Abstract
Activation-induced cytidine deaminase (AID) is a B-cell specific enzyme that targets immunoglobulin (Ig) genes to initiate class switch recombination (CSR) and somatic hypermutation (SHM)1. Through off-target activity, however, AID has a much broader impact on genomic instability by initiating oncogenic chromosomal translocations and mutations involved in lymphoma development and progression2. AID expression is tightly regulated in B cells and its overexpression leads to enhanced genomic instability and lymphoma formation3. The phosphatidylinositol 3-kinase (PI3K) δ pathway plays a key role in AID regulation by suppressing its expression in B cells4. Novel drugs for leukemia or lymphoma therapy such as idelalisib, duvelisib or ibrutinib block PI3Kδ activity directly or indirectly5–8, potentially affecting AID expression and, consequently, genomic stability in B cells. Here we show that treatment of primary mouse B cells with idelalisib or duvelisib, and to a lesser extent ibrutinib, enhanced the expression of AID and increased somatic hypermutation (SHM) and chromosomal translocation frequency to the Igh locus and to several AID off-target sites. Both these effects were completely abrogated in AID deficient B cells. PI3Kδ inhibitors or ibrutinib increased the formation of AID-dependent tumors in pristane-treated mice. Consistently, PI3Kδ inhibitors enhanced AID expression and translocation frequency to IgH and AID off-target sites in human chronic lymphocytic leukemia (CLL) and mantle cell lymphoma (MCL) cell lines, and patients treated with idelalisib, but not ibrutinib, showed increased SHM in AID off-targets. In summary, we show that PI3Kδ or BTK inhibitors increase genomic instability in normal and neoplastic B cells by an AID-dependent mechanism, an effect that should be carefully considered as such inhibitors are administered for years to patients.
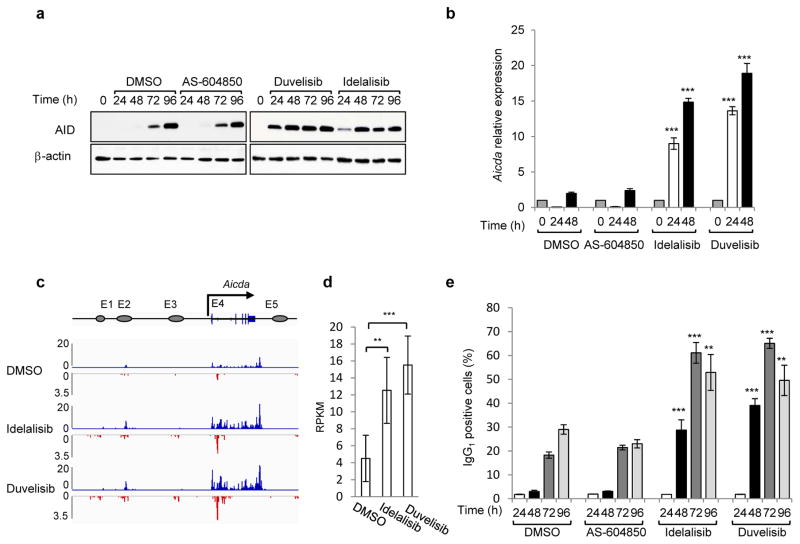
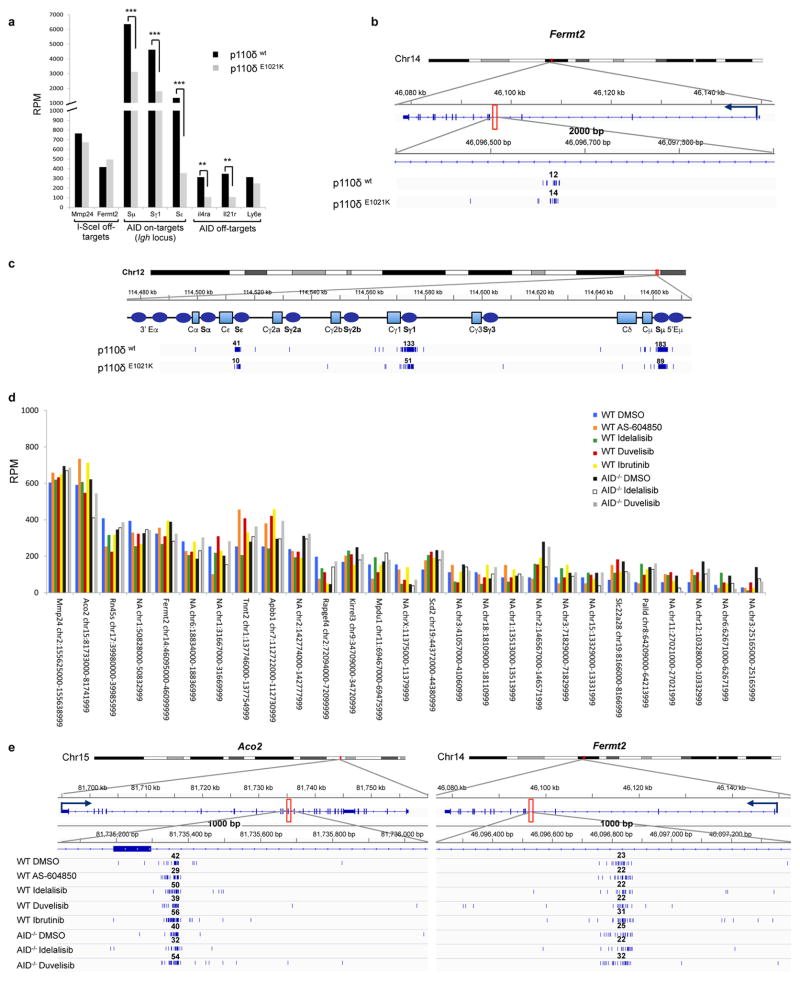
We first tested the effects of PI3Kδ blockade in primary mouse B cells stimulated with anti-CD40 plus IL-4 to undergo CSR9. In these cells, the PI3Kδ inhibitor idelalisib or the dual PI3Kγδ inhibitor duvelisib accelerated and increased AID induction whereas the PI3Kγ inhibitor AS-604850 did not affect AID abundance (Fig. 1a). Consistently, AID mRNA levels were significantly enhanced by either idelalisib or duvelisib (Fig. 1b). To more precisely define transcription changes of AID in activated mouse B cells treated with PI3Kδ inhibitors, we performed GRO-Seq analysis9. Of the 5 enhancers associated with the Aicda gene, we found a substantial increase in both sense and antisense transcription in the E4 enhancer downstream of the TSS (Fig. 1c,d), consistent with the pattern of AID expression we described in CSR-activated and germinal center (GC) B cells10. As a result of the enhanced AID expression, idelalisib and duvelisib increased CSR to IgG1 in vitro in activated B cells (Fig. 1e) as well as in vivo in GC B cells (Extended Data Fig. 1a–c). The effects were significant at doses ranging from 0.1 μM to 1 μM, which encompass the plasma concentration of these drugs observed in patients7,11 (Fig. 1e, Extended Data Fig. 1d–f). Idelalisib and duvelisib reduced B-cell proliferation, whereas AS-604850 did not (Extended Data Fig. 1g), demonstrating that PI3Kδ blockade enhanced AID expression and CSR despite an inhibition of B-cell proliferation12. In a reverse genetic experiment, B cells expressing a PI3Kδ gain-of-function mutant (PI3KδE1021K) recently discovered in patients with immunodeficiency and impaired CSR13,14, showed decreased AID mRNA and protein levels as well as CSR (Extended Data Fig. 1h–j).
Figure 1. Phosphatidylinositol 3-Kinase (PI3K)δ blockade increases AID expression and CSR in activated mouse B cells.
a, Western blot for AID protein from B cells treated with the indicated inhibitors (1 μM) (n = 3 biological replicates). For gel source data, see Supplementary Figure 1. b, Aicda mRNA levels were analyzed by qRT-PCR. Data are expressed as mean ± s.d. (n = 3 biological replicates).. ** P ≤ 0.01, *** P ≤ 0.001, two-tailed Student’s t-test from idelalisib or duvelisib vs DMSO-treated B cells. c, GRO-Seq profiles of Aicda gene in B cells at 48 h after activation (n = 2 biological replicates). Blue profiles: sense transcription, Red profiles: antisense transcription. d, Quantification of GRO-Seq sense and antisense reads per kilobase per million mapped reads (RPKM) in the Aicda gene, **P ≤ 0.01,***P ≤ 0.001, multiple test adjusted. e, IgG1 CSR in activated B cells. Data are expressed as mean ± s.d. (n = 3 biological replicates). ** P ≤ 0.01, *** P ≤ 0.001, two-tailed Student’s t-test from idelalisib or duvelisib vs DMSO treated B cells
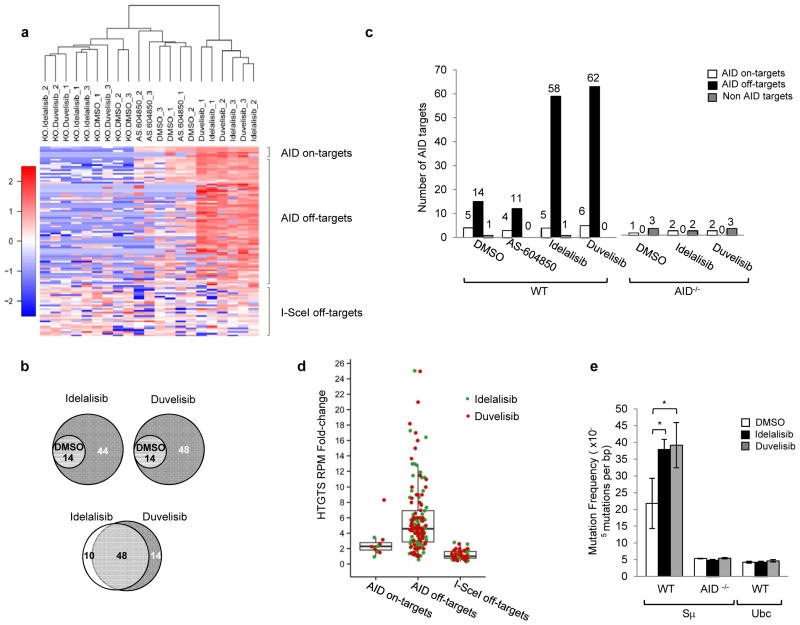
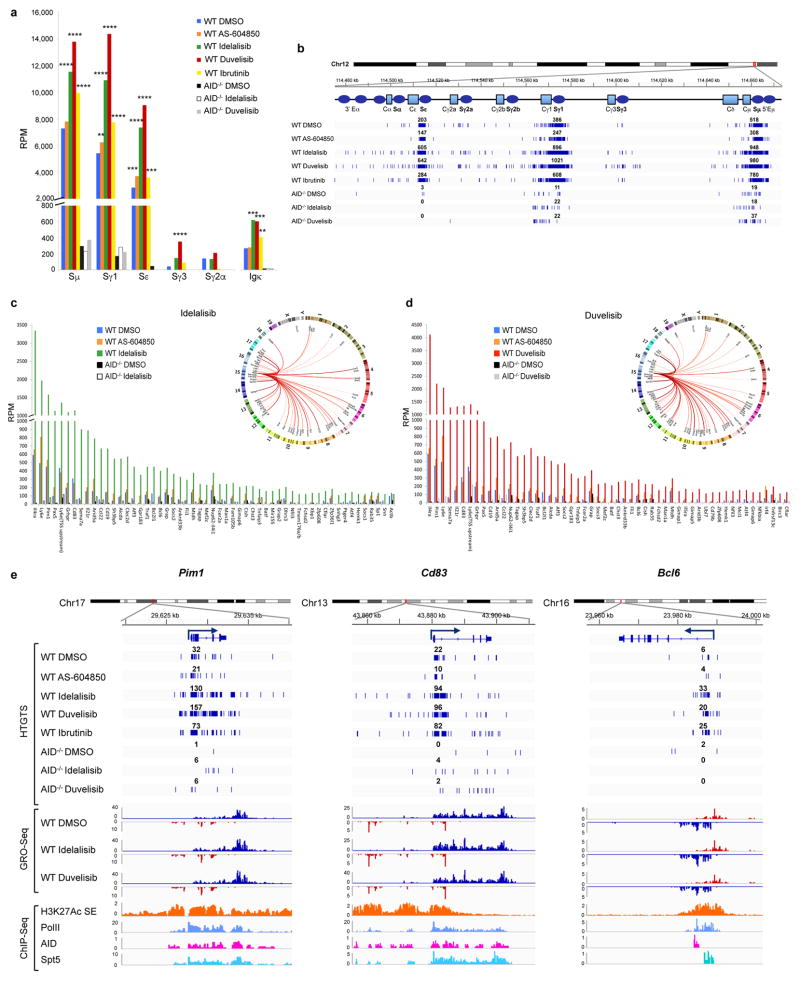
Because AID induces DNA damage and chromosomal translocations at defined on-target (Igh locus) and off-target (non-Igh locus) genomic sites1,9,15, we next analyzed whether the enhanced AID expression induced by PI3Kδ blockade would result in increased genome instability. We applied high-throughput genome-wide translocation sequencing (HTGTS)9 in order to generate genomic maps of chromosomal translocations in activated mouse B cells treated with idelalisib or duvelisib. By this approach, we sequenced thousands of translocation junctions between endogenous DSBs and a c-myc DSB initiated by the I-SceI nuclease9 (Supplementary Table 1). Overall, idelalisib or duvelisib similarly increased the formation of translocation junctions between c-myc and AID on target sites in the Igh locus or AID off-target sites in the genome (Fig. 2a–c). In the Igh locus, translocations junctions increased and clustered in the Sμ, Sγ1 and Sε regions as previously described9,15 (Extended Data Fig. 2a,b). In addition, the number of AID off-target sites in idelalisib or duvelisib, but not AS-604850, treated B cells was substantially increased (Fig. 2c, Supplementary Table 2). AID off-target sites induced by PI3Kδ blockade were widely dispersed across the genome (Extended Data Fig. 2c,d), were largely overlapping with AID off-target sites previously identified by HTGTS16, TC-Seq 15, AID ChIP-Seq 17 and RPA-ChIP 18 (Supplementary Table 2), and included Aicda itself as well as several genes involved in recurrent translocations in human lymphomas, such as Pax5, Pim1, Bcl6, Il21r and Bcl2l1 (Extended Data Fig. 2c–e). The median fold change translocation frequency was 2.0 and 2.6 for AID on-target sites and 4.3 and 4.7 for AID off-target sites for idelalisib or duvelisib, respectively, with 95 percentiles up to 12.9 and 17.0 fold changes for idelalisib or duvelisib (Fig. 2d). Both on-target and off-target translocation junctions were almost completely abrogated in AID-deficient B cells thus demonstrating that the increased genomic instability induced by PI3Kδ blockade was AID-dependent (Fig. 2c, Extended Data Fig. 2). Conversely, on-target and off-target translocation junctions were significantly reduced in B cells expressing the PI3KδE1021K active mutant (Extended Data Fig. 3a–c). As control, translocations to sites generated by the off-target activity of I-SceI that are independent of AID9 showed no changes in B cells treated with idelalisib or duvelisib (Extended Data Fig. 3d,e).
Figure 2. PI3Kδ blockade increases the frequency of genome-wide chromosomal translocations in activated mouse B cells.
a, Hierarchical clustering of translocation frequency analysis of HTGTS libraries generated from WT and AID−/− B cells treated with the indicated inhibitors. AID on-target (Igh and Igk loci) and off-target are listed in Supplementary Table 2. I-SceI off-targets are listed in Extended Data Fig. 3d. b, Venn diagrams showing the fraction of shared AID off-targets. c Numbers of AID on-target (Igh and Igk loci) and off-target hot spots from WT and AID−/− B cells. Data pooled from at least n = 3 biological replicates (Supplementary Table 1). d, Fold change of HTGTS junction frequency in B cells treated with idelalisib or duvelisib compared to DMSO. I-SceI off-targets are used as an internal control for non-AID mediated translocations. Whiskers extend to a maximum of 1.5X interquartile range (IQR) beyond the box. e, Mutation frequency of the immunoglobulin Sμ region in WT and AID−/− B cells. Cumulative frequency of C-to-T or G-to-A transition mutations is indicated. Data are expressed as mean ± s.d. (n = 3 biological replicates). * P < 0.05, two-tailed Student’s t-test.
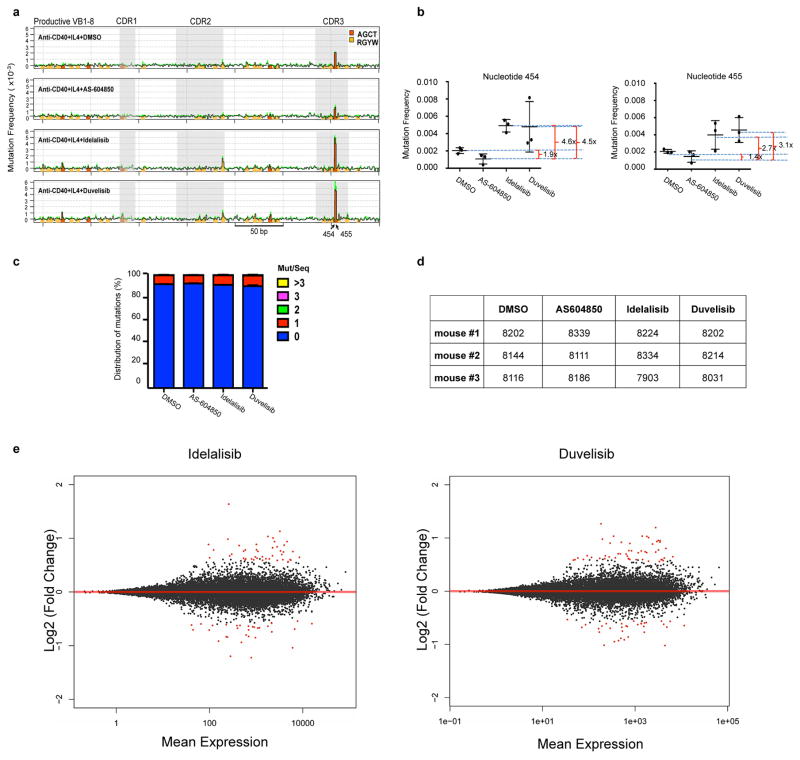
Next, we tested whether PI3Kδ blockade would also increase AID-mediated SHM. Treatment with idelalisib or duvelisib significantly increased SHM in the Sμ region in WT but not AID-deficient activated B cells and had no effect on the Ubc promoter, a site previously shown to lack AID-mediated SHM18 (Fig. 2e). Furthermore, PI3Kδ blockade increased 2.7 to 4.6 fold the mutation frequency at the V exon in activated B cells from mice harboring a VB1-8 exon in the productive allele19 (Extended Data Fig. 4a–d). Therefore, we concluded that PI3Kδ blockade increased both AID-dependent chromosomal translocation formation and SHM in activated mouse B cells.
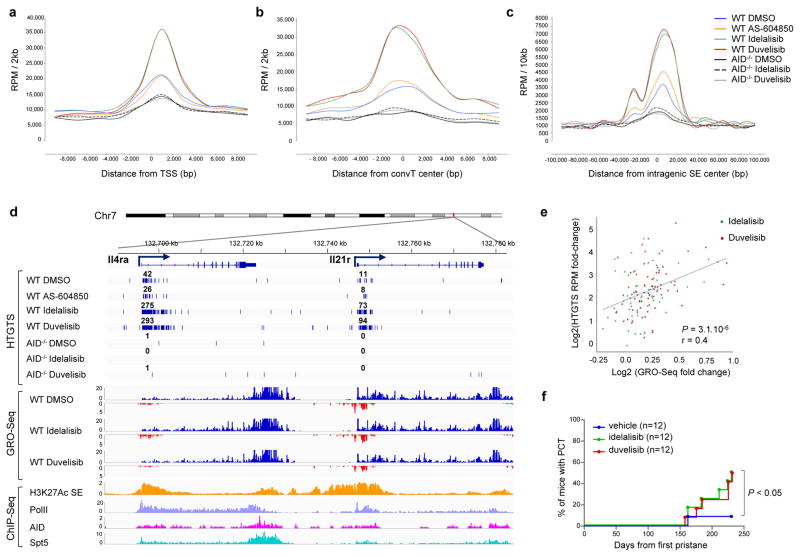
The current consensus is that AID preferentially targets genomic regions associated with high transcriptional activity, more frequently embedded within super-enhancers (SEs)10,18. SE regions typically are associated with events that direct AID binding and activity, such as RNA polymerase II (Pol II) transcriptional stalling with Spt5 accumulation20, foci of convergent transcription (ConvT)10 and non-coding RNA transcription21. In activated B cells treated with idelalisib or duvelisib, increased translocation junctions clustered 1–2 kb downstream of the transcription start site (TSS) (Fig. 3a), peaked around ConvT regions (Fig. 3b) and intragenic SEs (Fig. 3c), and were associated with AID, Pol II and Spt5 binding sites (Fig. 3d, Extended Data Fig. 2e). Overall these data show that PI3Kδ blockade increased the formation of translocations but did not change their pattern of distribution in the genome.
Figure 3. Enhanced genomic instability in mouse B cells by PI3Kδ blockade.
Genome-wide distribution of translocation junctions relative to transcription starting site (TSS) regions (a), convergent transcription (ConvT) regions (b) and intragenic super-enhancers (SEs) (c) in WT and AID−/− B cells treated with the indicated inhibitors. d, Detailed distribution of HTGTS junctions and GRO-Seq reads in representative gene examples. Numbers of translocation junctions in focal clusters are indicated in bold. Corresponding PolII, AID, Spt5 and H3K27Ac/SE ChIP-Seq profiles are shown. e, Fold change HTGTS junctions plotted against GRO-Seq transcription levels of AID targets in B cells treated with idelalisib or duvelisib over DMSO. Pearson’s correlation coefficient; P value calculated by two-tailed F-test. f, Development of PC tumor in mice induced with pristane and treated with idelalisib or duvelisib is plotted over time (n = 12 for each group in 2 independent cohorts of 6 mice). P values calculated by Log-rank (Mantel-Cox) test.
We recently showed that high RNA transcription levels are associated with increased AID off-target activity in B cells10. We analyzed GRO-Seq data in B cells treated with PI3Kδ inhibitors (Extended Data Fig. 4e, Supplementary Table 3) and found that translocation junction frequency positively correlated with transcription of AID targets (Fig. 3e), indicating that concomitant transcriptional changes induced by PI3Kδ blockade could influence translocation frequency in specific AID target sites.
To study the effect of PI3Kδ blockade on genomic instability in vivo, first we showed that both idelalisib and duvelisib significantly increased AID off-target activity at the endogenous c-myc locus in GC B cells (Extended Data Fig. 5a,b). Next, we investigated the effects of PI3Kδ blockade in the pristane-induced plasma cell (PC) tumor model where tumor formation is driven largely by AID-dependent Igh/c-myc translocations22. In this model, treatment with idelalisib or duvelisib significantly increased the frequency of PC tumor formation (Fig. 3f, Extended Data Fig. 5c–f). Overall, these data indicate that PI3Kδ blockade enhances AID-mediated genomic instability and tumor formation in vivo in mice.
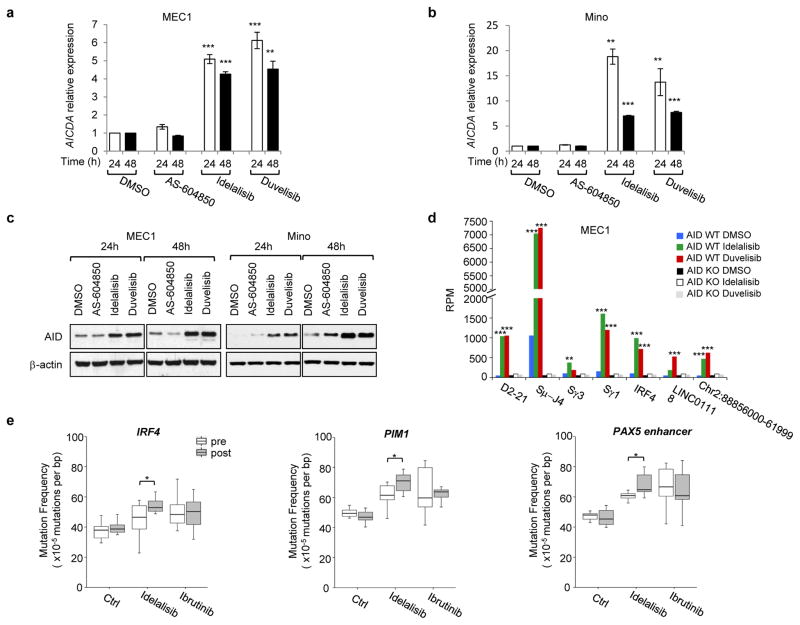
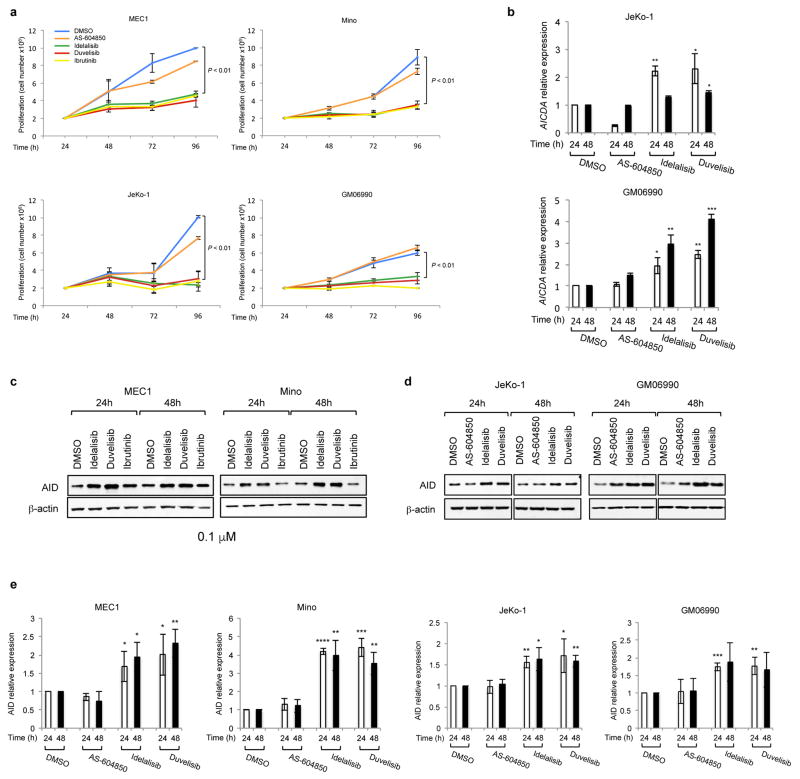
Since PI3Kδ inhibitors have been recently approved for the treatment of B cell malignancies, we next investigated whether PI3Kδ blockade also enhanced AID expression in human leukemia/lymphoma B cells. Idelalisib and duvelisib, but not AS-604850, blocked the proliferation of CLL (MEC1), MCL (Mino and JeKo-1) or B lymphoblastoid (GM06990) cell lines (Extended Data Fig. 6a), yet induced a significant increase in AID mRNA (Fig. 4a–b, Extended Data Fig. 6b) and protein expression (Fig. 4c, Extended Data Fig. 6c–e). To test whether enhanced AID expression by PI3Kδ blockade was sufficient to increase genomic instability, we introduced DSBs in the human c-MYC locus via CRISPR/Cas9 technology and adapted HTGTS to human B cells (Extended Data Fig. 7a,b). Translocation junctions induced by off-target Cas9 activity were easily identified as unique for each sgRNA sequence in MEC1 and JeKo-1 cell lines, and were excluded from the analysis (Extended Data Fig. 7c–h). HTGTS not only identified the IgH locus as a target for recurrent translocations in both cell lines, but also revealed translocations to recently identified AID off-targets in CLL and human lymphoma, such as PIM1, IRF4, miR142, and CXCR4 23–25. Remarkably, translocation to IgH as well as to AID off-targets were significantly increased by idelalisib and duvelisib treatment (Fig. 4d, Extended Data Fig. 8a–d, Supplementary Table 4) whereas no significant changes were observed in Cas9 off-targets that were used as internal control (Extended Data Fig. 7c–h). When we knocked-out AID in MEC1 cells by CRISPR/Cas9, chromosomal translocations to AID on- and off-targets were completely abrogated (Fig. 4d, Extended Data Fig. 8a,b,e). Thus, as in mouse B cells PI3Kδ blockade enhanced genomic instability by increasing AID expression also in malignant human B cells.
Figure 4. PI3Kδ inhibitors increase AID expression and genomic instability in human B cells.
a,b AICDA mRNA relative expression in the human MEC1 or Mino B cell lines treated with the indicated inhibitors (1 μM). Data are mean ± s.d. (n=3 technical replicates, n = 3 biological replicates). ** P ≤ 0.01, *** P ≤ 0.001; two-tailed Student’s t-test. c, Western blot for AID protein expression in MEC1 and Mino B cell lines (n = 4 biological replicates). For gel source data, see Supplementary Figure 1. d, Translocation junction frequency in AID on-target (D2-21 and Sμ-J4 region in the IGH locus) and AID off-target sites in MEC1 B cell line treated with 1 μM DMSO, idelalisib or duvelisib. AID knock-out (KO) MEC1 cells were generated by CRISPR/Cas9 mediated deletion (Extended Data Fig. 8e). Data are from pooled HTGTS libraries from 3 independent experiments. ** FDR ≤ 0.01, *** FDR ≤ 0.001. Statistical analysis in Methods. e, Mutation frequency calculated on the regions downstream TSS for IRF4 and PIM1, and the PAX5 enhancer region in CLL patients treated with idelalisib or ibrutinib. Untreated CLL patients are used as control. Boxplots indicate cumulative frequencies of C-to-T or G-to-A transition mutation in DNA samples collected before (pre) and after (post) treatment in each patient (control n=8, idelalisib n=10, ibrutinib n=10, Supplementary Table 6). Whiskers extend to a maximum of 1.5X interquartile range (IQR) beyond the box. * P < 0.05, paired samples two-tailed Student’s t-test.
Together with PI3Kδ inhibitors, drugs that block the Bruton tyrosine kinase (BTK) activity, such as ibrutinib, have been approved for the treatment of CLL and MCL5,26. Because BTK modulates PI3K signaling27 as shown by the block of AKT phosphorylation induced by ibrutinib in B cells (Extended Data Fig. 9a,b), we asked whether also AID expression and genomic instability were increased by ibrutinib. In CSR-activated mouse B cells, ibrutinib reduced cell proliferation, yet enhanced AID mRNA and protein levels as well as CSR (Extended Data Fig. 9c–f). Importantly, ibrutinib treatment increased translocation junctions to Igh and AID off-targets similarly although less potently than PI3Kδ inhibitors (Extended Data Fig. 2a,e and 9g,h, Supplementary Table 5) and increased the frequency of PC tumor formation in pristane-treated mice (Extended Data Fig. 5). In human B cells, ibrutinib reduced cell proliferation (Extended Data Fig. 6a), yet significantly enhanced AID expression (Extended Data Fig. 10a,b) as well as the frequency of translocation junctions to AID on-target and off-target sites (Extended Data Fig. 10c, Supplementary Table 4).
Finally, we asked whether evidence for an increased AID activity could be found in patients treated with idelalisib or ibrutinib. We collected samples of CLL patients treated for several months with either idelalisib or ibrutinib and analyzed the frequency of SHM in known AID off-target genes by paired comparison of samples before and after treatment. A significant increase of SHM frequency was found in the regions downstream the TSS of IRF4 and PIM1 genes and in the PAX5 enhancer region, all known AID off-targets24, in patients treated with idelalisib, but not in untreated or ibrutinib treated patients as well as in a series of control genes (Fig. 4e, Extended Data Fig. 10d, Supplementary Table 6).
Overall, we provide evidence that PI3Kδ blockade enhances genomic instability in normal and malignant B cells by increasing AID levels. These findings have several potential implications. First, increased AID levels induced by PI3Kδ inhibitors could facilitate secondary oncogenic mutations or translocations in normal or malignant B cells in patients. In this context, ibrutinib appears to have more limited effects, likely because of an indirect inhibition of the PI3K pathway. Since AID activity is thought to have an important role in CLL biology as mutational signatures consistent with AID activity can be tracked in the evolution of CLL clones28 and high AID expression in CLL is a poor prognostic factor29, long-term monitoring for clonal evolution over time in patients on these drugs is essential, particularly given that current follow-up is very short. Furthermore, increased AID expression could accelerate resistance to targeted therapy through an increased mutational rate, as described for targeted therapies against BCR-ABL30. Finally, this work demonstrates the general principle that drugs considered non DNA damaging may actually induce genomic instability, and represents the first application of a genome-wide translocation assay to demonstrate the genotoxic effects of drugs previously considered non DNA damaging.
Online Content. Methods, along with any additional Extended Data display items and Source Data, are available in the online version of the paper; references unique to these sections appear only in the online paper.
METHODS (online-only)
PI3K inhibitors
Idelalisib (CAL-101, GS-1101; PI3Kδ inhibitor), duvelisib (IPI-145, INK1197; PI3Kγδ dual inhibitor), AS-604850 (PI3Kγ inhibitor) and ibrutinib (BTK inhibitor) were purchased from Selleckchem (Houston, TX) and all used at 1 μM concentration in most experiments. In some experiments inhibitors were used at 0.1 μM or 0.5 μM concentrations, as specifically indicated in the corresponding figure legend.
Mice
Wild type (WT) mice, c-myc25xI-SceI and c-myc25xI-SceI;AID−/− in the 129S2 mice background. All mice carrying the 25xI-SceI cassette were heterozygous for the modified c-myc allele containing the I-SceI cassette and were previously described9,31. At least 3 independent mice of same sex (females) and similar age (8–12 weeks) were used for each experiment with B cells. No mice were excluded from the analysis and no randomization or blinding method was used. Animal experiments were performed under protocols approved by the Institutional Animal Care and Use Committee (IACUC) of Boston Children’s Hospital (Protocol 16-01-3093R) or by the Italian Ministry of Health for the University of Torino (approval n. 143/2013-B). They were housed and maintained in the specific pathogen-free facility at Boston Children’s Hospital.
Human leukemia/lymphoma cell lines
Human leukemia/lymphoma cell lines MEC1 (Chronic Lymphocytic Leukemia), JeKo-1 and Mino (Mantle Cell Lymphoma), and GM06990 (EBV-immortalized lymphoblastoid B cell line) were cultured in RPMI 1640 medium (Thermo Fisher Scientific, USA) supplemented with 10% fetal bovine serum (FBS), penicillin-streptomycin (100 units/ml) and L-Glutamine (2 mM). All cell lines tested negative for Mycoplasma contamination. Cell lines were authenticated as they were purchased from ATCC (JeKo-1, Mino), DSMZ (MEC1) or the Coriell Institute (GM06990).
For experiments with PI3K inhibitors, cells were plated in 6 well plates at a concentration of 5×105 cells/ml. Cells were collected at the indicated time points for RNA or protein isolation or for flow cytometry analysis or after 4 days of treatment to isolate genomic DNA for HTGTS libraries.
Samples from CLL patients
DNA before and after therapy from CLL patients (untreated n=8; idelalisib n=10; ibrutinib n=10; total 56 samples) was extracted from peripheral blood samples. Samples from idelalisib treated patients were collected in the 99-224 CLL Repository approved by the Dana-Farber Cancer Institute (DFCI) Institutional Review Board (IRB). Ibrutinib treated patients were enrolled on a phase 2, open-label, single-center, investigator-initiated study approved by the National Heart, Lung, and Blood Institutional Review Board at the National Institutes of Health (registered at www.clinicaltrials.gov NCT01500733). All patients provided written informed consent. All cases were diagnosed according to the International guidelines and consented according to internal protocols. Details of treatment and sample collection for each patient are summarized in Supplementary Table 6.
B cell purification and ex-vivo activation
Splenic mouse B cells were isolated from mice by immunomagnetic depletion with anti-CD43 MicroBeads (Invitrogen) as previously described9. Briefly, all the non-B cells were depleted with anti-CD43 MicroBeads combined with Dynabeads Biotin Binder (Invitrogen); naive B cells were cultured at a concentration of 5×105 cells/ml in RPMI medium supplemented with 15% FBS, penicillin-streptomycin (100 units/ml), L-Glutamine (2 mM), anti-CD40 (1 μg/ml, eBioscience) and recombinant mouse IL-4 (20 ng/ml; PeproTech). The purity of B cell population was typically 96–98% in all experiments, as documented by flow cytometric analysis of B220 expression in enriched cells. Cells were collected after 4 days of treatment with inhibitors to isolate genomic DNA for HTGTS libraries and targeted re-sequencing experiments. For RNA and protein extraction cells were collected at the indicated time points. Class switch recombination (CSR) was measured by staining with PE-labeled anti-mouse IgG1 (BD Biosciences) and Cy5-PE-labeled anti-mouse B220 (eBiosciences). Data acquisition was performed using a FACSVerse flow cytometer (BD biosciences).
Mouse immunization and purification of germinal center B cells
For immunization, sheep blood Alsevers (BD) were washed with PBS and re-suspended in PBS at a concentration of 1×109 sheep red blood cells (SRBCs) per ml. 8 to 12-week old mice were immunized by intraperitoneal injection of 2×108 SRBCs in a 200 ml volume. After 5 days, a booster injection was performed using 5-fold more SRBCs. On day 6 and for 7 consecutive days, animals were daily administered vehicle (0.5 % carboxymethylcellulose, 0.05 % Tween 80 in ultra-pure water) or idelalisib or duvelisib (10 mg/kg q.d.) by oral gavage. Spleens were collected at the end of treatment, placed on ice, washed in PBS to remove residual blood, cut into small pieces, crushed and physically dissociated using a Falcon cell strainer, and subjected to hypotonic lysis of erythrocytes. Mouse germinal center (GC) B cells were isolated from the spleens of immunized mice by immunomagnetic depletion: first non-B cells were depleted with anti-CD43 MicroBeads; next enriched B cells were incubated with a cocktail of biotinylated antibodies specific for CD11c (eBiosciences) and IgD (eBiosciences) to remove dendritic cells and mature naive B cells, respectively, as previously reported32. Enrichment of the GC B cells was evaluated with PE-labeled anti-mouse GL7 (eBiosciences) and Cy5-PE-labeled anti-mouse B220 (eBiosciences).
Induction of plasma cell (PC) tumor in mice
Eight weeks old female BALB/cAnNCrl mice were purchased from Charles River and housed in the University of Torino mouse facility under protocol approved by the Italian Ministry of Health. Commercial pristane (2,6,10,14-tetramethylpentadecane) was purchased from Sigma, St Louis, MO, USA. Pristane was administered by two 0.5 ml i.p. injections given 70 days apart, as previously described22. The mice were divided into 4 different groups: vehicle group (0.5 % carboxymethylcellulose, 0.05 % Tween 80 in ultra-pure water) and idelalisib or duvelisib or ibrutinib groups. Drugs were administered by oral gavage (10 mg/kg q.d.) for 70 days (5 days a week). Mice were followed-up for the development of ascites and were sacrificed when they reached a point of distress. Several tissues, including peritoneal tumor nodules, inflammatory granuloma, liver, spleen, intestine, were processed for histologic analysis. For histology, tissues and tumor nodules were fixed in 10% formalin over-night and transferred to 70% ethanol and embedded in paraffin. Four-μm-thick sections were stained with hematoxylin and eosin to evaluate the distribution of clusters of atypical plasma cells. PC tumors were diagnosed by finding clusters of 10 or more hyperchromatic, atypical plasma cells in hystology specimens, as previously reported22.
PCR amplification of junction fragments in pristane experiment
PCR for Igh/c-myc translocations was performed on 500 ng of genomic DNA extracted from ascites by adapting protocols previously described33,34. Briefly, we performed two rounds of PCR with Phusion High-Fidelity DNA Polymerase (Thermo Fisher Scientific) using primers listed in Supplementary Table 7. All PCR reactions were performed with appropriate positive controls (genomic DNA obtained from mouse B cells activated in vitro and treated with PI3Kδ inhibitors) and negative controls (DNA from AID−/− mouse B cells). PCR conditions were 98°C for 30s followed by 25 cycles (98°C, 10s; 62°C, 30s; 72°C 4 min) for both the first and second round. PCR amplicons were purified and sequenced to confirm Igh/c-myc translocations.
Protein extraction and Western blot analysis
Whole cell extracts were obtained from purified mouse B cells or cell lines treated with 1 μM PI3K inhibitors using GST-FISH buffer (10 mM MgCl2, 150 mM NaCl, 1% NP-40, 2% Glycerol, 1 mM EDTA, 25 mM HEPES (pH 7.5)) supplemented with protease inhibitors (Roche), 1 mM phenylmethanesulfonylfluoride (PMSF), 10 mM NaF and 1 mM Na3VO4. Extracts were cleared by centrifugation at 12,000 r.p.m. for 15 min. The supernatants were collected and assayed for protein concentration using the Bio-Rad protein assay method. 20 μg of proteins were loaded on 12% Mini-PROTEIN TGX gels (BIO-RAD), transferred on nitrocellulose membrane (GE Healthcare), blocked with 5% Skim milk (BIO-RAD). Primary antibodies for immunoblotting included: rat monoclonal anti-mouse AID (mAID-2 clone, eBioScience, CAT NO #14-5959-82), mouse monoclonal anti-human AID (ZA001, Life Technologies, CAT NO # 39-2500), rabbit monoclonal anti-PI3K p110δ (Y387, Abcam, CAT NO #32401), rabbit polyclonal anti-β–actin (Sigma, CAT NO #A2066), rabbit monoclonal anti-Phospho-AKT (S473) (D9E, Cell Signaling Technology, CAT NO #4060), rabbit monoclonal anti-AKT (pan) (C67E7, Cell Signaling Technology, CAT NO #4691). Membranes were developed with ECL solution (GE Healthcare). AID protein abundance was measured by ImageJ software and normalized for the β-actin intensity of the corresponding lane.
Quantitative RT-PCR analysis
Total RNA was isolated from primary mouse B cells and human lymphoma cells by TRIzol (Life Technologies). Before cDNA synthesis, 1 μg of total RNA was treated with 5 U/μl RNase-free recombinant DNase I (Roche). cDNA was transcribed using iScript cDNA synthesis kit following the manufacturer’s instructions (Biorad). All quantitative RT-PCR experiments were performed in triplicate on ICycler iQ Real-Time PCR Detection System (Bio-Rad Laboratories) with SYBR green dye. Primer pairs are listed in Supplementary Table 7. Expression levels for individual transcripts were normalized against β-actin for murine samples or HuPO for human samples. Fold change in transcript levels were calculated as fold change over not treated cells.
Retroviral transduction of mouse primary B cells
Retroviral supernatants were prepared from Phoenix packaging cells transfected with retroviral vectors. The pMX-I-SceI vector has been previously described9, PI3Kδ retroviruses (PI3K p110δ WT and PI3K p110δE1021K) were kindly provided by Drs. Klaus Okkenhaug and Fabien Garcon (The Babraham Institute, UK)13.
Briefly, Phoenix-ECO cells, a second-generation retrovirus producer cell line, were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), penicillin-streptomycin (100 units/ml) and L-Glutamine (2 mM). To generate retroviral particle, 3.5×106 Phoenix-ECO cells were plated per 10 cm dish. The following day, cells were transfected by calcium phosphate transfection method with 10 μg of each plasmid and 5 μg of pCL-Eco retrovirus packaging plasmid. The media was changed 8 h post-transfection. The viral supernatant was collected 48 h post-transfection, passed through a 0.45 μm filter, pooled and used either fresh or snap frozen.
For transduction, one volume of viral supernatant with polybrene (6 μg/ml) was added to mouse B cells after 24 h of activation with anti-CD40 plus IL-4, as previously described9. Plates were spin for 1.5 h at 2,400 r.p.m. and incubated overnight. Cells were washed and plated at a concentration of 5×105 cells/ml. At day 4 of stimulation, transduction efficiency was evaluated by measuring the percentage of transduced cells by EGFP expression (typical range was 50% to 85% of transduced cells). PI3K inhibitors were added at time of transduction and then maintained for the whole duration of the activation. CSR was evaluated by staining with Cy5-PE-labeled anti-mouse B220 (eBiosciences) and PE-labeled anti-mouse IgG1 (BD Biosciences). Data acquisition was performed using a FACSVerse flow cytometer (BD biosciences). CSR ranged between 15% and 40% for retrovirus- transduced B cells. DNA was isolated from cells at day 4 of culture according to standard methods for HTGTS libraries.
CRISPR/Cas9 sgRNA design and cloning
For SpCas9 expression and generation of single guide RNA (sgRNA), the 20-nt target sequences were selected to precede a 5′-NGG protospacer-adjacent motif (PAM) sequence. The two c-MYC sgRNAs (#1 and #2) and the AICDA sgRNA were designed with the CRISPR design tool from F. Zhang laboratory (http://crispr.mit.edu/). Oligonucleotides were purchased from Integrated DNA technology (IDT), annealed and cloned into the BsmbI-BsmBI sites downstream from the human U6 promoter in LentiCRISPR v2 plasmid (Addgene #52961). Oligonucleotides used in this study for cloning are listed in Supplementary Table 7.
Lenti-viral particles production
HEK293FT cells (Invitrogen) were maintained in 10% FBS-containing DMEM. To generate lentiviral particles, 5.5×106 HEK293FT cells were plated per 10 cm dish. The following day, cells were transfected by calcium phosphate transfection method with 7.2 μg of lentiCRISPR v2 plasmid, 3.6 μg of VSVG, 3.6 μg of RSV-REV, and 3.6 μg of PMDLg/pPRE. The media was changed 8 h post-transfection. The viral supernatant was collected 36 h post-transfection, passed through a 0.45 μm filter, pooled and used either fresh or snap frozen.
Human cell lines transduction with CRISPR/Cas9 lentiviruses
For transduction of JeKo-1 and MEC1 with c-MYC CRISPR/Cas9 lentiviruses, a total number of 4×105 human neoplastic cells were plated into six-well plates, at a concentration of 2×105 cells/ml. Lentiviral transduction was performed adding lentiviral supernatant, spinning for 1.5 h at 2,400 r.p.m. in the presence of polybrene (6 μg/ml). The viral supernatant was exchanged for fresh medium 8 h later. PI3K inhibitors were added 8 h before the infection and after washing. After 48 h, cells were selected with 0.2 μg/ml of puromycin to enrich for transduced cells. The cells were collected after 3 days from the puromycin addition. Genomic DNA was extracted as previously described for HTGTS libraries.
Generation of AID KO cell line clones
To generate the AID knock-out (KO) MEC-1 cell line, MEC-1 cells were transduced with AID CRISPR/Cas9 lentivirus according to the protocol described above. After 48 hours from transduction cells were selected with 0.2 μg/ml of puromycin for 3 days. The selected cells were seeded as single colonies in 96-well plates by serial dilutions. After 3–4 weeks of culture, cells derived from each colony were used to assess AID knock-out by western blotting and genomic sequencing of the sgRNA target region.
Surveyor assay
The genomic region flanking the CRISPR target sites was PCR amplified (Surveyor primers are listed in Supplementary Table 7), and products were purified using PCR purification kit (QIAGEN) following the manufacturer’s protocol. 400 ng total of the purified PCR products were mixed with 2 μl 10X Taq DNA Polymerase PCR buffer (Life Technologies) and ultrapure water to a final volume of 20 μl, and subjected to a re-annealing process to enable heteroduplex formation: 95°C for 10 min, 95°C to 85°C ramping at −2°C/s, 85°C to 25°C at −0.25°C/s, and 25°C hold for 1 min. After re-annealing, products were treated with SURVEYOR nuclease and SURVEYOR enhancer S (Transgenomics) following the manufacturer’s recommended protocol, and analyzed on 2% high-resolution agarose gel (Sigma Aldrich). Gels were stained with ethidium bromide (Sigma Aldrich) and imaged with a Gel Doc gel imaging system (Bio-rad). Quantification was based on relative band intensities. Indel percentage was determined by the formula, 100 X (1 − (1 − (b + c)/(a + b + c))1/2), where a is the integrated intensity of the undigested PCR product, and b and c are the integrated intensities of each cleavage products.
Generation of high-throughput genome-wide translocation sequencing (HTGTS) libraries
DNA was prepared from mouse and human B cells at day 4 of culture using rapid lysis buffer containing 10 μg/ml Proteinase K and incubation at 56°C overnight, followed by standard isopropanol extraction, wash in ethanol 70% and resuspension in TE buffer. HTGTS libraries were generated by emulsion-mediated PCR (EM–PCR) methods as previously described9. In brief, genomic DNA was digested overnight with HaeIII frequent cutter enzyme. HaeIII-generated blunt ends were A-tailed with Klenow polymerase (3′-5′ exo-; New England Biolabs). An asymmetric adapter (composed of an upper linker and a lower 3′-modified linker) was then ligated to fragmented DNA. To remove the unrearranged I-SceI cassettes or the unrearranged endogenous c-myc locus, ligation reactions were digested with both EcoRV and XbaI. In the first round of PCR, DNA was amplified using a biotinylated forward primer and an adapter-specific reverse primer and Phusion polymerase (Thermo-Scientific). 20 PCR cycles were performed in the following conditions: 98°C for 10 seconds, 58°C for 30 seconds, and 72°C for 30 seconds. Multiple reactions were performed in generating large-scale libraries. Biotinylated PCR products were isolated using the Dynabeads MyOne Streptavidin C1 kit (Invitrogen), followed by an additional 2 hour-digestion with blocking enzymes was performed. PCR products were eluted from the beads via a 30 minutes incubation at 65°C in 95% formamide/10 mM EDTA and purified. The purified products were then amplified in a second round with em-PCR in an oil-surfactant mixture. The emulsion mixture was divided into individual aliquots and PCR was performed using the following conditions: 20 cycles of 94°C for 30 seconds, 60°C for 30 seconds, and 72°C for 1 minute. Following PCR, the products were pooled and centrifuged in a table-top centrifuge for 5 minutes at 14,000 r.p.m. to separate the phases and the oil layer was removed. The sample was then extracted three times with diethyl ether and DNA was re-purified. The third, non-emulsion, round of PCR (10 cycles) was performed with the same primers as in round 2, but with the addition of linkers and barcodes for Illumina MiSeq sequencing. PCR products were size-fractionated for DNA fragments between 300 and 800 base pairs on a 1% agarose gel, column purified (QIAGEN) before loading onto Illumina Miseq machine for sequencing.
Nucleotide sequences of junctions were generated by Mi-Seq (Illumina NS500 SE150) sequencing at the Molecular Biology Core Facilities of the Dana-Farber Cancer Institute. At least three independent libraries were generated and analyzed for each experimental condition (Supplementary Table 1). Oligonucleotide primers used for mouse and human libraries preparation are listed in Supplementary Table 7.
HTGTS data analysis
Data process
First, we applied prinseq 0.235 to remove sequences with exact PCR duplicates, mean quality score < 20 and length < 50. Next, reads for each experimental condition were demultiplexed by designed barcode, and then filtered by the presence of the primer plus additional 5 downstream bases as bait portion. Barcodes and primers used are listed in Supplementary Table 1 and 7. Lastly, the barcode, primer and bait portion of the remained sequences were masked for alignment analysis.
Alignment and Filtering
The sequences for each experiment were aligned and filtered as previously described9. Briefly, we aligned sequences to the mouse reference genome (GRCm37/mm9) or human genome (GRCh38/hg38) using BLAT, and then filtered artificial junctions by removing PCR repeats (reads with same junction position in alignment to the reference genome and a start position in the read less than 3 bp apart), invalid alignments (including alignment scores < 30, reads with multiple alignments having a score difference < 4 and alignments having 10-nucleotide gaps) and ligation artifacts (for example, random HaeIII restriction sites ligated to bait breaksite). Translocation junction position was determined based on the genomic position of the 5′ end of the aligned read.
Hotspot Identification
Translocation junctions data from similar size biological replicates were pooled for hotspots identification. First, we employed SICER 1.136 to identify candidate regions where HTGTS junctions were significantly enriched against genome-wide background. The parameters used were as follows: window size, 1000; gap size, 2000; e, 0.000001; redundancy, 1; effective genome fraction, 0.77 for mouse or 0.74 for human. Next, we eliminated from analysis the following hotspots: (1) hotspots in the region ± 4Mb around Myc bait breaksite including the Pvt1 gene as previously described9; (2) hotspots with junctions number less than 5; (3) hotspots with strand bias. We used the following entropy formula to measure strand bias as S=−P×log2(P)−(1−P)×log2(1−P), where P is the percentage of junctions from the plus stand, and 1−P is the percentage of junctions from the minus strand. If P or 1−P were <10% (entropy S < 0.47) we eliminated the hotspot for a strand bias; (4) hotspots without significant enrichment against the local background. The local background p-value was calculated by Poisson distribution against the region that surrounds the hotspot (± 3 times the size of the hotspot). Bonferroni correction was used to adjust p-value for multiple tests. We set adjusted P = 0.01 as significance level. For JeKo-1, due to its complex karyotype37 which increases the local noise level, we set more stringent criteria for hotspot identification, including adjust P < 0.00001 and region size < 30 kb. Hotspots from different experiments that partially overlapped were merged to define common hotspot regions that were used as reference to compare junction frequency between different experiments.
Junction frequency calculation and representation
Translocation junction frequencies in hotspots were normalized to reads per million (RPM). In boxplot for fold-change comparison, to avoid “division by zero” error, 0 was replaced with 1, and then normalized to corresponding RPM in library. For clustering heatmap, the RPM was transformed in a log2 value, and then median centered. The genome-wide translocation circle plots were made using Circos tool38. Translocation junction distributions were visualized by IGV 2.3.639. For translocation frequency distribution around ConvT or SE centers, centers were defined as the central bp position of the ConvT or SE region, as we previously defined10. Regions ±4 Mb around the I-SceI c-myc breaksite on chr. 15 and the IgH S regions on chr. 12 were excluded in the analysis of junctions around TSS, ConvT or SE centers. For SE analysis, hotspots embedded within two adjacent SEs with center-to-center distance < 100kb were excluded because it was not possible to univocally assign them with one of the two SEs. All ChIP-Seq data used in this study were obtained from previously published data including SE10, AID17, Spt5 and Pol II20.
Statistical comparison of junction frequency in hotspots
Statistical significance of differential junction frequency in hotspots were performed using SICER 1.136 with the following parameters: window size = 1000, gap size = 2000, E-value = 0.000001, effective genome fraction=0.77 (mouse) or 0.74 (human) and FDR = 0.01 or FDR = 0.1.
GRO-Seq assay and analysis
Nuclei were isolated at day 2 from B cells activated with anti-CD40 plus IL-4 and treated with PI3K inhibitors, as previously described9. GRO-Seq libraries were sequenced on the Hi-Seq 2000 platform with single-end reads and analyzed as follows: GRO-seq data were aligned using Bowtie software40 mouse reference genome (GRCm37/mm9). Uniquely mapped, nonredundant sequence reads were retained. Next, we used HOMER software to count reads and calculate the nascent RNA expression levels as RPKM (reads per 1000bp per million mapped reads) in whole genes or in focal translocation clusters, and to identify transcripts from both strands of chromosomes41. The ConvT region was defined as sense and antisense transcription overlaps that were longer than 100 bp10. Statistical analysis for differential expression and log fold-change calculation were performed using DESeq242 in whole genes or in focal translocation clusters. The MA Plot of log fold-changes against mean of normalized counts were generated by function plotMA in R package DESeq242.
Amplification and targeted re-sequencing
Phusion High Fidelity DNA polymerase (Thermo-Scientific) was used to amplify selected regions from template genomic DNA. Oligonucleotide primers are listed in Supplementary Table 7: amplification conditions for each gene are available on request. Amplification products were purified using PCR purification kit (QIAGEN) and GEL extraction kit (QIAGEN) following the manufacturer’s protocol and sequenced bi-directionally in a Mi-Seq (Illumina NS500) sequencing platform at the Molecular Biology Core Facilities of the Dana-Farber Cancer Institute.
SHM data analysis
For SHM calculations, mouse and human intragenic and intergenic regions were targeted re-sequenced with primers indicated in Supplementary Table 7. Sequences with mean quality score < 20 and length < 50 were removed. Samples with less than 100 reads were excluded from analysis. The remained sequences were used to calculate mutation rate. Sequences obtained from each designed region were aligned to the reference sequence using BLASTN with alignment length > 200. Mutations were calculated after filtering steps, as previously described43. Briefly, mutations first had to pass a Neighbourhood Quality Standard (NQS) criteria requiring a minimum Phred score of 30 for the mutation itself, and 20 for the five adjacent bases on either side. Mutations that were within five bases of more than two additional mutations were excluded. Mutations within two bases of a deletion or insertion were also excluded. In addition, bases with mutation rate > 0.01 were excluded as a result of overwhelming influence of sequence error or SNP, of which bases with mutation rate > 0.2 were further regarded as SNP and were excluded. Finally, the average base mutation rate of 1–200 bp passing the above criteria were calculated from forward sequence, as well as reverse sequences if applicable. For average base mutation rates of C-to-T or G-to-A transitions, only C or G bp sites we counted.
Mutations on the VB1-8 productive allele were performed and analyzed as recently described19.
Code availability
Source code for genomic event analysis tools (GEAT) developed in our laboratory to perform the analysis is available at https://github.com/geatools/geat.
Data availability statement
All sequencing data has been deposited in the Gene Expression Omnibus database under accession number GSE77788. Source data for figures are provided with the paper.
Extended Data
Extended Data Figure 1. Phosphatidylinositol 3-Kinase (PI3K)δ blockade increases AID expression and CSR in activated mouse B cells.
a, Schematic representation of the in vivo experiment in WT mice immunized with sheep red blood cells (SRBCs) and treated with the indicated inhibitors (n = 6 biological replicates). b, Representative dot plots of IgG1 switched GL7+/B220+ germinal center (GC) B cells from mice treated with vehicle or the PI3Kδ inhibitors. c, Mean IgG1 CSR analyzed by flow cytometry in control and idelalisib or duvelisib treated mice. Data are expressed as mean ± s.d. (n=3). ** P ≤ 0.01, two-tailed Student’s t-test. d,e, IgG1 CSR analyzed by flow cytometry in activated B cells treated with 0.5 μM (d) or 0.1 μM (e) of the indicated drugs. Data are expressed as mean ± s.d. (n=3). * P < 0.05, ** P ≤ 0.01, *** P ≤ 0.001, two-tailed Student’s t-test f, Western blot for AID protein expression in activated B cells treated with the indicated inhibitors (0.1 μM) (n = 3 biological replicates). For gel source data, see Supplementary Figure 1. g, Viable cells were counted at the indicated time points by Trypan Blue exclusion in activated B cells treated with the indicated inhibitors (1 μM). Data are expressed as mean ± s.d. (n=3). P values calculated by two-tailed Student’s t-test. h, Representative western blot for AID and PI3K p110δ protein expression in mouse B cells transduced with retrovirus expressing PI3K p110δWT or the p110δE1021K active mutant (n = 4 biological replicates). i, Aicda mRNA levels analyzed by qRT-PCR in activated B cells transduced with PI3K p110δWT or the p110δE1021K active mutant. Data are expressed as mean ± s.d. (n=3). P values calculated by two-tailed Student’s t-test by comparing PI3K p110δE1021K vs PI3K p110δWT cells. j, IgG1 CSR analyzed by flow cytometry in activated B cells expressing PI3K p110δWT or the p110δE1021K active mutant. Data are expressed as mean ± s.d. (n=3). * P < 0.05, ** P ≤ 0.01, two-tailed Student’s t-test
Extended Data Figure 2. Frequency of AID mediated translocation junctions in activated mouse B cells is increased by PI3Kδ blockade.
a, Frequency of chromosomal translocations between the c-myc and the Igh and Igk loci, represented as reads per million (RPM) in WT and AID−/− activated B cells treated with the indicated inhibitors. Significance is calculated as false discovery rate (FDR) by comparing AS-604850, idelalisib or duvelisib to DMSO treated B cells as indicated in the Methods. ** FDR ≤ 0.01, *** FDR ≤ 0.001, **** FDR ≤ 1×10−10. b, Detailed view of the distribution of translocation junctions in the Igh locus from WT and AID−/− mouse B cells treated with the indicated inhibitors. Numbers of translocation junctions in focal clusters are indicated in bold. c, Histograms showing the AID off-targets with significantly increased frequency of HTGTS junctions (FDR < 0.001) induced by idelalisib treatment as compared to DMSO (54 out of 59 off-target sites, 91.5%) in WT and AID−/− activated B cells. Significance is calculated as indicated in the Methods. Circos plots show the overview of genome-wide translocation distribution. Individual translocations from c-myc to AID target sites are represented as arcs originating from c-myc DSB breaks on chromosome 15. Thickness and red color intensity of the arcs represent the fold increase of translocation frequency in idelalisib vs DMSO treated cells. d, Histograms showing the AID off-targets with significantly increased frequency of translocation junctions (FDR < 0.001) induced by duvelisib treatment as compared to DMSO (55 out of 63 off-target sites, 87.3%) in WT and AID−/− activated B cells. Significance is calculated as indicated in the Methods. Circos plots show the overview of genome-wide translocation distribution. e, Translocation junction distribution (top), GRO-Seq (middle) and ChIP-Seq (bottom) profiles in three representative AID off-target genes (Pim1, Cd83, Bcl6) in WT and AID−/− activated B cells treated with idelalisib, duvelisib or ibrutinib. GRO-Seq sense and antisense transcription is displayed in blue and red profiles. Numbers of translocation junctions in focal clusters are indicated in bold.
Extended Data Figure 3. Frequency of AID on-target and off-target translocation junctions is reduced in activated mouse B cells transduced with the PI3K p110δE1021K active mutant.
a, Histograms showing junction frequency in activated mouse B cells transduced with retrovirus expressing PI3K p110δWT or the p110δE1021K active mutant. Significance is calculated as FDR by comparing PI3K p110δWT to p110δE1021K transduced B cells as indicated in the Methods. ** FDR ≤ 0.01, *** FDR ≤ 0.001. b Detailed view of the distribution of translocation junctions in one representative I-Scel off-target (Fermt2). c, Detailed view of the distribution of translocation junctions in the Igh locus. Numbers of translocation junctions in focal clusters are indicated in bold. d, Histograms showing frequency of translocation junctions as RPM for I-Scel off-targets in WT and AID−/− activated B cells treated with the indicated inhibitors. e, Detailed view of the distribution of translocation junctions in two representative I-Scel off-target sites (Aco2 and Fermt2). Numbers of translocation junctions in focal clusters are indicated in bold.
Extended Data Figure 4. PI3Kδ blockade increases somatic hypermutation (SHM) frequency in Igh V region in activated mouse B cells and transcription of genes.
a, Map of mutations on the VB1-8 productive allele sequence in activated B cells treated with the indicated inhibitors. The y-axis indicates the mutation frequency at each nucleotide in sequences that have 0 to 2 mutations per sequence at day 4 of stimulation after subtraction of mutation frequency at day 0 of stimulation. Green shading indicates top s.e.m. from three independent mice. Orange and yellow bars mark the positions of AGCT and other RGYW motif that is not AGCT, respectively. One experiment is represented out of three biological replicates with similar results. b, Mutation frequency of nucleotide 454 (top) and 455 (bottom) of the VB1-8 productive allele at day 4 after subtraction of mutation frequency at day 0. Data represent mean of three independent stimulations of B cells from three independent mice. Error bars indicate s.d. Fold changes between each mean are indicated. c, Stacking bar chart shows percentage of sequences that have the indicated number of mutations per sequence. Data are displayed as the mean from three independent stimulations of B cells derived from three mice. Error bars indicate s.e.m. d, The number of reads from individual mice for the data are shown in (a). e, MA plots of log fold-changes generated by DESeq2 in R package against mean expression (normalized counts) from GRO-Seq data for idelalisib or duvelisib over DMSO. Red dots indicate significantly up-regulated genes (n=39 in idelalisib, n=47 in duvelisib) or down-regulated genes (n=32 in idelalisib, n=33 in duvelisib) with multiple test adjusted P value < 0.1 (Supplementary Table 3).
Extended Data Figure 5. PI3Kδ inhibitors and ibrutinib increase c-myc DSB formation and the incidence of plasma cell (PC) tumor in mice.
a, Detailed view of the distribution of rearrangements (deletions or inversions) in the c-myc locus in mice treated as above. Numbers of translocation junctions in focal clusters are indicated in bold. b, RPM Frequency of rearrangements (deletions or inversions) in the c-myc locus in GC B cell from mice treated in vivo with the idelalisib or duvelisib. Junctions within ±300 bp of primer region were excluded. Significance is calculated as false discovery rate (FDR) by comparing idelalisib or duvelisib to DMSO treated mouse as indicated in the Methods. ** FDR ≤ 0.01. c, Schematic representation of the experimental outline of pristane-induced PCT in mice treated with PI3Kδ inhibitors and ibrutinib. The mice were treated in two independent biological experimental replicates, each consisting of 6 mice per group. d, Direct PCR assay for Igh/c-myc translocation in mice with PC tumors. Translocations from c-myc to the IgHα locus are shown. Translocations for the only mouse in the vehicle group and from three example mice from treated groups are shown. Bands were purified from gels and were sequenced to confirm the Igh/c-myc translocation junction. For gel source data, see Supplementary Figure 1. e, Development of PC tumor in mice induced with pristane and treated with idelalisib, duvelisib or ibrutinib is plotted over time. The presence of PC tumors was confirmed by histology (n = 12 for each treatment in 2 independent cohorts of 6 mice). P values calculated by Log-rank (Mantel-Cox) test. f, Example histology of PC tumors in mice induced with pristane and treated with the indicated drugs. Magnification 40x; Scale bar = 50μm; Insets: high magnification image of clusters of atypical plasma cells.
Extended Data Fig 6. PI3Kδ blockade increases AID expression in human B cell lines.
a, Viable cells were counted at the indicated time points by Trypan Blue exclusion in MEC1, Mino, JeKo-1 and GM06990 cell lines treated with the indicated inhibitors (1 μM). Data are expressed as mean ± s.d. (n=3). P values calculated by two-tailed Student’s t-test. b, Histograms showing the AICDA mRNA relative expression in JeKo-1 and GM06990 cell lines treated with 1 μM inhibitors. Data are expressed as mean ± s.d. (n = 3 technical replicates, n = 3 biological replicates). * P < 0.05, ** P ≤ 0.01, *** P ≤ 0.001, two-tailed Student’s t-test. c, Western blot for AID protein expression in MEC1 and Mino B cell lines treated with 0.1 μM inhibitors (n = 2 biological replicates). d, Western blot for AID protein expression in JeKo-1 and GM06990 B cell lines treated with 1 μM inhibitors (n = 4 biological replicates). For gel source data, see Supplementary Figure 1. e, Histograms showing the AID protein relative expression at the indicated time points in MEC1, Mino, JeKo-1 and GM06990 cell lines treated with 1 μM inhibitors. AID abundance was measured by ImageJ software and normalized for the β-actin intensity of the corresponding lane. Data are expressed as mean ± s.d. (n=3 biological replicates). * P < 0.05, ** P ≤ 0.01, *** P ≤ 0.001, **** P ≤ 0.0001, two-tailed Student’s t-test
Extended Data Fig 7. Strategy to generate chromosomal translocations in human B cells from DSBs introduced in the c-MYC locus by CRISPR/Cas9 technology.
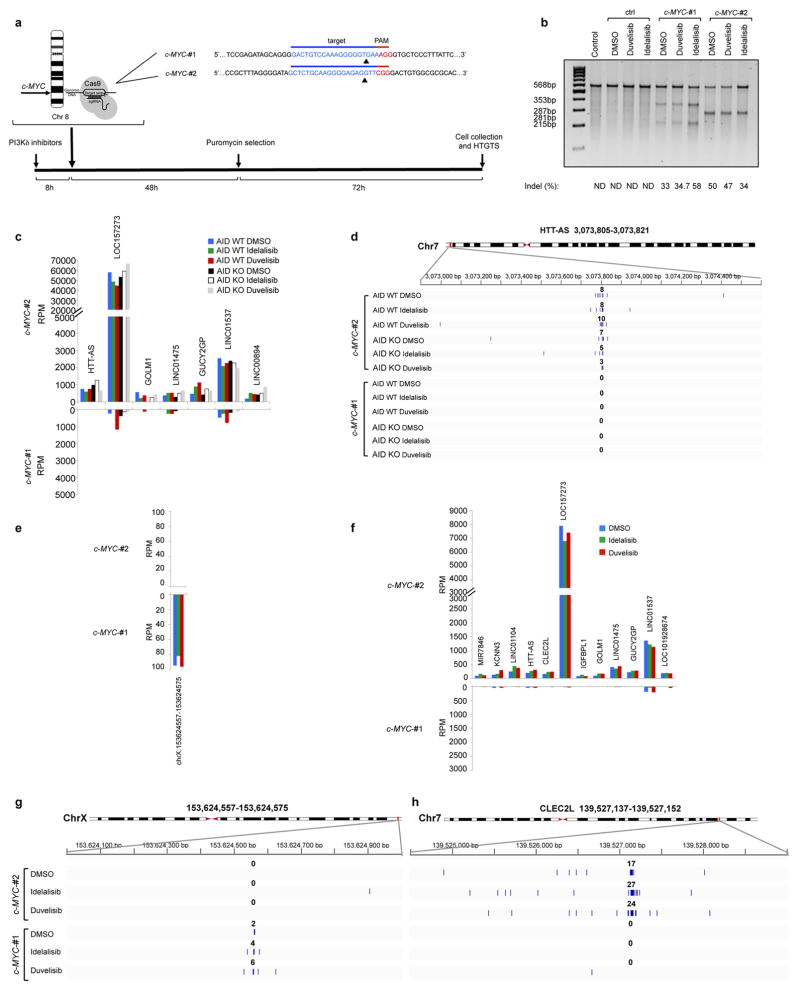
a, Schematic representation of the experimental strategy employed to generate chromosomal translocation from the c-MYC locus by introducing DSBs in c-MYC intron 1 by two different sgRNAs (c-MYC-#1 or c-MYC-#2). Arrowheads (black) indicate the cleavage sites introduced by Cas9 and PAM sequence is in red. b, Surveyor Assay to measure the cutting efficiency of the c-MYC gene targeted in JeKo-1 cell line. Locus modification efficiencies are analyzed 5 days after transduction of c-MYC-#1 or c-MYC-#2 Cas9 lentivirus. Estimated indel formation is indicated below each target. Black arrowheads indicate the size of the observed bands. N.D.: not detectable. One representative experiment out of three performed with comparable results is shown. For gel source data, see Supplementary Figure 1. c, Histograms showing frequency of translocation junctions in Cas9 off-target sites for c-MYC-#2 sgRNAs in MEC1 cell line treated with idelalisib or duvelisib. AID knock-out (KO) MEC1 cells were generated by CRISPR/Cas9 mediated deletion (Extended Data Fig. 8e). For each treatment, data are from pooled HTGTS libraries of similar size from independent experiments as indicated in Supplementary Table 1. d, Detailed view of the distribution of translocation junctions in representative Cas9 off-target site for c-MYC-#2. Numbers of translocation junctions in focal clusters are indicated in bold. e,f Histograms showing translocation junctions frequency in Cas9 off-targets sites for (e) c-MYC-#1 or (f) c-MYC-#2 sgRNAs in JeKo-1 cell line treated with idelalisib or duvelisib. For each treatment, data are from pooled HTGTS libraries of similar size as indicated in Supplementary Table 1 g,h, Detailed view of the distribution of translocation junctions in representative Cas9 off-target sites for c-MYC-#1 (g) or c-MYC-#2 (h). Numbers of translocation junctions in focal clusters are indicated in bold.
Extended Data Fig 8. Translocations to AID off-targets are increased by idelalisib and duvelisib treatment in MEC1 and JeKo-1 cell line.
Distribution of translocation junctions in the IgH locus (a) and in the IRF4 AID off-target gene (b) in MEC1 cells. AID knock-out (KO) MEC1 cells were generated by CRISPR/Cas9 mediated deletion. Numbers of translocation junctions in focal clusters are indicated in bold. c, Translocation junction frequency in AID on-target and AID off-target sites in JeKo-1 B cell line treated with DMSO, idelalisib or duvelisib (1 μM). Data are from pooled HTGTS libraries of similar size (Supplementary Table 1 and 4) from 3 independent experiments. Statistical analysis in Methods. * FDR ≤ 0.1, ** FDR ≤ 0.01. d, Distribution of translocation junctions in the PIM1 AID off-target gene in JeKo-1 B cell line. Numbers of translocation junctions in focal clusters are indicated in bold. e, AID knock-out (KO) MEC1 cells were generated by CRISPR/Cas9 mediated deletion, cloned and validated by indel sequencing of the Cas9 target site and by AID protein expression. Western Blot for AID showing the parental cells (WT), two AID KO clones (#4 and #10) and one AID WT clone (#6). AID KO clone #4 was treated with 1 μM DMSO, idelalisib or duvelisib (right panel) (n = 2 biological replicates). For gel source data, see Supplementary Figure 1.
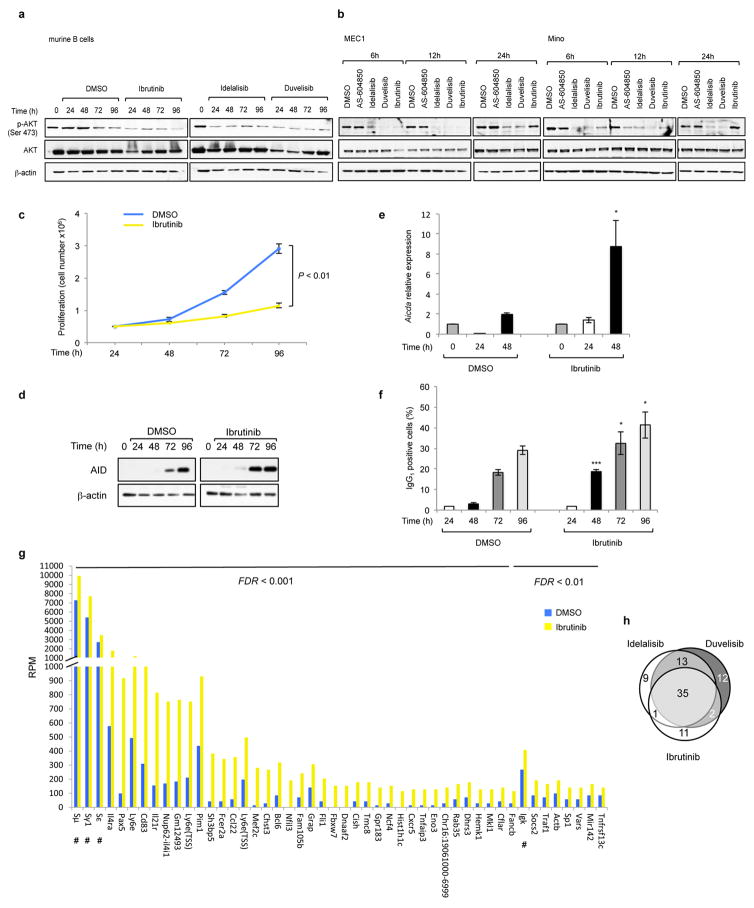
Extended Data Fig 9. Ibrutinib increases AID expression and the frequency of translocations to AID on- and off-target sites in mouse activated B cells.
a, AKT phosphorylation was detected by Western Blot in mouse activated B cells treated with DMSO, idelalisib, duvelisib or ibrutinib (1 μM) for the indicated time points (n = 2 biological replicates). For gel source data, see Supplementary Figure 1. b, MEC1 and Mino human lymphoma cells were treated with the indicated inhibitors (1 μM) and AKT phosphorylation was evaluated by Western Blot (n = 3 biological replicates). c, Viable cells were counted at the indicated time points by Trypan Blue exclusion in activated B cells treated with DMSO or ibrutinib (1 μM). Data are expressed as mean ± s.d. (n=3). P values calculated by two-tailed Student’s t-test. d, Western blot for AID protein expression in activated B cells treated with 1 μM ibrutinib. The DMSO panel from Fig. 1a is shown for comparison (n=3 biological replicates). e, Aicda mRNA levels analyzed by qRT-PCR in activated B cells treated with DMSO or ibrutinib (1 μM). Data are expressed as mean ± s.d. (n = 3 technical replicates, n = 3 biological replicates). * P < 0.05, two-tailed Student’s t-test. f, IgG1 CSR in activated B cells analyzed by flow cytometry. Data are expressed as mean ± s.d. (n = 3 biological replicates). * P < 0.05, *** P ≤ 0.001, two-tailed Student’s t-test. g, Histograms showing translocation junction frequency to AID on-target (Igh and Igk loci indicated with a #) and off-target sites in activated B cells treated with ibrutinib. Targets are divided based on the statistical significance of increased junctions frequency compared to DMSO treatment (FDR < 0.001 on the left; FDR < 0.01 on the right). Statistical analysis is indicated in the Method. For each treatment, data are from pooled HTGTS libraries of similar size from independent experiments as indicated in Supplementary Table 1 and 5. h, Venn diagrams showing the fraction of AID off-target sites shared in activated mouse B cells treated with ibrutinib, idelalisib or duvelisib.
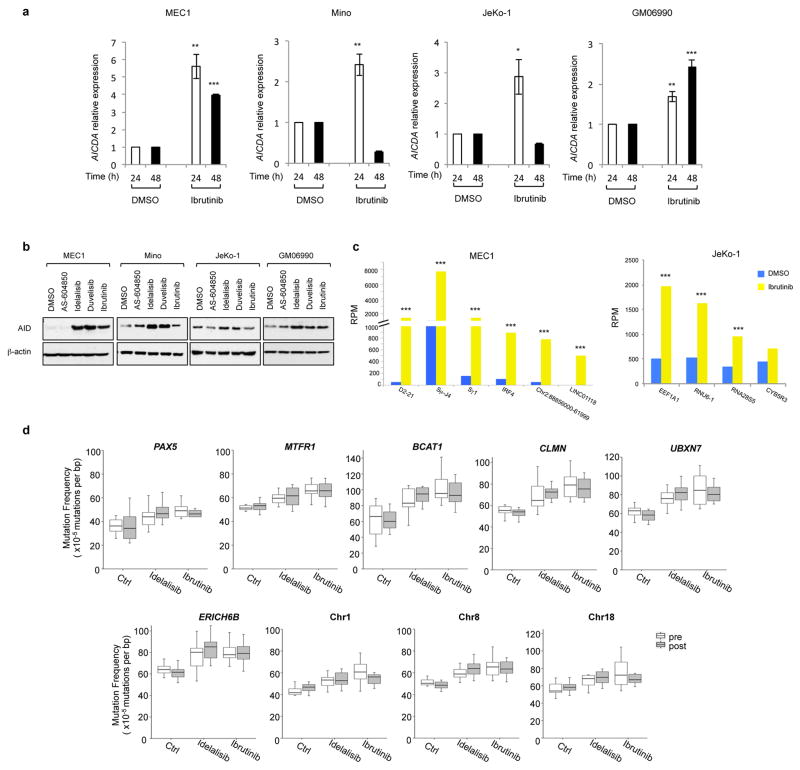
Extended Data Fig 10. Ibrutinib increases AID expression and the frequency of translocations to AID on- and off-target sites in human B cells.
a, AICDA mRNA levels analyzed by qRT-PCR in MEC1, Mino, JeKo-1 and GM06990 cell lines after treatment with DMSO or ibrutinib. Data are expressed as mean ± s.d. (n = 3 technical replicates, n = 3 biological replicates). * P < 0.05, ** P ≤ 0.01, *** P ≤ 0.001, two-tailed Student’s t-test. b, Western blot for AID protein expression in MEC1, Mino, JeKo-1 and GM06990 B cell lines treated with the indicated inhibitors (1 μM) for 48 h. For comparison, Mino and GM06990 panels correspond to the panel shown in Fig. 4c and Extended Data Fig. 6d, respectively, with the addition of the ibrutinib lane (n = 3 biological replicates). For gel source data, see Supplementary Figure 1. c, Histograms showing translocation junctions frequency to AID on-target and off-target sites in MEC1 and JeKo-1 B cell lines treated with ibrutinib. For each treatment, data are from pooled HTGTS libraries of similar size from three independent experiments (Supplementary Table 1 and 4). Significance is calculated as FDR in ibrutinib over DMSO treated human B cells. Statistical analysis is indicated in the Method. *** FDR ≤ 0.001. d, Mutation frequency of control non AID off-targets: intragenic regions for PAX5, MTFR1, BCAT1, CLMN, UBXN7, ERICH6B, and random intergenic regions on Chr. 1, Chr. 8, and Chr. 18 were targeted re-sequenced in CLL patients untreated or treated with idelalisib and ibrutinib. Boxplots indicate cumulative frequencies of C-to-T or G-to-A transition mutation in DNA samples collected before (pre) and after (post) treatment in each patient (control n=8, idelalisib n=10, ibrutinib n=10, Supplementary Table 6). Whiskers extend to a maximum of 1.5X interquartile range (IQR) beyond the box. P values calculated by paired samples two-tailed Student’s t-test.
Supplementary Material
Acknowledgments
We thank Drs. Klaus Okkenhaug and Fabien Garcon (The Babraham Institute, UK) for kindly providing WT and mutated PI3Kδ constructs, Dr. Feng Zhang (Broad Institute and MIT, Boston) for providing CRISPR/Cas9 plasmids. We thank Dr. Mark Fleming and Mark M. Awad for critically reading the manuscript.
This work was supported by NIH grants R01 CA196703-01 to R.C.; 1U10CA180861-01 to C.J.W., R01AI077595 to F.W.A; Associazione Italiana per la Ricerca sul Cancro grant IG-12023 to R.C. and MFAG to M.C.; Worldwide Cancer Research grant 12-0216 to R.C.; American Cancer Society Grant RSG-13-002-01-CCE to J.R.B.; T.C.C is supported by a National Research Foundation of Korea(NRF) fellowship; L.S.Y. was a Cancer Research Institute postdoctoral fellow; F.M. was a Lymphoma Research Foundation postdoctoral fellow; J.S. is a recipient of a PhRMA Foundation Research Fellowship; C.J.W. is Scholar of the Leukemia and Lymphoma Society; F.W.A. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Author Contributions
M.C. and C.P. planned and performed most experiments. Q.W. performed analysis of the data. T-C.C., F.L., T.P., C.A., C.V., M.G. performed experiments. E.K. and R.B contributed to CRISPR/Cas9 experiments. F.M. performed GRO-Seq experiments. L-S.Y. performed the mutational analysis on VB1-8 exon. A.W. S.N.K, J.R.B, J.S., C.J.W. provided clinical samples. F.W.A contributed to the design and interpretation of the experiments, and contributed to writing the manuscript. R.C. conceived and designed all the experiments, analyzed data, and wrote the manuscript with the help of M.C., Q.W. and C.P. All authors discussed the results and commented on the manuscript.
All sequencing data has been deposited in the Gene Expression Omnibus database under accession number GSE77788.
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
Readers are welcome to comment on the online version of the paper.
References
- 1.Alt FW, Zhang Y, Meng FL, Guo C, Schwer B. Mechanisms of programmed DNA lesions and genomic instability in the immune system. Cell. 2013;152:417–429. doi: 10.1016/j.cell.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nussenzweig A, Nussenzweig MC. Origin of chromosomal translocations in lymphoid cancer. Cell. 2010;141:27–38. doi: 10.1016/j.cell.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robbiani DF, et al. AID Produces DNA Double-Strand Breaks in Non-Ig Genes and Mature B Cell Lymphomas with Reciprocal Chromosome Translocations. Molecular cell. 2009;36:631–641. doi: 10.1016/j.molcel.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Omori SA, et al. Regulation of class-switch recombination and plasma cell differentiation by phosphatidylinositol 3-kinase signaling. Immunity. 2006;25:545–557. doi: 10.1016/j.immuni.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 5.Byrd JC, et al. Targeting BTK with ibrutinib in relapsed chronic lymphocytic leukemia. The New England journal of medicine. 2013;369:32–42. doi: 10.1056/NEJMoa1215637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gopal AK, et al. PI3Kdelta inhibition by idelalisib in patients with relapsed indolent lymphoma. The New England journal of medicine. 2014;370:1008–1018. doi: 10.1056/NEJMoa1314583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown JR, et al. Idelalisib, an inhibitor of phosphatidylinositol 3-kinase p110delta, for relapsed/refractory chronic lymphocytic leukemia. Blood. 2014;123:3390–3397. doi: 10.1182/blood-2013-11-535047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dong S, et al. IPI-145 antagonizes intrinsic and extrinsic survival signals in chronic lymphocytic leukemia cells. Blood. 2014;124:3583–3586. doi: 10.1182/blood-2014-07-587279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiarle R, et al. Genome-wide translocation sequencing reveals mechanisms of chromosome breaks and rearrangements in B cells. Cell. 2011;147:107–119. doi: 10.1016/j.cell.2011.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meng FL, et al. Convergent transcription at intragenic super-enhancers targets AID-initiated genomic instability. Cell. 2014;159:1538–1548. doi: 10.1016/j.cell.2014.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Advani RH, et al. Bruton tyrosine kinase inhibitor ibrutinib (PCI-32765) has significant activity in patients with relapsed/refractory B-cell malignancies. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2013;31:88–94. doi: 10.1200/JCO.2012.42.7906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rush JS, Liu M, Odegard VH, Unniraman S, Schatz DG. Expression of activation-induced cytidine deaminase is regulated by cell division, providing a mechanistic basis for division-linked class switch recombination. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:13242–13247. doi: 10.1073/pnas.0502779102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Angulo I, et al. Phosphoinositide 3-kinase delta gene mutation predisposes to respiratory infection and airway damage. Science. 2013;342:866–871. doi: 10.1126/science.1243292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lucas CL, et al. Dominant-activating germline mutations in the gene encoding the PI(3)K catalytic subunit p110delta result in T cell senescence and human immunodeficiency. Nature immunology. 2014;15:88–97. doi: 10.1038/ni.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klein IA, et al. Translocation-capture sequencing reveals the extent and nature of chromosomal rearrangements in B lymphocytes. Cell. 2011;147:95–106. doi: 10.1016/j.cell.2011.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu J, Tepsuporn S, Meyers RM, Gostissa M, Alt FW. Developmental propagation of V(D)J recombination-associated DNA breaks and translocations in mature B cells via dicentric chromosomes. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:10269–10274. doi: 10.1073/pnas.1410112111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamane A, et al. Deep-sequencing identification of the genomic targets of the cytidine deaminase AID and its cofactor RPA in B lymphocytes. Nature immunology. 2011;12:62–69. doi: 10.1038/ni.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qian J, et al. B cell super-enhancers and regulatory clusters recruit AID tumorigenic activity. Cell. 2014;159:1524–1537. doi: 10.1016/j.cell.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yeap LS, et al. Sequence-Intrinsic Mechanisms that Target AID Mutational Outcomes on Antibody Genes. Cell. 2015;163:1124–1137. doi: 10.1016/j.cell.2015.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pavri R, et al. Activation-induced cytidine deaminase targets DNA at sites of RNA polymerase II stalling by interaction with Spt5. Cell. 2010;143:122–133. doi: 10.1016/j.cell.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pefanis E, et al. Noncoding RNA transcription targets AID to divergently transcribed loci in B cells. Nature. 2014;514:389–393. doi: 10.1038/nature13580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Potter M. Neoplastic development in plasma cells. Immunological reviews. 2003;194:177–195. doi: 10.1034/j.1600-065x.2003.00061.x. [DOI] [PubMed] [Google Scholar]
- 23.Pasqualucci L, et al. Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature. 2001;412:341–346. doi: 10.1038/35085588. [DOI] [PubMed] [Google Scholar]
- 24.Puente XS, et al. Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nature. 2015 doi: 10.1038/nature14666. [DOI] [PubMed] [Google Scholar]
- 25.Landau DA, et al. Mutations driving CLL and their evolution in progression and relapse. Nature. 2015;526:525–530. doi: 10.1038/nature15395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang ML, et al. Targeting BTK with ibrutinib in relapsed or refractory mantle-cell lymphoma. The New England journal of medicine. 2013;369:507–516. doi: 10.1056/NEJMoa1306220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burger JA, Chiorazzi N. B cell receptor signaling in chronic lymphocytic leukemia. Trends in immunology. 2013;34:592–601. doi: 10.1016/j.it.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kasar S, et al. Whole-genome sequencing reveals activation-induced cytidine deaminase signatures during indolent chronic lymphocytic leukaemia evolution. Nature communications. 2015;6:8866. doi: 10.1038/ncomms9866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palacios F, et al. High expression of AID and active class switch recombination might account for a more aggressive disease in unmutated CLL patients: link with an activated microenvironment in CLL disease. Blood. 2010;115:4488–4496. doi: 10.1182/blood-2009-12-257758. [DOI] [PubMed] [Google Scholar]
- 30.Klemm L, et al. The B cell mutator AID promotes B lymphoid blast crisis and drug resistance in chronic myeloid leukemia. Cancer cell. 2009;16:232–245. doi: 10.1016/j.ccr.2009.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muramatsu M, et al. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 32.Cato MH, Yau IW, Rickert RC. Magnetic-based purification of untouched mouse germinal center B cells for ex vivo manipulation and biochemical analysis. Nature protocols. 2011;6:953–960. doi: 10.1038/nprot.2011.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kovalchuk AL, Müller JR, Janz S. Deletional remodeling of c-myc-deregulating chromosomal translocations. Oncogene. 1997;15:2369–2377. doi: 10.1038/sj.onc.1201409. [DOI] [PubMed] [Google Scholar]
- 34.Ramiro AR, et al. AID is required for c-myc/IgH chromosome translocations in vivo. Cell. 2004;118:431–438. doi: 10.1016/j.cell.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 35.Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zang C, et al. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 2009;25:1952–1958. doi: 10.1093/bioinformatics/btp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Camps J, et al. Genomic imbalances and patterns of karyotypic variability in mantle-cell lymphoma cell lines. Leukemia research. 2006;30:923–934. doi: 10.1016/j.leukres.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 38.Krzywinski M, et al. Circos: an information aesthetic for comparative genomics. Genome research. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robinson JT, et al. Integrative genomics viewer. Nature biotechnology. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Molecular cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu M, et al. Two levels of protection for the B cell genome during somatic hypermutation. Nature. 2008;451:841–845. doi: 10.1038/nature06547. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data has been deposited in the Gene Expression Omnibus database under accession number GSE77788. Source data for figures are provided with the paper.