Abstract
The translocation and assembly module (TAM) in bacteria consists of TamA and TamB that form a complex to control the transport and secretion of outer membrane proteins. Herein, we demonstrated that the DR_1462-DR_1461-DR_1460 gene loci on chromosome 1 of Deinococcus radiodurans, which lacks tamA homologs, is a tamB homolog (DR_146T) with two tamB motifs and a DUF490 motif. Mutation of DR_146T resulted in cell envelope peeling and a decrease in resistance to shear stress and osmotic pressure, as well as an increase in oxidative stress resistance, consistent with the phenotype of a surface layer (S-layer) protein SlpA (DR_2577) mutant, demonstrating the involvement of DR_146T in maintenance of cell envelope integrity. The 123 kDa SlpA was absent and only its fragments were present in the cell envelope of DR_146T mutant, suggesting that DR_146T might be involved in maintenance of the S-layer. A mutant lacking the DUF490 motif displayed only a slight alteration in phenotype compared with the wild type, suggesting DUF490 is less important than tamB motif for the function of DR_146T. These findings enhance our understanding of the properties of the multilayered envelope in extremophilic D. radiodurans, as well as the diversity and functions of TAMs in bacteria.
All Gram-negative and some Gram-positive bacteria are encased by two layers of membrane, with the outer membrane (OM) acting as a hydrophobic barrier against environmental stresses to maintain internal homeostasis and facilitate cell division1,2. The outer leaflet of many bacterial OMs is covered by a two-dimensional array of proteinaceous surface layer (S-layer)3. In Gram-positive bacteria and archaea, S-layers adhere to the peptidoglycan or pseudomurein, while in Gram-negative bacteria, S-layers are attached to the lipopolysaccharide (LPS) of the OM3,4,5. The S-layer functions as a protective barrier, but it also traps ions and is involved in cell fission3,4,6,7. S-layer proteins are secretory proteins with an N-terminal signal peptide3,4. Generally, secretory proteins are synthesized in the cytoplasm and transported through the cytoplasmic membrane by the Sec translocon system2,8,9, after which the signal peptide is cleaved by signal peptidase, and further transportation of the mature protein is achieved by outer membrane transportation systems including the β-barrel assembly machinery (BAM), the two-partner secretion system (TPSS) or the translocation and assembly module (TAM)1. The TAM consists of two components, TamA and TamB, that form a recently identified protein complex10 that is crucial for the assembly of outer membrane proteins10,11,12,13,14, as well as the virulence and colonization of pathogenic bacteria, but is not essential for viability in organisms studied thus far10,14,15. TamA belongs to the Omp85 protein family1,10,15 originally identified in Neisseria meningitidis2. TamB is an evolutionarily ancient and essential subunit of TAM10,11,15. Interestingly, the distribution of TamB is much broader than that of TamA, and TAM systems without TamA are found in some bacteria15,16. The properties and functions of TamB in cell envelope assembly and integrity remain to be determined.
D. radiodurans serves as an ideal model for studying the cell envelope and stress resistance since its initial isolation from γ-ray-sterilized canned meat17. To date, more than 50 species of the Deinococcus genus have been identified in a variety of environments18. Deinococcus are distinguished by their extraordinary tolerance to a number of lethal agents including ionizing radiation, hydrogen peroxide, osmotic pressure, desiccation, UV radiation and mitomycin C (MMC)19,20,21,22. It is widely accepted that efficient DNA repair systems and cell defence systems including an unusual cell envelope and small molecule antioxidants (e.g. Mn2+ and carotenoids) contribute to the survival of D. radiodurans under various stresses20,23,24,25,26,27,28. However, the mechanisms underpinning the extreme resistance of D. radiodurans have never been fully explained. D. radiodurans has an unusual multilayered cell envelope that includes a thick peptidoglycan cell wall, an outer membrane-like lipid layer and a S-layer, and it reacts positively with Gram stain despite sharing some characteristics with Gram-negative bacteria24,29,30. The SlpA (DR_2577) is the pivotal component of the S-layer of D. radiodurans. Knockout of SlpA results in the dissociation of the S-layer core structure and consequently leads to cell envelope damage28,31,32. Acosta and colleagues reported that SlpA of Thermus thermophilus, a bacterium evolutionally close to D. radiodurans, was assembled with assistance from the BAM complex33. However, the factors controlling S-layer proteins and cell envelope integrity are far from understood.
Herein, we identified a tamB (DR_146T) with two tamB motifs, even though a tamA homolog is not present in the genome of D. radiodurans15. Gene mutation, survival assays and cell envelope proteome analysis were performed to investigate the roles of DR_146T in cell envelope integrity with respect to SlpA and stress resistance.
Results
Identification of a tamB-containing locus in D. radiodurans
Two neighbouring sequences (DR_1462 and DR_1461) each containing a tamB motif were identified in the D. radiodurans genome using BLASTP, with each sequence sharing 28% and 29% amino acid sequence identity with Escherichia coli (E. coli) TamB encoded by b4221, respectively. However, a TamA homolog was not detected in D. radiodurans using the E. coli TamA as the query sequence, suggesting D. radiodurans might express a specific TAM lacking TamA.
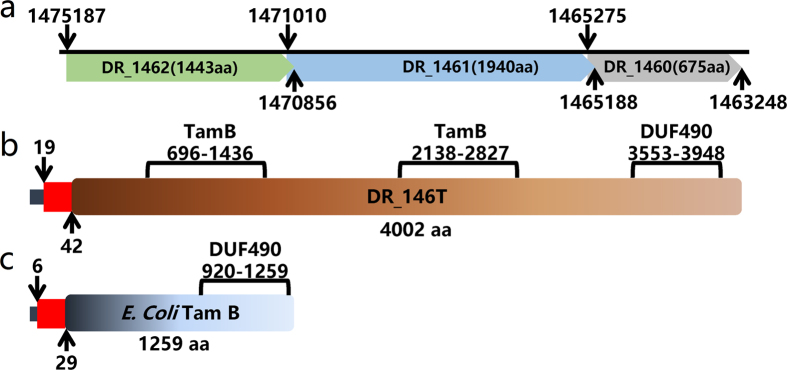
The current D. radiodurans genome sequence (NCBI accession: NC_001263.1) contains three consecutive annotated ORFs (DR_1462, DR_1461 and DR_1460) in the negative chain of D. radiodurans chromosome 1 (Fig. 1a). By re-sequencing these loci, we found five gaps and four base errors in the DNA sequence between 1463248 and 1475251 (Supplementary Fig. S1). The gaps introduced invalid start and/or stop codons in the predicted DR_1462, DR_1461 and DR_1460 genes (Supplementary Figs S1 and S2). Therefore, DR_1462, DR_1461 and DR_1460 actually form a single intact ORF, which we refer to as DR_146T (data submitted to NCBI, GenBank accession number is KY352801). This ORF was predicted to encode a TamB homolog of 4002 amino acid residues containing two TamB domains (PFAM signature: PF04357.11) and a C-terminal DUF490 domain, on contrast with E. coli TamB that has one TamB domain and one DUF490 domain (Fig. 1b and c). A signal peptide of 34 amino acids and a transmembrane helix spanning residues 20–42 were predicted using the SignalIP 3.0 Server and the TMHMM Server v. 2.0, respectively (Fig. 1b). Moreover, homologs of DR_146T are also present as intact hypothetical genes in other sequenced Deinococcus bacteria (Supplementary Table S3). Together these features imply that the DR_146T is a tamB homolog15.
Figure 1. Schematic representation of D. radiodurans DR_146T and E. coli TamB.
(a) The predicted DR_1462, DR_1461 and DR_1460 are annotated as found in the NCBI database. (b) The DR_146T containing two TamB motifs and one DUF490 motif was obtained from re-sequencing. (c) E. coli TamB encoded by b4221. Both DR_146T and E. coli TamB contain an N-terminal signal peptide (dark blue rectangle), which overlaps with the transmembrane segment (red rectangle).
DR_146T is involved in cell growth in D. radiodurans
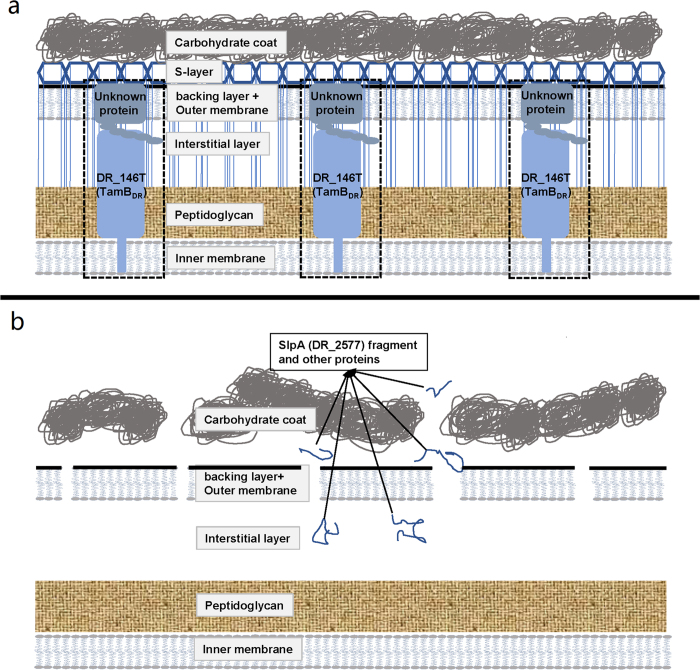
The growth of the DR_146T mutant (∆DR_146T) and the mutant deficient in DUF490 (∆DR_146T-DUF490) were slower than that of the wild type (Fig. 2). The DR_146T mutant grew twice as slowly as the wild type, while ∆DR_146T-DUF490 had only a slightly longer doubling time than the wild type. These results indicate that DR_146T might play an important role in cell growth or cell division. Moreover, the growth defect phenotype of ∆DR_146T is similar to that of the SlpA (DR_2577) mutant (Fig. 2), suggesting that DR_146T might contribute to cell envelope integrity along with the S-layer components.
Figure 2. Growth of D. radiodurans wild type and mutant strains.
The growth of DR_146T mutant (∆DR_146T, black) and SlpA mutant (∆DR_2577, purple) were much slower than that of the wild type (DRWT, blue), while the growth of the mutant deficient in the DUF490 motif of DR_146T (∆DR_146T-DUF490, green) was only slightly slower than that of the wild type. Cell growth was monitored by measuring the OD600 of cell cultures.
DR_146T is important for cell envelope integrity in D. radiodurans
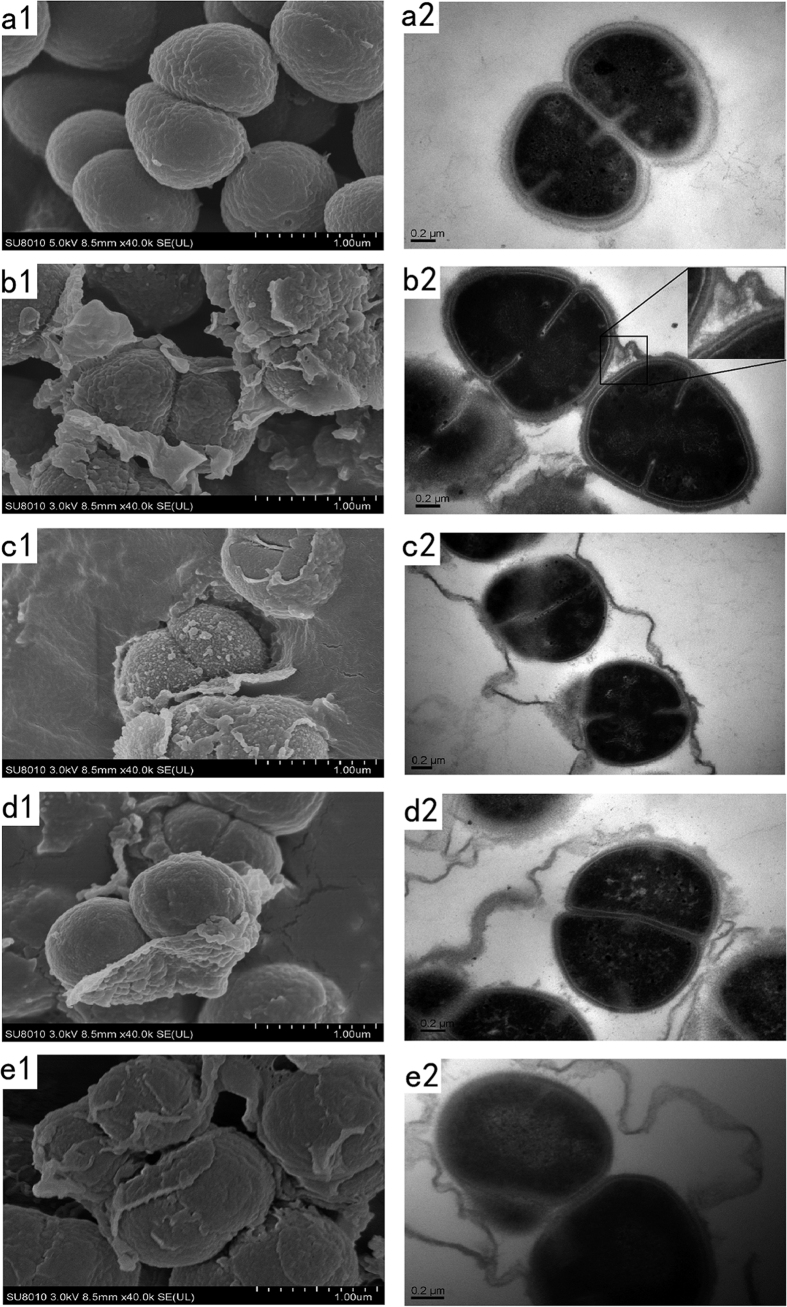
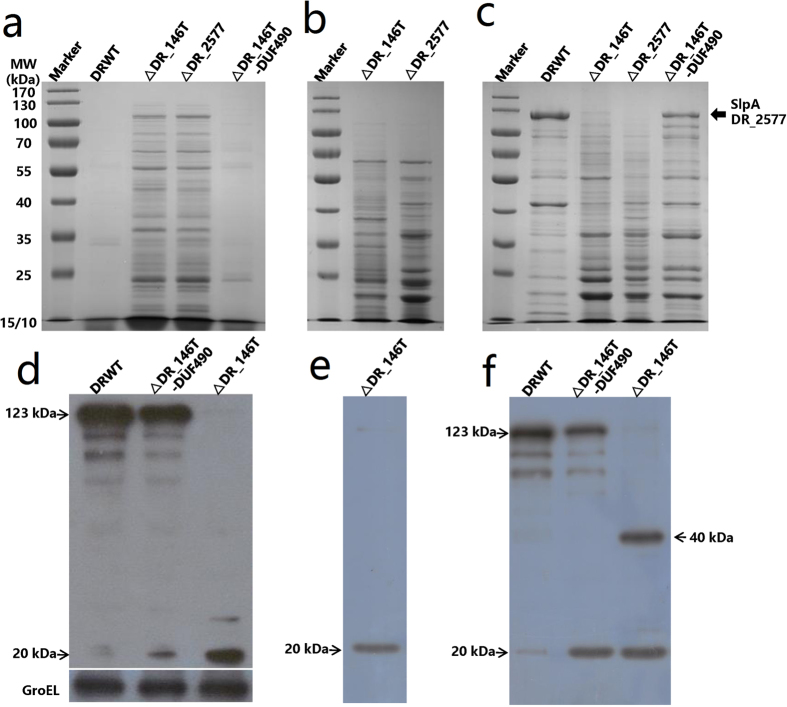
The wild type colonies were circular and smooth, while the colonies of DR_146T mutant were ring-shaped and rugose similar to those of SlpA mutant (Supplementary Fig. S3). In liquid media, mutant cells tended to aggregate and settle more easily than wild type cells (Supplementary Fig. S4). Scanning electron microscopy (SEM) images showed that wild type cells were elliptical and grouped into diplococci or tetracocci (Fig. 3a1), whereas mutant cells displayed surface variation and shedding of the outer layer (Fig. 3b1–e1). Meanwhile, the results of transmission electron microscopy (TEM) demonstrated that the ultrastructure of the cell envelope of the mutants was different from that of wild type cells (Fig. 3a2–e2). The cell envelope of ∆DR_146T exhibited the most severe damage with parts of the outer layer peeling off, and the inner layer exposed to the environment (Fig. 3d). Damage to the cell envelope of ∆DR_146T-TamB2nd-DUF490 that is mutated in the second TamB and DUF490 motifs (Fig. 3c) was similar to that of the ∆DR_146T cells (Fig. 3d), but more severe than that of ∆DR_146T-DUF490 cells (Fig. 3b), indicating that TamB motif appeared to be more important than the DUF490 motif. SEM and TEM images revealed that the outmost layer peeled in the SlpA mutant (∆DR_2577) (Fig. 3e). Together with the cell growth phenotype, these results indicated that DR_146T might play an important role in maintaining cell envelope integrity. The cytoplasmic membrane (inner membrane) and peptidoglycan layer could still be formed during cell division, but formation of the outer layers was clearly inhibited in the mutant.
Figure 3. SEM and TEM images of D. radiodurans wild type and mutant strains.
The cell envelope of ∆DR_146T exhibited severe damage with parts of the outer layer peeling off, and the inner layer exposed to the environment compared with that of the wild type. Images represent the SEM and TEM results for D. radiodurans wild type (a1–a2), ∆DR_146T-DUF490 (b1–b2), ∆DR_146T-TamB2nd -DUF490 (c1–c2), ∆DR_146T (d1–d2) and ∆DR_2577 (e1–e2), respectively. The inset diagram in (b2) shows an amplified region of the cell envelope. Scale bars indicate the corresponding length.
Mutation of DR_146T alters stress resistance in D. radiodurans
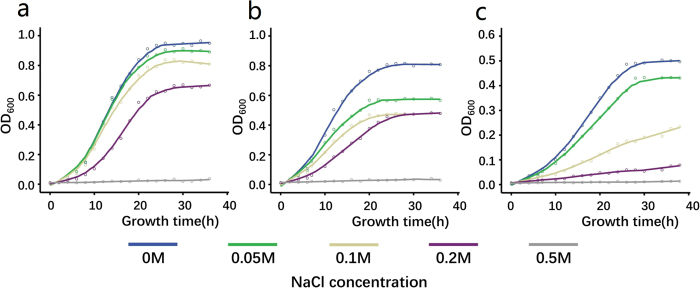
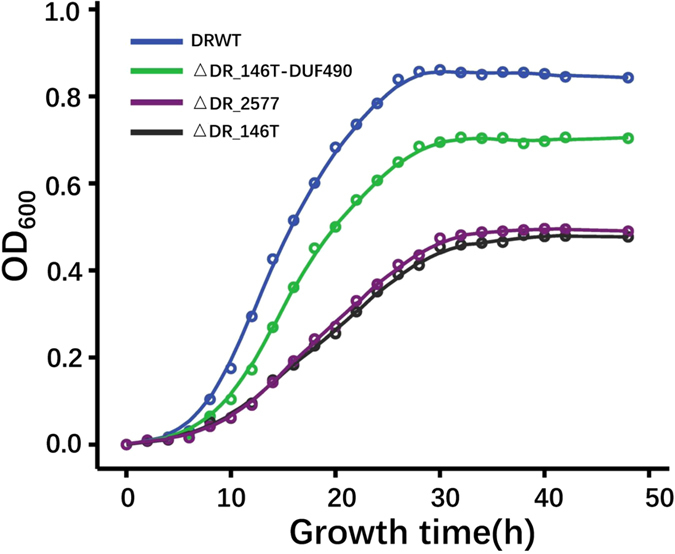
Under continuous vortexing, the survival fraction of ∆DR_146T was substantially lower than that of the wild type (Fig. 4), consistent with the survival phenotype of the SlpA mutant28. The survival of ∆DR_146T-DUF490 was slightly lower than that of the wild type. Fig. 5 shows that the cell resistance of ∆DR_146T and ∆DR_146T-DUF490 to osmotic pressure was lower than that of the wild type, indicating that mutation-induced deficiency in cell envelope integrity led to higher sensitivity to osmotic pressure. These suggest that DR_146T contributes to stress resistance, and the DUF490 motif might be less important than the TamB motif for the function of DR_146T.
Figure 4. Survival of D. radiodurans wild type and mutant strains to shear stress.
The survival fraction of ∆DR_146T was substantially lower than that for the wild type and ∆DR_146T-DUF490 under shear stress. DRWT, D. radiodurans wild type; ∆DR_146T-DUF490, mutant deficient in the DUF490 motif of DR_146T; ∆DR_146T, mutant deficient in DR_146T.
Figure 5. Growth of D. radiodurans wild type and mutant strains in TGY broth supplemented with different concentrations of NaCl.
The mutants ∆DR_146T and ∆DR_146T-DUF490 were more sensitive to osmotic pressure than the wild type. (a) DRWT, D. radiodurans wild type; (b) ∆DR_146T-DUF490, mutant deficient in the DUF490 motif of DR_146T; (c) ∆DR_146T, mutant deficient in DR_146T. Growth of the bacteria was monitored by measuring the OD600 of cell cultures.
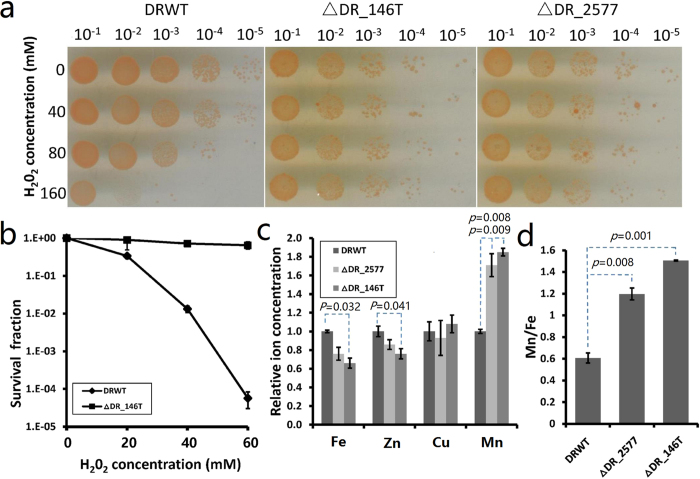
The resistance of ∆DR_146T and the SlpA mutant (∆DR_2577) to oxidative stress was much higher than that of wild type cells (Fig. 6a and b). The ∆DR_146T strain could survive high concentrations of H2O2 (160 mM) without any obvious decline in viability compared with controls (0 mM H2O2), indicating that disruption of DR_146T leads to an increase in resistance to oxidative stress. Previous studies demonstrated that accumulation of manganese ions (Mn2+) and a higher intracellular Mn/Fe ratio contribute to oxidative stress resistance in D. radiodurans through Mn complex-mediated ROS scavenging25,26,34. Thus, we measured the metal ion content in mutant and wild type cells by inductively-coupled plasma-mass spectrometry (ICP-MS) (Fig. 6c and d). The Mn ion content in ∆DR_146T and ∆DR_2577 was almost twice that of wild type cells, while the Fe and Zn ion content were slightly reduced in the mutants. Therefore, the H2O2 resistance of ∆DR_146T might be attributed to the increased Mn/Fe ratio in the mutant cells.
Figure 6. Survival of D. radiodurans wild type and mutant strains exposed to H2O2 treatment.
The ∆DR_146T and ∆DR_2577 showed higher resistance to H2O2 and increased Mn/Fe ratio than D. radiodurans wild type. (a) Comparison of the sensitivity of wild-type (DRWT), ∆DR_146T and ∆DR_2577 strains under H2O2 treatment. Cells (107 CFU ml−1) were dripped onto TGY plates following H2O2 treatment for 30 min and dilution with sterile phosphate buffer (0.1 M, pH 7.4). Different dilutions of cell cultures are indicated in the figure. (b) Survival assays of D. radiodurans wild type and ∆DR_146T strains. Cells were suspended in phosphate buffer (107 CFU ml−1), treated with H2O2 for 30 min, and the survival fraction was measured by counting bacterial colonies of treated compared with the untreated samples (0 mM H2O2). (c) ICP-MS analysis of the relative metal ion (Fe, Zn, Cu, Mn) content in wild type and mutant strains. (d) Ratio of Mn and Fe ion content in wild type and mutant strains. Experiments were independently performed three times. P-values indicate the significance compared with wild type cells.
Altered proteins in cell envelope, peeling fraction and culture’s supernatant
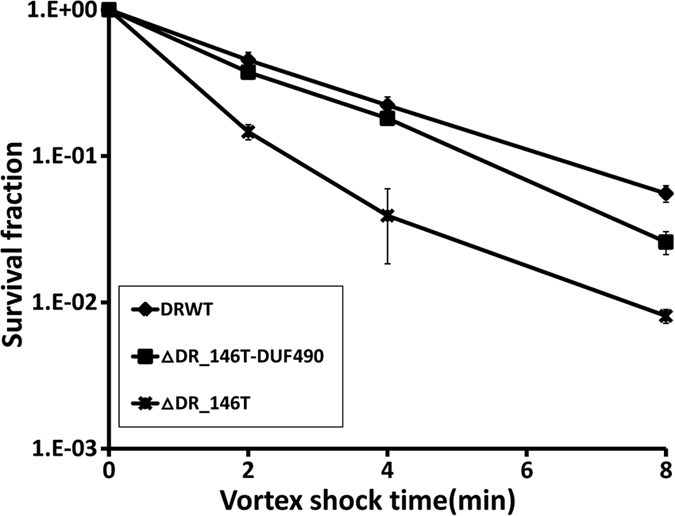
Each of the obtained whole cell envelope, culture’s supernatant and peeling cell envelope fractions was divided in half: half was used for mass spectrometry (MS) analysis, and the other half was for SDS-PAGE analysis on protein patterns of the wild type and the mutants. More than 80 proteins including DR_146T (matched to the predicted DR_1462, DR_1461 and DR_1460 from the NCBI database) were detected in the cell envelope of D. radiodurans wild type using MS analysis (Supplementary Table S4). Furthermore, the detection of peptides matched to the DR_146T from the cell envelope of D. radiodurans wild type cells by MS (Supplementary Fig. S5) suggests that DR_146T is a component of the cell envelope.
SDS-PAGE results revealed some extra protein bands in the culture’s supernatant of ∆DR_146T and ∆DR_2577, however, fewer proteins were detected in the culture’s supernatant of wild type and ∆DR_146T-DUF490 cells (Fig. 7a). The protein band containing SlpA with a molecular weight of 123 kDa (Fig. 7c) was subjected to MS analysis (Supplementary Table S5). Compared with wild type cells, many proteins including the 123 kDa SlpA were absent from the cell envelope fraction of ∆DR_146T (Fig. 7b and c), suggesting that DR_146T might be involved in maintenance of the surface layer. However, the cell envelope protein profile in ∆DR_146T-DUF490 was similar to that of the wild type, confirming that DUF490 may be not a pivotal factor in maintenance of the cell surface layer.
Figure 7.
SDS-PAGE analysis of proteins (a–c) and western blotting of SlpA (d–f) in the wild type and the mutants. Compared with wild type cells, the 123 kDa SlpA (indicated by an arrow) was absent from the cell envelope and only its fragments were present in the peeling envelope fraction and whole cell envelope of ∆DR_146T. (a) Cell culture’s supernatant; (b) peeling cell envelope; (c) whole cell envelope separated from bacterial high pressure homogenate. (d) Whole cell extract; (e) peeling cell envelope of DR_146T; (f) whole cell envelope separated from bacterial high pressure homogenate. DRWT, D. radiodurans wild type; ∆DR_146T, mutant deficient in DR_146T; ∆DR_2577, mutant deficient in DR_2577; ∆DR_146T-DUF490, mutant deficient in the DUF490 motif of DR_146T. GroEL was used as a loading control.
To further probe the roles of DR_146T in cell envelope assembly and integrity, proteins in the peeling cell envelope and culture’s supernatant were identified by MS, respectively. More than 30 proteins were identified in the culture’s supernatant of ∆DR_146T (Table 1), while only a few extracellular nucleases and proteases were detected in the culture’s supernatant of wild type cells (data not shown). Among these, surface layer proteins including Hpi (DR_2508), putative S-layer-like array-related protein (DR_1115, DR_1185, DR_0383 and DR_1124) and putative periplasm-located proteins (DR_1571, DR_1027, DR_0363 and DR_A0210) were detected in the culture’s supernatant of ∆DR_146T, suggesting some cell envelope proteins were released into the supernatant following mutation of DR_146T. More than 50 proteins were identified in the peeling envelope of ∆DR_146T (Table 2), and a number of proteins detected in the culture’s supernatant were also found in the peeling envelope fraction. However, some proteins were exclusive to the peeling envelope fraction, including SlpA, the putative outer membrane protein BamA (DR_0379) and the secretin (DR_0774)31. SlpA was one of the most abundant proteins in the peeling fraction but was absent in the culture’s supernatant, possibly due to the presence of its SLH domain which keeps it anchored to the outer membrane32,35. Since the 123 kDa SlpA was not detected in the cell envelope of ∆DR_146T (Fig. 7c) and proteins identified by MS were based on peptide matches following enzyme digestion, the SlpA detected in the peeling fraction could be fragments of SlpA. These results indicated that many periplasmic, outer membrane and S-layer proteins or their fragments dissociated from cells and were released into the culture in the absence of DR_146T. Proper assembly of the S-layer might be inhibited by mutation of this special TamB homolog in D. radiodurans.
Table 1. Proteins identified in the culture’s supernatant of ∆DR_146T by MS.
| Gene Number | Protein Description | Relative Abundance Indexa |
|---|---|---|
| DR_1115 | S-layer-like array-like protein | ++ |
| DR_0383 | S-layer-like array-like protein | + |
| DR_1185 | S-layer-like array-like protein | + |
| DR_2508 | hexagonally packed intermediate-layer surface protein | ++ |
| DR_1124 | SLH family protein | + |
| DR_A0210 | peptide ABC transporter, periplasmic peptide-binding protein | + |
| DR_0363 | peptide ABC transporter periplasmic peptide-binding protein | ++ |
| DR_1027 | amino acid ABC transporter, periplasmic amino acid-binding protein | +++ |
| DR_1571 | peptide ABC transporter periplasmic peptide-binding protein | ++++ |
| DR_0986 | extracellular solute-binding protein | ++ |
| DR_1551 | carboxyl-terminal protease | + |
| DR_1649 | immunogenic protein | + |
| DR_1893 | cyclophilin-type peptidyl-prolyl cis-trans isomerase | ++ |
| DR_1998 | catalase | + |
| DR_2070 | membrane lipoprotein | + |
| DR_2095 | c-type cytochrome | + |
| DR_2221 | tellurium resistance protein TerD | + |
| DR_2487 | cytochrome C4 | ++ |
| DR_2542 | cyclophilin-type peptidyl-prolyl cis-trans isomerase | +++ |
| DR_A0255 | aculeacin A acylase | + |
| DR_A0283 | serine protease | + |
| DR_0115 | hypothetical protein | + |
| DR_0459 | hypothetical protein | +++ |
| DR_0574 | hypothetical protein | + |
| DR_0685 | hypothetical protein | + |
| DR_0691 | hypothetical protein | + |
| DR_0969 | hypothetical protein | ++ |
| DR_0972 | hypothetical protein | +++ |
| DR_1306 | hypothetical protein | +++ |
| DR_1406 | hypothetical protein | + |
| DR_1805 | hypothetical protein | + |
| DR_1940 | hypothetical protein | ++ |
| DR_2319 | hypothetical protein | + |
| DR_2320 | hypothetical protein | + |
| DR_B0037 | hypothetical protein | ++ |
aRelative protein abundance was indicated by the index value (i = unweighted spectrum count of each protein/mass). +, i ≤ 0.20; ++, 0.20 < i ≤ 0.40; +++, 0.40 < i ≤ 0.60; ++++, 0.60 < i ≤ 0.80; +++++, i > 0.80.
Table 2. Proteins identified in the peeling cell envelope of ∆DR_146T by MS.
| Gene Number | Protein Description | Relative Abundance Indexa |
|---|---|---|
| DR_2577 | S-layer protein SlpA | +++ |
| DR_1185 | S-layer-like array-like protein | ++++ |
| DR_1115 | S-layer-like array-like protein | +++ |
| DR_0383 | S-layer-like array-like protein | +++ |
| DR_2508 | hexagonally packed intermediate-layer surface protein | ++ |
| DR_1124 | SLH family protein | ++ |
| DR_0379 | outer membrane protein | +++ |
| DR_0774 | general secretion pathway protein D | ++ |
| DR_0631 | cell division protein FtsZ | + |
| DR_0986 | extracellular solute-binding protein | + |
| DR_1290 | extracellular solute-binding protein | + |
| DR_1712 | extracellular solute-binding protein | + |
| DR_1955 | extracellular solute-binding protein | + |
| DR_A0246 | extracellular solute-binding protein | + |
| DR_1571 | peptide ABC transporter periplasmic peptide-binding protein | ++ |
| DR_0363 | peptide ABC transporter periplasmic peptide-binding protein | ++ |
| DR_0561 | maltose ABC transporter periplasmic maltose-binding protein | ++ |
| DR_1038 | branched-chain amino acid ABC transporter periplasmic amino acid-binding protein | ++ |
| DR_0788 | branched-chain amino acid ABC transporter periplasmic amino acid-binding protein | ++ |
| DR_0564 | amino acid ABC transporter periplasmic amino acid-binding protein | ++ |
| DR_B0014 | hemin ABC transporter, periplasmic hemin-binding protein | ++ |
| DR_2278 | amino acid ABC transporter periplasmic amino acid-binding protein | + |
| DR_A0210 | peptide ABC transporter, periplasmic peptide-binding protein | + |
| DR_1756 | periplasmic serine protease Do | + |
| DR_0327 | periplasmic serine protease | + |
| DR_1027 | amino acid ABC transporter, periplasmic amino acid-binding protein | + |
| DR_1277 | ABC transporter periplasmic substrate-binding protein | + |
| DR_2154 | amino acid ABC transporter periplasmic amino acid-binding protein | + |
| DR_2588 | iron ABC transporter periplasmic substrate-binding protein | + |
| DR_A0263 | branched-chain amino acid ABC transporter periplasmic amino acid-binding protein | + |
| DR_B0125 | iron ABC transporter substrate-binding protein | ++ |
| DR_2070 | membrane lipoprotein | ++ |
| DR_1536 | serine protease | ++ |
| DR_1459 | serine protease | + |
| DR_A0064 | serine protease | + |
| DR_1937 | serine protease | + |
| DR_B0007 | metal binding protein | + |
| DR_0479 | penicillin-binding protein 1 | + |
| DR_1232 | pilin, type IV | + |
| DR_0972 | hypothetical protein | ++++ |
| DR_1021 | hypothetical protein | ++++ |
| DR_2319 | hypothetical protein | +++ |
| DR_2517 | hypothetical protein | +++ |
| DR_1842 | hypothetical protein | +++ |
| DR_0116 | hypothetical protein | +++ |
| DR_1140 | hypothetical protein | +++ |
| DR_0025 | hypothetical protein | ++ |
| DR_1623 | hypothetical protein | ++ |
| DR_0560 | hypothetical protein | ++ |
| DR_1388 | hypothetical protein | ++ |
| DR_0486 | hypothetical protein | ++ |
| DR_0581 | hypothetical protein | ++ |
| DR_1818 | hypothetical protein | ++ |
aRelative protein abundance was indicated by the index value (i = unweighted spectrum count of each protein/mass). +, i ≤ 0.20; ++, 0.20 < i ≤ 0.40; +++, 0.40 < i ≤ 0.60; ++++, 0.60 < i ≤ 0.80; +++++, i > 0.80.
Relationship between DR_146T and S-layer protein SlpA
SDS-PAGE revealed that a protein band of 123 kDa corresponding to SlpA was missing from the envelope of ∆DR_146T (Fig. 7c). The expression level and location of SlpA were investigated using western blotting. The 123 kDa SlpA was detected in whole-cell extracts and cell envelope fractions of the wild type and the ∆DR_146T-DUF490 cells (Fig. 7d and f). By contrast, SlpA was present only as 20 kDa and/or 40 kDa fragments in the peeling fraction and whole cell envelope of ∆DR_146T (Fig. 7e and f).
Discussion
We identified the gene DR_146T as a tamB homolog in the extremophilic bacterium D. radiodurans, which is known for its multilayered cell envelope and exceptional stress resistance. Detection of homologs of DR_146T in other Deinococcus species indicates that the gene is conserved in this genus.
Evidence from several lines suggests that DR_146T is an intact functional gene. Firstly, we re-sequenced the gene loci containing the putative tamB homologs and found five gaps and four base errors that led to a frame shift and introduced invalid start and/or stop codons in the predicted DR_1462, DR_1461 and DR_1460 sequences. Analysis of the corrected sequences confirmed the presence of an intact ORF. Secondly, homologs of DR_146T are also present as intact ORFs in other sequenced Deinococcus bacteria. Thirdly, and conclusively, peptides of DR_146T were detected in the cell envelope of D. radiodurans wild type cells by MS analysis (Supplementary Fig. S5).
A schematic diagram of the multilayered cell envelope of D. radiodurans is proposed in Fig. 8, adapted from previous reports28,31,36,37. DR_146T might function as a TamB-like protein, anchored in the inner membrane with its N-terminal transmembrane helix and spanning the peptidoglycan and periplasm, based on the topology of TamB in E. coli11. However, the unusual DR_146T containing two TamB domains is larger than typical bacterial TamB which contains only a single TamB domain15, such as TamB in E. coli, Citrobacter rodentium and Salmonella enterica10. TamB in the inner membrane is reported to cooperate with TamA in the outer membrane to form a hetero-oligomeric TAM complex10,11. However, there does not appear to be a TamA homolog in D. radiodurans, indicating that DR_146T might act as a solo TamB. The presence of TamB in other bacteria lacking TamA has been reported, and the TAM system is suggested to have evolved from an original combination of TamB and BamA, which evolved into TamA in Proteobacteria by a later gene duplication event15,29,38. Whether there was any interaction between TamB and BamA in D. radiodurans is not clear. We did not rule out the possibility that other genes with no or little homology to tamA might have similar functions to tamA in D. radiodurans. Considering that the thickness of the cell envelope of D. radiodurans is at least twice that of E. coli39, the two TamB domains might be needed for DR_146T to span the thicker peptidoglycan and periplasm in D. radiodurans.
Figure 8. Proposed schematic diagram of the D. radioduranscell envelope, adapted from previous reports11,28,31,36,37.
(a) Wild type strain. (b) DR_146T knockout strain. The TAM complex containing the DR_146T and an unknown cooperating protein was indicated by dashed box. In wild type cells, the S-layer assembles properly and acts as a scaffold for the cell envelope. In the DR_146T mutant, SlpA (DR_2577) is present as fragments in the peeling cell envelope. Components in the carbohydrate coat, outer membrane and periplasm dissociate from the cell.
In DR_146T mutant cells, the cell envelope was disrupted and some periplasmic and S-layer proteins were released, suggesting DR_146T is involved in maintaining the integrity of the cell envelope. A number of periplasmic and S-layer proteins were detected at altered relative abundance in the peeling cell envelope of DR_146T mutants, indicating that the proteome integrity of the cell envelope was altered by the mutation. Recently, Smith et al. reported that the abundance of 12 proteins, including protein quality control systems and protein secretion, were altered in the membrane of the TamB homolog (MorC) mutant strain of Aggregatibacter actinomycetemcomitans compared with the wild type strain40. The main functions of the S-layer are maintaining cell shape, acting as a mechanical barrier3,4,7, providing a binding scaffold for large molecules and ions41, and mediating bacterial adhesion42,43. DR_146T might facilitate the assembly of the S-layer, because its mutation results in the loss of the 123 kDa SlpA, which is involved in formation and attachment of the surface layer to the inner cell envelope28. In addition, the S-layer functions as a sheath surrounding groups of cells, and forming on the surface of daughter cells when they separate44. In the absence of DR_146T, detachment of the S-layer sheath from the cell surface was observed in SEM and TEM images (Fig. 3). The S-layer protein of Thermus thermophilus, an ancient bacterial lineage closely related to Deinococcus, was assembled under the assistance of the BAM complex33. The S-layer of D. radiodurans may be assembled under the assistance of TamB encoded by DR_146T.
We demonstrated that deficiencies in the cell envelope following mutation of DR_146T led to increased sensitivity to shear stress and osmotic pressure due to a decrease in rigidity of the cell envelope, consistent with the SlpA mutant. Meanwhile, DR_146T and SlpA mutants showed increased resistance to oxidative stress, as well as an increased intracellular Mn2+ content and Mn/Fe ion ratio, both of which contribute to oxidative stress resistance through non-enzymic ROS scavenging25,26,45. An increase in the permeability of the cell envelope in the mutant strains might facilitate the penetration of Mn ions across the cell wall. Furthermore, we cannot eliminate the possibility that mutation of DR_146T affected metal ion transporters directly. Indeed, an iron ABC transporter substrate-binding protein (DR_B0125) was detected in the peeling cell envelope of the DR_146T mutant, suggesting that it was detached, and this could explain the observed decrease in Fe ion content in the mutants. The mechanism by which DR_146T is involved in intracellular Mn accumulation remains to be explained by further studies.
In conclusion, tamB homolog DR_146T in D. radiodurans consists of two tamB motifs and one DUF490 motif. DR_146T might be involved in the maintenance of cell envelope integrity and stress resistance through its impact on SlpA. Further investigations are necessary to elucidate protein conformation, structure-activity relationships and details of the mechanism of this unusual TamB homolog in cell envelope assembly. Given the important roles of the TamB homolog in cell envelope integrity, these findings are not only useful for understanding the mechanism of TAM in this and other organisms, but also valuable for screening new antibiotics that target TamB.
Methods
Bacterial strains and growth conditions
All bacterial strains, plasmids, and primers used in this study are listed in Supplementary Tables S1 and S2, respectively. D. radiodurans was cultured in TGY liquid medium (0.5% tryptone, 0.3% yeast extract, 0.1% D-glucose) or on TGY plates (TGY liquid medium supplemented with 1% agar) at 30 °C. E. coli was grown in LB broth (1% tryptone, 0.5% yeast extract, 1% NaCl) or on LB plates (LB liquid medium supplemented with 1% agar) at 37 °C. Appropriate antibiotics were added into the medium where required.
Sequencing analysis of gene loci containing tamB motifs
Gene loci on chromosome 1 (NC_001263.1) containing tamB sequences were searched using BLAST (NCBI, http://blast.ncbi.nlm.nih.gov/Blast.cgi) with tamB from E. coli as the query sequence. Target DNA was sequenced using a 3730xl DNA Analyzer (ABI, USA). ORFs were predicted using OFR finder (https://www.ncbi.nlm.nih.gov/orffinder/). DNA and protein sequence analyses were performed using BLAST. Signal peptide analysis was conducted using the SignalIP 3.0 Server (http://www.cbs.dtu.dk/services/SignalP-3.0/). Transmembrane helices were analysed using the TMHMM Server v. 2.0 (http://www.cbs.dtu.dk/services/TMHMM-2.0/).
Construction of mutant strains
Tripartite ligation and double-crossover recombination methods were used as described previously46,47 with some modifications. Briefly, DNA fragments upstream and downstream of the targeted sequence were amplified and digested with BamHI and HindIII, respectively. The streptomycin resistance fragment was amplified from vector pMD18-T and digested with BamHI and HindIII. Upstream, downstream and streptomycin resistance fragments were ligated with T4 DNA ligase, and ligation products were transformed into competent D. radiodurans cells using the CaCl2 method. The homozygous mutant strain was confirmed by PCR and DNA sequencing.
Growth curves of mutant and wild type strains
Wild type and mutant strains were cultivated to an OD600 of ~1.0, inoculated into fresh TGY medium at a ratio of 1:50 (v/v) and incubated at 30 °C. Bacterial growth was monitored by measuring the OD600 value using a SpectraMax M5 microplate reader (Molecular Devices, USA). Experiments were performed independently in triplicate.
Scanning electron microscopy (SEM) and transmission electron microscopy (TEM) analyses
All experimental bacterial strains were cultured to an OD600 of ~1.0. For SEM analysis, harvested cells were double fixed and dehydrated as described by Li48, and dehydrated cells were coated with gold-palladium and observed using a SU8010 ultra-high resolution SEM (Hitachi, Japan).
For TEM analysis, specimens were processed as described by Li48, cut into ultrathin (70–90 nm) sections using microtome, stained with uranyl acetate and alkaline lead citrate for 15 min, and observed using a Hitachi H-7650 TEM (Hitachi, Japan).
Survival assays under shear stress, osmotic pressure and oxidative stress
Bacterial survival assays under shear stress were performed as described previously28 with some modifications. Briefly, bacterial cultures at an OD600 of ~1.0 were diluted with fresh TGY medium to OD600 = 0.1, mixed with sterile zirconia ceramic beads, and exposed to shear stress using a tissue homogenizer (RETSCH MM301, Germany) at 30 Hz for different treatment time (0, 2, 4 or 8 min). Suspensions were diluted and spread onto TGY plates. Bacterial colonies were counted and survival fraction was measured. For the osmotic pressure experiment, bacterial cultures at an OD600 of ~1.0 were inoculated into fresh TGY medium to which NaCl (0.05 M, 0.1 M, 0.2 M and 0.5 M) was added. Cultures were grown with shaking at 30 °C and the OD600 was measured using a SpectraMax M5 microplate reader. For the oxidative stress experiment, bacteria were treated with H2O2 in sterile phosphate buffer (0.01 M, pH 7.4) for 30 min, diluted and spread onto TGY plates. For the dripping test, 6 μl of cells was dripped onto TGY plates. All experiments were independently performed in triplicate.
Measurement of intracellular metal ion concentration by inductively-coupled plasma-mass spectrometry (ICP-MS)
ICP-MS assays were performed as described previously by Sun34 with some modifications. D. radiodurans wild type and mutant strains were cultured to an OD600 of ~1.0. Cells were collected and washed three times with 2 mM EDTA in 0.01 M phosphate buffer (pH 7.4), rinsed three times with phosphate buffer without EDTA, dried and treated with nitric acid (1 M) for further metal ion concentration assays using ICP-MS (ELAN DRC-e, PerkinElmer, USA).
Separation of cell envelope, peeling and culture’s supernatant fractions
Whole cell envelope fractions were prepared according to previously described methods32 with some modifications. Bacterial cultures at an OD600 of ~1.0 were centrifuged at 2000 g for 10 min. Cells were washed and suspended in 0.01 M phosphate buffer (pH 7.4), then disrupted at 4 °C using an ultra-high pressure continuous flow cell disrupter (JN-3000 PLUS, JNBIO, China). Undisrupted cells were removed by centrifugation (2000 g for 10 min). The supernatant was centrifuged again (20000 g for 20 min), and the envelope precipitate was washed three times in 0.01 M phosphate buffer (pH 7.4).
For separation of culture’s supernatant and peeling cell envelope fractions, bacterial cultures at an OD600 of ~1.0 were centrifuged at 2000 g for 10 min. The obtained supernatant was centrifuged again at 20000 g for 20 min to separate peeling and supernatant fractions. Supernatant fractions were concentrated by vacuum evaporation. Peeling fractions were washed three times with 0.01 M phosphate buffer (pH 7.4). During sample preparation, protease inhibitor cocktail (Selleckchem, USA) was added to protect proteins against proteolytic degradation.
Each of the obtained whole cell envelope, culture’s supernatant and peeling cell envelope fractions was divided in half: half was used for MS analysis, and the other half was for SDS-PAGE analyses on protein patterns of the wild type and the mutants. For electrophoresis, 6–10% (w/v) separating gels and 5% (w/v) stacking gels were used. The gels were stained with Coomassie Brilliant Blue G250.
Identification of proteins by MS
Protein samples in solution were denatured by RapiGest SF (Waters, USA), reduced with TRIS-(2-carboxyethyl)-phosphine and alkylated with 20 mM iodoacetamide for 45 min at room temperature in darkness. Trypsin was added at 1:50 trypsin-to-protein mass ratio for digestion overnight and 1:100 trypsin-to-protein mass ratio for a second 4 h-digestion. The peptides were then desalted and dried by vacuum evaporation. For proteins in selected gel slice, the gel was diced into small pieces and reduced with TRIS-(2-carboxyethyl)-phosphine, then alkylated with 20 mM iodoacetamide for 45 min at room temperature in darkness. The obtained samples were placed into 0.65 mL siliconized tubes, then washed with 100 μl 50% acetonitrile/25 mM NH4HCO3 for 3 times and dried by vacuum evaporation. After overnight trypsin digestion, 5 μl of 5% formic acid was added to the sample to stop the reaction. The digested solution was transferred into a clean 0.65 ml siliconized tube. 50 μl 50% acetonitrile/5% formic acid was added to wash the peptides for 3 times, and the peptide supernatant was dried by vacuum evaporation. Then the peptides were diluted with 0.1% formic acid.
The peptides were analysed by HPLC-MS/MS (Triple TOF 5600 + LC/MS/MS system, AB SCIEX, USA) as described previously28,31 with some modifications. Samples were loaded on a prepacked column (200 μm × 500 mm, Chrome XP C18-CL, 3 μm, 300 Å) to desalinate for 10 min at a flow rate of 4 μl/min. Subsequently, they were eluted at 0.3 μl/min with a C18 HPLC reversed-phase column (75 μm × 150 mm, Chrome XP C18-CL, 3 μm, 300 Å) using a mobile phase of solution A (5% acetonitrile and 0.1% formic acid) and solution B (95% acetonitrile and 0.1% formic acid). During elution, solution B was increased linearly from 5% to 40% over 60 min. Eluted peptides were introduced directly into the mass spectrometer and peptide identification was performed using the D. radiodurans protein sequence database (NCBI). The unweighted spectrum count of each protein was divided by its mass, and the resulting index was used as an indicator of relative protein abundance in each sample31.
Western blotting
Protein expression levels were confirmed using western blotting as described previously49. A 6× His tag was fused to the C-terminal of SlpA (DR_2577), and monoclonal anti-6× His mouse antibody (Protein tech, USA) was used to detect SlpA-6× His. Horseradish peroxidase-conjugated goat anti-mouse and anti-rabbit IgG were added as secondary antibodies. The expression level of GroEL served as an internal loading control, and was detected using rabbit anti-GroEL polyclonal antibody (Sigma, USA).
Statistical analysis
Data were processed using SPSS 18.0 statistical software (SPSS, USA) and are presented as means ± SD. F-tests and independent sample T-tests were used to assess the significance of differences between results. P < 0.05 was considered statistically significant.
Additional Information
How to cite this article: Yu, J. et al. A tamB homolog is involved in maintenance of cell envelope integrity and stress resistance of Deinococcus radiodurans. Sci. Rep. 7, 45929; doi: 10.1038/srep45929 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (31370119, 31670083, 31210103904, 31370102).
Footnotes
The authors declare no competing financial interests.
Author Contributions J.Y. and B.T. designed the experiments. J.Y., T.L., S.D., Y.W., J.L. and Q.L. performed the experiments. J.Y., B.T., T.L., S.D., Y.W., J.L., Q.L., H.X. and Y.H. analysed the data. B.T. and H.X. supervised experimental work and evaluated data. J.Y. and B.T. wrote manuscript.
References
- Webb C. T., Heinz E. & Lithgow T. Evolution of the beta-barrel assembly machinery. Trends in microbiology 20, 612–620 (2012). [DOI] [PubMed] [Google Scholar]
- Voulhoux R., Bos M. P., Geurtsen J., Mols M. & Tommassen J. Role of a Highly Conserved Bacterial Protein in Outer Membrane Protein Assembly. Science Bulletin 299, 265–265 (2003). [DOI] [PubMed] [Google Scholar]
- Sleytr U. B., Schuster B., Egelseer E. M. & Pum D. S-layers: principles and applications. FEMS microbiology reviews 38, 823–864 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleytr U. B. & Beveridge T. J. Bacterial S-layers. Trends Microbiol. 7, 253–260 (1999). [DOI] [PubMed] [Google Scholar]
- Olabarría G., Carrascosa J. L., De Pedro M. A. & Berenguer J. A Conserved Motif in S-Layer Proteins Is Involved in Peptidoglycan Binding in Thermus thermophilus. J Bacteriol. 178, 4765–4772 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pum D., Toca-Herrera J. L. & Sleytr U. B. S-layer protein self-assembly. International journal of molecular sciences 14, 2484–2501 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbino E., Carasi P., Mobili P., Serradell M. A. & Gomez-Zavaglia A. Role of S-layer proteins in bacteria. World journal of microbiology & biotechnology 31, 1877–1887 (2015). [DOI] [PubMed] [Google Scholar]
- Manting E. H. & Driessen A. J. Escherichia coli translocase the unravelling of a molecular machine. Molecular microbiology 37, 226–238 (2000). [DOI] [PubMed] [Google Scholar]
- Jeeves M. & Knowles T. J. A novel pathway for outer membrane protein biogenesis in Gram-negative bacteria. Molecular microbiology 97, 607–611 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selkrig J. et al. Discovery of an archetypal protein transport system in bacterial outer membranes. Nature structural & molecular biology 19, 506–510 (2012). [DOI] [PubMed] [Google Scholar]
- Shen H. H. et al. Reconstitution of a nanomachine driving the assembly of proteins into bacterial outer membranes. Nature communications 5, 5078, 10.1038/ncomms6078 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubenrauch C. et al. Effective assembly of fimbriae in Escherichia coli depends on the translocation assembly module nanomachine. Nat Microbiol 1, 8, 16064, 10.1038/nmicrobiol (2016). [DOI] [PubMed] [Google Scholar]
- Heinz E. et al. Conserved Features in the Structure, Mechanism, and Biogenesis of the Inverse Autotransporter Protein Family. Genome biology and evolution 8, 1690–1705 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal H., Kenedy M. R., Lybecker M. & Akins D. R. The TamB ortholog of Borrelia burgdorferi interacts with the beta-barrel assembly machine (BAM) complex protein BamA. Molecular microbiology 102, 757–774 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz E., Selkrig J., Belousoff M. J. & Lithgow T. Evolution of the Translocation and Assembly Module (TAM). Genome biology and evolution 7, 1628–1643 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubenrauch C., Grinter R. & Lithgow T. The modular nature of the beta-barrel assembly machinery, illustrated in Borrelia burgdorferi. Molecular microbiology 102, 753–756 (2016). [DOI] [PubMed] [Google Scholar]
- Andeson A., Nordan H., Cain R., Parrish G. & Duggan D. Studies on a radio resistant micrococcus. Isolation, morphology, cultural characteristics, and resistance to gamma radiation. Food Technology 10, 575–577 (1956). [Google Scholar]
- Masters C. I., Murray R. G., Moseley B. E. & Minton K. W. DNA polymorphisms in new isolates of Deinococcus radiopugnans. Journal of general microbiology 137, 1459–1469 (1991). [DOI] [PubMed] [Google Scholar]
- Rajpurohit Y. S. & Misra H. S. DR1769, a Protein with N-Terminal Beta Propeller Repeats and a Low-Complexity Hydrophilic Tail, Plays a Role in Desiccation Tolerance of Deinococcus radiodurans. Journal of bacteriology 195, 3888–3896 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minton K. W. DNA Repair in the Extremely Radioresistant Bacterium Deinococcus-Radiodurans. Molecular microbiology 13, 9–15 (1994). [DOI] [PubMed] [Google Scholar]
- Zhang C. et al. The Site-Directed A184S Mutation in the HTH Domain of the Global Regulator IrrE Enhances Deinococcus radiodurans R1 Tolerance to UV Radiation and MMC Shock. J Microbiol Biotechnol 25, 2125–2134 (2015). [DOI] [PubMed] [Google Scholar]
- Farci D., Slavov C., Tramontano E. & Piano D. The S-layer Protein DR_2577 Binds Deinoxanthin and under Desiccation Conditions Protects against UV-Radiation in Deinococcus radiodurans. Frontiers in microbiology 7, 155, 10.3389/fmicb (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H. et al. DR2539 is a novel DtxR-like regulator of Mn/Fe ion homeostasis and antioxidant enzyme in Deinococcus radiodurans. Biochemical and biophysical research communications 396, 413–418 (2010). [DOI] [PubMed] [Google Scholar]
- Battista J. R. Against all odds: The survival strategies of Deinococcus radiodurans. Annual review of microbiology 51, 203–224 (1997). [DOI] [PubMed] [Google Scholar]
- Daly M. J. et al. Accumulation of Mn(II) in Deinococcus radiodurans Facilitates Gamma-Radiation Resistance. Science 306, 1025–1028 (2004). [DOI] [PubMed] [Google Scholar]
- Daly M. J. et al. Small-molecule antioxidant proteome-shields in Deinococcus radiodurans. PloS one 5, e12570, 10.1371/journal.pone.0012570 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosal D. et al. How radiation kills cells: survival of Deinococcus radiodurans and Shewanella oneidensis under oxidative stress. FEMS microbiology reviews 29, 361–375 (2005). [DOI] [PubMed] [Google Scholar]
- Rothfuss H., Lara J. C., Schmid A. K. & Lidstrom M. E. Involvement of the S-layer proteins Hpi and SlpA in the maintenance of cell envelope integrity in Deinococcus radiodurans R1. Microbiology 152, 2779–2787 (2006). [DOI] [PubMed] [Google Scholar]
- Nesper J. et al. Omp85(Tt) from Thermus thermophilus HB27: an ancestral type of the Omp85 protein family. Journal of bacteriology 190, 4568–4575 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- White O. et al. Genome Sequence of the Radioresistant Bacterium Deinococcus radiodurans R1. Science 286, 319–327 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farci D. et al. New features of the cell wall of the radio-resistant bacterium Deinococcus radiodurans. Biochimica et biophysica acta 1838, 1978–1984 (2014). [DOI] [PubMed] [Google Scholar]
- Farci D. et al. Purification and characterization of DR_2577 (SlpA) a major S-layer protein from Deinococcus radiodurans. Frontiers in microbiology 6, 414, 10.3389/fmicb (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acosta F., Ferreras E. & Berenguer J. The β-barrel assembly machinery (BAM) is required for the assembly of a primitive S-layer protein in the ancient outer membrane of Thermus thermophilus. Extremophiles: life under extreme conditions 16, 853–861 (2012). [DOI] [PubMed] [Google Scholar]
- Sun H. et al. Identification and evaluation of the role of the manganese efflux protein in Deinococcus radiodurans. BMC microbiology 10, 319, 10.1186/1471-2180-10-319 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra C. S., Basu B. & Apte S. K. Surface (S)-layer proteins of Deinococcus radiodurans and their utility as vehicles for surface localization of functional proteins. Biochimica et biophysica acta 1848, 3181–3187 (2015). [DOI] [PubMed] [Google Scholar]
- BG. T. & RG. M. Isolation and characterization of the plasma membrane and the outer membrane of Deinococcus radiodurans strain Sark. Canadian journal of microbiology 27, 729–734 (1981). [DOI] [PubMed] [Google Scholar]
- Makarova K. S. et al. Genome of the extremely radiation-resistant bacterium Deinococcus radiodurans viewed from the perspective of comparative genomics. Microbiology and molecular biology reviews 65, 44–79 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada Mallarino L. et al. TtOmp85, a beta-barrel assembly protein, functions by barrel augmentation. Biochemistry 54, 844–852 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishimoto-Okada A. et al. Comparison of the envelope architecture of E. coli using two methods: CEMOVIS and cryo-electron tomography. Journal of electron microscopy 59, 419–426 (2010). [DOI] [PubMed] [Google Scholar]
- Smith K. P., Voogt R. D., Ruiz T. & Mintz K. P. The conserved carboxyl domain of MorC, an inner membrane protein of Aggregatibacter actinomycetemcomitans, is essential for membrane function. Molecular oral microbiology 31, 43–58 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbino E., Carasi P., Araujo-Andrade C., Tymczyszyn E. E. & Gomez-Zavaglia A. Role of S-layer proteins in the biosorption capacity of lead by Lactobacillus kefir. World journal of microbiology & biotechnology 31, 583–592 (2015). [DOI] [PubMed] [Google Scholar]
- Hynonen U. & Palva A. Lactobacillus surface layer proteins: structure, function and applications. Applied microbiology and biotechnology 97, 5225–5243 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara J. et al. Loss of adherence ability to human gingival epithelial cells in S-layer protein-deficient mutants of Tannerella forsythensis. Microbiology 153, 866–876 (2007). [DOI] [PubMed] [Google Scholar]
- Pum D., Messner P. & Sleytr U. Role of the S Layer in Morphogenesis and Cell Division of the Archaebacterium Methanocorpusculum sinense. Journal of bacteriology 173, 6865–6873 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H. et al. Regulation of MntH by a dual Mn(II)- and Fe(II)-dependent transcriptional repressor (DR2539) in Deinococcus radiodurans. PloS one 7, e35057, 10.1371/journal.pone.0035057 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. et al. Protease activity of PprI facilitates DNA damage response: Mn2+-dependence and substrate sequence-specificity of the proteolytic reaction. PloS one 10, e0122071, 10.1371/journal.pone.0122071 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. et al. DrRRA: a novel response regulator essential for the extreme radioresistance of Deinococcus radiodurans. Molecular microbiology 67, 1211–1222 (2008). [DOI] [PubMed] [Google Scholar]
- Li J. et al. Biosynthesis of gold nanoparticles by the extreme bacterium Deinococcus radiodurans and an evaluation of their antibacterial properties. International Journal of Nanomedicine 11, 5931–5944 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L. et al. DqsIR quorum sensing-mediated gene regulation of the extremophilic bacterium Deinococcus radiodurans in response to oxidative stress. Molecular microbiology 100, 527–541 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.