Abstract
Osteoblasts (OBs) play an important role in bone fracture healing, yet the extreme adverse microenvironment in fracture sites has a negative impact on the survival of OBs. Therefore, it is important to study how OBs behave in the complex fracture microenvironment. Studies have shown that autophagy plays a pivotal role in maintaining cellular homeostasis and defending the cell against adverse microenvironments. In this study we found the induction of autophagy in OBs at femoral bone fracture sites, which may be a result of ischemia, oxidative stress and hypoxia within the local area. At fracture sites a low pH environment also developed. Until now it has been unclear whether the induction of autophagy in osteoblasts is triggered by the acidic pH environment. Therefore, we cultured OBs in vitro in media of different pH values, and found both autophagy and apoptosis increased in OBs in acidic conditions. However, when autophagy inhibitor chloroquine (CQ) was used, apoptosis increased significantly compared with that without CQ. Thus indicating that inhibition of autophagy may promote apoptosis in OBs in an acidic environment, which may provide a new therapeutic strategy to decrease cell apoptosis in OBs through the use of drugs that modulate the autophagic state.
Physical trauma including bone fractures are accountable for an increasing proportion of the global disease burden1,2 posing a major socioeconomic, clinical, and scientific challenge. Survival of patients after severe tissue trauma depends on an efficient molecular and cellular responses and effective regeneration of the damaged tissue. Bone fracture healing is a complex process, initially the soft (cartilage) callus forms, followed by formation of the hard (bone) callus. Although most fractures heal routinely, about 5–10% of fractures fail to heal normally and are described as displaying bone nonunion3. Many factors contribute to the pathogenesis of a delayed union or bone nonunion. It is known that osteoblasts (OBs), the cells responsible for bone formation, play a significant role3. The extreme adverse microenvironment in fracture sites has a negative effect on the survival of OBs, which may be a key cause for bone nonunion and segmental bone loss after fractures. As such, it is of clinical significance to study how the osteoblasts react in the complex fracture environment.
Autophagy is a catabolic process of eukaryotic cells in which cellular components, such as damaged macromolecules and organelles, are degraded by the lysosome in order to maintain cellular homeostasis4,5,6. Three major forms of autophagy are observed in mammalian cells, namely: chaperone-mediated autophagy, microautophagy, and macroautophagy. Macroautophagy (referred to hereafter as autophagy), the most widely studied form, acts in concert with the ubiquitin-proteasome system (UPS) to maintain cellular homeostasis7. The cytoplasmic material including damaged organelles, intracellular pathogens and protein aggregates are enclosed by double-membraned vesicles known as autophagosomes. Autophagosomes are delivered to lysosomes where upon their contents are degraded after fusion. These catabolic products can re-enter bioenergetic and/or anabolic process8.
During physiological conditions, autophagy plays an important role in removing defective organelles or protein aggregates. However, when cells are exposed to stressful conditions, autophagy is induced to degrade cellular components in order to provide an energy source5,6. Some reports have shown that hypoxia, serum deprivation and oxidative stress induce autophagy and promote the survival of mesenchymal stem cells or OBs9,10,11,12,13. However, acidic pH environments also develop in fracture sites14,15 and the role of these acidic environments in the induction of autophagy in OB’s is still unclear.
After bone fracture, the accumulation of lactic acid may occur as the result of an interruption in blood supply in the local environment. Both hypoxic and ischemic conditions can cause extracellular acidosis with low pH, consequently an acidic local environment can develop at the fracture site14,15. The application of tissue engineering can also lead to local acidosis during the induction of ossification16,17. This local acidosis can lead to the cell death of osteoblasts, which results in bone loss18,19,20,21,22. Osteoblasts may persist initially in a low pH environment before undergoing cell death, yet how the osteoblasts react in this environment is still not well understood. Therefore, we hypothesized that autophagy may be directly induced in OBs when challenged by an acidic environment and autophagy may play a pivotal role in the protection of OBs from apoptosis in such conditions.
In this study, autophagy was first detected at bone fracture sites in vivo. Subsequent in vitro study was conducted to investigate autophagy and apoptosis of OBs in cultured in media of different acidities. It is proposed that modulation of the autophagy state may provide a new therapeutic strategy to promote bone healing.
Results
Autophagy is induced in OBs at fracture sites
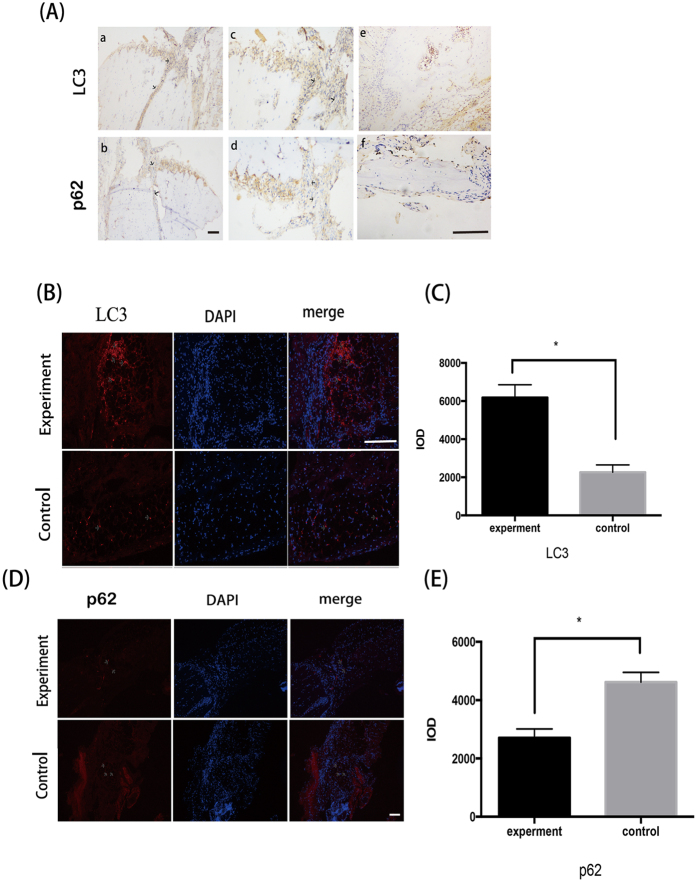
Positive immunohistological or immunofluorescent staining was not found in group A(6 h) and group B(24 h). Immunohistological examination of femora 36 hours post fracture in group C, showed that LC3 immunoreactivity could be detected in a punctate pattern in OBs at fracture sites, which was not observed in un-fractured controls (Fig. 1A). Similarly, in group C immunofluorescent staining of LC3 also showed high expression in OBs around the bone marrow cavity in the fractured areas compared with the control femora, suggesting that autophagy was induced in OBs at fracture sites (Fig. 1B).
Figure 1. Autophagy is induced in OBs near fracture sites in the group 36 h after fracture.
Longitudinal sections of rat fractured bones were detected. (A) Immunohistological staining showed the expression of LC3 and P62 in OBs at fractured sites (arrows). Size bar = 100 μm. As showed in figures e and f were control groups, a and b were experiment groups. a and c were from the same section with different magnification which showed the expression of LC3. b and d were from the same section with different magnification which showed the expression of P62. (B,D) Immunofluorescence analysis noted the distinct punctate pattern of LC3 and P62 expression (arrows) in OBs near fracture sites after 36 h. Size bar = 100 μm. (C,E) Quantified analysis of LC3 and P62 expression in osteoblasts in bones from the control and fracture group. *P < 0.05 vs.control. Values are the mean ± sd, *P < 0.05 in comparison to the control by one-way ANOVA.
Low pH media decreases cell viability and induces apoptosis in OBs
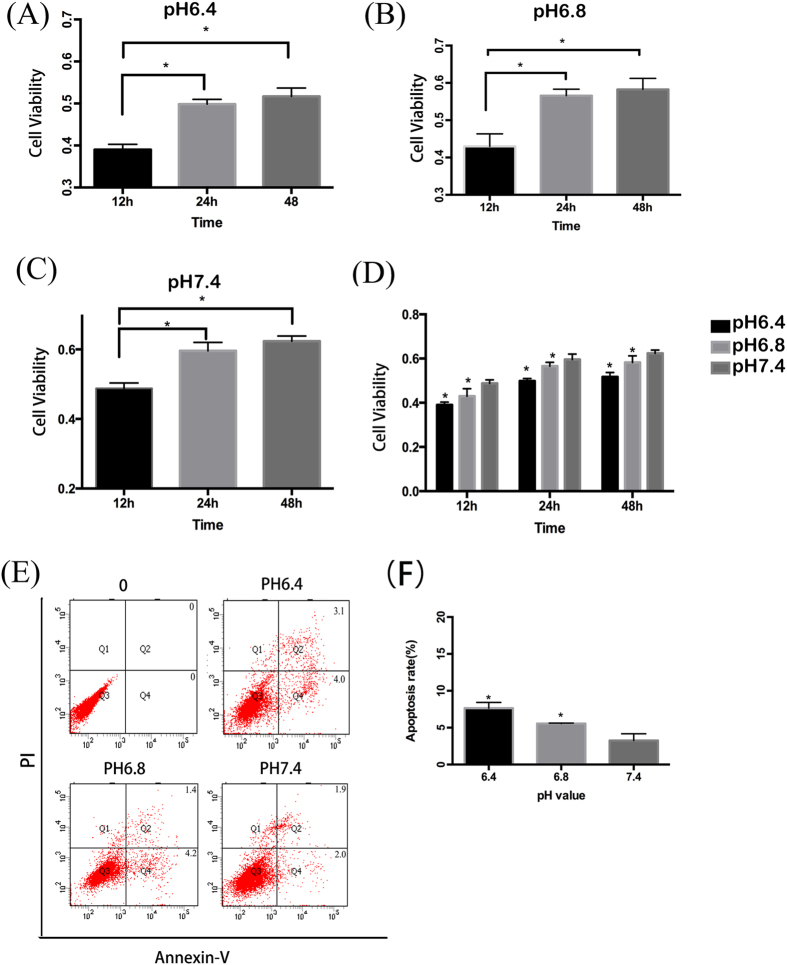
Viability of osteoblastic cell line, MC3T3-E1, was markedly influenced by the acidity of the culture media in which they are grown. Both pH values of 6.4 and 6.8 induced a significant decrease in cell viability compared with that of pH 7.4 (P < 0.05). While all cells died when cultured in media at pH 6.0. Furthermore, a pH- and time-dependent decrease in cell viability was observed, with a significant difference in viability between pH value of 6.4 and 6.8 (Fig. 2A–D). Levels of apoptosis were determined by flow cytometry utilising Annexin-V and PI dual labelling. Cells in the Q2 and Q4 quadrants represent apoptotic cells. Increased rate of primarily early apoptosis (but also some late apoptosis) was detected in MC3T3-E1 cells in media with pH 6.8 and 6.4 after 48 h of exposure. Compared with cells cultured at pH 7.4, those at pH 6.4 and 6.8 exhibited statistically significantly increased levels of apoptosis (P < 0.05). (Fig. 2E,F). Comparison between the levels of apoptosis between the two acidic culture conditions showed increased apoptosis at pH6.4, however there is no statistically significant difference (P > 0.05).
Figure 2. Acidic pH environment has a negative effect on the survival of OBs.
OBs were cultured in media with different pH values for 12, 24, 48 h and cell viability was measured by MTT. Apoptosis was determined by flow cytometry. (A–D) MC3T3-E1 cells was cultured in media with pH6.4, 6.8 and 7.4 for 12, 24, 48 h, and cell viability was detected. *P < 0.05 vs.12 h with pH6.4 and 6.8, and *P < 0.05 vs.6 h with pH7.4. (E) Cell apoptosis increased in media with pH 6.8 and 6.4 at the time of 48 h including both early apoptosis and late apoptosis. (F) Analysis of apoptotic rate with pH 6.4, 6.8 and 7.4 at the time of 48 h. *P < 0.05 vs.pH 7.4. Values are the mean ± sd, *P < 0.05 in comparison to the control by one-way ANOVA.
Low pH media induces autophagy in OBs
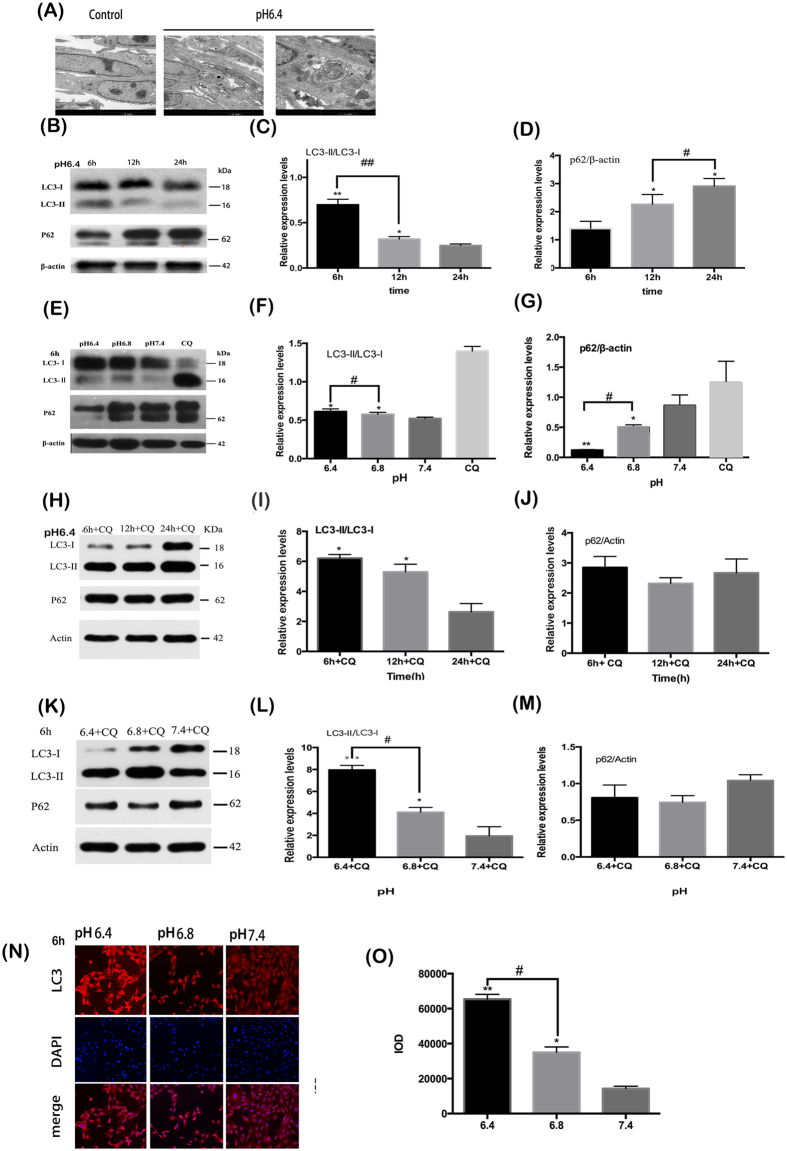
Transmission electron microscope (TEM), is the gold standard for the detection of autophagy. TEM was carried out to confirm the existence of autophagy in the osteoblastic cells cultured in acidic media. As shown in Fig. 3A, the morphology of the cells was changed after exposure to low pH media, with more autophagosomes observed in the pH 6.4 and 6.8 conditions. The typical double-membrane structures surrounded with cytoplasm and mitochondria were found primarily in the cells cultured in an acid environment.
Figure 3. Acidic pH environment induces autophagy in murine osteoblastic cell line MC3T3-E1.
(A) More autophagosomes were observed in the media of pH 6.4 with typical double-membrane structures surrounded with cytoplasm and mitochondria. (B)The expression of LC3-II and P62 in OBs as the treatment time in pH 6.4 is prolonged. (C) Densitometric analysis of LC3-II/LC3-I at 6 h, 12 h and 24 h. **P < 0.01 vs.24 h, *P < 0.05 vs.24 h. (D) Densitometric analysis of P62/β-actin at 6 h, 12 h and 24 h. *P < 0.05 vs.6 h. (E) The expression of LC3-II and P62 in OBs as the pH value decreased for 6 h. (F,G) Densitometric analysis of LC3-II/LC3-I and P62/β-actin with pH 6.4, 6.8 and 7.4 for 6 h. *P < 0.05 vs. pH 7.4. (H) The expression of LC3-II/LC3-I and P62 in OBs in pH 6.4 with CQ as the treatment time was prolonged. (I,J) Densitometric analysis of LC3-II/LC3-I and P62/β-actin at 6 h, 12 h and 24 h in pH 6.4. *P < 0.05 vs.24 h + CQ. (K) The expression of LC3-II/LC3-I and P62 in OBs as the pH value decreased for 6 h. (L,M) Densitometric analysis of LC3-II/LC3-I and P62/β-actin among 6.4 + CQ, 6.8 + CQ and 7.4 + CQ for 6 h. **P < 0.01 vs.pH 7.4 + CQ, *P < 0.05 vs. pH 7.4 + CQ. (N) Immunofluorescence staining of LC3 were labelled red and nuclei were stained with blue. Size bar = 100 μm. The brightness of red around nuclei increased with the decrease of pH value in media. (O) Quantified analysis of immunofluorescence staining of LC3. **P < 0.01 vs.pH 7.4, *P < 0.05 vs. pH 7.4. Values are the mean ± sd, *P < 0.05.**p < 0.01, in comparison to the control by one-way ANOVA.
In order to determine whether an acidic environment increases autophagy in osteoblasts, expression level analysis of autophagy marker protein LC3, which can be converted from LC3-I to LC3-II, was undertaken. Under prolonged exposure to acidic conditions, the ratio of LC3-II/LC3-I in OBs decreased (Fig. 3B,C). Western blot analysis showed that after 6 h of cell culture, an inverse correlation was observed between the increasing expression of LC3-II in OBs and decreasing pH of the media(Fig. 3E,F). Autophagy is a highly dynamic process; detection of P62, the substrate of autophagy, is utilised to monitor autophagy flux. Contrary to the LC3-II, P62 expression increased gradually with treatment time prolonged and the increase of pH(Fig. 3B,D,E,G). Our results suggested that reduction of environmental pH increased the expression of LC3-II and reduced the levels of P62 (Fig. 3B–G). Furthermore, OBs cells exposed to low pH and treated with CQ were also studied. The ratio of LC3-II/LC3-I in pH 6.4 with CQ decreased as the treatment time is prolonged and the increse of pH (Fig. 3H–M). In addition this study has shown the cellular localisation of LC3 is restricted to in the cytoplasm of osteoblasts under acid environment (Fig. 3N,O).
Suppression of autophagy promotes apoptosis in OBs
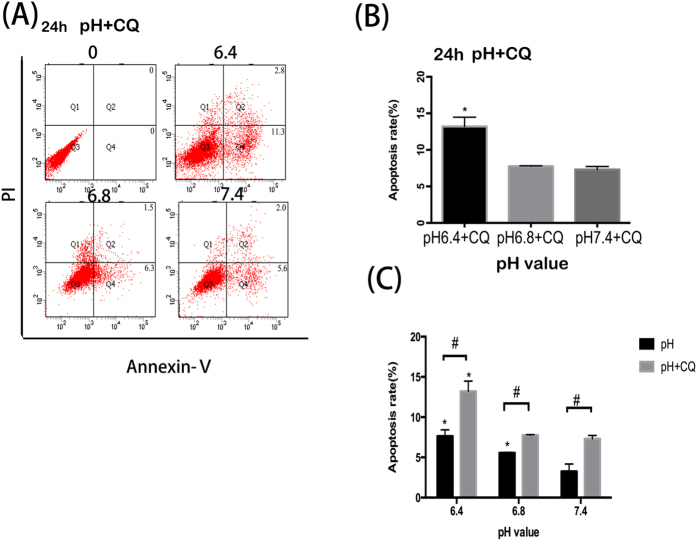
In order to study the relationship between apoptosis and autophagy, MC3T3-E1 cells were exposed to autophagy inhibitor CQ. The rate of cellular apoptosis increased when cells were treated with CQ in parallel with exposure to acidic conditions (Fig. 4A,B). Moreover, a larger proportion of apoptotic cells were observed when the cells were treated with CQ at all three experimental pH levels (7.4,6.8 and 6.4) (Fig. 4C).
Figure 4. Suppression of autophagy promotes apoptosis in OBs.
(A) Cell apoptosis increased in media with CQ with the decrease of pH value at the time of 24 h. (B) Cell apoptosis rate with 6.4 + CQ, 6.8 + CQ and 7.4 + CQ for 24 h. *P < 0.05 vs. pH 6.8 + CQ(or pH 7.4 + CQ). (C) Moreover, the rate of apoptosis in OBs with CQ is higher than that without CQ. Values are the mean ± sd, *P < 0.05 in comparison to the control by one-way ANOVA.
Discussion
Low tissue pH and locally acidic environments develops at bone fracture sites or in tumours, as a result of hypoxic or ischemic conditions, and poses a deleterious environmental stress. The induction of autophagy has been increasingly observed in stem cells under serum deprivation, oxidative or hypoxic stress and radiation insult4,9,10,11. However, whether autophagy is directly induced in low acidic situations is still unknown and the connections between apoptosis and autophagy in OBs is still unclear.
Previous study has shown osteocytes can display a punctate distribution of LC3 in both rats and human cortical bone. However, particulate LC3 fluorescence has not been observed in bone-lining cells or osteoblasts. Furthermore, autophagic osteocytes were primarily observed residing at a distance from the haversian canal23,24. Our study found the expression of LC3 in osteoblasts which indicated that osteoblasts may undergo autophagy at bone fracture sites. Given the acidic environment caused by hypoxic and ischemic conditions at the bone fracture site we postulated that osteoblasts may be induced to autophagy through extracellular acidosis and low pH.
Thus, we designed in vitro experiments to investigate the levels of autophagy in OBs cultured in media of a variety of pH. In the present study, autophagosomes were observed in OBs in media of pH 6.4 and 6.8 using TEM. Autophagy decrease was detected with prolonged exposure to low pH, by examining the autophagy-reliable marker LC3. Meanwhile, the expression of LC3-II in OBs increased when cells were grown in decreasing pH conditions. Furthermore, LC3-II accumulated greatly with the inhibition of lysosomal degradation and autophagic flux, through the administration of CQ25,26. Autophagy is a highly dynamic process. The amount of LC3-II detected by western blot (WB) during the process of autophagy, reflects synthesis as well as consumption of the protein by the lysosomal compartment. In addition to protein levels of LC3, P62 is an important marker to study autophagic flux27,28,29,30. P62 is exclusively degraded after being bound directly to LC3 by the autophagy-lysosome pathway31,32. Therefore, P62 decreases after autophagy is induced and accumulation of P62 protein level can be detected when autophagy is inhibited27,28. Thus, in this study P62 was also examined to detect the dynamic autophagic flux. Our data shows that P62 detection by western blot exhibits two bands which may result from phosphorylation of P62.
Paradoxically, autophagy is closely linked to both cell survival as well as the induction of the death process. However, the relationship between autophagy and apoptosis is still not well understood33,34. Many studies have reported the complex relationship between apoptosis, autophagy and necrosis with different conclusions35. There is not a consensus over the relation between autophagy and apoptosis36,37,38,39,40. Autophagy is a highly controlled catabolic mechanism for the removal of dysfunctional or unwanted proteins and organelles, which plays an important role in protective process in various stress environment41,42. However, this study found that apoptosis was promoted and cell viability decreased with increasing acidity. Furthermore, levels of apoptosis also increased when autophagy inhibitor CQ was used, which indicates inhibition of autophagy may promote apoptosis in OBs when in acidic environment. Autophagy plays many roles in regulating cell survival and cell death. It can convert from a defensive mechanism against environmental stimuli, to a cell death pathway when conditions change43,44. Our findings suggest that culture of OBs in low pH media may activate the intrinsic protective autophagic process, as such the cells will be more tolerant to acidic stress. However, if the low pH is far beyond the tolerance of OBs or the induced autophagy is supressed, the cells are more easily to undergo apoptosis.
In summary, we found for the first time that autophagy is induced in bone fracture sites in vivo and that acidic culture conditions enhanced autophagy in OBs in vitro. We also present that co-treatment with autophagy inhibitor CQ renders cells more sensitive to apoptosis. However, in future additional experiments are needed to confirm that autophagy plays the protective role in helping OBs survive in an acidic environment. These results put forward a new understanding of low pH environments and the process of autophagy in OB. These findings may provide a new strategy to promote bone fracture healing in low pH tissue environments through the use of drugs that modulate the autophagic state.
Methods
Animal studies
Six-week-old male KM rats were used in this experiment. The body weights ranged from 30 g to 40 g at the beginning of the experiment. Twenty-seven KM rats underwent fracture of the right femora and were then randomly assigned into 3 groups of 9 (group A,B and C). The rats were treated with anaesthesia by intraperitoneal injection of 10% chloral hydrate. All animals receive no treatment after the bone fracture models were established. The animals from group A, B and C were humanely killed with sodium pentothal injections 6 h, 24 h and 36 h post-fracture, respectively. All fractured femora were harvested and the left femora were also extracted to act as a non-fractured control. The specimens were fixed with 4% paraformaldehyde and decalcified in 10% EDTA solution which changed every 4 days at 4 °C, then subjected to graded ethanol dehydration with xylene, and embedded in paraffin wax. We conducted immunohistochemical and immunofluorescence staining for LC3 and P62. Samples were observed and imaged by microscope (Carl Zeiss Canada).
All animal studies were carried out under the guide of the Care and Use of Laboratory Animals of the Chinese Science and Technology Ministry. This experiment was approved by the Committee on the Ethics of Animal Experiments of the Second Hospital of Shandong University (Permit Number: KYLL-2016LW-0016).
In vitro cell culture
Murine osteoblastic cell line, MC3T3-E1, was cultured in α-minimal essential media (α-MEM), supplemented with 10% fetal bovine serum (FBS), containing 100 units/ml penicillin and 100 μg/mL streptomycin, and maintained at 37 °C humidified atmosphere of 5% CO2. The media was changed every 3 days. The pH value of media was adjusted by adding 5 N HCL and 5 N NaHCO, and 20 m M HEPES were added to α-MEM3 in order to avoid pH variations. Before each experiment, the acidic pH media was measured and filtered to ensure consistency of the experimental conditions.
MTT assay for cell viability
In order to determine whether the acidic environment has a negative impact on cell proliferation, cells were exposed to different pH media for 12, 24, 48 h, before cell viability was measured by MTT assay as previously described45. Osteoblasts were seeded into 96-well plates with a density of 5 × 104 cells/well. After being incubated for 24 h, the cells were replaced with media of different pH (6.0,6.4,6.8, and 7.4) at indicated times. Then 10 μL of MTT stock solution (5 mg/ml) was added into the wells. After being incubated in the dark at 37 °C for 4 h, 100 μL DMSO was added into the well. The absorbance was measured at 490 nm with a bio-tek 311 microplate plate reader (elx 800, bio-tek, Winooski, vt).
Apoptosis detection by flow cytometry
Annexin V (a sensitive indicator for detection of early apoptosis) and propidium iodide (PI) (a sensitive indicator for detection of late apoptosis), were used to detect apoptosis in obteoblasts. The experiment was divided into two groups (A and B). In group A, osteoblasts were cultured in media of different pH values (6.4,6.8 and 7.4). In group B before cells were exposed to media with same pH values as group A, CQ was added to inhibit autophagy. Cellular apoptosis was measured by the Annexin V-FITC Apoptosis Detection Kit. Cells were plated in 6-well plates and maintained at 37 °C for 24 h, and then treated with media of different pH value (6.4,6.8, and 7.4). Cells were harvested and washed with cold PBS gently for three times after 48 h, and then resuspended in 100 μL Binging Buffer. Annexin V-FITC and PI were added and the cells were incubated for 15 min in the dark. 400 μL Binging Buffer was then added and the cells were analysed by flow cytometry (Accuri C6 Cytometer, BD Biosciences, San Jose, USA).
Transmission electronic microscopy
After being treated with media of different pH (6.4,6.8 and 7.4), the cells were prefixed in 2.5% glutaraldehyde for 2 h at 4 °C, then harvested by centrifugation and washed with 0.1 M PBS. The samples were then postfixed in 1% osmium tetroxide through a series of ethanol dehydration. Finally, the samples were embedded in Durcopan ACM, stained, and imaged by transmission electron microscope (JEM-2000EX; JEOL Co; Japan).
Immunofluorescence staining for LC3
After being seeded in 24-well plates and treated with different pH (6.4,6.8 and 7.4) for 6 h, cells were rinsed with phosphate-buffered saline (PBS) for three times and fixed with 4% paraformaldehyde for 20 min at room temperature. The samples were then permeabilized with 0.5% TritonX-100 for 20 min and washed three times. Subsequently the samples were incubated with goat serum for 30 min at room temperature. Antibody raised against LC3 was added to every sample and samples were maintained at 4 °C overnight. After being washed with PBST three times, fluorescein-conjugated secondary antibody was added and incubated in dark place for 1 h. DAPI was used to counterstain cell nuclei, and then visualized by microscope.
Western blot analysis
After OBs were treated with pH 6.4 for 6 h, 12 h and 24 h autophagy marker protein LC3 was detected by Western blot. Cells were washed with cold PBS and lysed with a lysis buffer on the ice. After a series of centrifugations and sample heating, the protein concentrations were measured using the BCA assay. The protein extracts were separated by 15% SDS-polyacrylamide gel and transferred to polyvinylidene difluoride (PVDF) membranes, which had been cut according to the protein band. The membranes were blotted in 5% skim milk for 1 h, then washed and incubated in specific primary antibody LC3 (1:800 dilution) and P62 (1:1000 dilution) overnight at 4 °C. Finally the membrane was put into secondary antibody (1:1000 dilution) for 1 h at room temperature and the protein band was visualized by labwork software.
Materials
The murine osteoblastic cell line MC3T3-E1 was purchased from Boster (Boster, Wuhan, China). α-minimal essential media (α-MEM) purchased from Hyclone (Logan, Utah, USA). Fetal bovine serum (FBS) was purchased from Gibco (Grand Island, NY, USA). 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolinum bromide (MTT), 4,6-diamidino-2-phenylindole (DAPI), penicillin, streptomycin, Hepes, chloroquine (CQ), were purchased from Solarbio (Beijing Solarbio Science & Technology, Beijing, China). The primary antibodies against P62, LC3 were purchased from Sigma (St. Louis, MO, USA), β-actin was purchased from ZSGB (ZSGB-BIO, Beijing, China). The Annexin V-FITC apoptosis detection kit was purchased from BD Biosciences (San Diego, CA, USA).
Statistical analysis
Data are presented as mean ± standard deviation (SD) of three replicates. One-way ANOVA test and t test were employed to compare the difference between experimental groups. SPSS 17.0 software was used to analyse the data. The results were considered significant at P < 0.05.
Additional Information
How to cite this article: Zhang, Z. et al. Acidic pH environment induces autophagy in osteoblasts. Sci. Rep. 7, 46161; doi: 10.1038/srep46161 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Acknowledgments
This study was supported by the Distinguished Middle-Aged and Young Scientist Encourage and Reward Foundation of Shandong Province (BS2013YY050), and Science and Technology Program of Shandong Province (No. 2014GSF118145), and Independent Innovation Program of Jinan Universities and Colleges (26010205081417). The authors would like to thank Jue Wang and Bing Hu for helpful discussions and technical assistance.
Footnotes
The authors declare no competing financial interests.
Author Contributions Study design: Qingguo Lai. Animal study:Zhichao Zhang, Yanan Li, Xiaopeng Tang, Jiangbo Ci, and Yan Li. In vitro experiments: Zhichao Zhang and Chao Xu. Data collection and interpretation: Zhichao Zhang, Qingguo Lai, Shaolong Sun, Bingbing Xu. Manuscript writing: Qingguo Lai and Zhichao Zhang. All the authors discussed the results and reviewed the manuscript.
References
- Lim S. S. et al. A Comparative Risk Assessment of Burden of Disease and Injury Attributable to 67 Risk Factors and Risk Factor Clusters in 21 regions, 1990–2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet, 380(9859), 2224–60 (2012). [DOI] [PMC free article] [PubMed]
- Murray C. J. et al. Disability-Adjusted Life Years (DALYs) for 291 Diseases and Injuries in 21 Regions, 1990–2010: A Systematic Analysis for the Global Burden of Disease Study 2010. Lancet. 380, 2197–2223 (2012). [DOI] [PubMed] [Google Scholar]
- Wang T., Zhang X. & Bikle D. D. Osteogenic Differentiation of Periosteal Cells During Fracture Healing. J Cell Physiol. (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z. et al. [X-ray Induces Autophagy in Human Mesenchymal Stem Cells]. Zhonghua Xue Ye Xue Za Zhi. 32, 602–605 (2011). [PubMed] [Google Scholar]
- Levine B. & Kroemer G. Autophagy in the Pathogenesis of Disease. CELL. 132, 27–42 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Levine B., Cuervo A. M. & Klionsky D. J. Autophagy Fights Disease through Cellular Self-Digestion. Nature. 451, 1069–1075 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong E. & Cuervo A. M. Integration of Clearance Mechanisms: The Proteasome and Autophagy. Cold Spring Harb Perspect Biol. 2, a6734 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong A. S., Cheung Z. H. & Ip N. Y. Molecular Machinery of Macroautophagy and its Deregulation in Diseases. Biochim Biophys Acta. 1812, 1490–1497 (2011). [DOI] [PubMed] [Google Scholar]
- Lee Y. et al. Increased SCF/c-kit by Hypoxia Promotes Autophagy of Human Placental Chorionic Plate-Derived Mesenchymal Stem Cells Via Regulating the Phosphorylation of mTOR. J Cell Biochem. 114, 79–88 (2013). [DOI] [PubMed] [Google Scholar]
- Zhang Q. et al. Autophagy Activation: A Novel Mechanism of Atorvastatin to Protect Mesenchymal Stem Cells From Hypoxia and Serum Deprivation Via AMP-activated Protein Kinase/Mammalian Target of Rapamycin Pathway. Stem Cells Dev. 21, 1321–1332 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Puerto M. C. et al. Activation of Autophagy by FOXO3 Regulates Redox Homeostasis During Osteogenic Differentiation. Autophagy 12, 1804–1816 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nollet M. et al. Autophagy in Osteoblasts is Involved in Mineralization and Bone Homeostasis. Autophagy 10, 1965–1977 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirigian L. S. et al. Osteoblast Malfunction Caused by Cell Stress Response to Procollagen Misfolding in alpha2(I)-G610C Mouse Model of Osteogenesis Imperfecta. J Bone Miner Res. 31, 1608–1616 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman R. J., Duthie R. B. & Francis M. J. Nuclear Magnetic Resonance Studies of Fracture Repair. Clin Orthop Relat Res. 297–303 (1985). [PubMed] [Google Scholar]
- Newman R. J., Francis M. J. & Duthie R. B. Nuclear Magnetic Resonance Studies of Experimentally Induced Delayed Fracture Union. Clin Orthop Relat Res. 253–261 (1987). [PubMed] [Google Scholar]
- Beck-Broichsitter B. E. et al. Endocultivation: Metabolism During Heterotopic Osteoinduction in Vivo-Monitoring with Fiber Optic Detection Devices. Tissue Eng Part C Methods 18, 740–746 (2012). [DOI] [PubMed] [Google Scholar]
- Beck-Broichsitter B. E. et al. Using Eddy Currents for Noninvasive in Vivo pH Monitoring for Bone Tissue Engineering. Oral Maxillofac Surg. 19, 55–60 (2015). [DOI] [PubMed] [Google Scholar]
- Arnett T. R. Acidosis, Hypoxia and Bone. Arch Biochem Biophys. 503, 103–109 (2010). [DOI] [PubMed] [Google Scholar]
- Kaunitz J. D. & Yamaguchi D. T. TNAP, TrAP, Ecto-Purinergic Signaling, and Bone Remodeling. J Cell Biochem. 105, 655–662 (2008). [DOI] [PubMed] [Google Scholar]
- Han S. H. et al. Acidic pH Environments Increase the Expression of Cathepsin B in Osteoblasts: The Significance of ER Stress in Bone Physiology. Immunopharmacol Immunotoxicol 31, 428–431 (2009). [DOI] [PubMed] [Google Scholar]
- Jilka R. L., Weinstein R. S., Parfitt A. M. & Manolagas S. C. Quantifying Osteoblast and Osteocyte Apoptosis: Challenges and Rewards. J Bone Miner Res. 22, 1492–1501 (2007). [DOI] [PubMed] [Google Scholar]
- Burckhardt P. The Role of Low Acid Load in Vegetarian Diet On Bone Health: A Narrative Review. Swiss Med Wkly. 146, w14277 (2016). [DOI] [PubMed] [Google Scholar]
- Zahm A. M., Bohensky J., Adams C. S., Shapiro I. M. & Srinivas V. Bone Cell Autophagy is Regulated by Environmental Factors. Cells Tissues Organs 194, 274–278 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas V., Bohensky J., Zahm A. M. & Shapiro I. M. Autophagy in Mineralizing Tissues: Microenvironmental Perspectives. Cell Cycle 8, 391–393 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D. J. et al. Guidelines for the Use and Interpretation of Assays for Monitoring Autophagy. Autophagy 8, 445–544 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaf M. B., Keulers T. G., Vooijs M. A. & Rouschop K. M. LC3/GABARAP Family Proteins: Autophagy-(Un)related Functions. Faseb J. (2016). [DOI] [PubMed] [Google Scholar]
- Bjorkoy G. et al. Monitoring Autophagic Degradation of p62/SQSTM1. Methods Enzymol. 452, 181–197 (2009). [DOI] [PubMed] [Google Scholar]
- Katsuragi Y., Ichimura Y. & Komatsu M. P62/SQSTM1 Functions as a Signaling Hub and an Autophagy Adaptor. Febs J. 282, 4672–4678 (2015). [DOI] [PubMed] [Google Scholar]
- Moscat J., Karin M. & Diaz-Meco M. T. P62 in Cancer: Signaling Adaptor Beyond Autophagy. Cell. 167, 606–609 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley J. H. & Nogales E. Next-Generation Electron Microscopy in Autophagy Research. Curr Opin Struct Biol. 41, 211–216 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M. et al. Homeostatic Levels of P62 Control Cytoplasmic Inclusion Body Formation in Autophagy-Deficient Mice. Cell. 131, 1149–1163 (2007). [DOI] [PubMed] [Google Scholar]
- Zhan J. et al. Crosstalk Between the Autophagy-Lysosome Pathway and the Ubiquitin-Proteasome Pathway in Retinal Pigment Epithelial Cells. Curr Mol Med. 16, 487–495 (2016). [DOI] [PubMed] [Google Scholar]
- Vergilio C. S. & de Melo E. J. Autophagy, Apoptosis and Organelle Features During Cell Exposure to Cadmium. Biocell 37, 45–54 (2013). [PubMed] [Google Scholar]
- Chiarelli R., Agnello M., Bosco L. & Roccheri M. C. Sea Urchin Embryos Exposed to Cadmium as an Experimental Model for Studying the Relationship Between Autophagy and Apoptosis. Mar Environ Res. 93, 47–55 (2014). [DOI] [PubMed] [Google Scholar]
- Maiuri M. C., Zalckvar E., Kimchi A. & Kroemer G. Self-Eating and Self-Killing: Crosstalk Between Autophagy and Apoptosis. Nat Rev Mol Cell Biol. 8, 741–752 (2007). [DOI] [PubMed] [Google Scholar]
- Amaravadi R. K. et al. Autophagy Inhibition Enhances Therapy-Induced Apoptosis in a Myc-induced Model of Lymphoma. J Clin Invest. 117, 326–336 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boya P. et al. Inhibition of Macroautophagy Triggers Apoptosis. Mol Cell Biol. 25, 1025–1040 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh C. H., Hsu S. P., Yang C. C., Chien C. T. & Wang N. P. Hypoxic Preconditioning Reinforces HIF-alpha-dependent HSP70 Signaling to Reduce Ischemic Renal Failure-Induced Renal Tubular Apoptosis and Autophagy. Life Sci. 86, 115–123 (2010). [DOI] [PubMed] [Google Scholar]
- Domart M. C. et al. Concurrent Induction of Necrosis, Apoptosis, and Autophagy in Ischemic Preconditioned Human Livers Formerly Treated by Chemotherapy. J Hepatol. 51, 881–889 (2009). [DOI] [PubMed] [Google Scholar]
- Gonzalez-Polo R. A. et al. The Apoptosis/Autophagy Paradox: Autophagic Vacuolization Before Apoptotic Death. J Cell Sci. 118, 3091–3102 (2005). [DOI] [PubMed] [Google Scholar]
- Ravikumar B. et al. Regulation of Mammalian Autophagy in Physiology and Pathophysiology. Physiol Rev. 90, 1383–1435 (2010). [DOI] [PubMed] [Google Scholar]
- Klionsky D. J. & Emr S. D. Autophagy as a Regulated Pathway of Cellular Degradation. Science 290, 1717–1721 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones R. G. The Roles, Mechanisms, and Controversies of Autophagy in Mammalian Biology. F1000 Biol Rep. 1, 68 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piemontese M. et al. Low Bone Mass and Changes in the Osteocyte Network in Mice Lacking Autophagy in the Osteoblast Lineage. Sci Rep. 6, 24262 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M. B., Nielsen S. E. & Berg K. Re-Examination and Further Development of a Precise and Rapid Dye Method for Measuring Cell Growth/Cell Kill. J Immunol Methods 119, 203–210 (1989). [DOI] [PubMed] [Google Scholar]