Summary
Porphyromonas gingivalis, an asaccharolytic bacterium, utilizes amino acids as energy and carbon sources. Since amino acids are incorporated into the bacterial cells mainly as di- and tri-peptides, exopeptidases including dipeptidyl-peptidase (DPP) and tripeptidyl-peptidase are considered to be prerequisite components for their metabolism. We recently discovered DPP11, DPP5, and acylpeptidyl oligopeptidase in addition to previously reported DPP4, DPP7, and prolyl tripeptidyl peptidase A. DPP11 is a novel enzyme specific for acidic P1 residues (Asp and Glu) and distributed ubiquitously in eubacteria, while DPP5 is preferential for the hydrophobic P1 residue and the first entity identified in prokaryotes. Recently, acylpeptidyl oligopeptidase with a preference for hydrophobic P1 residues was found to release N-terminally blocked di- and tri-peptides. Furthermore, we also demonstrated that gingipains R and K contribute to P1-basic dipeptide production. These observations implicate that most, if not all, combinations of di- and tri-peptides are produced from extracellular oligopeptides even with an N-terminal modification. Here, we review P. gingivalis exopeptidases mainly in regard to their enzymatic characteristics. These exopeptidases with various substrate specificities benefit P. gingivalis for obtaining energy and carbon sources from the nutritionally limited subgingival environment.
Abbreviations: DPP, dipeptidyl peptidase; PtpA, prolyl tripeptidyl peptidase A; AOP, acylpeptidyl oligopeptidase; Rgp, gingipain R; Kgp, gingipain K; MCA, 4-methycoumaryl-7-amide; Z-, benzyloxycarbonyl-
Keywords: Periodontitis, Amino acid metabolism, Dipeptidyl peptidase (DPP), Exopeptidase, Porphyromonas gingivalis
1. Introduction
Porphyromonas gingivalis, a Gram-negative black-pigmented anaerobe, is a major causative agent of chronic periodontitis [1], which leads to permanent tooth loss [2]. Recently, much attention has been paid to this bacterium and other periodontopathic ‘red complex species’ (Tannerella forsythensis, Treponema denticola) [3], because of their close relationships to systemic diseases, such as atherosclerotic cardiovascular disorder [4], [5], [6], decreased kidney function [7], and rheumatoid arthritis [8].
A common feature among these bacteria is that they do not ferment glucose or sucrose (asaccharolytic), but rather utilize amino acids as energy and carbon sources [9], [10], [11]. In P. gingivalis, nutritional extracellular proteins are initially degraded to oligopeptides by potent cysteine endopeptidases, i.e., gingipains R (Rgp) and K (Kgp) [12], [13], [14], then oligopeptides are degraded to di- and tri-peptides, the main incorporated forms in P. gingivalis [15], [16]. As for the amino acid transport system, an analysis of the P. gingivalis genome indicated the existence of two types of oligopeptide transporters [10], which are considered to mediate di- and tri-peptide incorporation. In addition, a sodium ion-driven serine/threonine transporter with a sequence similar to that of the Escherichia coli serine transporter has been reported [17]. In this context, exopeptidases consisting of dipeptidyl peptidases (DPPs), tripeptidyl peptidase, and acylpeptidyl oligopeptidase (AOP) producing di- and tri-peptides from oligopeptides are viewed as important for P. gingivalis to acquire proteinaceous nutrition from the mixed-species environment of the subgingival sulcus.
Initially, DPP4, DPP7, and prolyl tripeptidyl peptidase A (PtpA) were the only exopeptidases identified in P. gingivalis. These share substrates according to their altered specificities, as DPP4 is highly specific for Pro at the penultimate position from the N-terminus (P1 position), though it accepts Ala to a lesser extent [18], [19], DPP7 is preferential to P1 hydrophobic amino acids [20], and PtpA liberates tripeptides with P1-position Pro, an activity that is able to compensate DPP4 and DPP7, which are unable to accept oligopeptides with Pro at the third position [21]. Although these three exopeptidases could not sufficiently explain the entire metabolism of extracellular oligopeptides, no other members were added to the list of P. gingivalis exopeptidases for a period of 10 years. Even though Asp and Glu are located in central routes of metabolism in P. gingivalis [10], [15], [22] (Fig. 1), oligopeptides with acidic amino acid residues do not seem to be efficiently produced. Moreover, none of DPPs [19], [21], [23] and PtpA [20] are incapable of utilizing N-terminally blocked polypeptides, such as the various serum proteins present in gingival crevicular fluid.
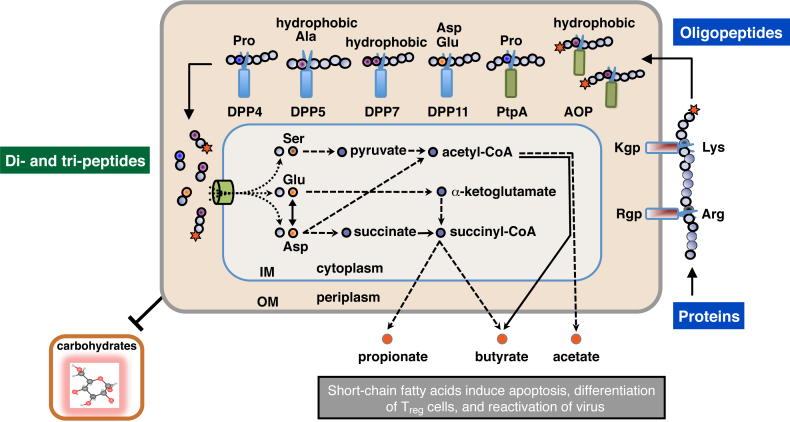
Figure 1.
Schematic illustration of extracellular oligopeptide metabolism in P. gingivalis. The metabolic pathway of P. gingivalis from the extracellular polypeptides, di- and tri-peptide incorporation, amino acid metabolism, and excretion as short-chain fatty acids, are schematically illustrated [10], [15]. Amino acids, except for Ser and Thr, are mainly transported as di- and tri-peptides via oligopeptide transporters [10]. Rgp and Kgp are mainly localized on the outer membrane (OM), while DPPs, PtpA, and AOP are located in periplasmic space [23], [24], [25]. Scissors indicate peptidases, which cleave peptide bonds at specific positions. Stars represent acylaminoacyl groups at the N-terminus. IM, inner membrane.
The majority of these disadvantages have been overcome by recent discoveries such as a novel Asp/Glu-specific DPP, DPP11 [23], DPP5, which is specific for hydrophobic P1 amino acid [24], and AOP in P. gingivalis [25]. Furthermore, we demonstrated that gingipains are able to produce dipeptides from oligopeptides [24]. These peptidases with various substrate specificities benefit P. gingivalis for colonization in nutrition-limited subgingival environments.
2. Peptidases involved in degradation of extracellular proteinaceous nutrients
2.1. Gingipains
P. gingivalis produces substantial quantities of cysteine endopeptidases that cleave peptide substrates with basic residues at the P1 position [12], [13], [14]. These peptidases, termed Rgp and Kgp, have been shown to be major virulence factors of the species. Gingipains are able to degrade many human proteins including complement system proteins, cytokines, and integrin. Their potent activities are implicated in most phases of the pathogenesis of periodontal disease, from adherence and colonization through nutrient acquisition and neutralization of host defense.
The gingipain null (rgpA-, rppB-, kgp-) mutant KDP136 was reported unable to grow in defined medium with human albumin as the sole carbon source [26]. Comparative studies of the virulence of isogenic mutants lacking RgpA, RgpB, and Kgp indicated that Kgp contributed more to the pathogenicity of P. gingivalis in murine lesion and periodontal models [27], though Rgp activity was always found to be at a level higher than Kgp activity [28]. The roles of gingipains as virulence factors have been the focus point of many reviews [29], [30], [31], and will not be further described in this article.
2.1.1. Rgp
Rgp (EC 3.4.22.37) is classified in MEROPS as clan CD, family C25, peptidase C25.001 [32]. Its specificity is highly limited to the Arg-|-Xaa bond with no particular preference for P2 and P1′ sites [14], [33]. Two closely related genes, rgpA and rgpB, are present in the P. gingivalis genome, and possibly developed by gene duplication and rearrangement. The rgpA gene translation product, RgpA, is composed of 1703 amino acids, consisting of the propeptide, a catalytic domain, four adhesin domains, and a C-terminal domain. RgpB, encoded by rgpB, is composed of 736 amino acids, consisting of the propeptide, and catalytic and C-terminal domains. Heterogeneous molecules with Arg-specific activity have been observed in both P. gingivalis bacterial cells and culture supernatant samples containing vesicles. The heterogeneity in the Rgp molecules was implicated in the existence of the two genes, auto-proteolysis occurring at the domain junctions, complex formation between the catalytic and adhesion domains, and LPS association [34].
2.1.2. Kgp
Kgp (EC 3.4.22.47), classified as clan CD, family C25, peptidase C25.002, is specific for the P1 Lys residue [35], [36] and encoded by a genomic single gene, kgp [37]. The translation product of kgp is composed of 1723 amino acids, producing a multi-domain protein similar to RgpA, and consists of the propeptide, a caspase-like catalytic domain, four adhesin domains, and a C-terminal domain. Kgp possesses an additional putative immunoglobulin-like domain located between the caspase-like and first adhesion domains. Soluble and cell-associated forms of Kgp are detected in the same manner as those of Rgp [33].
2.2. Exopeptidases
Extracellular oligopeptides produced by gingipains are converted into di- or tri-peptides in the periplasm space (Fig. 1), then incorporated into bacterial cell across the inner membrane. These peptides are finally hydrolyzed in the cytoplasm into single amino acids by dipeptidase, aminopeptidase, and carboxyl peptidase, and further converted into metabolic end-products, such as ammonia and butyrate [15], [16], which are considered to be virulence factors that cause host tissue damage [38], [39], [40]. Accordingly, periplasmic exopeptidases, i.e., DPP4, DPP5, DPP7, DPP11, and AOP that produce di- and tri-peptides are considered to play critical roles in both cell growth and pathogenicity.
2.2.1. DPP4
The serine exopeptidase DPP4 hydrolyzes the peptide bond at the carboxyl side of Pro (NH2-Xaa1-Pro2-|-Xaa3-) and is classified as clan SC, family S9, subfamily S9B, peptidase S09.013. Pro2 residue can be substituted by Ala or hydroxyproline, though the rate of hydrolysis is substantially lower than that of Pro2. P. gingivalis DPP4 is composed of 723 amino acids with calculated masses of 82,018 (precursor) and 80,235 (mature form) [41]. The growth of dpp4-, dpp7, or ptpA single-knockout strains was shown to be comparable with that of wild-type strain W83, while that of the triple-knockout mutant was retarded [42]. P. gingivalis DPP4 also hydrolyzes biologically active peptides that include substance P, fibrin inhibitory peptide, and β-casomorphin [19], and binds to fibronectin, thus it mediates bacterial adhesion to host cells [43]. A recent study also demonstrated that DPP4 was closely associated with biofilm formation in a murine subcutaneous abscess model [44].
2.2.2. DPP5
DPP5, classified as peptidase S9.012, was first isolated from Aspergillus fumigatus culture supernatant samples [45] and, until our discovery of P. gingivalis DPP5, species carrying this exopeptidase were restricted to the fungi A. fumigatus, Aspergillus orizae (koji fungus, used for fermentation of soybeans), and Microsporum canis, which causes the skin disease dermatophytosis [46]. A. fumigatus DPP5 has been reported to be frequently detected in mycelium of patients with invasive aspergillosis and aspergilloma, and was reported to be one of the major antigens in immunocompetent patients with aspergilloma [47].
In a report published in 2014, we noted that a P. gingivalis ATCC 33277 dpp4-dpp7-dpp11 triple-knockout strain (NDP511) still possessed some hydrolyzing activities for Met-Leu-, Lys-Ala-, Gly-Phe-, and Ser-Tyr-4-methylcoumaryl-7-amide (MCA), indicating the expression of an unidentified DPP with a preference for the hydrophobic P1 residue [24]. In order to identify this speculated DPP, we searched for candidates among the 2090 protein-coding sequences of this strain. Consequently, it was found that PGN_0756, tentatively annotated as prolyl oligopeptidase, exhibited DPP activity preferential for Ala and hydrophobic residues at the P1 position. Its amino acid sequence was 28.5% identical to A. fumigatus DPP5. Indeed, the substrate specificity of PGN_0756 was similar to that of fungal DPP5. Hence, we concluded that PGN_0756 represents P. gingivalis DPP5 and should be classified as clan SC, family S9, subfamily C, S09.075 [32]. The discovery of bacterial type DPP5 revealed that this enzyme is distributed from eubacteria, archaea members, and eukaryotes including fungi, as well as higher animals and plants [25].
Although DPP5 and DPP7 showed a hydrophobic P1 preference, the former has no apparent amino acid preference at the P2 position (N-terminus) [25], in contrast to the hydrophobic P2 preference of DPP7 [48]. Thus, DPP5 and DPP7 share substrates based on the differences in their P2 position specificities. For example, DPP7 is able to hydrolyze dipeptidyl MCA substrates harboring non-hydrophobic residues at the P1 position to some extent, such as Leu-Gln- and Leu-Arg-MCA, which are scarcely hydrolyzed by DPP5. In contrast, Lys-Ala- and Gly-Phe-MCA are predominantly hydrolyzed by DPP5.
We previously reported that Porphyromonas endodontalis, a bacterium frequently detected in infected root canals of patients with acute symptoms, showed Lys-Ala-MCA-hydrolyzing activity much greater than that of P. gingivalis [49]. Also, results of a biochemical study that used recombinant peptidases demonstrated that this difference was mainly caused by an 18-fold increase in kcat/Km of P. endodontalis DPP5 as compared to that of P. gingivalis DPP5 [50].
2.2.3. DPP7
DPP7 is classified in MEROPS as clan SC, family S46, subfamily S9B, peptidase S46.001. P. gingivalis DPP7 was the first S46-family peptidase reported that hydrolyzes the peptide bond at the carboxyl side of aliphatic or aromatic residues (Yaa) (NH2-Xaa1-Yaa2-|-Xaa3-), but does not cleave substrates with a blocked N-terminus and long oligopeptides, such as insulin B chain, azocasein, and type I collagen [20]. We recently found that P. gingivalis DPP7 is also preferential for hydrophobic residues at the P2 as well as P1 positions, which are mainly mediated by Phe664 [51].
Unlike DPP4 and DPP5, the species distribution of DPP7 as well as DPP11 seems to be limited to the bacterial kingdom, and DPP7 proteins of P. gingivalis [20], [51], P. endodontalis [49], and Pseudoxanthomonas mexicana WO24 [52] have been biochemically characterized to date. Recently, Ogasawara's group was first to report the crystal structure of P. mexicana DPP7 (named DAPBII). They demonstrated that Gly675 of DAPBII is located at the bottom of a wall of the S1 subsite, which is in accordance with our finding that Gly666 of P. gingivalis DPP7, equivalent to Gly675, is critical for hydrophobic P1 specificity [51].
Because of confusion in nomenclature, human and mouse DPP7 (also known as QPP and DPPII, respectively) are post-Pro aminopeptidase belonging to clan SC, S28.002 [53], [54], and hence, essentially distinct from P. gingivalis DPP7 [20], [51].
2.2.4. DPP11
In P. gingivalis, the gene coding DPP11 (PGN_0607 [55]/PG_1283 [10]) was reported to be an isoform of authentic DPP7 (PGN_1479) [20] or only described as a hypothetical protein [54]. However, we revealed that the gene encodes a novel DPP that hydrolyzes the peptide bond on the carboxyl side of acidic residues (NH2-Xaa1-Asp/Glu2-|-Xaa3-) [23] and newly classified it separately from DPP7 as peptidase S46.002 in clan SC, family S46, subfamily S9B [32].
DPP11 was unexpectedly discovered during our attempt to identify the Lys-Ala-MCA-hydrolyzing entity of P. endodontalis most similar to DPP7 [23]. Accordingly, degenerated PCR cloning was performed with primers designed from the nucleotide sequences of PGN_1479, PGN_0607 and their homologues from other species [20]. As a result, a gene with an open-reading frame encoding 717 amino acids (uploaded as AB610284) was cloned, and its amino acid sequence was shown to be 38.3% and 57.9% identical to that of authentic P. gingivalis DPP7 and the DPP7 isoform, respectively. However, the AB610284 and PGN_0607 recombinant proteins did not hydrolyze any of the 80 commercially available dipeptidyl-MCA substrates examined. Finally, we determined that they were novel enzymes, designated as DPP11, with preference for acidic amino acid P1 residues (Asp and Glu) [23]. Also, hydrophobic residues were found to be preferable at the N-terminus (P2 position). DPP11 does not hydrolyze N-terminally modified substrates, such as acetyl-Leu-Asp-MCA, the same as DPP4 and DPP7. DPP11 orthologues are widely distributed throughout the bacterial kingdom [51].
The acidic P1-position preference of P. gingivalis DPP11 is primarily mediated by Arg673. Asp at the P1 position is more preferential than Glu [23]. In some species, such as Shewanella putrefaciens, Arg673 is substituted by Ser, in which the Asp preference is converted toward Glu [51]. Arg673 of DPP11 is replaced by Gly666 in DPP7 of all species, which allows acceptance of a large hydrophobic residue at the P1 position [23].
We found that a dpp11-knockout strain lost most of its Asp/Glu-dependent DPP activity, while growth was partially (30%) retarded [23]. Because orthologues of peptidase family S46 comprising DPP11 as well as DPP7 are not present in humans, DPP11 may be a potential drug target for chronic periodontitis.
2.2.5. Prolyl tripeptidyl peptidase A (PtpA)
Different from DPPs, which are unable to hydrolyze oligopeptides with Pro at the third position from the N-terminus, PtpA, classified as Clan SC, family S9, subfamily B; S09.017, hydrolyzes the Pro3-Xaa4 bond and liberates an N-terminal tripeptide (NH2-Xaa1-Xaa2-Pro3) [21]. An unblocked N-terminus is absolutely required for cleavage and no cleavage occurs with substrates with Pro at the P2 position [56]. Synthetic substrates Ala-Ala-Pro- and Gly-Ala-Pro-p-nitroanilide are suitable for measurement of the activity [21]. PtpA is possibly related to degradation of type I collagen carrying the tri-peptidyl repeat (-Gly-Xaa-Pro-)n. Reflecting the similarity of the substrate specificities, the amino acid sequence of P. gingivalis PtpA is partially homologous (23.5%) to that of P. gingivalis DPP4. The crystal structure of P. gingivalis PtpA was previously reported [56].
2.2.6. Acylpeptidyl oligopeptidase (AOP)
The dpp4-dpp5-dpp7-triple knockout strain NDP211 was found to have lost most DPP activities except for that of DPP11 and a small level of Met-Leu-MCA-hydrolyzing activity. However, further disruption of dpp11 in NDP211, termed NDP212, reversed that Met-Leu-MCA-hydrolyzing activity [24], which strongly suggested induction of an unidentified DPP under emergent conditions. Our search revealed that an S9-family peptidase, PGN_1349, was responsible for hydrolysis of Met-Leu-MCA in NDP212 [25]. Maximal activity of PGN_1349 was achieved with benzyloxycarbonyl- (Z-)VKM-MCA. Thus, the Met-Leu-MCA-hydrolyzing activity observed in the quatro-dpp knockout strain NDP212 was not mediated by DPP, but rather by the non-DPP peptidase PGN_1349. Since PGN_1349 preferentially liberates di- and tri-peptides from acylated oligopeptides, we designated it acylpeptidyl oligopeptidase (AOP). Details of the biochemical and enzymatic properties of AOP will be reported elsewhere. Expression of AOP is likely beneficial for the organism, because it provides N-terminally unblocked serum proteins toward DPPs and PtpA.
2.3. Absence of exopeptidases with preference to hydrophilic residues
P. gingivalis exopeptidases recognize P1-position Pro, acidic, or hydrophobic residues in their substrate peptides. Hydrophobic interaction is also involved in the P2-position preference for DPP7 and DPP11 [48]. In contrast, P. gingivalis does not seem to possess a DPP to liberate N-terminal dipeptides composed of hydrophilic residues, since it does not hydrolyze Thr-Ser- or Gly-Gly-MCA [24]. In fact, no DPP for hydrophilic amino acids has been found in any organisms to date [32]. The hydrophilic interaction between DPP and the dipeptidyl portion of a substrate may be not sufficient to form an enzyme–substrate complex, because the interaction is likely weakened by surrounding water and hydrophilic macromolecules.
3. Subcellular localization and comparison of enzymatic properties among exopeptidases
All four DPP and AOP activities have been detected in P. gingivalis cells, though not in culture medium, indicating that they are present as so-called cell-associated forms. Furthermore, subcellular fractionation demonstrated that DPP5 and DPP11 are localized in the periplasmic space of the cell (Fig. 1) [23], [24]. Indeed, DPPs do not have the C-terminal domain, which is required for outer membrane localization of Rgp and Kgp mediated by the type IX secretion system [57]. Periplasmic localization of P. gingivalis DPPs, PtpA, and AOP has also been suggested based on proteome analysis [58]. The differing subcellular localization between DPPs and gingipains seems to be compatible with the process of extracellular polypeptide processing.
We determined and compared the enzymatic parameters of recombinant forms of DPPs and AOP (Table 1). The kcat/Km values for the best substrates for their respective peptidases were the highest in DPP4 followed by DPP11, while DPP5 and DPP7 possessed moderate values, and AOP had the lowest. Interestingly, this order was in parallel with substrate confinement in these exopeptidases, from Pro-specific DPP4, to Asp/Glu-specific DPP11, hydrophobic P1- and P2-specific DPP7, hydrophobic P1-specific DPP5, and the most relaxed hydrophobic P1-specific AOP. This coincidence is reasonable, because DPP4 may fold into the most specific steric structures for its substrate, whereas AOP, which is able to liberate di- and tri-peptides with and without N-terminal modification, may fold into a more open structure that is able to accept several types of substrates.
Table 1.
Enzymatic parameters of P. gingivalis exopeptidases.
| Enzyme | Peptidyl MCAa | kcat (s−1) | Km (μM) | kcat/Km (μM−1 s−1) | Reference |
|---|---|---|---|---|---|
| DPP4 | Gly-Pro- | 6,917 ± 1253 | 100.9 ± 20.1 | 66.8 ± 2.0 | This study |
| DPP5 | Lys-Ala- | 7,577 ± 637 | 687.6 ± 11.0 | 11.0 | [24] |
| DPP7 | Met-Leu- | 394 ± 79 | 39.6 ± 16.0 | 10.6 ± 2.5 | [51] |
| DPP11 | Leu-Asp- | 10,707 ± 140 | 19.5 ± 0.4 | 547.4 ± 6.3 | This study |
| AOP | Z-VKM- | 592 ± 40 | 4.9 ± 1.0 | 123.3 ± 17.3 | This study |
The most suitable peptidyl MCA is presented. Mean ± S.D.
4. Exopeptidases as potential virulence factors of P. gingivalis
All four DPP activities have been detected in P. gingivalis strains ATCC 33277, ATCC 49417, W83, W50, HW24D1, HNA99, HG1690, and 16-1 [50], and the exopeptidases discussed in this article contribute to bacterial growth. The findings presented here suggest that the activities of these exopeptidases influence both bacterial colonization in the oral cavity and clearance from the cardiovascular system after entering the blood circulation from a periodontal lesion.
It has been reported that P. gingivalis DPP4 promotes the degradation of collagen and gelatin mediated by host proteases, and also inhibits binding between gingival fibroblasts and fibronectin [43]. In addition, DPP4 is involved in biofilm formation by P. gingivalis, which is closely related to bacterial virulence [44]. Hence, it is reasonable to postulate that other DPPs also contribute to biofilm formation and mixed-species colonization in subgingival plaque. Furthermore, evaluation of the roles of P. gingivalis DPPs in systemic diseases, especially type-2 diabetes mellitus and cardiovascular diseases, would be interesting. Our preliminary data indicate that the human peptide hormone incretin GLP-1, which induces secretion of insulin from the pancreas, is degraded by P. gingivalis bacterial cells and DPP4 in vitro. This is now an important point of focus for further studies.
5. Conclusion
P. gingivalis expresses various exopeptidases, i.e., DPP4, DPP5, DPP7, DPP11, PtpA, and AOP, in periplasmic space, which produce di- and tri-peptides from most oligopeptides. This oligopeptide processing step is important as an extracellular event in the metabolism of asaccharolytic P. gingivalis. Rgp and Kgp are also involved in dipeptide production. An organized subcellular localization of various exopeptidases and gingipains is a rational explanation for processing of proteinaceous nutrients present in the subgingival environment, thus providing a means of efficient survival for the bacterium.
Additional note
To activate studies of DPPs of P. gingivalis and other oral bacteria, we contacted the Peptide Institute (Osaka, Japan) to produce DPP substrates. As a result, Leu-Asp- and Met-Leu-MCA, substrates for DPP11 and DPP7, respectively, have recently become available (The Peptide Institute, Supplemental Product List 28-3, 2015).
Conflict of interest
The authors have no conflict of interest to declare.
Acknowledgements
We thank Dr. R. Kamijyo (Showa University) for providing us the opportunity to present this article. This study was conducted in collaboration with Drs. T. Ono, T.T. Baba, and T. Kobayakawa (Nagasaki University), Dr. S.M.A. Rouf (Faculty of Applied Sciences, Islamic University, Bangladesh), and Drs. Y. Shimoyama and S. Kimura (Iwate Medical University). This study was supported by Grants-in-aid for Scientific Research from the Japan Society for the Promotion of Science (to T.K.N. and Y.O.-N.), a grant from the Institute for Fermentation, Osaka (to T.K.N.), and grants from the Joint Research Promotion Project of Nagasaki University Graduate School of Biomedical Sciences in 2013 and 2014 (to Y.O.-N.).
References
- 1.Slots J., Listgarten M.A. Bacteroides gingivalis, Bacteroides intermedius and Actinobacillus actinomycetemcomitans in human periodontal diseases. J Clin Periodontol. 1988;15:85–93. doi: 10.1111/j.1600-051x.1988.tb00999.x. [DOI] [PubMed] [Google Scholar]
- 2.White D., Mayrand D. Association of oral Bacteroides with gingivitis and adult periodontitis. J Periodontal Res. 1981;16:259–265. doi: 10.1111/j.1600-0765.1981.tb00974.x. [DOI] [PubMed] [Google Scholar]
- 3.Socransky S.S., Haffajee A.D. Periodontal microbial ecology. Periodontol 2000. 2002;38:135–187. doi: 10.1111/j.1600-0757.2005.00107.x. [DOI] [PubMed] [Google Scholar]
- 4.Iwai T., Inoue Y., Umeda M., Huang Y., Kurihara N., Koike M. Oral bacteria in the occluded arteries of patients with Buerger disease. J Vasc Surg. 2005;42:107–115. [Google Scholar]
- 5.Genco R.J., Van Dyke T.E. Prevention: reducing the risk of CVD in patients with periodontitis. Nat Rev Cardiol. 2010;7:479–480. doi: 10.1038/nrcardio.2010.120. [DOI] [PubMed] [Google Scholar]
- 6.Tabeta K., Yoshie H., Yamazaki K. Current evidence and biological plausibility linking periodontitis to atherosclerotic cardiovascular disease. Jpn Dent Sci Rev. 2014;50:55–62. [Google Scholar]
- 7.Kshirsagar A.V., Offenbacher S., Moss K.L., Barros S.P., Beck J.D. Antibodies to periodontal organisms are associated with decreased kidney function: the dental atherosclerosis risk in communities study. Blood Purif. 2007;25:125–132. doi: 10.1159/000096411. [DOI] [PubMed] [Google Scholar]
- 8.Detert J., Pischon N., Burmester G.R., Buttgereit F. The association between rheumatoid arthritis and periodontal disease. Arthritis Res Ther. 2010;12:218. doi: 10.1186/ar3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Citron D.M., Poxton I.R., Baron E. 9th ed. vol. 1. ASM Press; Washington, DC: 2007. Bacteroides, Porphyromonas, Prevotella, Fusobacterium, and other anaerobic Gram-negative rods; pp. 911–932. (Manual of clinical microbiology). [Google Scholar]
- 10.Nelson K.E., Fleischmann R.D., DeBoy R.T., Paulsen I.T., Fouts D.E., Eisen J.A. Complete genome sequence of the oral pathogenic bacterium Porphyromonas gingivalis strain W83. J Bacteriol. 2003;185:5591–5601. doi: 10.1128/JB.185.18.5591-5601.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seshadri R., Myers G.S., Tetelin H., Eisen J.A., Heidelberg J.F., Dodson R.J. Comparison of the genome of the oral pathogen Treponema denticols with other spirochete genomes. Proc Natl Acad Sci USA. 2004;101:5646–5651. doi: 10.1073/pnas.0307639101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mayrand D., Holt S.C. Biology of asaccharolytic black-pigmented Bacteroides species. Microbiol Rev. 1988;52:134–152. doi: 10.1128/mr.52.1.134-152.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shah H.N., Gharbia S.E., Kowlessur D., Wilkie E., Brocklehurst K. Isolation and characterization of gingivain, a cysteine proteinase from Porphyromonas gingivalis strain W83. Biochem Soc Trans. 1990;18:578–579. doi: 10.1042/bst0180578. [DOI] [PubMed] [Google Scholar]
- 14.Chen Z., Potempa J., Polanowski A., Wikstrom M., Travis J. Molecular cloning and structural characterization of the Arg-gingipain proteinase of Porphyromonas gingivalis. Biosynthesis as a proteinase-adhesin polyprotein. J Biol Chem. 1992;267:18896–18901. [Google Scholar]
- 15.Takahashi N., Sato T., Yamada T. Metabolic pathways for cytotoxic end product formation from glutamate- and aspartate-containing peptides by Porphyromonas gingivalis. J Bacteriol. 2000;182:4704–4710. doi: 10.1128/jb.182.17.4704-4710.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takahashi N., Sato T. Preferential utilization of dipeptides by Porphyromonas gingivalis. J Dent Res. 2001;80:1425–1429. doi: 10.1177/00220345010800050801. [DOI] [PubMed] [Google Scholar]
- 17.Dashper S.G., Brownfield L., Slakeski N., Zilm P.S., Rogers A.H., Reynolds E.C. Sodium ion-driven serine/threonine transport in Porphyromonas gingivalis. J Bacteriol. 2001;183:4142–4148. doi: 10.1128/JB.183.14.4142-4148.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abiko Y., Hayakawa M., Murai S., Takiguchi H. Glycylprolyl dipeptidylaminopeptidase from Bacteroides gingivalis. J Dent Res. 1985;64:106–111. doi: 10.1177/00220345850640020201. [DOI] [PubMed] [Google Scholar]
- 19.Banbula A., Bugno M., Goldstein J., Yen J., Nelson D., Travis J. Emerging family of proline-specific peptidases of Porphyromonas gingivalis: purification and characterization of serine dipeptidyl peptidase, a structural and functional homologue of mammalian prolyl dipeptidyl peptidase IV. Infect Immun. 2000;68:1176–1182. doi: 10.1128/iai.68.3.1176-1182.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Banbula A., Yen J., Oleksy A., Mak P., Bugno M., Travis J. Porphyromonas gingivalis DPP-7 represents a novel type of dipeptidylpeptidase. J Biol Chem. 2001;276:6299–6305. doi: 10.1074/jbc.M008789200. [DOI] [PubMed] [Google Scholar]
- 21.Banbula A., Mak P., Bugno M., Silberring J., Dubin A., Nelson D. Prolyl tripeptidyl peptidase from Porphyromonas gingivalis. A novel enzyme with possible pathological implications for the development of periodontitis. J Biol Chem. 1999;274:9246–9252. doi: 10.1074/jbc.274.14.9246. [DOI] [PubMed] [Google Scholar]
- 22.Mazumdar V., Snitkin E.S., Amar S., Segre S. Metabolic network model of a human oral pathogen. J Bacteriol. 2009;191:74–90. doi: 10.1128/JB.01123-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohara-Nemoto Y., Shimoyama Y., Kimura S., Kon A., Haraga H., Ono T. Asp- and Glu-specific novel dipeptidyl peptidase 11 of Porphyromonas gingivalis ensures utilization of proteinaceous energy sources. J Biol Chem. 2011;286:38115–38127. doi: 10.1074/jbc.M111.278572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohara-Nemoto Y., Rouf S.M.A., Naito M., Yanase A., Tetsuo F., Ono T. Identification and characterization of prokaryotic dipeptidyl-peptidase 5 from Porphyromonas gingivalis. J Biol Chem. 2014;289:5436–5448. doi: 10.1074/jbc.M113.527333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nemoto T.K., Ono T., Shimoyama Y., Kimura S., Ohara-Nemoto Y. The 87th congress of Japanese Biochemical Society. 2014 Program 4T10p-07 (4P-117) 2014. Mechanism on oligomer–monomer transition of a novel peptidase from periodontopathic bacterium. [in Japanese] [Google Scholar]
- 26.Shi Y., Ratnayake D.B., Okamoto K., Abe N., Yamamoto K., Nakayama K. Genetic analyses of proteolysis, hemoglobin binding, and hemagglutination of Porphyromonas gingivalis. Construction of mutants with a combination of rgpA, rgpB, kgp, and hagA. J Biol Chem. 1999;274:17955–17960. doi: 10.1074/jbc.274.25.17955. [DOI] [PubMed] [Google Scholar]
- 27.O’Brien-Simpson N.M., Paolini R.A., Hoffmann B., Slakeski N., Dashper S.G., Reynolds E.C. Role of RgpA, RgpB, and Kgp proteinases in virulence of Porphyromonas gingivalis W50 in a murine lesion model. Infect Immun. 2001;69:7527–7534. doi: 10.1128/IAI.69.12.7527-7534.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Potempa J., Pike R., Travis J. Titration and mapping of the active site of cysteine proteases from Porphyromonas gingivalis (gingipains) using peptidyl chloromethanes. Biol Chem. 1997;378:223–230. doi: 10.1515/bchm.1997.378.3-4.223. [DOI] [PubMed] [Google Scholar]
- 29.Imamura T., Travis J., Potempa J. The biphasic virulence activities of gingipains: activation and inactivation of host proteins. Curr Protein Pept Sci. 2003;4:443–450. doi: 10.2174/1389203033487027. [DOI] [PubMed] [Google Scholar]
- 30.Kadowaki T., Takii R., Yamatake K., Kawakubo T., Tsukuba T., Yamamoto K. A role for gingipains in cellular responses and bacterial survival in Porphyromonas gingivalis-infected cells. Front Biosci. 2007;12:4800–4809. doi: 10.2741/2428. [DOI] [PubMed] [Google Scholar]
- 31.Fitzpatrick R.E., Wijeyewickrema L.C., Pike R.N. The gingipains: scissors and glue of the periodontal pathogen, Porphyromonas gingivalis. Future Microbiol. 2009;4:471–487. doi: 10.2217/fmb.09.18. [DOI] [PubMed] [Google Scholar]
- 32.Rawlings N.D., Waller M., Barrett A.J., Bateman A. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2014;42:D503–D509. doi: 10.1093/nar/gkt953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kadowaki T., Yoneda M., Okamoto K., Maeda K., Yamamoto K. Purification and characterization of a novel arginine-specific cysteine proteinase (argingipain) involved in the pathogenesis of periodontal disease from the culture supernatant of Porphyromonas gingivalis. J Biol Chem. 1994;269:21371–21378. [PubMed] [Google Scholar]
- 34.Potempa J., Pike R., Travis J. The multiple forms of trypsin-like activity present in various strains of Porphyromonas gingivalis are due to the presence of either Arg-gingipain or Lys-gingipain. Infect Immun. 1995;63:1176–1182. doi: 10.1128/iai.63.4.1176-1182.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fujimura S., Shibata Y., Nakamura T. Comparative studies of three proteases of Porphyromonas gingivalis. Oral Microbiol Immunol. 1992;7:212–217. doi: 10.1111/j.1399-302x.1992.tb00027.x. [DOI] [PubMed] [Google Scholar]
- 36.Scott C.F., Whitaker E.J., Hammond B.F., Colman R.J. Purification and characterization of a potent 70-kDa thiol lysyl-proteinase (Lys-gingipain) from Porphyromonas gingivalis that cleaves kininogens and fibrinogen. J Biol Chem. 1993;268:7935–7942. [PubMed] [Google Scholar]
- 37.Pavloff N., Pemberton P.A., Potempa J., Chen W.C., Pike R.N., Prochazka V. Molecular cloning and characterization of Porphyromonas gingivalis lysine-specific gingipain. A new member of an emerging family of pathogenic bacterial cysteine proteinases. J Biol Chem. 1997;272(3):1595–1600. doi: 10.1074/jbc.272.3.1595. [DOI] [PubMed] [Google Scholar]
- 38.Singer R.E., Buckner B.A. Butyrate and propionate: important components of toxic dental plaque extracts. Infect Immun. 1981;32:458–463. doi: 10.1128/iai.32.2.458-463.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ho Y.-C., Chang Y.-C. Effects of a bacterial lipid byproduct on human pulp fibroblasts in vitro. J Endod. 2007;33:437–441. doi: 10.1016/j.joen.2006.12.022. [DOI] [PubMed] [Google Scholar]
- 40.Kurita-Ochiai T., Seto S., Suzuki N., Yamamoto M., Otsuka K., Abe K. Butyric acid induces apoptosis in inflamed fibroblasts. J Dent Res. 2008;87:51–55. doi: 10.1177/154405910808700108. [DOI] [PubMed] [Google Scholar]
- 41.Kiyama M., Hayakawa M., Shiroza T., Nakamura S., Takeuchi A., Masamoto Y. Sequence analysis of the Porphyromonas gingivalis dipeptidyl peptidase IV gene. Biochim Biophys Acta. 1998;1396:39–46. doi: 10.1016/s0167-4781(97)00225-x. [DOI] [PubMed] [Google Scholar]
- 42.Oda H., Saiki K., Tonosaki M., Yajima A., Konishi K. Participation of the secreted dipeptidyl and tripeptidyl aminopeptidases in asaccharolytic growth of Porphyromonas gingivalis. J Priodontal Res. 2009;44:362–367. doi: 10.1111/j.1600-0765.2008.01117.x. [DOI] [PubMed] [Google Scholar]
- 43.Kumagai Y., Yagishita H., Yajima A., Okamoto T., Konishi K. Molecular mechanism for connective tissue destruction by dipeptidyl aminopeptidase IV produced by the periodontal pathogen Porphyromonas gingivalis. Infect Immun. 2005;73:2655–2664. doi: 10.1128/IAI.73.5.2655-2664.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clais S., Boulet G., Kerstens M., Horemans T., Teughels W., Quirynen M. Importance of biofilm formation and dipeptidyl peptidase IV for the pathogenicity of clinical Porphyromonas gingivalis isolates. Pathog Dis. 2014;70:408–413. doi: 10.1111/2049-632X.12156. [DOI] [PubMed] [Google Scholar]
- 45.Beauvais A., Monod M., Debeaupuis J.P., Diaquin M., Kobayashi H., Latgé J.P. Biochemical and antigenic characterization of a new dipeptidyl-peptidase isolated from Aspergillus fumigatus. J Biol Chem. 1997;7(272):6238–6244. doi: 10.1074/jbc.272.10.6238. [DOI] [PubMed] [Google Scholar]
- 46.Monod M., Beauvais A. Dipeptidyl-peptidases IV and V of Aspergillus. In: Rawlings N.D., Salvesen G.S., editors. Handbook of proteolytic enzymes. 3rd ed. Elsevier; Amsterdam: 2012. pp. 3392–3394. [Google Scholar]
- 47.Loussert C., Schmitt C., Prevost M.C., Balloy V., Fadel E., Philippe B. In vivo biofilm composition of Aspergillus fumigatus. Cell Microbiol. 2010;12:405–410. doi: 10.1111/j.1462-5822.2009.01409.x. [DOI] [PubMed] [Google Scholar]
- 48.Rouf S.M.A., Ohara-Nemoto Y., Ono T., Shimoyama Y., Kimura S., Nemoto T.K. Phenylalanine664 of dipeptidyl peptidase (DPP) 7 and phenylalanine671 of DPP11 mediate preference for P2-position hydrophobic residues of a substrate. FEBS Open Bio. 2013;3:177–181. doi: 10.1016/j.fob.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kon A. Pathogenic factors of Porphyromonas endodontalis. Dent J Iwate Med Univ. 2002;27:187–196. [in Japanese with English abstract] [Google Scholar]
- 50.Nishimata H., Ohara-Nemoto Y., Baba T.T., Hoshino T., Fujiwara T., Shimoyama Y. Identification of dipeptidyl-peptidase (DPP) 5 and DPP7 in Porphyromonas endodontalis, distinct from those in Porphyromonas gingivalis. PLOS ONE. 2014;9:e114221. doi: 10.1371/journal.pone.0114221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rouf S.M.A., Ohara-Nemoto Y., Hoshino T., Fujiwara T., Ono T., Nemoto T.K. Discrimination based on Gly and Arg/Ser at Position 673 between dipeptidyl-peptidase (DPP) 7 and DPP11, widely distributed DPPs in pathogenic and environmental Gram-negative bacteria. Biochimie. 2013;95:824–832. doi: 10.1016/j.biochi.2012.11.019. [DOI] [PubMed] [Google Scholar]
- 52.Sakamoto Y., Suzuki Y., Iizuka I., Tateoka C., Roppongi S., Fujimoto M. S46 peptidases are the first exopeptidases to be members of clan PA. Sci Rep. 2014;15:4977. doi: 10.1038/srep04977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Underwood R., Chiravuri M., Lee H., Schmitz T., Kabcenell A.K., Yardley K. Sequence, purification, and cloning of an intracellular serine protease, quiescent cell proline dipeptidase. J Biol Chem. 1999;274:34053–34058. doi: 10.1074/jbc.274.48.34053. [DOI] [PubMed] [Google Scholar]
- 54.Bezerra G.A., Dobrovetsky E., Dong A., Seitova A., Crombett L., Shewchuk L.M. Structures of human DPP7 reveal the molecular basis of specific inhibition and the architectural diversity of proline-specific peptidases. PLoS ONE. 2012;7:e43019. doi: 10.1371/journal.pone.0043019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Naito M., Hirakawa H., Yamashita A., Ohara N., Shoji M., Yukitake H. Determination of the genome sequence of Porphyromonas gingivalis strain ATCC 33277 and genomic comparison with strain W83 revealed extensive genome rearrangements in P. gingivalis. DNA Res. 2008;15:215–225. doi: 10.1093/dnares/dsn013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ito K., Nakajima Y., Xu Y., Yamada N., Onohara Y., Ito T. Crystal structure and mechanism of tripeptidyl activity of prolyl tripeptidyl aminopeptidase from Porphyromonas gingivalis. J Mol Biol. 2006;362:228–240. doi: 10.1016/j.jmb.2006.06.083. [DOI] [PubMed] [Google Scholar]
- 57.Sato K., Naito M., Yukitake H., Hirakawa H., Shoji M., McBride M.J. A protein secretion system linked to bacteroidete gliding motility and pathogenesis. Proc Natl Acad Sci USA. 2010;107:276–281. doi: 10.1073/pnas.0912010107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Veith P.D., Chen Y.Y., Gorasia D.G., Chen D., Glew M.D., O’Brien-Simpson N.M. Porphyromonas gingivalis outer membrane vesicles exclusively contain outer membrane and periplasmic proteins and carry a cargo enriched with virulence factors. J Proteome Res. 2014;13:2420–2432. doi: 10.1021/pr401227e. [DOI] [PubMed] [Google Scholar]