Summary
During eukaryotic translation initiation, eIF3 binds the solvent-accessible side of the 40S ribosome and recruits the gate-keeper protein eIF1 and eIF5 to the decoding center. This is largely mediated by the N-terminal domain (NTD) of eIF3c, which can be divided into three parts: 3c0, 3c1 and 3c2. The N-terminal part, 3c0, binds eIF5 strongly, but only weakly to the ribosome-binding surface of eIF1, whereas 3c1 and 3c2 form a stoichiometric complex with eIF1. 3c1 contacts eIF1 through Arg-53 and Leu-96, while 3c2 faces 40S protein uS15/S13, to anchor eIF1 to the scanning pre-initiation complex (PIC). We propose that the 3c0:eIF1 interaction diminishes eIF1 binding to the 40S, whereas 3c0:eIF5 interaction stabilizes the scanning PIC by precluding this inhibitory interaction. Upon start codon recognition, interactions involving eIF5, and ultimately 3c0:eIF1 association facilitate eIF1 release. Our results reveal intricate molecular interactions within the PIC, programmed for rapid scanning-arrest at the start codon.
eTOC Blurb
During translation initiation, eIF3 binds the solvent-accessible side of the 40S ribosome. Obayashi et al. propose that the N-terminal domain of eIF3c reaches into the decoding center, to not only anchor the gate-keeper eIF1, but facilitate eIF1 release on AUG selection. eIF5 appears to play a role in this regulation.
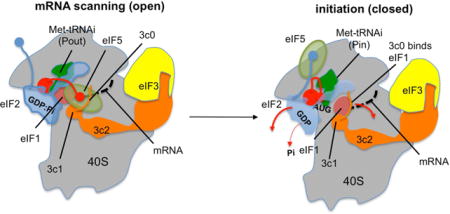
Ribosomes initiate translation with levels of stringency varying between bacteria (low) and eukaryotes (high) (Asano, 2014). The high accuracy of initiation in eukaryotes results from suppressing initiation from non-AUG codons like GUG and UUG. This stringency is imposed partly by eukaryotic initiation factors (eIFs) that bind the small (40S) ribosomal subunit in the 43S preinitiation complex (PIC), i. e. eIF1A, eIF1, eIF2, eIF3 and eIF5 (Asano, 2014; Hinnebusch, 2014). Like its bacterial counterpart IF1, eIF1A binds the 40S A-site. The other four factors engage in numerous mutual interactions to form the multifactor complex (MFC) with Met-tRNAiMet bound to eIF2-GTP in the ternary complex, whereby MFC can be isolated free of ribosomes from various eukaryotes (Asano et al., 2000; Dennis et al., 2009; Meleppattu et al., 2015; Sokabe et al., 2012). eIF4F, comprised of m7G-cap binding subunit eIF4E, RNA helicase eIF4A and scaffold eIF4G, mediates attachment of the mRNA 5′ end to the PIC in its open, scanning-competent conformation (Kumar et al., 2016). A key event in start codon selection is dissociation from the 40S of eIF1, a gatekeeper molecule that maintains the open conformation of the PIC (Pestova and Kolupaeva, 2002; Saini et al., 2010). During scanning, the eIF1 physically opposes full accommodation of tRNAi in the P-site, keeping it in the POUT conformation (Lomakin and Steitz, 2013; Rabl et al., 2011; Weisser et al., 2013). Once tRNAi base pairs to the AUG start codon, eIF1 is released, Met-tRNAi is fully accommodated in the P-site (PIN state), and the PIC adopts the closed conformation incompatible with scanning. The resulting 40S initiation complex is ready for subsequent 60S subunit joining.
In this work, we examine the structural role of the NTD of the eIF3c-subunit of eIF3, a crucial binding partner of eIF1 and eIF5 in the MFC, and key regulator of start codon selection (Asano et al., 2000; Asano et al., 2001a; Karásková et al., 2012; Phan et al., 1998; Valásek et al., 2004). eIF3 is a multisubunit complex (Asano et al., 1997) that binds the solvent-accessible side of the 40S (Srivastava et al., 1992). Cross-linking and integrated modeling studies suggest that eIF3c-NTD extends into the 40S decoding center proximal to eIF1 (Erzberger et al., 2014). eIF5 is the GTPase activating protein for eIF2 (Asano et al., 2001b; Huang et al., 1997). Independently of the catalytic NTD, the eIF5 C-terminal domain (CTD) interacts with eIF1A, eIF2β, eIF3c, and eIF4G at various stages of initiation (Luna et al., 2013; Luna et al., 2012; Reibarkh et al., 2008; Singh et al., 2012; Yamamoto et al., 2005). While an initial cryoEM study revealed density potentially corresponding to eIF5-CTD facing eIF1 and eIF2 in the PIC (Hussain et al., 2014), this was not observed in more recent PIC structures (Llacer et al., 2015). Thus, the location and structural role of eIF5-CTD in the PIC also remains unclear.
Genetic studies have revealed that eIF3c-NTD contains two distinct elements with opposing roles in initiation accuracy. Box12 is required for accurate initiation, and substitution mutations in this element increase non-AUG initiation (for the Sui− or suppressor of initiation codon mutation phenotype). The Box6 element is required for initiation at non-AUG codons, and substitutions in Box 6 suppress effects conferred by a Sui− mutation (for the Ssu− or suppressor of Sui phenotype) (Karásková et al., 2012; Valásek et al., 2004). Henceforth, Box6 and Box12 are designated as an Ssu+ (Box6Ssu+), and a Sui+ element (Box12Sui+), respectively. Certain Box6 or Box12 mutations decrease eIF1 binding to the eIF3c-NTD, suggesting that the eIF3c-NTD helps to stabilize eIF1 in the PIC not only during mRNA scanning, but also during the switch to the closed state upon start codon selection. Herein, we employed a battery of biophysical methods including nuclear magnetic resonance (NMR) spectroscopy to dissect eIF3c-NTD into three units, 3c0, 3c1 and 3c2, and locate the latter two within the recently solved cryo-EM PIC structure (Erzberger et al., 2014). Based on physical interaction studies involving eIF1, eIF3c-NTD and eIF5, we propose that, by interacting with the N-terminal unit 3c0, eIF5 modulates the ability of eIF3c-NTD to either anchor or release eIF1. Our model explains distinct contributions of eIF3c Box 6Ssu+ and Box 12Sui+ to the accuracy of start codon selection in vivo.
Results
Functional dissection of eIF1-binding elements in eIF3c-NTD
To map eIF1 binding sites in the eIF3c-NTD, we divided the latter into three regions: 3c0 encompassing amino acids (aa) 1–58, including the conserved N-terminus required for eIF5 binding (Karásková et al., 2012) and most of Box6Ssu+; 3c1 encompassing aa 59–87, which contains a conserved hydrophobic segment; and 3c2 comprising aa 88–163, including predicted α-helices (http://bioinf.cs.ucl.ac.uk/psipred/) and Box12Sui+ (Fig. 1A and S1A). GST fusions to eIF3c-NTD fragments with different combinations of these regions (eIF3c-A to -G, Fig. 1A) were tested for eIF1 binding using GST pulldown assays. Fragment eIF3c-D58–163 essentially covers the previously determined minimal eIF1-binding site (aa. 60–137) (Karásková et al., 2012).
Fig. 1. Functional dissection of the eIF1-binding site within eIF3c-NTD.
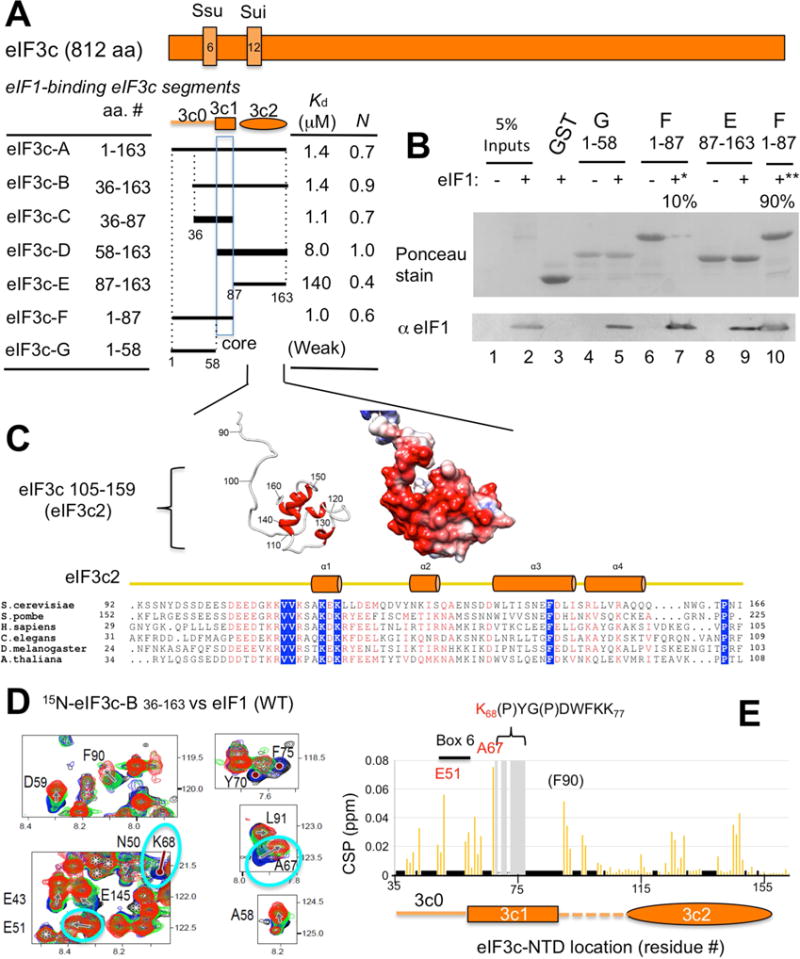
(A) Location of eIF1-binding site in eIF3c primary structure (orange rectangle), highlighting regions of Ssu and Sui mutation sites, Box6 and Box12 (boxes with numbers). Orange schematics below indicate functional elements identified in eIF3c-NTD, 3c0, 3c1 and 3c2. The lines further beneath depict eIF3c deletion constructs used in this study. Dotted lines define the boundaries of eIF3c-NTD regions i to iv (Fig. S1A). Table summarizes the results of ITC analysis for eIF1 binding, Kd and N (stoichiometry; number of eIF3c molecules bound to a eIF1 molecule) (See Fig. S2). “(Weak)”, the weakest binding observed with eIF3c-G in GST pulldown. (B) GST pulldown assay. ~5 μg of indicated GST-eIF3c fusion proteins (0. 15–0. 2 nmol) were allowed to bind 70 μg of recombinant eIF1 (~5 nmol; ~25 μM) in E. coli lysates (lanes labeled “+”) and the protein complexes pulled down and analyzed with 5% in-put amounts of lysates by SDS-PAGE, followed by immunoblotting with anti-yeast eIF1 (bottom) and Ponceau staining (top). “−”, uninduced E. coli lysates were used as a negative control. In lanes 7 (*) and 10 (**), 10 % and 90% of the GST-eIF3c-F complex were analyzed, respectively. (C) Solution structure of eIF3c 105–159 found within yeast eIF3c-NTD (36–163), determined by NMR spectroscopy (see Supplementary text and Table S3). Ribbon diagram is shown to the left. Right, electrostatic potential distribution (negative in red, neutral in white, and positive in blue) calculated according to Coulomb’s law. Bottom, α-helices are aligned with amino acid sequences of eIF3c_N (Pfam). (D–E) NMR CSP studies on interaction between 15N-eIF3c-B36–163 and eIF1. (D) Close-up views of 1H-15N HSQC spectra of 15N-eIF3c eIF3c-B in the absence (black) or presence of WT eIF1 (panel 1) (1:0. 3 molar ratio in blue, 1:0. 6 in green, 1:1 in red) See Fig. S3B for the entire spectrum of 15N-eIF3c eIF3c-B with or without WT eIF1. (E) Chemical shift perturbation, Δδ, was computed as described in Supplementary methods and presented for each assigned amino acid. “P”, proline. Short black bar, unassigned. Shaded, residue with line broadening by eIF1. Three amino acids showing largest Δδ are labeled. Labeled in red are amino acids whose CSP were resolved by eIF1 mutations defective in eIF3c binding. See also Figures S1, S2, S3, S4 and Tables S1 and S3.
The strongest eIF1 binding was observed with eIF3c-A1–163, -B36–163, -C36–87 and –F1–87, which all include the C-terminal half of 3c0 and the entire 3c1 (Fig. S1B, lanes 2, 3, 6 and 8 and Fig. S1C). Isothermal titration calorimetry (ITC) assays demonstrated apparent Kd values of ~1μM for these constructs (Fig. 1A and S2A) with standard deviations of <15% (n=3, Fig. S2B). eIF3c-D58–163, containing regions 3c1 and 3c2, exhibited weaker association with eIF1 (Fig. S1B, lane 4) with an apparent Kd of ~8μM (Fig. 1A and S2A–B). eIF3c-D58–163 regions therefore bind eIF1 with a significantly lower affinity than the constructs with regions 3c0–3c1 (p<0. 006, n=3). In contrast, the two NTD segments lacking 3c1, eIF3c-E87–163 (3c2) and eIF3c-G1–58 (3c0), did not appear to interact with eIF1 in GST pulldown assays (Fig. S1B, lanes 5 and 7), but displayed appreciable binding when the eIF1 concentration was increased ~50-fold to ~30μM (Fig. 1B, lanes 5 and 9, eIF1 detected by anti-eIF1; Fig. S1C, lanes 3 and 7, eIF1 indicated by arrowheads in Coomassie staining). Note that in Fig. 1B, amounts of eIF1 bound to GST-eIF3c-E87–163 (lane 9) and eIF3c-G1–58 (lane 5) are <10% of that bound to GST-eIF3c-F1–87 with 3c0 and 3c1 (lanes 7 and 10; where 10 and 90% of the pulldown fraction were loaded. Anti-eIF1 signal in lane 10 is saturated due to overloading). Consistent with the pulldown results, the Kd for eIF3c-E87–163 binding to eIF1 is >100 μM (Fig. 1A).
The ITC assay revealed that eIF3c-D58–163 forms a stoichiometric complex with eIF1 (N=1.0, Fig. 1A and S2B), while other segments containing 3c1 and 3c0 (A1–163, B36–163, C36–87 and F1–87) display N values (number of eIF3c molecules bound per eIF1 molecule) significantly less than 1.0 (p<0.03, n=3). These results suggest that eIF1 has more than one binding site for eIF3c regions 3c0 and 3c1.
Based on these results, we identify 3c1 as the core eIF1-binding site in eIF3c-NTD. Low-affinity eIF1 binding by flanking region 3c0 containing Box6Ssu+ contributes to the high-affinity eIF1 binding (~1μM) by fragments containing 3c1 and 3c0, likely through interaction with more than one site on eIF1. Because we failed to generate an eIF3c segment containing only 3c1, the contribution of C-terminal flanking 3c2 remained unclear. However, based on the low-affinity eIF1 binding to eIF3c-E87–163, 3c2 containing Box12Sui+ likely contributes to the relatively high-affinity binding (~8μM) observed for eIF3c-D58–163.
CSP mapping with 15N-eIF3c-NTD identifies amino acids involved in eIF1 binding
Next, we used NMR chemical shift perturbation (CSP) mapping to delineate eIF3c residues directly involved in eIF1 binding. We first determined the structure of eIF3c-NTD by NMR spectroscopy using [13C, 15N] eIF3c-B36–163 segment (see Supplementary text and Table S3 for details), which demonstrated that the region covering most of 3c2 (residues 105–159) folds into α-helical globule (Fig. 1C). The eIF3c backbone resonance assignments were then used for CSP studies. As shown in Fig. S3, CSPs induced by eIF1 binding are nearly identical between 15N-eIF3c-A1–163 and –B36–163, which is consistent with our GST-pull down and ITC studies (Fig. 1A, S1B–C and S2) (Karásková et al., 2012). We further observed large CSPs for A67 (circled in blue in Fig. 1D; indicated by arrow in Fig. S3 lower panels), and E51 residues (circled in blue in Fig. 1D) accompanied by the strong resonance line broadening in the stretch: K68(P)YG(P)DWFKK77 (K68, Y70 and F75 highlighted in Fig. 1D; others highlighted in Fig. S3A–B; prolines are in parenthesis). In contrast, all CSPs in 3c2 were minor (<0.04 ppm) (Fig. 1E) except for F90, which was considered spurious inasmuch as it was not eliminated by an eIF1 mutation that abolishes interaction with eIF3c (shown below in Fig. S4, panel 1). Collectively, these results indicate that the eIF1-binding site on eIF3c NTD resides in the area covering Box6Ssu+ of 3c0 (containing E51) and core region 3c1 (aa 58–87, contains A67K(P)YG(P)DWFKK77) (Fig. 1A and S1A).
Structure of eIF3c-NTD105–159 and integrated modeling of eIF3:eIF1:40S complex structure define two globular units within eIF3c-NTD
In a recent cryo-EM study of the eIF1/eIF3/40S complex, which integrated extensive crosslinking information, it was proposed that the eIF3c-NTD projects from the solvent side along the 40S subunit into the decoding center, where eIF1 is bound (Erzberger et al., 2014). However, structural information for the eIF3c-NTD was lacking. We therefore incorporated NMR structure of eIF3c segment 105–159 (Fig. 1C) into the integrated modeling platform and calculated a new localization for the whole eIF3 complex (Fig. S5). The resulting localization densities for eIF3c-NTD had a resolution of 18 Å (Fig. 2A, left), guided by 4 high-confidence crosslinks (Fig. 2A, right), which is a clear improvement from the 38 Å precision in our previous model (Erzberger et al., 2014). The eIF3c-NTD is resolved into two globular units that span the ~60Å distance between eIF1 and rpS13/uS15 (Fig. 2A and S5). The one is located near rpS13/uS15 and was assigned as α-helical globular structure in 3c2 (aa 105–159) shown in Fig. 1C. The other is adjacent to eIF1 and, thus, was assigned as the core eIF1-binding region 3c1 (aa 59–87) (Fig. 1A). Indeed, recent medium-resolution cryo-EM reconstructions (Aylett et al., 2015; Llacer et al., 2015) reveal densities consistent with the positions of 3c1 and 3c2 (Fig. 2B). The N-terminal region 3c0 (Fig. 1A) was not localized in the eIF3:eIF1:40S structure, presumably because it cannot bind eIF1 when eIF1 is bound to the 40S subunit (as discussed below). Therefore, integrative modeling that incorporates the NMR structure of eIF3c105–159 pinpointed the locations of the 3c1 and 3c2 elements within the PIC, with 3c1 directly contacting eIF1.
Fig. 2. Location of eIF3c2 105–159 globule within the eIF1:eIF3:40S structure.
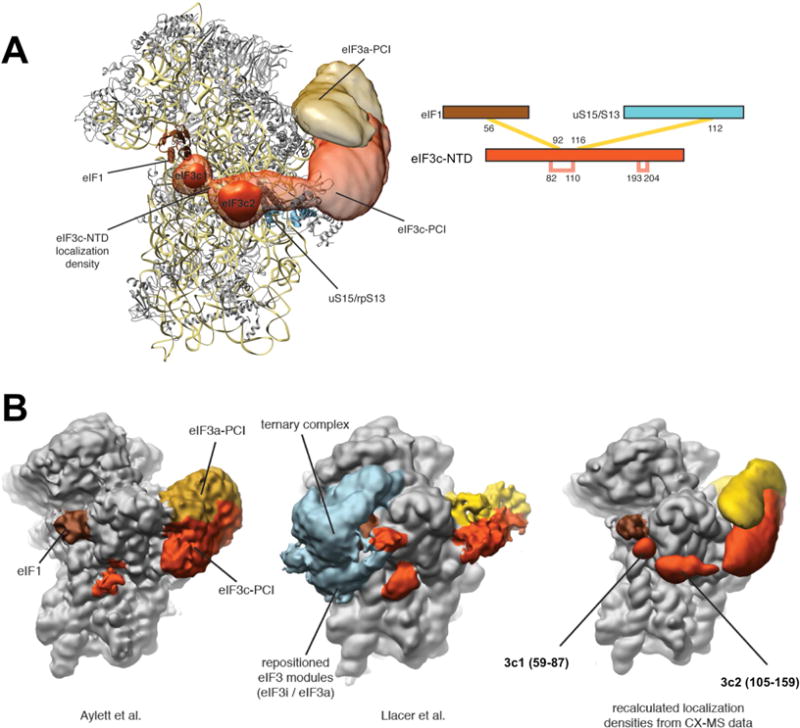
(A) Recalculated integrated modeling localization densities (Erzberger et al., 2014) incorporating the eIF3c-NTD NMR structure. Left, 40S subunit shown as a ribbon diagram with overlayed localization densities for eIF3a (gold) and eIF3c (orange-red). A higher contour level of the eIF3c-NTD is used to emphasize the predicted two-domain architecture. Right, eIF3c-NTD-specific cross-links that anchor the globular domain of eIF3c-NTD. Interstrand crosslinks in yellow, intrastrand crosslinks in pink. (B) Comparison of recent cryo-EM reconstructions of eIF3 complexes [Left – (Aylett et al., 2015), Center – (Llacer et al., 2015)] with the current localization densities derived from integrative modeling superposed on a 40S-eIF1 structure (right). Densities for eIF3a and eIF3c are colored as in panel A, eIF1 is shown in brown and additional densities present in the Llacer et al structure shown in light blue. In the Erzberger structure, 3c1 is defined as eIF3c aa. 58–87, based on the NMR studies in Fig. 3–4. See also Figure S5.
NMR evidence that eIF3c-NTD segments 3c1–3c2 interact with a limited surface of eIF1 compatible with 40S binding
eIF1 is comprised of an unstructured N-terminal tail (NTT) and a globular domain with a β1-β2-α1-β3-β4-α2-β5 fold (Fletcher et al., 1999; Reibarkh et al., 2008) (Fig. 3A and Table S4). To determine the eIF1 residues contacted by the 3c1–3c2 units in the complex formed with eIF3c D58–163, we performed CSP experiments using 15N-eIF1. As shown in Fig. 3B and summarized in Fig. 3A, strong CSPs were observed for R53, K56, I93 and L96 residues on eIF1 thereby indicating that these residues on eIF1 direct its interaction with eIF3c D58–163. In contrast, resonances corresponding to residues within or nearby the two eIF1 ribosome-binding sites (Martin-Marcos et al., 2013; Rabl et al., 2011), including K60 at the α1 C-terminus and T40/T41 near the β1-β2 loop (loop-1), were only marginally affected (Fig. 3A–B). As summarized in Fig. 4A, the eIF3c-D58–163-binding site on eIF1 is comprised of the N-terminal and central portions of α1 and the adjacent hydrophobic area containing I93 (residues painted red or orange). In agreement with this, the previous EM study showed that K56 on eIF1 crosslinks with K92 on eIF3c which is located in the vicinity of 3c1 region (Fig. 2A). Because eIF1 interacts with the ribosome via residues K59 and K60 at the C-terminus of α1, and R36 in loop-1 (residues painted cyan in Fig. 4A), stoichiometric eIF1 binding to the 3c1-3c2 segment of eIF3c appears to be compatible with eIF1:ribosome association.
Fig. 3. NMR CSP mapping of eIF3c binding site on eIF1.
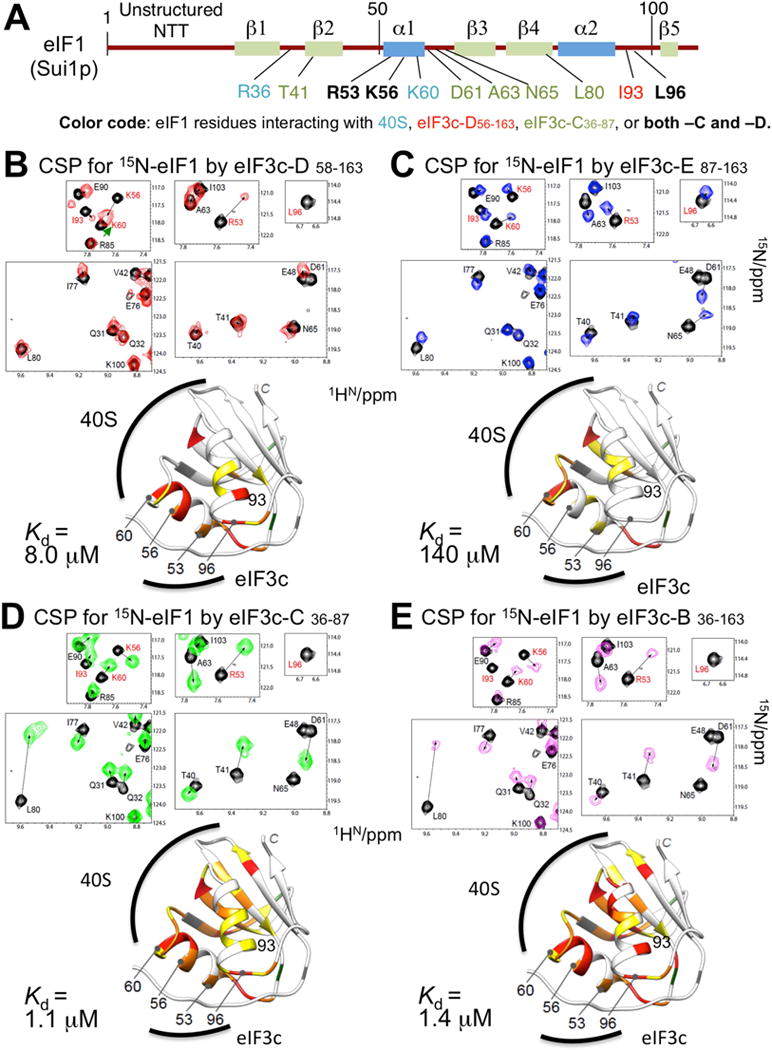
(A) Primary structure of yeast eIF1 (brown horizontal line) with boxes indicating secondary structure elements. Amino acids whose resonance was shifted due to addition of distinct eIF3c fragments are shown in colors based on the code on the bottom. (B)–(E) Top, CSP of 15N-eIF1 resonances caused by eIF3c-D58–163 (Panel B), eIF3c-E78–163 (Panel C), eIF3c-C36–87 (Panel D), and eIF3c-B36–163 (Panel E) were highlighted with arrows in the specified areas of 1H-15N HSQC spectra. The spectra taken in the presence and absence of eIF3c fragments (1:1.2) are shown in color and gray, respectively. The eIF1 residues assigned to the resonances are shown with their amino acid numbers. Amino acids of high relevance (R53, K56, K60, I93, L96) are highlighted in red. Bottom, the eIF1 residues affected by each eIF3c segment are painted orange or yellow for strong or moderate CSP of >0. 1 ppm or 0. 05~0. 1 ppm, respectively, in the ribbon diagram of yeast eIF1 structure. The eIF1 residues whose resonances caused line broadening were painted red. Locations of amino acids of high relevance are indicated. Prolines (11, 46, and 72) and unassigned residues (23, 34–36, 66, and 107) are painted green and gray, respectively. In panel B, note that, upon eIF3c-D58–163 addition, the cross peak for K60 was shifted only slightly (green arrowhead in the spectrum), overlapping with that for K56, which shifted a greater distance (long black arrow). See also Tables S1 and S4.
Fig. 4. Summary of eIF3c:eIF1 interaction models.
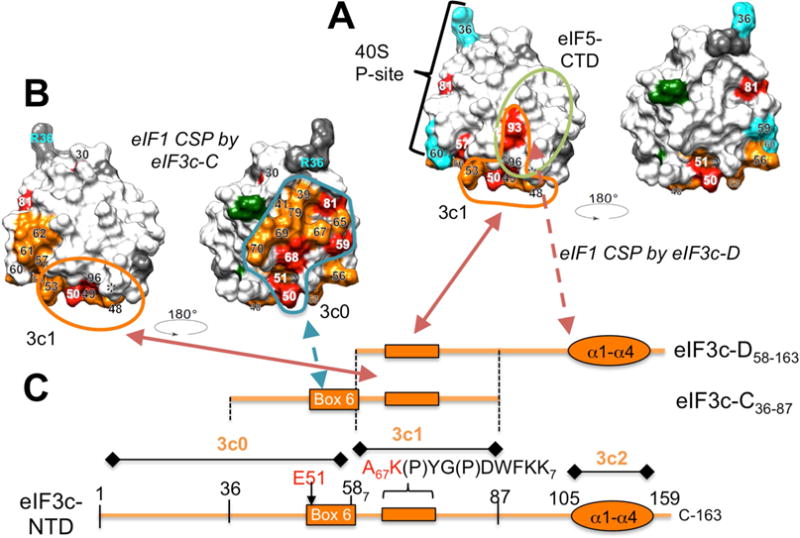
(A) and (B) Left, the eIF1 residues showing strong CSP or line broadening by eIF3c-D58–163 (Panel A) or eIF3c-C36–87 (Panel B) are presented (with unassigned and proline residues) by the same colors on the space-filled model viewed from the same angle as in Fig. 3B and D, respectively. For simplicity, however, residues showing moderate CSP (yellow in Fig. 3) are not presented. Residues 36, 59, and 60 known to contact the ribosome are shown in cyan (Rabl et al., 2011). 3c1- and eIF5-binding sites implicated in the scanning PIC are circled with orange and dark green lines, respectively. Right, the space-filled model of eIF1 rotated 180° relative to the model to the left. In (B), 3c0-binding site is indicated by blue line. (C) Schematic on the bottom (horizontal orange line) describes the primary structure of eIF3c-NTD (aa. 1–163) with orange boxes indicating the locations of Box6 and the area of line-broadening by eIF1. Orange oval, α-helical globule of 3c2. Lines above denote the locations of eIF3c segments, D58–163 and C36–87, used for the CSP studies on 15N-eIF1. Arrows indicate the proposed interactions between defined areas of eIF3c and eIF1.
By comparing CSPs between eIF3c-D58–163 (Fig. 3B) vs. eIF3c-E87–163, containing only 3c2 (Fig. 3C), it is clear that eliminating the 3c1 core eIF1-binding region dramatically reduces affinity of eIF3C NTD for eIF1 (Fig. 1A and S1 & S2). However, weak/moderate CSPs (0.05<ppm<0.1) were observed in the N-terminal half of α1 and β4 of eIF1 (Fig. 3C), while stronger CSPs were located in the C-terminus of α1 (K60) and the α1-β3 loop (N65). Considering the small N value (0.4) observed for E87–163 in ITC assays indicating multiple binding sites on eIF1 (Fig. 1A and S2B), we suggest that eliminating 3c1 disrupts the stoichiometric binding to eIF1 seen for eIF3c-D58–163, which allows isolated 3c2 (E87–163) to engage in weak and likely non-physiological interactions with multiple surfaces on eIF1.
In conclusion, eIF3c-D58–163 containing 3c1 and 3c2, but not 3c2 alone binds eIF1 in a manner compatible with eIF1 binding to the ribosome. Thus, the role of 3c2 in stimulating eIF1 binding to eIF3c-NTD, if any, appears to be indirect.
NMR evidence that segment 3c0-Box6Ssu+ interacts with the ribosome-binding surface of eIF1
Relative to 3c1–3c2 segment D58–163, fragment C36–87, containing 3c1 and part of 3c0, displayed CSPs of greater intensity for a larger number of 15N-eIF1 resonance peaks (Fig. 3D). Herein, in addition to R53 and L96 eIF1 residues, extensive CSPs were also observed for D61, A63 and N65, which are localized in the α1-β3 loop, T41 in β2 near loop-1, and L80 (Fig. 3D, summarized in Fig. 3A). This suggest that C36–87 fragment binds an entire side of β-sheets 1–4 of eIF1 that is adjacent to K60 at the α1 C-terminus, the 40S contact site, and is likely to overlap with the second 40S contact site in loop-1, R36 (cyan lettering in Fig. 4B). Interestingly, the resonance corresponding to I93 was slightly shifted in the presence of C36–87 without attenuation of its signal (Fig. 3D, yellow for weak/moderate interaction), but did not disappear (line broadening) as observed for D58–163 (red in Fig. 3B). As summarized in Fig. 4B, this pattern suggests that C36–87 still retains interaction with R53 and L96 of eIF1 through the core element, 3c1, while its interaction with eIF1-I93 is diminished due to lack of 3c2. This supports an indirect stimulatory role for 3c2 in eIF1 binding to eIF3c-NTD (dotted line in Fig. 4A).
Importantly, these data also suggest that the presence of the C-terminal half of 3c0 in C36–87 confers more extensive interactions with the ribosome-binding surfaces of eIF1 (Fig. 4B). This is in agreement with its N value in ITC experiments of 0.7, indicating >1 binding sites on eIF1 (Fig. 1A and S2B). Hence, we propose that 3c0 does not engage eIF1 in the scanning PIC because its binding site on eIF1 overlaps with the 40S-binding surface.
The conclusion that the C-terminal half of 3c0 containing Box6Ssu+ engages ribosome-binding surface of eIF1 is further supported by ITC analysis indicating that the K37E substitution in eIF1 loop-1 reduces eIF1 binding to eIF3c-NTD by 4-fold (Fig. 5B and S6A). Importantly, 3c0:eIF1 interaction may explain the Ssu− phenotype of the Box6R mutation (Valásek et al., 2004). Notwithstanding that due to the relatively low affinity of eIF1 for eIF3c-NTD (Kd=1 μM), 3c0 is unlikely to displace eIF1 from PIC (eIF1:40S subunit Kd=1–10 nM) (Martin-Marcos et al., 2013), by competing with the eIF1:40S subunit interaction 3c0 may increase the chance that eIF1 is inappropriately released from the 40S subunit at a non-AUG codon. By disrupting this competition, the Box6R mutation of 3c0 is expected to stabilize the scanning PIC and diminish non-AUG initiation (Ssu− phenotype). Thus, combined with the genetic findings (Valásek et al., 2004), the CSP study in Fig. 3D suggests that 3c0:eIF1 interaction impedes eIF1 binding to the ribosome.
Fig. 5. Effect of eIF1 mutations on interaction with eIF3c-NTD, eIF5-CTD and the 40S subunit.
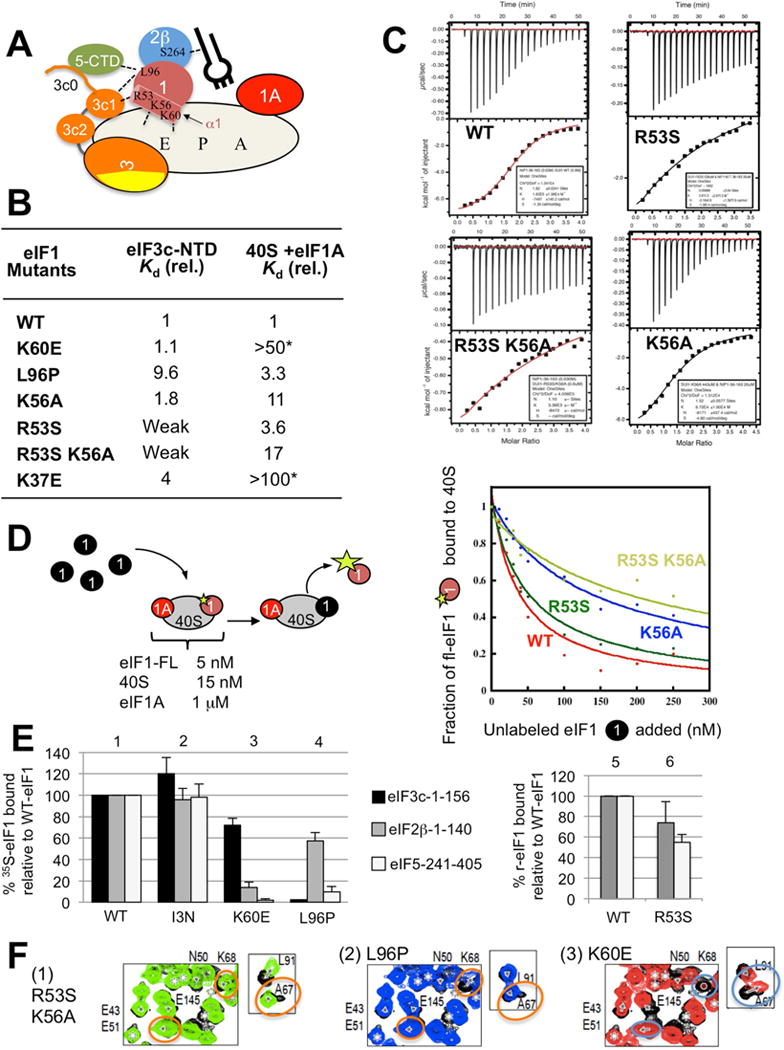
(A) Interactions described here to stabilize the open PIC are presented by dotted lines. Circles indicate eIFs, its subunits or domain. eIF1 or eIF2β amino acids involved in the interactions are presented within the circles. The cylinder attached to eIF1 is its α1. Plug, Met-tRNAi. The largest oval, 40S subunit with tRNA-binding sites (A, P and E). (B) Affinity of eIF1 or its mutants for 40S or eIF3c-NTD. Shown are relative Kd values compared to the value obtained with WT eIF1. Original values are shown in Fig. S6. Asterisk, values based on a previous study (Martin-Marcos et al., 2013). (C) ITC thermograms for eIF3c-B binding to eIF1 and its mutants indicated. (D) 40S binding assay by eIF1 mutants. (Left) Schematics illustrating experimental schemes. Binding of fluorescently labeled eIF1 (brown circle) to 40S (oval):eIF1A (red circle) complex was monitored by FA in the presence of given concentrations of indicated unlabeled eIF1 forms (filled circle). The size of the star indicates the degree of FA. (Right) Fraction of 40S bound to the labeled eIF1 was plotted against the concentration of unlabeled eIF1 species. (E) GST-pulldown assay. The values for the binding of eIF1 mutants to GST-fusion proteins indicated on top are presented relative to those obtained with WT eIF1. eIF1-I3N was used as a negative control. (F) Close-up views of 1H-15N HSQC spectra of 15N-eIF3c eIF3c-B in the absence (black) or presence of indicated mutant eIF1 protein species (panels 1–3) (1:1. 2 in color). See also Figures S6, S7 and Table S1.
The largest fragment examined, eIF3c-B36–163, containing the C-terminal half of 3c0, and full 3c1 and 3c2, induced a combination of CSPs observed for both eIF3c-D58–163 (3c1+3c2) and eIF3c-C36–87 (3c0+3c1) (Fig. 3E). These included major perturbations in the following eIF1 residues: R53 and L96 (due to 3c1), I93 (due to 3c2), and residues in the proximity of both eIF1 ribosome-binding surfaces (attributed to 3c0). Altogether, these results suggest that 3c0 and 3c2, flanking the core eIF1 binding element 3c1, may differentially modulate interaction of eIF3c-NTD with eIF1.
Arg-53 and Leu-96 of eIF1 make critical connections to the eIF3c-NTD within the scanning PIC
CSP analysis implicated eIF1 residues R53 and L96, in the N-terminal end of α1 and nearby hydrophobic patch, in interaction with all three eIF3c-NTD constructs that bind eIF1 with strong affinity (Fig. 3B, D, and E). Accordingly, we tested the effect of substituting these residues on eIF1 binding to eIF3c-B36–163 in vitro. As controls, we examined eIF1 substitutions K56A and K60E, which are involved in 40S binding (see Fig. 3A and 5A for eIF1 residues altered). In the ITC assay, R53S substitution reduced eIF3c binding below the detection limit, whereas L96P substitution reduced the affinity by 10-fold (Fig. 5B–C). In contrast K56A and K60E exerted little effect on eIF3c-NTD:eIF1 binding (Fig. 5B and S6A), which is consistent with NMR data. These results were verified by CSP experiments (Fig. 5F and S4). These results also agree with our previous GST pulldown assays indicating that simultaneous substitution of eIF1 residues K52, R53, K56, K59 and K60 distributed along α1 (sui1-M5) (Reibarkh et al., 2008) and I93, L96 and G97 in the hydrophobic patch (sui1-93-97) (Cheung et al., 2007) reduces eIF1 binding to eIF3c (italicized are amino acids whose single substitution was found here to reduce the interaction).
The eIF1 substitutions L96P and K60E are known to allow mis-initiation from UUG codons in vivo (Sui− phenotype) (Martin-Marcos et al., 2013), which we verified using a UUG-his4 allele and UUG-lacZ reporter (Fig. 6A, rows 1–3). The K60E substitution strongly impairs 40S binding in vitro (Martin-Marcos et al., 2013). Since the L96P substitution reduces eIF1 interaction with eIF3c-NTD (Fig. 5B), its strong Sui− phenotype (Fig. 6A) could be attributed to defective interaction with eIF3c. However, neither eIF1 R53S nor K56A elevate UUG initiation (Fig. 6A, rows 4–5), even though R53S had a greater effect than L96P on eIF3c-NTD binding (Fig. 5B). Thus, the dramatic reduction in initiation stringency conferred by L96P likely results from disrupting eIF1 interactions with other components of the scanning PIC besides eIF3c-NTD.
Fig. 6. Effect of eIF1 mutations on stringent translation initiation in vivo.
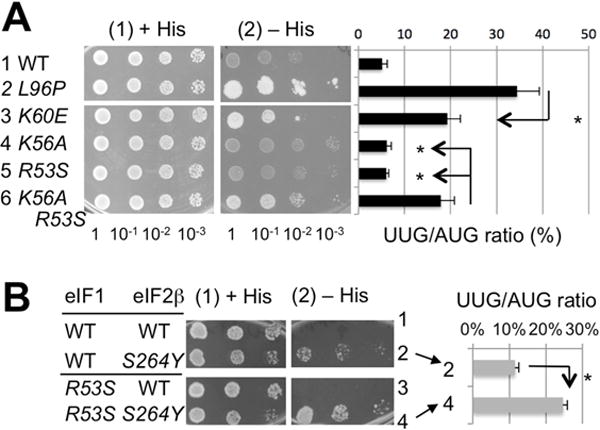
(A) and (B) Sui− phenotype tests. Indicated yeast eIF1 mutants are assayed for his4-UUG expression (His+ test, panel 2 with − His plate; panel 1 with + His plate shown as a loading control) or UUG/AUG initiation ratio (graph to the left). *, p<0. 05 (Panel A, n=5; Panel B, n=4). See Supplementary text for details. See also Tables S1 and S2.
To test this tenet, we examined the effects of L96P on eIF1 interactions with its other known binding partners: the 40S subunit, the eIF2β-NTT, and the eIF5-CTD (Fig. 5A). We determined the Kd for the 40S·eIF1 complex by measuring changes in fluorescence anisotropy (FA) of fluorescently labeled eIF1 in the presence of increasing 40S concentration (Maag et al., 2005). By this approach, we showed that K60E (Martin-Marcos et al., 2013) essentially eliminated, while L96P (this study) modestly reduced eIF1 affinity for 40S, respectively (Fig. 5B, column 3). In contrast, L96P strongly reduced eIF1 interaction with the eIF5-CTD in GST pulldown assays (Fig. 5E, column 4). This is consistent with our previous CSP and spin-labeling studies identifying the hydrophobic patch harboring L96 as the eIF5-CTD binding site (Luna et al., 2012; Reibarkh et al., 2008) (Fig. 4A). However, L96P only slightly reduced eIF1 binding to the eIF2β-NTT (Fig. 5E; see Fig. 5A for summary of interaction involving eIF1-L96). Thus, the strong Sui− phenotype of L96P (Fig. 6A) likely arises from combined defects of reduced eIF1 binding to the eIF3c-NTD, eIF5-CTD (Fig. 5E), and perhaps the 40S subunit (Fig. 5B).
Despite the fact that eIF1 substitution R53S essentially abolishes binding to the eIF3c-NTD (Fig. 5B), it has no effect on initiation accuracy (Fig. 6A), implying that eIF1-R53S retains other interactions with the PIC that compensate for impaired interaction with eIF3c. Employing a variation of the FA assay in which excess unlabeled eIF1 competes with WT labeled eIF1 for ribosome binding (Fig. 5D, left), we found that R53S has only a slight effect on 40S binding (Fig. 5B and D, green curve). Moreover, GST pulldown assays revealed only modest effects of R53S on binding to the eIF2β-NTT and eIF5-CTD (Fig. 5E, right). The CSP assay with 15N-eIF1-R53S also demonstrates robust eIF5-CTD interaction with this mutant, as observed with WT 15N-eIF1 (Fig. S7) (Reibarkh et al., 2008). Thus, R53S specifically abolishes eIF1 interaction with the eIF3c-NTD (Fig. 5B), which is not sufficient to impair accuracy of start site selection in vivo.
To demonstrate a role for eIF1-R53 in stabilizing the scanning PIC in vivo, we generated double mutants. Combining R53S and K56A in eIF1 did not alter the defect in eIF3c-NTD:eIF1 binding seen for R53S alone (Fig. 5B), and conferred only a moderate decrease in 40S:eIF1 binding affinity beyond the 11-fold reduction in KD induced by K56A alone (Fig. 5B and D, blue and light green curves). Nevertheless, the R53S,K56A double mutant displayed a marked increase in UUG initiation that was not observed for single mutants (Fig. 6A, row 6). Since K56A has no effect on eIF1 binding to eIF2β-NTT and eIF5-CTD when combined with four other eIF1 substitutions in α1 (M5 mutation) (Reibarkh et al., 2008), we conclude that the synthetic Sui− phenotype of the R53S,K56A substitution (Fig. 6A) results from the combined loss of eIF1 interaction with eIF3c-NTD conferred by R53S and weakened 40S binding conferred by K56A (Fig. 5B). As shown in Fig. 6B (panel 2, row 2 vs. 4), the eIF1-R53S substitution also exacerbates the elevated UUG initiation caused by the eIF2β-S254Y variant (encoded by SUI3-2), previously attributed to increased GTP hydrolysis (Huang et al, 1997) and stabilizing the PIN conformation of Met-tRNAi at UUG codons (Martin-Marcos et al., 2014). Our findings imply that the defective stabilization of the closed/PIN conformation at UUG codons conferred by eIF2β-S254Y is normally mitigated by the eIF1/eIF3c-NTD interaction (disrupted by eIF1-R53S) to diminish acceptance of codon-anticodon mismatches in the P site.
In conclusion, these results show that eIF1-R53 and –L96 are key eIF3c-NTD interaction sites in vivo. Within the scanning PIC, eIF3c-NTD appears to be the sole binding partner of eIF1-R53, whereas eIF1-L96 appears to engage both eIF3c-NTD and eIF5-CTD. Thus, multiple interactions between eIF3c, eIF5 and the ribosome collaborate in retaining eIF1 within the scanning PIC (Fig. 5A; also see Supplementary Results).
eIF5 regulates eIF3c-NTD interactions with eIF1
We speculated that the 3c0:eIF1 interaction competes with 40S:eIF1 interaction to favor eIF1 dissociation from the PIC at the start codon. We therefore addressed whether the known interaction of eIF5-CTD with 3c0 (Karásková et al., 2012) can preclude this destabilizing effect of 3c0 on the scanning PIC. As shown in Fig. S8, GST pulldown assays demonstrated that GST-eIF3c-A1–163, -F1–87, and –G1–58, but not GST-eIF3c-E87–163 associates with eIF5, supporting the idea that the minimal eIF5-binding segment in eIF3-NTD spans across 3c0 residues 1–46 (Karásková et al., 2012). To examine competition between eIF1 and eIF5 for 3c0 binding, we used GST-eIF3c-F1–87 and eIF3c-A1–163, which both exhibit high affinity for eIF1 (Fig. 1A), and monitored their binding to the full-length eIF5 (in 1:1 stoichiometry) in the presence of >10-fold molar excess of eIF1. As shown in Fig. 7A, eIF1 inhibited eIF5 binding by fragment F1–87 to 51±0.8% (p<0. 0001, n=4) (lanes 6 vs 7, red arrow), supporting the idea that eIF1 competes with eIF5 for 3c0 binding. Importantly, this inhibition was not observed with A1–163 fragment (lanes 9 vs 10), indicating that the presence of 3c2 in fragment A1–163 allows eIF1 and eIF5 to avoid competition for binding to eIF3c-NTD.
Fig. 7. GST pulldown and analytical ultracentrifugation (AUC) experiments characterizing interaction between eIF3c-NTD, eIF1 and eIF5.
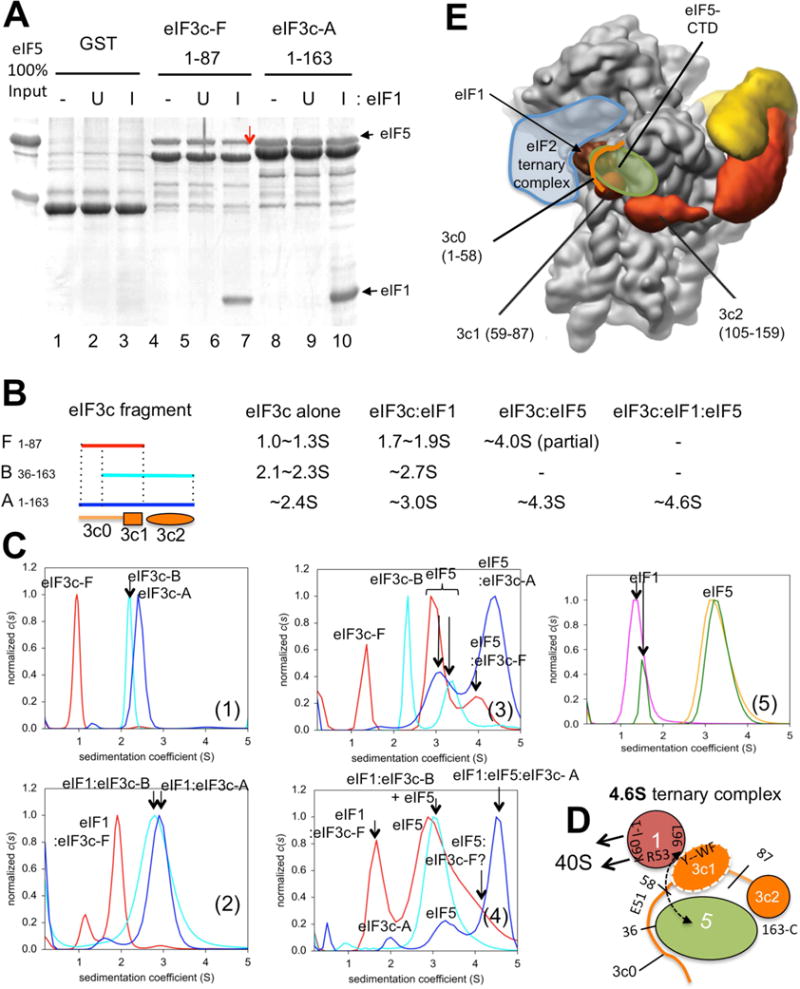
(A) GST pulldown assay demonstrating inhibition of eIF5 binding to eIF3c by excess eIF1. ~5 μg of indicated GST-eIF3c fusion proteins (~0. 2 nmol) were allowed to bind ~5 μg of eIF5 (~0. 1 nmol) in the presence of 70 μg of recombinant eIF1 (~5 nmol) present in induced (I) lysates and the complex analyzed by SDS-PAGE and Commassie staining. U and -, uninduced lysate or buffer, respectively, was added in place of induced lysates. (B) Summary of AUC interaction studies. Left, eIF3c-NTD fragments used are shown with bars indicating their relative locations in eIF3c primary structure. Second, third, forth and fifth columns list sizes of eIF3c species or complexes formed. -, no complex formation. (C) AUC analysis. The sedimentation coefficient (c(s)) distributions of reactions containing eIF3c-F (red), -B (cyan) and -A (blue), either alone (panel 1) or in the presence of eIF1 (panel 2), eIF5 (panel 3) or both (panel 4). Panel 5, eIF1 (pink), eIF5 (orange) and the mixture thereof (green). Proposed peak assignments are presented for each experiment. (D) Schematic illustration of the proposed 4. 6S trimeric complex. eIF3c-NTD is drawn as blue orange line representing unstructured segments, 3c0 (aa. 1–58), and orange circles representing 3c1 (aa. 59–87) and 3c2 (aa. 105–159), as found in cryo-EM models in Fig. 2 and redefined based on 15N-eIF3c-B CSP studies (Fig. 3). eIF5 (dark green circle) is depicted as contacting both ends of eIF3c-NTD. eIF1 (brown circle) is bound to 3c1 via R53 and L96 (labeled). K60 and loop-1 (l-1) of eIF1 are open for 40S binding (arrows). E51, showing CSP with eIF1; Y--WF; Y70, W74, F75, showing line broadening with eIF1 (Fig. 1E and S3). Numbers along eIF3c schematics indicate boundaries of eIF3c units. Dotted arrow indicates the interaction between eIF1-L96 and eIF5, suggested here to stabilize the trimeric complex, as it does in the scanning PIC (Luna et al., 2012; Reibarkh et al., 2008). (E) Locations of eIF2 ternary complex (blue drawing) (Llacer et al., 2015), eIF5-CTD (dark green circle, this study) and 3c0 (aa. 1–58) (orange line, this study) are superimposed onto the re-calculated cryo-EM structure, as shown in Fig. 2B, right. See also Figure S8 and Table S1.
To corroborate these findings, we used analytical ultracentrifugation (AUC) to examine the size and hence, stoichiometry, of complexes formed by eIF1, eIF5 and different eIF3c-NTD fragments. When tested alone, eIF3c fragments A1–163, F1–87, and B36–163 display single peaks (Fig. 7C, panel 1), ranging in size from 1.7S to 3.0S (Fig. 7B). Addition of eIF1 allowed formation of a dimeric complex with each eIF3c fragment (Fig. 7C, panel 2, and 7B), as expected from their high affinity for eIF1 (Fig. 1A). In assays where eIF5 fragments were included (Fig. 7C, panel 3), F1–87 bound eIF5 partially, whereas B36–163, lacking the eIF5 binding site in 3c0, did not bind eIF5 at all (red and cyan lines). Interestingly, eIF5 assembled into a 4.3S complex with A1–163, leaving no unbound fragment A1–163 (blue line). This strong interaction with eIF5 requires 3c2 devoid of F1–87 (red line).
As shown in Fig. 7C, panel 4, the AUC assay confirmed formation of a stable 4.6S trimeric complex comprised of eIF1, eIF5 and eIF3c-A1–163 (blue line) (Asano et al., 2000; Singh et al., 2004). In contrast, eIF1, eIF5 and eIF3c-F1–87 did not assemble into a trimeric complex (Fig. 7C, panel 4, red line), even though eIF1 and F1–87 formed a 1.7S complex. These results support competition by eIF1 and eIF5 for 3c0 binding, which can be relieved by 3c2 present in eIF3c-A1–163 but not eIF3c-F1–87.
When bound to eIF3c-B36–163 defective in eIF5-binding, eIF1 was unable to bind eIF5, and free eIF5 and the B36–163:eIF1 complex were found co-sedimenting at ~3S (Fig. 7C, panel 4, cyan line). Thus, forming the trimeric complex requires eIF5 interaction with the N-terminal region of 3c0 (aa 1–35). Because eIF1 and eIF5 did not form a complex in the absence of eIF3c fragments (Fig. 7C, panel 5), we conclude that the entire eIF3c-NTD (aa. 1–163) bridges these two proteins, with eIF1 bound to its C-terminal portion, as found in eIF3c-D58–163 (Fig. 1A) and (Karásková et al., 2012). The proposed interactions in the eIF5:eIF3c-NTD:eIF1 trimeric complex are depicted in Fig. 7D. Here, it should be noted that the eIF5-CTD:3c0 interaction precludes the 3c0:eIF1 interaction that otherwise competes with eIF1:40S association; and we propose that this stabilizes the scanning PIC. Based on these findings, we suggest approximate locations of the eIF5-CTD and eIF3c0 in the PIC (Fig. 7E) compatible with the proposed roles of these segments in regulating the transition from scanning to start codon recognition.
Discussion
The results of NMR and complementary quantitative binding assays presented in this work revealed two distinct eIF1 complexes formed with overlapping eIF3c-NTD segments that appear to function at different stages of the initiation pathway. The C-terminal segment of the eIF3c-NTD (fragment D59–163) contains the core eIF1-binding unit 3c1 (aa 59–87) and the adjacent globular domain 3c2 (aa 105–159), which bind to a limited surface on eIF1, including R53 and L96, in a manner compatible with eIF1 binding to the 40S subunit (Fig. 4A and C). We have assigned two densities projecting from the main body of eIF3 in the eIF1:eIF3:40S cryo-EM structure (Erzberger et al., 2014) as 3c1, which contacts eIF1, and 3c2 interacting with uS15 (Fig. 2). In contrast, eIF3c fragment C36–87, containing 3c1 and the C-terminal half of 3c0, interacts with a broader surface of eIF1 that includes R53 and L96 but additionally contains residues surrounding the two 40S binding sites at the C-terminus of α1 and loop-1 (Fig. 4C and B). Based on the Ssu− phenotype of a mutation in Box6 (aa 51–60) within 3c0, we propose that interaction of eIF1 with 3c0 occludes the 40S-binding surface in eIF1 and thus facilitates eIF1 dissociation at the start codon—the event diminished at UUG codons by the Box6R Ssu− mutation. This destabilizing effect is likely to be opposed in the scanning PIC through eIF5-CTD binding to 3c0, which shifts eIF1 interaction from eIF3c-NTD elements 3c0/3c1 to 3c1/3c2 and thereby eliminates occlusion of the 40S binding surface on eIF1 by segment 3c0 (Fig. 7D). Dissolving the eIF5-CTD:3c0 interaction thus emerges as a key step in the transition from the open to closed conformation of the PIC, and we propose a plausible mechanism for this rearrangement below.
In agreement with our proposal that the 3c1/3c2 segments of eIF3c-NTD cooperate to anchor eIF1 on the scanning PIC, eIF1 substitution L96P, which perturbs the interface with 3c1, reduces eIF1 binding to the eIF3c-NTD. By also impairing eIF1 binding to the eIF5-CTD, L96P dramatically elevates UUG initiation in the manner expected for destabilization of the scanning complex (Martin-Marcos et al., 2013). eIF1 substitution R53S, which affects the neighboring surface in helix α1, dramatically reduces eIF3c-NTD binding but does not substantially impair the eIF1:eIF5-CTD interaction. Because R53S elevates UUG initiation only when combined with the α1 substitution K56A, which weakens eIF1:40S interaction, we conclude that a network of eIF1 interactions with the eIF3c-NTD, eIF5-CTD, and 40S subunit cooperate to anchor eIF1 to the scanning PIC and block initiation at non-AUG codons (Fig. 5A). Based on the cryo-EM model in Fig. 2A, the role of 3c2 in anchoring eIF1 to the PIC appears to be indirect. Consistently, mutations altering Box12Sui+ (aa 111–120) within 3c2 can elevate UUG initiation by either increasing or decreasing eIF1 retention in native PICs (Karásková et al., 2012). This complexity may reflect dual role of 3c2 in promoting eIF1 binding to segment 3c1 and eIF5-CTD binding to 3c0 in the scanning PIC, while preventing the more stable eIF1 complex formed with 3c0/3c1 on AUG recognition. In addition, by directly contacting 40S protein uS15/S13, 3c2 is likely to stabilize eIF1 binding to the scanning PIC (Fig. 2A).
Recent studies reveal structural rearrangements within 43S/48S PICs between different steps of initiation (Hussain et al., 2014; Llacer et al., 2015; Simonetti et al., 2016). However, it is unclear exactly how start codon selection induces transition from the open to closed conformations of the PIC. Based on our findings and other work done using yeast S. cerevisiae as a model system, we propose that the eIF1A-NTT plays such a signaling role (Saini et al., 2010) (Fig. 8). During mRNA scanning, eIF1A-NTT interaction with the eIF5-CTD helps to retain eIF1 in the PIC (Luna et al., 2013) (Fig. 8a). Thus, in addition to binding the 3c0 element and eIF1, the eIF5-CTD binds the basic eIF1A-NTT through a distinct acidic surface. This interaction is also important as it antagonizes eIF5-CTD interaction with the positively charged eIF2β-NTT, which would otherwise promote eIF1 release (Luna et al., 2012; Nanda et al., 2013). On Met-tRNAiMet anticodon pairing to AUG, eIF1A-NTT binds to the codon:anticodon duplex in the P-site (Hussain et al., 2014) (Fig. 8b). This releases the eIF5-CTD for interaction with eIF2β-NTT, which in turn disrupts eIF5-CTD interaction with both eIF1 (Luna et al., 2012) and 3c0 (Fig. 8b–c). The 3c0 segment is now free to engage eIF1 and occlude its ribosome-binding surface, interfering with eIF1 re-association with the 40S subunit and thus allowing Met-tRNAiMet to remain stably anchored in the PIN state (Fig. 8d). These effects are expected to amplify the subtle distortion of eIF1 structure and perturbation of its 40S binding site that accompanies Met-tRNAiMet isomerization to the PIN state (Hussain et al., 2014). In this way, 3c0 ensures irreversible eIF1 release from the decoding center in response to AUG recognition and subsequent closure of the ribosome structure and formation of the 40S initiation complex.
Fig. 8. Model of MFC rearrangement during translation initiation.
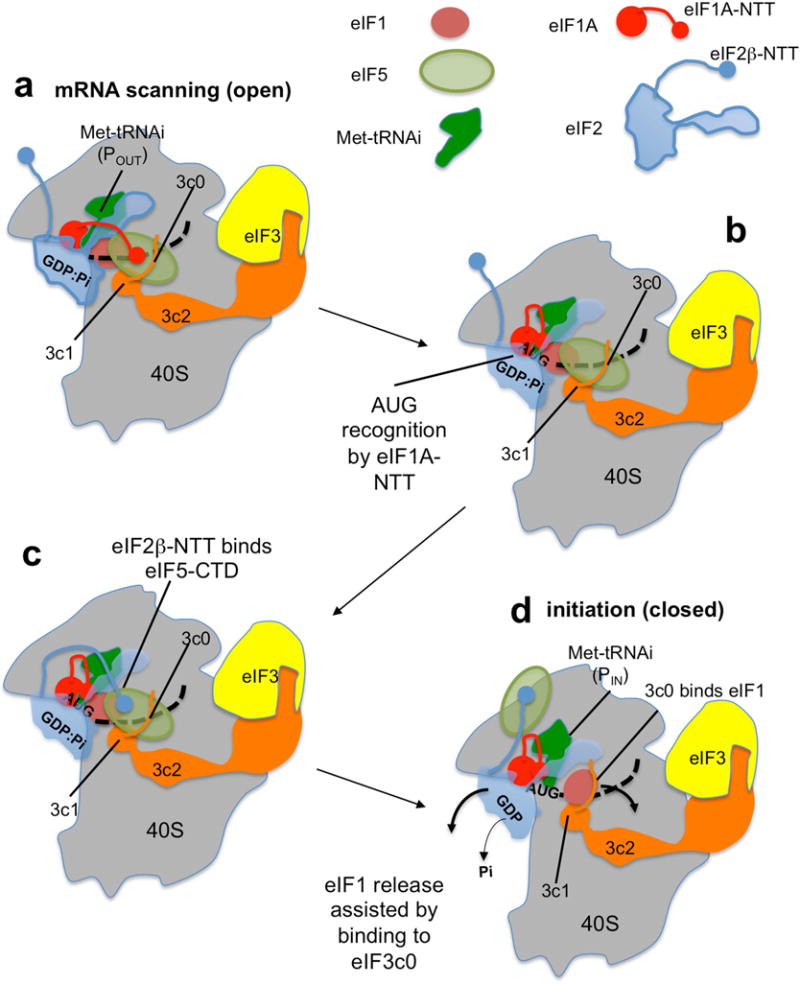
(a) During mRNA (dotted line) scanning, eIF5 and eIF3c-NTD play crucial roles in eIF1 anchoring. eIF5 and 3c1 directly bind eIF1 to anchor it to the PIC (this study). eIF1 maintains the PIC in the open conformation and prevents Met-tRNAi to accommodate in the P-site (POUT). eIF1A-NTT also binds eIF5 (Luna et al., 2013), preventing its binding by eIF2β-NTT. The binding partner of eIF2β-NTT at this stage may be eIF1 (Nanda et al., 2013) or alternatively, rRNA or mRNA, as it binds RNA (Singh et al., 2012). (b) Met-tRNAi base-pairing to the AUG codon causes a scanning arrest. This is enhanced by eIF1A-NTT binding to the codon:anticodon duplex, resulting in eIF1 distortion (Hussain et al., 2014; Llacer et al., 2015). eIF5 is now available for eIF2β-NTT binding. (c) eIF2β-NTT binds eIF5, resulting in disruption of eIF5 binding to eIF1 (Luna et al., 2012) and 3c0. (d) 3c0 assists eIF1 release by binding to its ribosome binding site (this study). tRNAiMet bound to the start codon positions in the P-site (PIN). eIF1 release is followed by Pi release from eIF2 (Algire et al., 2005), promoting ejection of eIF2:GDP in complex with eIF5 (Singh et al., 2006). The model that eIF1 remains associated with eIF3 after its release from the 40S decoding site was previously proposed (Karásková et al., 2012; Singh et al., 2012).
It is noteworthy that human eIF1 also binds eIF3c-NTD (Fletcher et al., 1999) and eIF5-CTD (Luna et al., 2012). While the eIF3c-NTD segments corresponding to 3c0 are shorter in animals and plants, they contain an acidic element similar to Box6, lying next to the conserved core region 3c1 (Boxed in Fig. S1B). Moreover, eIF3c-NTD in animals and plants is predicted to form an α-helical structure, as found in yeast 3c2 (Fig. 1C). Further work on the human and yeast systems is expected to reveal Eukarya-wide conservation of the MFC’s role in promoting scanning and AUG selection through the coordinated interactions of the eIF3c-NTD with eIF1, eIF5 and potentially other parts of eIF3 (Hussain et al., 2014; Simonetti et al., 2016; Valásek et al., 2003).
Experimental Procedures
Protein purification and yeast methods
Isotopically labeled or unlabeled proteins were expressed in E. coli transformants carrying appropriate plasmids (Table S1) and purified as described in Supplementary text. Yeast Saccharomyces cerevisiae strains used in this study are constructed as described in Supplementary text, and listed in Table S2. Standard yeast molecular biology methods including growth and β-galactosidase assays were used throughout (Lee et al., 2007) (see Supplemental Experimental Procedures for details).
Biophysical methods
ITC, NMR spectroscopy, FA and AUC are all performed as described in Supplemental Experimental Procedures. Detailed NMR data and structural statistics for eIF3c-B36–163 and eIF1 are summarized in Table S3 and S4, respectively. We re-ran integrative modeling prediction including the new information from the eIF3c-B NMR structure, with parameters and methods identical to those previously described (Erzberger et al., 2014).
Supplementary Material
Highlights.
eIF3c N-terminal domain is divided into three regions, 3c0, 3c1 and 3c2.
3c1 and eIF5 anchor eIF1 to the 40S ribosome during mRNA scanning.
On AUG, 3c0 binds eIF1 ribosome-binding site, facilitating eIF1 release.
eIF5 prevents 3c0 from binding eIF1 before AUG selection.
Acknowledgments
We thank Hiroshi Matsuo and Erin Adamson for comments, Ivan Topisirovic for proof reading and Michaela Flax and Speed Rogers for technical assistance. This work was supported by a grant from NIH (R01 GM64781), a pilot grant from University of Kansas COBRE-PSF (P30 GM 110761), and an Innovative Award from KSU Terry Johnson Cancer Center to KA; CA68262 and GM47467 to GW; an intramural grant from NICHD, NIH, to AGH; and JSPS KAKENHI 15H01634 and 26440026 to TN.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
EO, REL, TN, PMM, HH, CRS, JPE and KA designed, performed experiments and analyzed data. FZ, HA, and JM performed experiments and analyzed data. RP, SU, FS, and TU analyzed data. CM, IH, EP, HY, MLN, BT, EA, SH, ED, AN, and PG performed experiments. REL, AGH, GW and KA wrote the paper.
References
- Algire MA, Maag D, Lorsch JR. Pi release from eIF2, not GTP hydrolysis, is the step controlled by start-site selection during eukaryotic translation initiation. Mol Cell. 2005;20:251–262. doi: 10.1016/j.molcel.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Asano K. Why is start codon selection so precise in eukaryotes? Translation. 2014;2:e28387. doi: 10.4161/trla.28387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano K, Clayton J, Shalev A, Hinnebusch AG. A multifactor complex of eukaryotic initiation factors eIF1, eIF2, eIF3, eIF5, and initiator tRNAMet is an important translation initiation intermediate in vivo. Genes Dev. 2000;14:2534–2546. doi: 10.1101/gad.831800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano K, Phan L, Valasek L, Schoenfeld LW, Shalev A, Clayton J, Nielsen K, Donahue TF, Hinnebusch AG. A multifactor complex of eIF1, eIF2, eIF3, eIF5, and tRNAiMet promotes initiation complex assembly and couples GTP hydrolysis to AUG recognition. Cold Spring Harbor Symposia on Quantitative Biology. 2001a;66:403–415. doi: 10.1101/sqb.2001.66.403. [DOI] [PubMed] [Google Scholar]
- Asano K, Shalev A, Phan L, Nielsen K, Clayton J, Valasek L, Donahue TF, Hinnebusch AG. Multiple roles for the carboxyl terminal domain of eIF5 in translation initiation complex assembly and GTPase activation. EMBO Journal. 2001b;20:2326–2337. doi: 10.1093/emboj/20.9.2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano K, Vornlocher H-P, Richter-Cook NJ, Merrick WC, Hinnebusch AG, Hershey JWB. Structure of cDNAs encoding human eukaryotic initiation factor 3 subunits: possible roles in RNA binding and macromolecular assembly. J Biol Chem. 1997;272:27042–27052. doi: 10.1074/jbc.272.43.27042. [DOI] [PubMed] [Google Scholar]
- Aylett CHS, Boehringer D, Erzberger JP, Schaefer T, Ban N. Structure of a Yeast 40S–eIF1–eIF1A–eIF3–eIF3j initiation complex. Nat Struc Mol Biol. 2015;22:269–271. doi: 10.1038/nsmb.2963. [DOI] [PubMed] [Google Scholar]
- Cheung Y-N, Maag D, Mitchell SF, Fekete CA, Algire MA, Takacs JE, Shirokikh N, Pestova T, Lorsch JR, Hinnebusch A. Dissociation of eIF1 from the 40S ribosomal subunit is a key step in start codon selection in vivo. Genes Dev. 2007;21:1217–1230. doi: 10.1101/gad.1528307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis MD, Person MD, Browning KS. Phosphorylation of plant translation initiation factors by CK2 enhances the in vitro interaction of multifactor complex components. J Biol Chem. 2009;284:20615–20628. doi: 10.1074/jbc.M109.007658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erzberger JP, Stengel F, Pellarin R, Zhang S, Schaefer T, Aylett CHS, Cimermancic P, Boehringer D, Sali A, Aebersold R, et al. Molecular Architecture of the 40S,eIF1,eIF3 Translation Initiation Complex. Cell. 2014;158:1123–1135. doi: 10.1016/j.cell.2014.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher CM, Pestova TV, Hellen CUT, Wagner G. Structure and interactions of the translation initiation factor eIF1. EMBO Journal. 1999;18:2631–2639. doi: 10.1093/emboj/18.9.2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG. The Scanning Mechanism of Eukaryotic Translation Initiation. Annu Rev Biochem. 2014;83:779–812. doi: 10.1146/annurev-biochem-060713-035802. [DOI] [PubMed] [Google Scholar]
- Huang H, Yoon H, Hannig EM, Donahue TF. GTP hydrolysis controls stringent selection of the AUG start codon during translation initiation in Saccharomyces cerevisiae. Genes Dev. 1997;11:2396–2413. doi: 10.1101/gad.11.18.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain T, Llácer JL, Fernández IS, Munoz A, Martin-Marcos P, Savva CG, Lorsch JR, Hinnebusch AG, Ramakrishnan V. Structural changes enable start codon recognition by the eukaryotic translation initiation complex. Cell. 2014;159:597–607. doi: 10.1016/j.cell.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karásková M, Gunisˇová S, Herrmannová A, Wagner S, Munzarová V, Valásˇek LS. Functional Characterization of the Role of the N-terminal Domain of the c/Nip1 Subunit of Eukaryotic Initiation Factor 3 (eIF3) in AUG Recognition. J Biol Chem. 2012;287:28420–28434. doi: 10.1074/jbc.M112.386656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Hellen CUT, Pestova TV. Toward the mechanism of eIF4F-mediated ribosomal attachment to mammalian capped mRNAs. Genes Dev. 2016;30:1573–1588. doi: 10.1101/gad.282418.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B, Udagawa T, Singh CS, Asano K. Yeast phenotypic assays on translational control. Methods Enzymol. 2007;429:139–161. doi: 10.1016/S0076-6879(07)29006-8. [DOI] [PubMed] [Google Scholar]
- Llacer JL, Hussain T, Marler L, Aitken CE, Thakur A, Lorsch JR, Hinnebusch AG, Ramakrishnan V. Conformational Differences between Open and Closed States of the Eukaryotic Translation Initiation Complex. Mol Cell. 2015;59:399–412. doi: 10.1016/j.molcel.2015.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomakin IB, Steitz TA. The initiation of mammalian protein synthesis and mRNAscanningmechanism. Nature. 2013;500:307–311. doi: 10.1038/nature12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna RE, Arthanari H, Hiraishi H, Akabayov B, Tang L, Cox C, Markus MA, Luna LE, Ikeda Y, Watanabe R, et al. The interaction between eukaryotic initiation factor 1A and eIF5 retains eIF1 within scanning preinitiation complexes. Biochemitry. 2013;52:9510–9518. doi: 10.1021/bi4009775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna RE, Arthanari H, Hiraishi H, Nanda J, Martin-Marcos P, Markus M, Arabayov B, Milbradt A, Luna LE, Seo H-C, et al. The C-terminal domain of eukaryotic initiation factor 5 promotes start codon recognition by its dynamic interplay with eIF1 and eIF2β Cell Reports. 2012;1:689–702. doi: 10.1016/j.celrep.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maag D, Fekete CA, Gryczynski Z, Lorsch JR. A conformational change in the eukaryotic translation preinitiation complex and release of eIF1 signal recognition of the start codon. Molecular Cell. 2005;17:265–275. doi: 10.1016/j.molcel.2004.11.051. [DOI] [PubMed] [Google Scholar]
- Martin-Marcos P, Nanda J, Luna LE, Wagner G, Lorsch JR, Hinnebusch AG. β-hairpin loop of eIF1 mediates 40S ribosome binding to regulate initiator tRNAMet recruitment and accuracy of AUG selection in vivo. J Biol Chem. 2013;288:27546–27562. doi: 10.1074/jbc.M113.498642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Marcos P, Nanda JS, Luna RE, Zhang F, Saini AK, Cherkasova VA, Wagner G, Lorsch JR, Hinnebusch AG. Enhanced eIF1 binding to the 40S ribosome impedes conformational rearrangements of the preinitiation complex and elevates initiation accuracy. RNA. 2014;20:150–167. doi: 10.1261/rna.042069.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meleppattu S, Kamus-Elimeleh D, Zinoviev A, Cohen-Mor S, Orr I, Shapira M. The eIF3 complex of Leishmania-subunit composition and mode of recruitment to different cap-binding complexes. Nucl Acids Res. 2015;43:6222–6235. doi: 10.1093/nar/gkv564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanda J, Saini AK, Munoz AM, Hinnebusch AG, Lorsch JR. Coordinated Movements of Eukaryotic Translation Initiation Factors eIF1, eIF1A and eIF5 Trigger Phosphate Release from eIF2 in response to Start Codon Recognition by the Ribosomal Pre-initiation Complex. J Biol Chem. 2013;288:5316–5329. doi: 10.1074/jbc.M112.440693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Kolupaeva VG. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes and Development. 2002;16:2906–2922. doi: 10.1101/gad.1020902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan L, Zhang X, Asano K, Anderson J, Vornlocher HP, Greenberg JR, Qin J, Hinnebusch AG. Identification of a translation initiation factor 3 (eIF3) core complex, conserved in yeast and mammals, that interacts with eIF5. Mol Cell Biol. 1998;18:4935–4946. doi: 10.1128/mcb.18.8.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabl J, Leibundgut M, Ataide SF, Haag A, Ban N. Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiaiton factor 1. Science. 2011;331:730–736. doi: 10.1126/science.1198308. [DOI] [PubMed] [Google Scholar]
- Reibarkh M, Yamamoto Y, Singh CR, Rio Fd, Fahmy A, Lee B, Luna RE, Ii M, Wagner G, Asano K. Eukaryotic initiation factor (eIF) 1 carries two distinct eIF5-binding faces important for multifactor assembly and AUG selection. J Biol Chem. 2008;283:1094–1103. doi: 10.1074/jbc.M708155200. [DOI] [PubMed] [Google Scholar]
- Saini AK, Nanda JS, Lorsch JR, Hinnebusch AG. Regulatory elements in eIF1A control the fidelity of start codon selection by modulating tRNAiMet binding to the ribosome. Genes Dev. 2010;24:97–110. doi: 10.1101/gad.1871910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonetti A, Querido JB, Myasnikov AG, Mancera-Martinez E, Renaud A, Kuhn L, Hashem Y. eIF3 Peripheral Subunits Rearrangement after mRNA Binding and Start-Codon Recognition. Mol Cell. 2016;63:206–217. doi: 10.1016/j.molcel.2016.05.033. [DOI] [PubMed] [Google Scholar]
- Singh CR, Lee B, Udagawa T, Mohammad-Qureshi SS, Yamamoto Y, Pavitt GD, Asano K. An eIF5/eIF2 complex antagonizes guanine nucleotide exchange by eIF2B during translation initiation. EMBO J. 2006;25:4537–4546. doi: 10.1038/sj.emboj.7601339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh CR, Watanabe R, Chowdhury D, Hiraishi H, Murai MJ, Yamamoto Y, Miles D, Ikeda Y, Asano M, Asano K. Sequential eIF5 binding to the charged disordered segments of eIF4G and eIF2β stabilizes the 48S pre-initiation complex and promotes its shift to the initiation mode. Mol Cell Biol. 2012;32:3978–3989. doi: 10.1128/MCB.00376-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh CR, Yamamoto Y, Asano K. Physical association of eukaryotic initiation factor 5 (eIF5) carboxyl terminal domain with the lysine-rich eIF2b segment strongly enhances its binding to eIF3. Journal of Biological Chemistry. 2004;279:49644–49655. doi: 10.1074/jbc.M409609200. [DOI] [PubMed] [Google Scholar]
- Sokabe M, Fraser CS, Hershey JW. The Human Translation Initiation Multi-Factor Complex Promotes Methionyl-tRNAi Binding to the 40S Ribosomal Subunit. Nucl Acids Res. 2012;40:905–913. doi: 10.1093/nar/gkr772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava S, Verschoor A, Frank J. Eukaryotic initiation factor 3 does not prevent association through physical blockage of the ribosomal subunit-subunit interface. J Mol Biol. 1992;220:301–304. doi: 10.1016/0022-2836(92)90946-h. [DOI] [PubMed] [Google Scholar]
- Valásek L, Mathew AA, Shin BS, Nielsen KH, Szamecz B, Hinnebusch AG. The yeast eIF3 subunits TIF32/a, NIP1/c, and eIF5 make critical connections with the 40S ribosome in vivo. Genes and Development. 2003;17:786–799. doi: 10.1101/gad.1065403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valásek L, Nielsen KH, Zhang F, Fekete CA, Hinnebusch AG. Interaction of eIF3 subunit NIP1/c with eIF1 and eIF5 promote preinitiation complex assembly and regulate start codon selection. Molecular and Cellular Biology. 2004;24:9437–9455. doi: 10.1128/MCB.24.21.9437-9455.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisser M, Voigts-Hoffmann F, Rabl J, Leibundgut M, Ban N. The crystal structure of the eukaryotic 40S ribosomal subunit in complex with eIF1 and eIF1A. Nat Struct Mol Biol. 2013;20:1015–1017. doi: 10.1038/nsmb.2622. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Singh CR, Marintchev A, Hall NS, Hannig EM, Wagner G, Asano K. The eukaryotic initiation factor (eIF) 5 HEAT domain mediates multifactor assembly and scanning with distinct interfaces to eIF1, eIF2, eIF3 and eIF4G. Proc Natl Acad Sci USA. 2005;102:16164–16169. doi: 10.1073/pnas.0507960102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


