Robertson E. M. & Miall R. C. The resting human brain and motor learning. Current biology: CB 19, 1023–1027 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis C. M., Baldassarre A., Committeri G., Romani G. L. & Corbetta M. Learning sculpts the spontaneous activity of the resting human brain. Proceedings of the National Academy of Sciences of the United States of America 106, 17558–17563 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartzell J. F., Tobia M. J., Davis B., Cashdollar N. M. & Hasson U. Differential lateralization of hippocampal connectivity reflects features of recent context and ongoing demands: an examination of immediate post-task activity. Human brain mapping 36, 519–537, doi: 10.1002/hbm.22644 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett D. S. et al. Dynamic reconfiguration of human brain networks during learning. Proceedings of the National Academy of Sciences of the United States of America 108, 7641–7646, doi: 10.1073/pnas.1018985108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubert M., Lohmann G., Margulies D. S., Villringer A. & Ragert P. Long-term effects of motor training on resting-state networks and underlying brain structure. NeuroImage 57, 1492–1498, doi: 10.1016/j.neuroimage.2011.05.078 (2011). [DOI] [PubMed] [Google Scholar]
- Stevens W. D., Buckner R. L. & Schacter D. L. Correlated low-frequency BOLD fluctuations in the resting human brain are modulated by recent experience in category-preferential visual regions. Cerebral cortex (New York, N Y: 1991) 20, 1997–2006 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sami S., Robertson E. M. & Miall R. C. The Time Course of Task-Specific Memory Consolidation Effects in Resting State Networks. Journal of Neuroscience 34, 3982–3992, doi: 10.1523/Jneurosci.4341-13.2014 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin P. et al. Global and local brain network reorganization in attention-deficit/hyperactivity disorder. Brain imaging and behavior 8, 558–569, doi: 10.1007/s11682-013-9279-3 (2014). [DOI] [PubMed] [Google Scholar]
- Bullmore E. & Sporns O. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat Rev Neurosci 10, 186–198, doi: 10.1038/nrn2575 (2009). [DOI] [PubMed] [Google Scholar]
- Buckner R. L. et al. Cortical Hubs Revealed by Intrinsic Functional Connectivity: Mapping, Assessment of Stability, and Relation to Alzheimer’s Disease. Journal of Neuroscience 29, 1860–1873 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q. et al. Assessing dynamic brain graphs of time-varying connectivity in fMRI data: application to healthy controls and patients with schizophrenia. NeuroImage 107, 345–355, doi: 10.1016/j.neuroimage.2014.12.020 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossley N. A. et al. Cognitive relevance of the community structure of the human brain functional coactivation network. Proceedings of the National Academy of Sciences of the United States of America 110, 11583–11588, doi: 10.1073/pnas.1220826110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy K., Birn R. M., Handwerker D. A., Jones T. B. & Bandettini P. A. The impact of global signal regression on resting state correlations: Are anti-correlated networks introduced? Neuroimage 44, 893–905, doi: 10.1016/j.neuroimage.2008.09.036 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholvinck M. L., Maier A., Ye F. Q., Duyn J. H. & Leopold D. A. Neural basis of global resting-state fMRI activity. Proceedings of the National Academy of Sciences of the United States of America 107, 10238–10243 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power J. D., Barnes K. A., Snyder A. Z., Schlaggar B. L. & Petersen S. E. Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. Neuroimage 59, 2142–2154 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews-Hanna J. R. et al. Disruption of large-scale brain systems in advanced aging. Neuron 56, 924–935 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews-Hanna J. R., Reidler J. S., Sepulcre J., Poulin R. & Buckner R. L. Functional-anatomic fractionation of the brain’s default network. Neuron 65, 550–562, doi: 10.1016/j.neuron.2010.02.005 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornito A., Harrison B. J., Zalesky A. & Simons J. S. Competitive and cooperative dynamics of large-scale brain functional networks supporting recollection. Proc Natl Acad Sci USA 109, 12788–12793, doi: 10.1073/pnas.1204185109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnelle V. et al. Salience network integrity predicts default mode network function after traumatic brain injury. Proc Natl Acad Sci USA 109, 4690–4695, doi: 10.1073/pnas.1113455109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A. C. et al. Causal interactions between fronto-parietal central executive and default-mode networks in humans. Proceedings of the National Academy of Sciences of the United States of America 110, 19944–19949, doi: 10.1073/pnas.1311772110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison R. M., Womelsdorf T., Gati J. S., Everling S. & Menon R. S. Resting-state networks show dynamic functional connectivity in awake humans and anesthetized macaques. Human brain mapping 34, 2154–2177, doi: 10.1002/Hbm.22058 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonardi N. et al. Principal components of functional connectivity: a new approach to study dynamic brain connectivity during rest. NeuroImage 83, 937–950, doi: 10.1016/j.neuroimage.2013.07.019 (2013). [DOI] [PubMed] [Google Scholar]
- Lee H. L., Zahneisen B., Hugger T., Levan P. & Hennig J. Tracking dynamic resting-state networks at higher frequencies using MR-encephalography. NeuroImage 65, 216–222, doi: 10.1016/j.neuroimage.2012.10.015 (2013). [DOI] [PubMed] [Google Scholar]
- Allen E. A. et al. Tracking whole-brain connectivity dynamics in the resting state. Cereb Cortex 24, 663–676, doi: 10.1093/cercor/bhs352 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deco G., Jirsa V. K. & McIntosh A. R. Emerging concepts for the dynamical organization of resting-state activity in the brain. Nat Rev Neurosci 12, 43–56, doi: 10.1038/nrn2961 (2011). [DOI] [PubMed] [Google Scholar]
- de Pasquale F. et al. Temporal dynamics of spontaneous MEG activity in brain networks. Proc Natl Acad Sci USA 107, 6040–6045, doi: 10.1073/pnas.0913863107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pasquale F. et al. A cortical core for dynamic integration of functional networks in the resting human brain. Neuron 74, 753–764, doi: 10.1016/j.neuron.2012.03.031 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S. M. et al. Correspondence of the brain’s functional architecture during activation and rest. Proceedings of the National Academy of Sciences of the United States of America 106, 13040–13045 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utevsky A. V., Smith D. V. & Huettel S. A. Precuneus Is a Functional Core of the Default-Mode Network. Journal of Neuroscience 34, 932–940, doi: 10.1523/Jneurosci.4227-13.2014 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon V. & Uddin L. Q. Saliency, switching, attention and control: a network model of insula function. Brain structure & function 214, 655–667, doi: 10.1007/s00429-010-0262-0 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulden N. et al. The salience network is responsible for switching between the default mode network and the central executive network: replication from DCM. NeuroImage 99, 180–190, doi: 10.1016/j.neuroimage.2014.05.052 (2014). [DOI] [PubMed] [Google Scholar]
- Rubinov M. & Sporns O. Complex network measures of brain connectivity: uses and interpretations. NeuroImage 52, 1059–1069 (2010). [DOI] [PubMed] [Google Scholar]
- Barnes A., Bullmore E. T. & Suckling J. Endogenous human brain dynamics recover slowly following cognitive effort. PloS one 4, e6626, doi: 10.1371/journal.pone.0006626 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton A. T., Morgan V. L., Rogers B. P. & Gore J. C. Modulation of steady state functional connectivity in the default mode and working memory networks by cognitive load. Human brain mapping 32, 1649–1659, doi: 10.1002/hbm.21138 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasenkamp W., Wilson-Mendenhall C. D., Duncan E. & Barsalou L. W. Mind wandering and attention during focused meditation: a fine-grained temporal analysis of fluctuating cognitive states. NeuroImage 59, 750–760, doi: 10.1016/j.neuroimage.2011.07.008 (2012). [DOI] [PubMed] [Google Scholar]
- Park B. et al. Are brain networks stable during a 24-hour period? NeuroImage 59, 456–466, doi: 10.1016/j.neuroimage.2011.07.049 (2012). [DOI] [PubMed] [Google Scholar]
- Eichenlaub J. B. et al. Resting Brain Activity Varies with Dream Recall Frequency Between Subjects. Neuropsychopharmacology 39, 1594–1602, doi: 10.1038/Npp.2014.6 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kesteren M. T. R., Fernandez G., Norris D. G. & Hermans E. J. Persistent schema-dependent hippocampal-neocortical connectivity during memory encoding and postencoding rest in humans. Proceedings of the National Academy of Sciences of the United States of America 107, 7550–7555 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peigneux P. et al. Offline persistence of memory-related cerebral activity during active wakefulness. PLoS biology 4, e100, doi: 10.1371/journal.pbio.0040100 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker J. T. et al. Disruption of cortical association networks in schizophrenia and psychotic bipolar disorder. JAMA Psychiatry 71, 109–118, doi: 10.1001/jamapsychiatry.2013.3469 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.