Abstract
Discriminating between inherited and non-inherited sporadic hearing loss is challenging. Here, we attempted to delineate genetic inheritance in simplex cases of severe-to-profound congenital hearing loss in Korean children. Variations in SLC26A4 and GJB2 in 28 children with bilateral severe-to-profound non-syndromic hearing loss (NSHL) without familial history were analyzed using Sanger sequencing. Genetic analysis of individuals without mutations in SLC26A4 and GJB2 was performed by whole exome sequencing (WES). Bi-allelic mutations in SLC26A4 and GJB2 were identified in 12 and 3 subjects, respectively. Of the 13 individuals without mutations in SLC26A4 and GJB2, 2 and 1 carried compound heterozygous mutations in MYO15A and CDH23, respectively. Thus, 64.3% (18/28) of individuals with NSHL were determined to be genetically predisposed. Individuals with sporadic severe-to-profound NSHL were found to mostly exhibit an autosomal recessive inheritance pattern. Novel causative candidate genes for NSHL were identified by analysis of WES data of 10 families without mutations in known causative genes. Bi-allelic mutations predisposing to NSHL were identified in 64.3% of subjects with sporadic severe-to-profound NSHL. Given that several causative genes for NSHL are still unidentified, genetic inheritance of sporadic congenital hearing loss could be more common than that indicated by our results.
Hearing loss is a common sensorial disorder, with an incidence of 1 in 500–1000 among children1. At least half of these cases are attributable to genetic factors, and more than two-third of the cases in this subset are classified as non-syndromic hearing loss (NSHL)2, which is associated with autosomal dominant (AD) and recessive (AR), X-linked, and maternal inheritance patterns. Of the more than 100 genes (>140 loci) associated with NSHL, approximately 70% contribute to AR-NSHL (http://hereditaryhearingloss.org/)3.
Most individuals with AR-NSHL experience severe-to-profound congenital hearing loss3,4. Severe-to-profound congenital or prelingual hearing loss results in the delay of language and behavioural development at an early age5. Therefore, early diagnosis of hearing loss should be mandatory in screening procedures such as newborn hearing screening. Profound congenital hearing impairment detected in newborn screening is usually sporadic6. Over 90% of newborns with this condition are born of parents with normal hearing7, who will require genetic counseling for hearing rehabilitation and future family planning. Unfortunately, the genetic characteristics of individuals with sporadic hearing loss have not been sufficiently investigated till date6. Specifically, sporadic cases of severe-to-profound congenital hearing loss have been rarely weighted in previous targeted exon sequencing (TES) analyses. Moreover, genetic contribution is so heterogeneous that diversity among individuals of different ethnicities is great. For example, the most common genetic cause of hearing loss among Caucasians is GJB2 mutation, whereas that among East Asians is SLC26A4 mutation8,9. Therefore, it is necessary to delineate the characteristics of genetic predisposition patterns according to ethnicity.
Several studies have employed TES for identifying causative mutations of hearing loss6,10,11,12,13. Whole exome sequencing (WES) has also been successfully used for analysis of genetic factors for hearing loss5,14, Additionally, WES has facilitated the identification of novel genes associated with Mendelian disorders including hearing loss15,16,17. Recent studies have identified several causative genes for NSHL, including GPSM2, DNMT1, ELMOD3, GRXCR2, and ADCY1, by WES15,16,18,19,20.
In the present study, we aimed to establish a molecular diagnosis of sporadic congenital hearing loss in 28 individuals with severe-to-profound congenital NSHL (auditory hearing threshold >70 dB nHL) by genetic analysis by Sanger sequencing and WES.
Methods
Subjects
This study was approved by the institutional review board of the Severance Hospital, Yonsei University Health System (IRB#4-2015-0659). After obtaining informed consent, individuals with hearing loss were enrolled in the Yonsei University Hearing Loss (YUHL) cohort, and their clinical and pedigree data were recorded. The experimental methods were performed in accordance with the approved guidelines. For children, written informed consent was obtained from the parents. From the YUHL cohort, 28 individuals with bilateral severe-to-profound hearing loss without familial history were selected for this study, which was conducted between January 2013 and December 2015 at the Department of Otorhinolaryngology, Severance Hospital, Seoul, Korea. The selected individuals exhibited no other syndromic features except hearing loss and were referred to the Severance Hospital for cochlear implantation. Individuals with a history of non-inherited risk factors for hearing loss, such as perinatal cytomegalovirus infection, neonatal intensive care unit treatment, and middle ear infection, were excluded.
Diagnosis of hearing loss
Physical examination and otoscopy were administered by an otologist. All subjects underwent evaluation for auditory brainstem response (ABR), auditory steady state response (ASSR), and oto-acoustic emission (OAE) for diagnosis of hearing impairment. Subjects who exhibited thresholds <70 dB nHL for ABR were excluded. Average thresholds for ASSR were measured at frequencies of 0.5, 1, 2, and 4 kHz. None of the subjects suspicious for auditory neuropathy were unresponsive in ABR evaluation and presented normal OAE findings. All participants underwent temporal bone computed tomography (TBCT) and magnetic resonance imaging for evaluation of inner ear and temporal bone anomalies. In subjects with unilateral or bilateral enlarged vestibular aqueduct (EVA), all of the exons of SLC26A4 were sequenced using direct Sanger sequencing. In subjects without EVA, GJB2 was sequenced using direct Sanger sequencing. The primer sequences for SLC26A4 and GJB2 are listed in Table S1. The genotypes of individuals with no causative mutations in SLC26A4 or GJB2 were analyzed using WES. In case of subjects with cochlear implants, audiological outcomes at 6 months post-surgery were measured in terms of auditory performance (CAP) score and age equivalence based on the sequenced language scale for infants (SELSI)21,22.
DNA preparation, whole exome sequencing, sequence alignment, and variant calling
Whole blood (3 ml) was collected from the participants and, when available, their siblings and parents for segregation analysis. Genomic DNA was extracted from peripheral leukocytes using red blood cell and cell lysis solutions and a protein precipitation solution (QIAGEN). Whole exome capture was performed using the Agilent SureSelect V5 enrichment capture kit (Agilent Technologies). The enriched library was then sequenced using the HiSeq 2500 sequencing system (Illumina; 101-base paired-end sequencing). Image analysis and base calling were performed with the Pipeline software (Illumina) using default parameters. Sequence reads were mapped to the human reference genome assembly (GRCh37/hg19) using the CLC Genomic Workbench (version 9.0.1) software (QIAGEN). Mapping was performed using the “Map Reads to Reference” function of the CLC Genomic Workbench software with the following settings: mismatch cost, 2; insertion cost, 3; deletion cost, 3; length fraction, 0.5; similarity fraction, 0.9; and map to nonspecific reads, random. Nonspecific reads were ignored for count and coverage. All variants with a minimum coverage of 2 were called using the “Basic Variant Caller” function of the CLC Genomic Workbench and annotated.
Filtering and evaluation of variants
Variant ranking and calling for disease-causing mutations was performed according to the accepted standard in molecular diagnostics23,24. The variant filtering process is described in Fig. S1 and Table S2. In the first step, variants with minor allele frequencies >1% in the single nucleotide polymorphism (dbSNP; version 138) or 1000 genomes (2504 individuals; phase 3 data) databases were excluded. In the second step, variants present in the homozygous or hemizygous state in 32 healthy Korean individuals without hearing loss (internal control WES data) were excluded. In step 3, synonymous variants and intronic variants not located within splice site regions were excluded. In step 4, variants of all 72 genes known to be monogenic factors for NSHL were systematically evaluated25. In step 5, if there were no possible causative variant, a recessive inheritance pattern was assumed on the basis of the pedigree analysis results. Therefore, homozygous, bi-allelic, and de novo heterozygous variants were retained, while single heterozygous variants, except for de novo variants, were excluded from further evaluation. In case of male participants with hearing loss, hemizygous variants were also considered. In the final step, the remaining variants were ranked based on conservation of the mutated amino acid residue across species and their probable impact on the function of the encoded protein. Mutation calling was performed by geneticists and cell biologists with knowledge of clinical phenotypes and pedigree structure and experience with WES analysis. The remaining variants were confirmed in the original participant DNA samples by Sanger sequencing. Segregation analysis was performed whenever parental DNA was available.
Copy-number variant (CNV) analysis
Analysis of CNV was performed using the paired-end WES data using the EXCAVATOR version 2.226 and ExomeDepth version 1.1.1027 tools with default settings. The GRCh37/hg19 database was used as the reference assembly for calculation of GC content. The WES dataset of 32 internal control subjects was compared with that of the study participants. Copy number variations at specific target regions were estimated according to different CNV detection algorithms using the Agilent SureSelect V5 kit.
Results
Family recruitment and clinical assessment
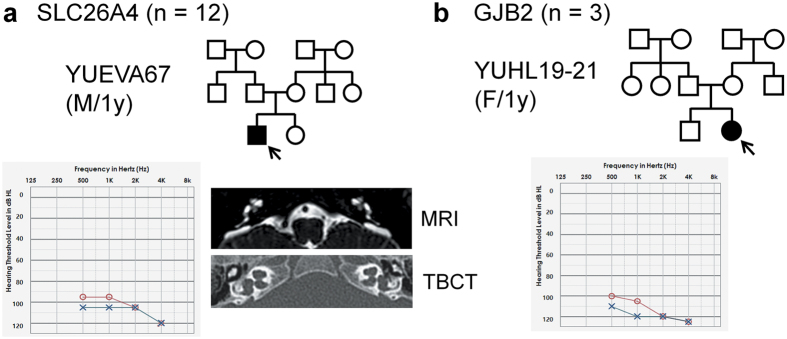
This study included 28 unrelated children (male, 15; female, 13; mean age, 2.6 ± 1.8 years; age range, 8–70 months) with sporadic severe-to-profound NSHL and without syndromic features or familial history of hearing impairment. The average thresholds for ASSR in the right and left ears were 103.0 ± 24.5 dB and 97.4 ± 23.0 dB, respectively. Of the 28 children, 12 exhibited EVA and/or Mondini’s deformity (cochlear anomaly where the cochlea has only 1.5 turns), representing DFNB4 hearing impairment with bi-allelic mutations in SLC26A4 (Fig. 1a). Direct Sanger sequencing for all of the exons of SLC26A4 revealed bi-allelic mutations in SLC26A4 in all 12 of these subjects (Table 1); one of the subjects, Yonsei University enlarged vestibular aqueduct (YUEVA)-65, exhibited a novel variant—a nonsense mutation of p.Y27*. Apart from EVA and/or Mondini’s deformity, these subjects exhibited no other anatomical inner/middle ear anomalies. Direct Sanger sequencing for GJB2 in the 16 children without EVA revealed pathogenic bi-allelic mutations in GJB2 in 3 subjects (Fig. 1b and Table 1).
Figure 1. Family pedigree and audiological phenotype of families with causative mutations in SLC26A4 or GJB2.
(a) The family pedigree of a representative patient with sporadic congenital hearing loss (YUEVA67) from among twelve patients with bi-allelic mutations of SLC26A4 is shown. The estimated thresholds for auditory steady-state response (ASSR) are >90 dB HL at 500, 1000, 2000, and 4000 Hz (red color, right ear; blue color, left ear). Internal auditory canal magnetic resonance imaging and temporal bone computed tomography images reveal bilateral enlarged vestibular aqueduct and endolymphatic sac. (b) The family pedigree and ASSR findings of a representative patient with sporadic congenital hearing loss (YUHL19-21) from among three patients with bi-allelic mutations of GJB2 are depicted. YUHL, Yonsei University hearing loss; YUEVA, Yonsei University enlarged vestibular aqueduct.
Table 1. Bi-allelic mutations of SLC26A4 and GJB2 in 15 families with sporadic congenital hearing loss.
| Patient | Sex | Age | Gene | Genomic change | Protein change | Location | Mutation type | HGMD |
|---|---|---|---|---|---|---|---|---|
| YUEVA 38 | M | 4y | SLC26A4 | c.2168 A > G | p.His723Arg | Exon 19 | Missense | yes |
| c.439 A > G | p.Met147Val | Exon 5 | Missense | yes | ||||
| YUEVA 59 | M | 2y | SLC26A4 | c.2168 A > G | p.His723Arg | Exon 19 | Missense | yes |
| c.919-2 A > G | Splicing | Intron 7 | Splicing variant | yes | ||||
| YUEVA 60 | M | 3y | SLC26A4 | c.2027_2028insT | p.Arg677Alafs*11 | Exon 17 | Insertion | yes |
| c.2168 A > G | p.His723Arg | Exon 19 | Missense | yes | ||||
| YUEVA 61 | F | 2y | SLC26A4 | c.2168 A > G | p.His723Arg | Exon 19 | Missense | yes |
| c.919-2 A > G | Splicing | Intron 7 | Splicing variant | yes | ||||
| YUEVA 65 | M | 2y | SLC26A4 | c.1229 C > T | p.Thr410Met | Exon 10 | Missense | yes |
| c.81 C > A | p.Tyr27* | Exon 2 | Nonsense | no | ||||
| YUEVA 67 | M | 11 m | SLC26A4 | c.2168 A > G | p.His723Arg | Exon 19 | Missense | yes |
| c.919-2 A > G | Splicing | Intron 7 | Splicing variant | yes | ||||
| YUEVA 69 | M | 2y | SLC26A4 | c.2168 A > G | p.His723Arg | Exon 19 | Missense | yes |
| c.439 A > G | p.Met147Val | Exon 5 | Missense | yes | ||||
| YUEVA 72 | M | 4y | SLC26A4 | c.2168 A > G | p.His723Arg | Exon 19 | Missense | yes |
| c.919-2 A > G | Splicing | Intron 7 | Splicing variant | yes | ||||
| YUEVA 73 | M | 2y | SLC26A4 | c.2168 A > G | p.His723Arg | Exon 19 | Missense | yes |
| c.439 A > G | p.Met147Val | Exon 5 | Missense | yes | ||||
| YUEVA 74 | F | 2y | SLC26A4 | c.2168 A > G | p.His723Arg | Exon 19 | Missense | yes |
| c.2168 A > G | p.His723Arg | Exon 19 | Missense | yes | ||||
| YUEVA 77 | F | 1y | SLC26A4 | c.2168 A > G | p.His723Arg | Exon 19 | Missense | yes |
| c.2168 A > G | p.His723Arg | Exon 19 | Missense | yes | ||||
| YUEVA 111 | M | 4y | SLC26A4 | c.916_917insG | p.Val306Glyfs*24 | Exon 7 | Insertion | yes |
| c.919-2 A > G | Splicing | Intron 7 | Splicing variant | yes | ||||
| YUHL 6–21 | F | 5y | GJB2 | c.605_606insAGAAG ACTGTCTTCACAG TGTTCATGATTGC AGTGTCTGGAATTTG | p.Cys202* | Exon2 | Inserion | yes |
| c.9 G > A | p.Trp3* | Exon2 | Nonsense | yes | ||||
| YUHL 11–21 | M | 10 m | GJB2 | c.235delC | p.Leu79Cysfs*3 | Exon2 | Deletion | yes |
| c.427 C > T | p.Arg143Trp | Exon2 | Missense | yes | ||||
| YUHL 19–21 | F | 11 m | GJB2 | c.299_300delAT | p.His100Rfs*14 | Exon2 | Deletion | yes |
| c.427 C > T | p.Arg143Trp | Exon2 | Missense | yes |
YUHL, Yonsei University hearing loss; YUEVA, Yonsei University enlarged vestibular aqueduct.
Identification of causative mutations by whole exome sequencing
The genetic characteristics of 13 individuals without causative mutations in GJB2 and SLC26A4 were further analyzed using WES. On an average, the WES datasets of each of the 13 individuals had 99.2%, 97.7%, and 92.4% of mappable bases represented by coverage of at least 1, 10, and 20 reads, respectively. The mean exome coverage was 71.6.
The 72 known NSHL-associated genes were first examined for the presence of variants (Table S3)25. Whole exome sequencing of these genes achieved an average coverage of 65.8 (Table S4). Variants that existed in homozygous or hemizygous states in 32 healthy Korean control subjects were excluded, as were common variants with minor allele frequencies >1%, as described in Materials and Methods. We also examined the most recent version of dbSNP (http://www.ncbi.nlm.nih.gov/SNP/), exome variant server (EVS; http://evs.gs.washington.edu/EVS/), exome aggregation consortium (ExAC; http://exac.broadinstitute.org/), and the National Biobank of Korea control databases (NBK) to evaluate the filtered variants. The remaining variants had minor allele frequencies <0.01, and were ranked based on conservation of the affected amino acid residue and the predicted pathogenicity. In addition, we investigated whether the surviving variants had minor allele frequencies <0.005 and 0.0005 for AR and AD genes, respectively; these thresholds were consistent with the minor allele frequency thresholds that Shearer et al. suggested for pathologic variants in NSHL28.
Compound-heterozygous mutations were identified in two known AR NSHL-linked genes, MYO15A (YUHL8-21 and YUHL13-21) and CDH23 (YUHL24-21), in three individuals with NSHL; in two of these families, where parental DNA available, segregation was confirmed by Sanger sequencing (Table 2 and Figs S2, S3). Thus, NSHL in these three individuals most likely resulted from these recessive mutations.
Table 2. Causative mutations detected by WES in three individuals with NSHL.
| Gene Symbol | Indiv-idual | Sex | Age of onset | Nucleotide changea | Amino acid change | Exon (zygosity, segrega-tion) | Amino acid sequence conservationb | Frequencies in the dbSNP databasec | Frequencies in the ExAC databased | Frequencies in the NBK databasee | PP2f | MTg | PRO-VEANh | SIFTi |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MYO15A | YUHL 8–21 | Fm | 2 yr | c.7990 C > A | p.Pro2664Thr | 42 (het, ND) | G. gallus | rs577657134 (MAF: A = 0.0002/1) rs564438028 | 12/118432 (no hom) | A = 0.00377834 | Bn (0.026) | PM (0.914) | Del (−2.55) | Dam (0.028) |
| c.9221 T > C | p.Met3074Thr | 53 (het, ND) | M. musculus | (MAF: C = 0.0002/1) | 1/120686 (no hom) | ND | Bn0.006 | DC(0.952) | Neu (−2.08) | Tol (0.184) | ||||
| MYO15A | YUHL 13–21 | Ml | 1 yr | c.4322 G > T | p.Gly1441Val | 11 (het, F) | D. rerio | rs772995303 (NA) | 1/116996 (no hom) | ND | PD (1.000) | DC (0.999) | Del (−8.66) | Dam (0.000) |
| c.1651 G > A | p.Ala551Thr | 1 (het, M) | M. musculus | rs747175448 (NA) | ND | ND | BN 0.004 | PM (0.999) | Neu (−0.82) | Dam (0.012) | ||||
| CDH23 | YUHL 24–21 | Fm | 1 mo | c.7145 G > A | p.Arg2382Gln | 49 (het, F) | D. rerio | rs759439688 (MAF: A = 0.00002/3)) | 3/120700 (no hom) | ND | PD (0.977) | N/ | Neu (−0.77) | Dam (0.001) |
| c.7361 C > T | p.Thr2454Met | 50 (het,ND) | D. rerio | rs772949926 (MAF: T = 0.00010/12)) | 12/120752 (no hom) | ND | PD (0.998) | No | Del (−3.18) | Dam (0.001) |
Bn, benign; Dam, damaging; DC, disease causing; Del, deleterious; ExAC, exome aggregation consortium; F, heterozygous mutation identified in father; Fm, female; het, heterozygous in affected individual; M, heterozygous mutation identified in mother; MAF, minor allele frequency; Ml, male; mo, month; NA, not applicable; ND, no data or DNA available; Neu, neutral; NSHL, nonsyndromic hearing loss; PD, probably damaging; PM, polymorphism; PP2, PolyPhen-2 prediction score Humvar; PROVEAN, protein variation effect analyzer; SIFT, sorting intolerant from tolerant; SNP, single nucleotide polymorphism; Tol, tolerant; WES, whole exome sequencing; yr, years; YUHL, Yonsei University hearing loss.
acDNA mutations are numbered according to human cDNA reference sequences NM_016239.3 (MYO15A) and NM_022124.5 (CDH23); +1 corresponds to A of the translation initiation codon ATG. bAmino acid residue is continually conserved throughout evolution, including in the indicated species. cdbSNP database (http://www.ncbi.nlm.nih.gov/SNP). dExAC browser (http://exac.broadinstitute.org/). eNational Biobank of Korea, Centers for Disease Control and Prevention. fPolyPhen-2 prediction score HumVar ranges from 0 to 1.0; 0 = benign, 1.0 = probably damaging (http://genetics.bwh.harvard.edu/pph2/). gMutation taster (http://www.mutationtaster.org/). hPROVEAN (http://provean.jcvi.org/index.php). hSIFT (http://sift.jcvi.org/).
Two individuals with profound congenital NSHL exhibited variants in two AD genes, MYO7A (YUHL26-21) and DFNA5 (YUHL44-21) (Table S5). Parental DNA was available in both cases, and segregation analysis by Sanger sequencing revealed that the unaffected mothers (YUHL26-12 and YUHL44-12, respectively) carried the variant in both families (Table S5 and Fig. S4), indicating that these variants were not segregated with the affected status.
As shown in Table S6, heterozygous variants in AR NSHL-linked genes were also identified in six individuals. However, these variants were probably not the causative factors for NSHL in these cases since bi-allelic mutations in AR genes are required to be in trans configuration in order to be causative.
Whole exome sequencing did not cover all 1340 coding exons of the 72 known NSHL-linked genes evaluated in this study. Approximately 2.8% of these exons (38/1340; 17 of 72 genes) were not covered (read depth <1; Table S4). Of the 38 uncovered exons, 24 were not targeted by the Agilent SureSelect V5 kit, while the remaining 14 were targeted but poorly covered. Since seven of the uncovered exons belonged to five AD genes, they were analyzed by Sanger sequencing. However, no causative mutations were identified in these exons in 13 individuals with NSHL. Of the 12 AR genes with uncovered exons, only those with heterozygous mutations detected by WES—PCDH15 in YUHL14-21, MYO3A in YUHL26-21, and TSPEAR in YUHL36-21 (Table S6)—were analyzed by Sanger sequencing of the uncovered exons. However, no additional mutations were detected, which left the genetic etiology of NSHL in these individuals unresolved.
Identification of candidate genes for non-syndromic hearing loss by whole exome sequencing
The WES data of 10 individuals who did not exhibit definite causative mutations in known NSHL-linked genes were analyzed for mutations in monogenic candidate genes. Since NSHL in these 10 individuals was sporadic and had congenital or prelingual onset, and the most frequent mode of inheritance of NSHL-linked mutations is AR inheritance, we assumed the following inheritance patterns: (1) bi-allelic variants in recessive genes, (2) hemizygous variants in X-chromosome genes in affected male subjects, and (3) de novo heterozygous variants in families in which trio analysis was possible. Variant filtering reduced the number of candidate genes to one to 13 in 10 families, as outlined in Fig. S1. In case of YUHL44-21 (Fig. S2c), variant filtering was begun with 170,215 variants from the normal reference sequence. This number was reduced to 929 upon exclusion of homozygous and hemizygous variants in healthy Korean individuals, common variants (minor allele frequencies >1% in public databases), and synonymous variants. Upon considering only those genes with de novo heterozygous variants, hemizygous variants or more than two variants in the same gene, the number of variants was further reduced to 35 variants (28 genes). Exclusion of artefacts by direct inspection of sequence alignment and exclusion of variants with minor allele frequencies <0.005 in public databases left nine variants in eight candidate genes — HEATR1, SLC25A14, SSX3, ADAMTSL3, CEND1, DOPEY2, SWI5, and CAND2. These variants were subsequently ranked based on their extent of amino acid conservation across species and predicted likelihood to be deleterious for the function of the encoded protein. The WES datasets of other individuals with NSHL in the present study were analyzed in the same manner to identify candidate genes (Table S7). All mutations in novel candidate genes were confirmed by Sanger sequencing of the DNA of the affected individuals and their parents and were found to segregate with the affected status.
In the 10 individuals without causative mutations in known NSHL-linked genes, the EXCAVATOR and ExomeDepth tools detected 126 and 158 CNVs, respectively (Table S8). We specifically focused on deletion or duplication of alleles in an AR pattern and identified a bi-allelic deletion in YUHL10-21 (Figs S2c and S5). The deleted region was located between 30,995,157 and 30,996,641 on chromosome 6 (hg19) and corresponded to a part of the coding sequence of the third exon of MUC22 (Fig. S5c). This deletion is expected to result in the truncation of the MUC22 protein. However, further study is required to determine whether this deletion is associated with hearing loss.
Audiological performance after cochlear implantation according to genotype
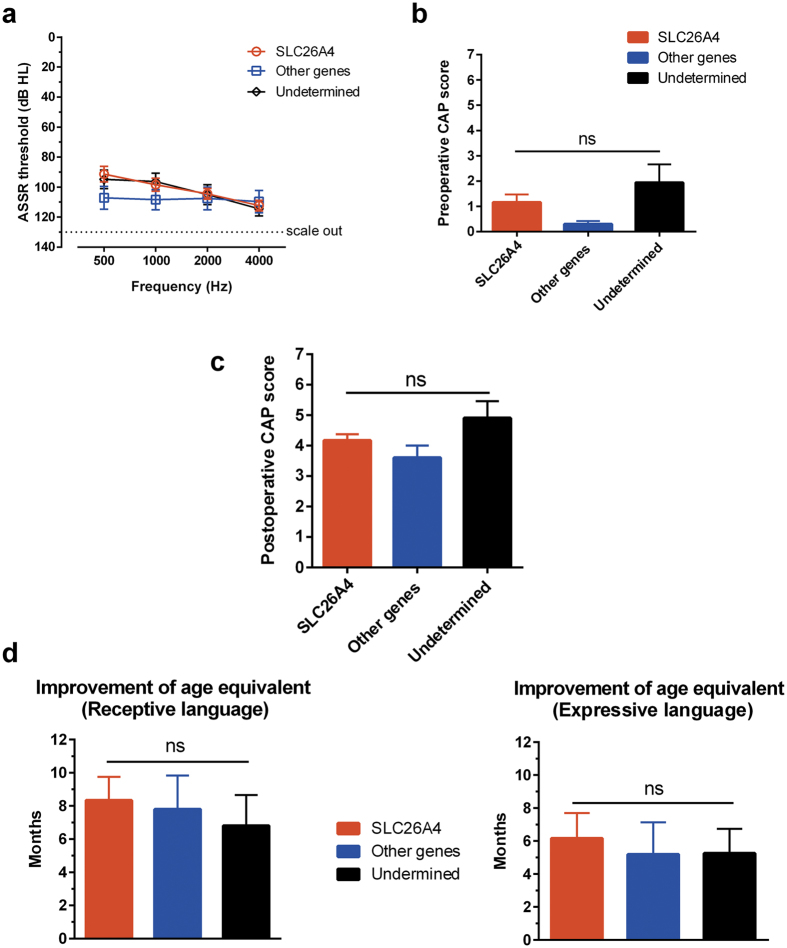
Of the 28 subjects included in the present study, 25 received cochlear implants for hearing rehabilitation. The mean age at operation was 26.0 ± 15.8 months. The type of implanted device was CI24RE (CochlearTM, Australia) or Concerto (Med-ElTM, Austria), depending on the decision of the patients. These 25 subjects were subdivided into three groups—the SLC26A4 mutation, other gene mutations (including GJB2, MYO15A, and CDH23), and unidentified etiology groups—according to genotype, and preoperative hearing thresholds for ASSR were compared among these groups (Fig. 2a). Although the group of individuals with mutations in other genes appeared to exhibit comparatively worse hearing thresholds at 500 and 1000 Hz, there were no significant differences in preoperative threshold among the three groups at any of the frequencies (two-way analysis of variance; P > 0.05). All three groups exhibited average preoperative CAP scores <2, where a score of 2 indicated response to spoken signals (e.g., “go” or “boo”). There were no statistically differences in the average preoperative or 6-month postoperative CAP scores among the groups (Fig. 2b and c)21. Age equivalence in terms of receptive and expressive language was evaluated according to the SELSI22. As shown in Fig. 2d, there were no significant differences in the improvement of receptive or expressive language among the groups.
Figure 2. Audiological performance of cochlear implantation.
Twenty-five children received cochlear implants. They were sub-divided into three groups—the SLC26A4 mutation, other gene mutations (including GJB2, MYO15A, and CDH23), and unidentified etiology groups—according to genotype. (a) Preoperative hearing thresholds for auditory steady-state response (ASSR). (b) Comparison of preoperative categories of auditory performance (CAP) scores among the three groups. (c) Comparison of postoperative CAP scores among the three groups. (d) Comparison of improvement in age equivalence in terms of receptive and expressive language among the three groups. ns, not significant (one-way analysis of variance).
Discussion
In the present study, we comprehensively evaluated the contribution of genetic predisposition to severe-to-profound sporadic congenital hearing loss by Sanger sequencing and WES. Of the 28 included subjects, 18 (64.3%) carried pathogenic bi-allelic mutations in 1 of 72 genes known to be associated with NSHL. This diagnostic rate is exceptionally higher compared with that reported in a previous study (37%) involving sporadic NSHL11. However, upon exclusion of subjects with SLC26A4 mutations from the present cohort, the diagnostic rate of NSHL in the present study decreases to 37.5% (6 of 16 subjects), which is comparable to that reported in the previous study11.
Interestingly, the frequency of SLC26A4 mutations in subjects with sporadic congenital hearing loss in the present study is higher compared that reported in previous studies. Pathogenic variants of SLC26A4 have been previously reported in approximately 10% of cases of severe-to-profound hearing loss in multi-ethnic cohorts11,29. Similarly, the frequency of SLC26A4 mutation in Japanese individuals with genetically diagnosed NSHL was reported to be 7.5%, which is comparable to that reported in the two previous studies30. In Korea, approximately 15–26% of cases of profound hearing loss were found to be attributable to SLC26A4 mutations regardless of familial history9,31. There are several possible reasons for the high frequency of SLC26A4 mutations observed in the present study. First, the present cohort only comprised individuals with congenital hearing loss, which usually results from genetic defects. Second, genetic evaluation is convincing and easily agreed to by patients in cases where TBCT findings reveal EVA, which might have resulted in patient selection. Nevertheless, the high rate of bi-allelic mutations in patients with sporadic profound hearing loss implies that sporadic congenital hearing loss is caused in a majority of cases in Korea by SLC26A4 mutations and not by GJB2 mutations. GJB2 mutations account for just 9% of the total frequency observed in this study. Therefore, Korean patients should first be evaluated for SLC26A4 mutations prior to comprehensive genetic analyses such as TES or WES. Because SLC26A4 comprises 21 exons and requires more than 20 primers for Sanger sequencing, TBCT can be more cost-effective for ruling out SLC26A4 mutations. Given that the frequency of SLC26A4 mutations in Korean patients with EVA is >90%, it is reasonable to perform TBCT for discriminating between patients with hearing loss due to SLC26A4 mutations and those with NSHL due to other AR mutations9.
Since the genetic factors associated with hearing loss are extremely heterogeneous, and mutations in nearly 100 genes have been identified as causative factors for NSHL, next-generation sequencing approaches such as TES and WES have been employed for molecular diagnosis of NSHL. Targeted exon sequencing has been used as a cost effective method for genetic diagnosis5,6,10,11,12,13. It provides greater coverage and depth and is less expensive compared with WES. Over the past few years, mutations in several novel genes have been identified as causative factors for NSHL by WES15,16,18,19,20,32,33, which highlights the necessity for the updating and optimization of TES for inclusion of these new genes. Considering the over 70 known genes associated with NSHL and the genes that will be identified in the near future, TES might not actually be much cheaper than WES. In this regard, WES has an advantage over TES because the former method can be employed for evaluation of any gene. However, WES also has a few limitations as a diagnostic tool. As shown in Table S4, WES fails to capture certain regions of the exome, which vary depending on the exome capture kit used. In addition, there are mitochondrial genes and microRNAs associated with NSHL25 and these are not generally covered by WES. Therefore, WES users need to know the type of capture kit used and consider and resolve any uncovered regions. Improvements in exome capture kits are expected to reduce the extent of uncovered regions with WES.
A previous study involving WES identified causative mutations of NSHL in 60.0% (12 of 20) of the included families14, which is higher than the frequency of causative mutations of NSHL observed among individuals evaluated by WES in the present study (23.1%; 3 of 13 individuals). This discrepancy might be attributable to the following factors. First, in the previous study, the authors pre-screened only GJB2, which is the most common genetic cause of NSHL in the Middle East; in contrast, the present study involved pre-screening for both GJB2 and SLC26A4 mutations. Second, the present cohort only comprised individuals with sporadic hearing loss, whereas the previous study included multiplex cases with at least two affected members. Third, the Korean population is generally outbred, whereas the previous study included only consanguineous families, which exhibit an increased prevalence of AR disorders34.
Another study employing WES identified causative mutations of NSHL in 5 of 11 (45.4%) Korean patients with sporadic mild-to-moderate NSHL; the authors also suggested various modes of inheritance, including digenic inheritance (GRP98/PDZ7)5. Similar to the present cohort, the previous study cohort did not include individuals with GJB2 mutations or EVA. Their reported frequency of causative mutations (45.4%) is higher compared with that observed in the present study (23.1%) despite the fact that mild-to-moderate hearing loss is less frequently diagnosed by molecular methods than severe-to-profound hearing loss. This discrepancy might be attributable to the relatively small sample sizes of both studies. In the present study, we identified two single heterozygous variants in two AR genes in three families — YUHL13-21, YUHL20-21, and YUHL44-21 (Table S6). Among these families, YUHL13-21 exhibited heterozygous variants in GIPC3 and LOXHD1 despite already possessing compound-heterozygous mutations in MYO15A. Therefore, the contribution of variants of GIPC3 and LOXHD1 to hearing loss is not clear. Heterozygous mutations in YUHL20-21 and YUHL44-21 corresponded to GRXCR2/LOXHD1 and FAM65B/KARS, respectively. It is not clear whether hearing loss in these cases was a result of digenic inheritance, and it is difficult to prove this hypothesis without any experimental evidence. Incidental findings of heterozygous variants in known NSHL genes have also been reported by Diaz-Horta et al.14.
Two families, YUHL26 and YUHL44, exhibited variants in AD genes (MYO7A and DFNA5, respectively) that did not segregate with the affected status in the family (Table S5). In both families, the unaffected mothers carried the same variant (Fig. S4), indicating that the variants are not de novo. Incomplete penetrance is observed in AD inheritance; however, it is not clear whether non-segregation in these cases resulted from incomplete penetrance.
Language development after cochlear implantation depends on conditions such as age at implantation, duration of implant use, and preoperative residual hearing35. Additionally, genetic predisposition might have an effect on post-implantation outcomes. While a few previous studies have reported better performance in individuals with GJB2 mutations compared with that in individuals with other genetic etiologies, a few others have demonstrated that individuals with SLC26A4 mutations exhibit better outcomes than those with an unknown aetiology36,37,38,39,40. Recently, Wu et al. reported that GJB2 and SLC26A4 mutations were associated with good long-term post-implant outcomes when implantation was done before the age of 3.5 years. However, we found no significant differences in post-operative auditory outcomes among groups with SLC26A4 mutations, mutation in other genes, and molecularly undetermined etiology, which might be because the present study only included short-term follow-up data. Thus, long-term outcomes may differ according to the genotype. Nevertheless, it is noteworthy that preoperative residual hearing was uniform across all groups in the present study (Fig. 2a). In previous studies that reported better post-implant outcomes in individuals with SLC26A4 mutations as well, preoperative hearing and auditory function in individuals with SLC26A4 mutations were better compared to those in control subjects36,40. This indicates that postoperative outcomes correlate with preoperative hearing function, and comparison between groups with dissimilar preoperative hearing functions is inappropriate. Thus, it is reasonable that, among individuals with similar preoperative hearing thresholds, specific genetic etiologies—at least, GJB2 or SLC26A4 mutations—do not result in better or worse postoperative auditory outcomes.
Sanger sequencing of top priority genes according to ethnicity renders the genetic evaluation process relatively cost-effective and simple. With the exception of SLC26A4 and GJB2 mutations, mutations in genes linked to hearing loss are extremely rare and can be effectively evaluated by WES31. Our findings indicate that direct sequencing of SLC26A4 and GJB2 followed by WES is a simple and precise protocol for molecular diagnosis of sporadic NSHL in a pediatric population.
Additional Information
How to cite this article: Jung, J. et al. Genetic Predisposition to Sporadic Congenital Hearing Loss in a Pediatric Population. Sci. Rep. 7, 45973; doi: 10.1038/srep45973 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This study was provided with bioresources from the National Biobank of Korea, Centers for Disease Control and Prevention, Republic of Korea (4845-301, 4851-302 and -307). This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education (2015R1D1A1A01056685 to H.Y.G.) and by the Korea government (MSIP) (2016R1A2B4007268 to C.J.Y. and 2014R1A2A1A01003385 to Y.J.). We are grateful to Alice Jaewon Lee for the scientific discussion and proofreading the manuscript.
Footnotes
The authors declare no competing financial interests.
Author Contributions J.J. and L.J.S. performed experiments, analyzed data and wrote the paper; C.K.J. and Y.S. performed experiments and collected data; Y.J., G.H.Y. and C.J.Y designed and wrote the paper.
References
- Xing G. et al. Identification of OSBPL2 as a novel candidate gene for progressive nonsyndromic hearing loss by whole-exome sequencing. Genet Med 17, 210–218 (2015). [DOI] [PubMed] [Google Scholar]
- Hilgert N., Smith R. J. & Van Camp G. Function and expression pattern of nonsyndromic deafness genes. Curr Mol Med 9, 546–564 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton C. C. & Nance W. E. Newborn hearing screening–a silent revolution. N Engl J Med 354, 2151–2164 (2006). [DOI] [PubMed] [Google Scholar]
- Smith R. J. & Hone S. Genetic screening for deafness. Pediatr Clin North Am 50, 315–329 (2003). [DOI] [PubMed] [Google Scholar]
- Kim N. K. et al. Whole-exome sequencing reveals diverse modes of inheritance in sporadic mild to moderate sensorineural hearing loss in a pediatric population. Genet Med 17, 901–911 (2015). [DOI] [PubMed] [Google Scholar]
- Gu X. et al. Genetic testing for sporadic hearing loss using targeted massively parallel sequencing identifies 10 novel mutations. Clin Genet 87, 588–593 (2015). [DOI] [PubMed] [Google Scholar]
- White K. R. Early hearing detection and intervention programs: opportunities for genetic services. Am J Med Genet A 130A, 29–36 (2004). [DOI] [PubMed] [Google Scholar]
- Kelsell D. P. et al. Connexin 26 mutations in hereditary non-syndromic sensorineural deafness. Nature 387, 80–83 (1997). [DOI] [PubMed] [Google Scholar]
- Shin J. W., Lee S. C., Lee H. K. & Park H. J. Genetic Screening of GJB2 and SLC26A4 in Korean Cochlear Implantees: Experience of Soree Ear Clinic. Clin Exp Otorhinolaryngol 5 Suppl 1, S10–13 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekin D. et al. A next-generation sequencing gene panel (MiamiOtoGenes) for comprehensive analysis of deafness genes. Hear Res 333, 179–184 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan-Heggen C. M. et al. Comprehensive genetic testing in the clinical evaluation of 1119 patients with hearing loss. Hum Genet 135, 441–450 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi B. Y. et al. Diagnostic application of targeted resequencing for familial nonsyndromic hearing loss. PLoS One 8, e68692 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagawa M. et al. Targeted exon sequencing successfully discovers rare causative genes and clarifies the molecular epidemiology of Japanese deafness patients. PLoS One 8, e71381 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Horta O. et al. Whole-Exome Sequencing Efficiently Detects Rare Mutations in Autosomal Recessive Nonsyndromic Hearing Loss. PLoS ONE 7, e50628 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein C. J. et al. Mutations in DNMT1 cause hereditary sensory neuropathy with dementia and hearing loss. Nat Genet 43, 595–600 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh T. et al. Whole exome sequencing and homozygosity mapping identify mutation in the cell polarity protein GPSM2 as the cause of nonsyndromic hearing loss DFNB82. Am J Hum Genet 87, 90–94 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Horta O. et al. Whole-exome sequencing efficiently detects rare mutations in autosomal recessive nonsyndromic hearing loss. PLoS One 7, e50628 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaworek T. J. et al. An alteration in ELMOD3, an Arl2 GTPase-activating protein, is associated with hearing impairment in humans. PLoS Genet 9, e1003774 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imtiaz A., Kohrman D. C. & Naz S. A frameshift mutation in GRXCR2 causes recessively inherited hearing loss. Hum Mutat 35, 618–624 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Cortez R. L. et al. Adenylate cyclase 1 (ADCY1) mutations cause recessive hearing impairment in humans and defects in hair cell function and hearing in zebrafish. Hum Mol Genet 23, 3289–3298 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archbold S., Lutman M. E. & Marshall D. H. Categories of Auditory Performance. Ann Otol Rhinol Laryngol Suppl 166, 312–314 (1995). [PubMed] [Google Scholar]
- Kim Y. T. Content and reliability analyses of the sequenced language scale for infants (SELSI). Commun Sci Disord 7, 1–23 (2002). [Google Scholar]
- MacArthur D. G. et al. Guidelines for investigating causality of sequence variants in human disease. Nature 508, 469–476 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y. et al. Clinical Whole-Exome Sequencing for the Diagnosis of Mendelian Disorders. New England Journal of Medicine 369, 1502–1511 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abou Tayoun A. N. et al. Improving hearing loss gene testing: a systematic review of gene evidence toward more efficient next-generation sequencing-based diagnostic testing and interpretation. Genet Med 18, 545–553 (2016). [DOI] [PubMed] [Google Scholar]
- Magi A. et al. EXCAVATOR: detecting copy number variants from whole-exome sequencing data. Genome Biol 14, R120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plagnol V. et al. A robust model for read count data in exome sequencing experiments and implications for copy number variant calling. Bioinformatics 28, 2747–2754 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearer A. E. et al. Utilizing ethnic-specific differences in minor allele frequency to recategorize reported pathogenic deafness variants. Am J Hum Genet 95, 445–453 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D. et al. Spectrum of DNA variants for non-syndromic deafness in a large cohort from multiple continents. Hum Genet, doi: 10.1007/s00439-016-1697-z. (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moteki H. et al. Comprehensive genetic testing with ethnic-specific filtering by allele frequency in a Japanese hearing-loss population. Clin Genet, doi: 10.1111/cge.12677. (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. H. et al. Exploration of molecular genetic etiology for Korean cochlear implantees with severe to profound hearing loss and its implication. Orphanet J Rare Dis 9, 167 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girotto G. et al. Linkage study and exome sequencing identify a BDP1 mutation associated with hereditary hearing loss. PLoS One 8, e80323 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. et al. Exome sequencing and linkage analysis identified tenascin-C (TNC) as a novel causative gene in nonsyndromic hearing loss. PLoS One 8, e69549 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamamy H. Consanguineous marriages: Preconception consultation in primary health care settings. Journal of Community Genetics 3, 185–192 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niparko J. K. et al. Spoken language development in children following cochlear implantation. JAMA 303, 1498–1506 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. C. et al. Genetic characteristics in children with cochlear implants and the corresponding auditory performance. Laryngoscope 121, 1287–1293 (2011). [DOI] [PubMed] [Google Scholar]
- Angeli S. I. et al. Influence of DFNB1 status on expressive language in deaf children with cochlear implants. Otol Neurotol 32, 1437–1443 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer P. W. et al. The effect of GJB2 allele variants on performance after cochlear implantation. Laryngoscope 113, 2135–2140 (2003). [DOI] [PubMed] [Google Scholar]
- Green G. E. et al. Performance of cochlear implant recipients with GJB2-related deafness. Am J Med Genet 109, 167–170 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y. J. et al. The effect of GJB2 and SLC26A4 gene mutations on rehabilitative outcomes in pediatric cochlear implant patients. Eur Arch Otorhinolaryngol 270, 2865–2870 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.