Essential tremor has a high heritability, but its molecular genetic determinants remain unknown. Müller et al. conduct a genome-wide association study in more than 2800 patients with essential tremor and more than 6800 controls of European descent, and identify three new loci associated with the disease.
Keywords: genome-wide association study, movement disorders, tremor, genetics, essential tremor
Abstract
We conducted a genome-wide association study of essential tremor, a common movement disorder characterized mainly by a postural and kinetic tremor of the upper extremities. Twin and family history studies show a high heritability for essential tremor. The molecular genetic determinants of essential tremor are unknown. We included 2807 patients and 6441 controls of European descent in our two-stage genome-wide association study. The 59 most significantly disease-associated markers of the discovery stage were genotyped in the replication stage. After Bonferroni correction two markers, one (rs10937625) located in the serine/threonine kinase STK32B and one (rs17590046) in the transcriptional coactivator PPARGC1A were associated with essential tremor. Three markers (rs12764057, rs10822974, rs7903491) in the cell-adhesion molecule CTNNA3 were significant in the combined analysis of both stages. The expression of STK32B was increased in the cerebellar cortex of patients and expression quantitative trait loci database mining showed association between the protective minor allele of rs10937625 and reduced expression in cerebellar cortex. We found no expression differences related to disease status or marker genotype for the other two genes. Replication of two lead single nucleotide polymorphisms of previous small genome-wide association studies (rs3794087 in SLC1A2, rs9652490 in LINGO1) did not confirm the association with essential tremor.
Introduction
Essential tremor, defined as a ‘bilateral, largely symmetric postural or kinetic tremor’ (Deuschl et al., 1998), is a common movement disorder with a reported prevalence of 0.9%, increasing to 4.6% in the population above the age of 65 (Louis and Ferreira, 2010). The disease is progressive and significant disabilities occur (Lorenz et al., 2011; Louis et al., 2011). Hands and arms are predominantly affected but head, voice and leg tremor occur. Twin studies estimated the heritable component of essential tremor between 45% and 90% (Kuhlenbäumer et al., 2014). Two small genome-wide association studies (GWAS) (452/436 cases per discovery stage) identified essential tremor associated single nucleotide polymorphisms (SNPs) in SLC1A2 (Thier et al., 2012), coding for the glutamate transporter EAAT2, and in LINGO1 (Stefansson et al., 2009), involved in oligodendrocyte differentiation and axonal regeneration. Replication studies yielded mixed results for both genes.
Materials and methods
Patient recruitment
Approval was obtained from all responsible ethics committees. All participants gave written consent. Movement disorder specialists at each recruitment site (Supplementary material) based the diagnosis mainly on presence of postural and action tremor in the arms exceeding the amplitude seen in enhanced physiological tremor and not attributable to other causes (Parkinson’s disease, dystonia, medication). Supplementary Table 1 lists the sample characteristics of the discovery and replication samples. Controls were ethnically matched to cases and either derived from biobanks PopGen (Krawczak et al., 2006) and KORA-gen (Wichmann et al., 2005) (Europe sites) or from North American sites. All patients and controls were of North-Western European (CEU) ancestry. Four hundred and thirty-six samples from Kiel, Tübingen and Innsbruck from the discovery stage of a previous GWAS of essential tremor (Thier et al., 2012) were genotyped anew.
Genotyping
All samples were processed on Axiom® Genome-Wide CEU 1 Array Plate by Affymetrix. Genotypes were called using the Affymetrix Power Tools (APT, version 1.15.1). Genotypes of case and control samples were called together in five batches.
Quality control
Quality control was performed at sample and marker level using pyGenClean (version 1.2.3) (Lemieux Perreault et al., 2013). Samples were excluded if the missingness rate exceeded 2% or sex information was discordant. We controlled all samples for deviating ethnicity (based on the ethnic classification of 1000 genome samples from CEU, YRI and JPT-CHB populations) and cryptic relatedness [identity by descent (IBD) > 0.125]. Markers failing the following thresholds: missingness rate (<2%), minor allele frequency rate (>5%) or Hardy Weinberg Equilibrium (P > 0.0001) were excluded. We examined markers for informative missingness and batch effects (Anderson et al., 2010). We report detailed procedures and numbers of excluded samples for individual quality control steps in Supplementary Table 2.
Imputation
The pruned dataset was pre-phased using SHAPEIT (version 2.12). We used IMPUTE2 (version 2.3.0) for imputation. In both steps 1000 Genomes Phase I integrated variant set release (v3) in NCBI b37 was used as reference panel. Only imputed variants with a minor allele frequency > 0.01 and an imputation quality score (INFO) above 0.3 were analysed further.
Tests for association
We used (i) a logistic additive model implemented in PLINK (version 1.07) (Purcell et al., 2007); and (ii) GEMMA’s linear mixed model (version 0.94) (Zhou and Stephens, 2012) for association testing. We adjusted for sex and population stratification using 10 dimensions of an IBD-derived multidimensional scaling matrix (PLINK) or using a GEMMA-generated relatedness matrix. The imputed dosage data were analysed using GEMMA.
Replication
The most promising SNPs (n = 59) in the finding stage were studied further. We selected SNPs in narrow regional bins in linkage disequilibrium (LD) and controlled their cluster plots. LD bins were defined as SNPs being located in a 500 kb window and with r2 > 0.5. We used two Sequenom® panels, processed all samples in one centre and tested for association using logistic models implemented in PLINK. Due to incomplete phenotype data, additional covariates could not be included without severely compromising sample size. We investigated follow-up SNPs in the combined discovery and replication data using logistic regression (PLINK).
Genotyping of variants associated with essential tremor in previous genome-wide association studies
We genotyped all available samples using TaqMan® probes for SNPs rs9652490 in LINGO1 and rs3794087 in SLC1A2 and called alleles using QuantStudioTM Real-Time PCR software v1.1 (Life Technologies).
Clinicopathological assessment of patients and tissue handling
Ethics review board approval and participants’ written consents were obtained. All patient brain samples are from individuals diagnosed with definite essential tremor (Deuschl et al., 1998). Control individuals were reported to be free of neurological disorders. Autopsy procedures have been described previously (Rajput et al., 2004). One hemisphere of all patient brains was examined histologically in detail by a neuropathologist. Controls underwent a standard neuropathological assessment. Nineteen cases with essential tremor and five control subjects were of Canadian origin, 12 control subjects were of German origin (Supplementary Table 3).
DNA/RNA extraction
We isolated total RNA from 30 mg lateral cerebellar cortex tissue from 19 essential tremor cases and 17 control subjects using AllPrep® DNA/RNA Mini Kit (Qiagen). RNA concentration was measured using NanoDrop® Spectrophotometer and RNA quality determined by gel electrophoresis.
Quantitative reverse transcription polymerase chain reaction
We prepared cDNA from 500 ng RNA for quantitative PCR (qPCR) using the RevertAid® H Minus Reverse Transcriptase kit (Thermo Fisher). RNA expression was analysed for the genes PPARGC1A, STK32B, CTNNA3, ACTB and the Purkinje cell housekeeping gene PCP2. Expression analysis was performed using a StepOnePlus® system and SYBR® Green PCR Master Mix (Thermo Fisher). We performed at least two technical replicates in one qPCR run (four replicates in two qPCR runs for ACTB, PCP2 and STK32B). We calculated expression using bioconductor packages ReadqPCR and NormqPCR. Technical replicates were averaged, then gene-expression was normalized using ACTB and PCP2 and expression differences assessed using Student’s t-test.
Results
Discovery stage
All discovery samples were genotyped under identical conditions. In the discovery stage 1778 patients with essential tremor and 5376 control samples were genotyped and 416 462 SNPs included after quality pruning. The estimated statistical power was ∼90% for an odds ratio (OR) of 1.3 (Supplementary Fig. 1). We first computed marker-wise P-values using the logistic additive model implemented in PLINK. QQ-plots showed a high level of genomic inflation (λ = 1.25) despite including 10 dimensions of an IBD-derived multidimensional scaling matrix, indicating incomplete correction for population stratification. Therefore, we also used GEMMA’s linear mixed model with adjustment for sex and population stratification, which reduced the genomic inflation to a very low level with λ = 1.01 (Zhou and Stephens, 2012). In addition, we analysed the imputed dosage data using GEMMA (Manhattan and QQ-Plots: Supplementary Figs 2–4).
Two variants successfully replicated in independent cohort
In the replication stage we genotyped an independent cohort of 1029 patients with essential tremor and 1065 control samples for 59 best markers selected from all three analyses (PLINK, GEMMA, GEMMA imputed data). We applied logistic regression analysis (PLINK) in the replication stage and in a combined analysis of both stages. Two variants, both selected from the GEMMA first stage analysis, met a Bonferroni-corrected significance threshold of P = 8.47 × 10−4 (Tables 1 and 2). The first variant rs10937625 [P = 7.36 × 10−4, OR = 0.77, 95% confidence interval (CI) 0.66–0.90] locates to an intronic region of STK32B (Fig. 1A), coding for a serine/threonine kinase. The second variant rs17590046 [P = 6.81 × 10−4, OR = 0.75 (95% CI 0.64–0.89)] lies in an intron of PPARGC1A (Fig. 1B), a transcriptional coactivator. LD analysis showed no additional associated markers in neighbouring genes (Fig. 1A and B). None of the SNPs chosen based on the PLINK analysis of the discovery stage or the imputed data was successfully replicated. Therefore, we do not report any imputed data. Combined analysis of first and second stage data revealed a region on chromosome 10q21.3 containing variants with small P-values in PLINK as well as GEMMA analysis, including three intronic variants in the adhesion molecule gene CTNNA3[rs7903491, P = 2.49 × 10−7, OR = 1.10 (95% CI 1.03–1.18); rs12764057, P = 1.19 × 10−8, OR = 1.17 (95% CI 1.09–1.26) and rs10822974, 1.65 × 10−7, OR = 1.16 (95% CI 1.08–1.24)] (Fig. 1C). The variants are located close to each other but are not in strong LD (r2 < 0.8) and testing for independent haplotypic effects using PLINK showed no correlation (Fig. 1C). Association results for individual stages and combined analysis of the 20 most significant markers are summarized in Supplementary Table 4. The minor allele frequencies of successfully replicated markers rs10937652 and rs1750046 were highly consistent between different recruiting centres and discovery/replication samples while higher deviations between stages were observed for markers in CTNNA3 (Supplementary Table 5 and Supplementary Fig. 5). Heterogeneity metrics of associated markers between different stages and recruitment sites are displayed in Table 1 and forest plots in Supplementary Figs 6–10. In many studies, including this one, the age at onset data follow a bimodal distribution (Supplementary Fig. 11). Stratification into early (<25 years) and late (>50 years) age at onset groups did not reveal large differences in the allele frequencies of the top 20 markers between both groups (Supplementary Table 6 and Supplementary Fig. 12). We do not present subgroup analyses because these did not provide additional insights.
Table 1.
Association results for the three associated loci
| Discovery stage | Replication stage | Combined stages | Heterogeneity | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | CHR | BP | Minor allele (a) | Major allele (A) | P | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | I2 | Tau2 |
| rs10937625 | 4 | 5 128 159 | C | T | 4.42 × 10−6 | 0.80 (0.73–0.88) | 7.36 × 10−4 | 0.77 (0.66–0.90) | 5.21 × 10−10 | 0.80 (0.74–0.87) | 39.6% | 0.0049 |
| rs17590046 | 4 | 24 362 541 | C | T | 3.39 × 10−6 | 0.80 (0.73–0.88) | 6.81 × 10−4 | 0.75 (0.64–0.89) | 1.39 × 10−9 | 0.79 (0.72–0.86) | 47.6% | 0.0082 |
| rs12764057 | 10 | 68 845 715 | G | T | 3.99 × 10−5 | 1.28 (1.19–1.39) | 0.275 | 1.07 (0.94–1.22) | 1.19 × 10−8 | 1.17 (1.09–1.26) | 61.6% | 0.0089 |
| rs10822974 | 10 | 68 850 419 | G | A | 3.02 × 10−5 | 1.27 (1.18–1.37) | 0.796 | 0.98 (0.86–1.13) | 1.65 × 10−7 | 1.16 (1.08–1.24) | 67.3% | 0.0118 |
| rs7903491 | 10 | 68 917 164 | A | G | 1.10 × 10−6 | 1.20 (1.11–1.30) | 0.407 | 1.05 (0.93–1.20) | 2.49 × 10−7 | 1.10 (1.03–1.18) | 92.4% | 0.0676 |
SNP = dbSNP ID of the marker; CHR = chromosome; BP = chromosomal position (GRCh37), OR (95% CI) = odds ratio with 95% confidence interval; I2 = ratio of true heterogeneity to total observed variation; tau2 = between studies variance. Heterogeneity metrics were calculated between samples included in: discovery stage with European descent, discovery stage with North American descent, replication stage with European descent and replication stage with North American descent.
Table 2.
Genotype counts and minor allele frequencies for the three associated loci
| Genotype counts cases | Genotype counts controls | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | CHR | BP | Minor Allele (a) | Major Allele (A) | aa | aA | AA | MAF | aa | aA | AA | MAF |
| rs10937625 | 4 | 5 128 159 | C | T | 113 | 942 | 1622 | 0.2182 | 403 | 2492 | 3410 | 0.2615 |
| rs17590046 | 4 | 24 362 541 | C | T | 68 | 775 | 1888 | 0.1668 | 263 | 2112 | 4047 | 0.2054 |
| rs12764057 | 10 | 68 845 715 | G | T | 485 | 1283 | 930 | 0.4175 | 898 | 2974 | 2537 | 0.3721 |
| rs10822974 | 10 | 68 850 419 | G | A | 625 | 1271 | 637 | 0.4976 | 1827 | 3116 | 1312 | 0.5412 |
| rs7903491 | 10 | 68 917 164 | A | G | 453 | 1311 | 881 | 0.4191 | 1347 | 3208 | 1848 | 0.4609 |
MAF = minor allele frequency; SNP = dbSNP ID of the marker; CHR = chromosome; BP = chromosomal position (GRCh37).
Figure 1.
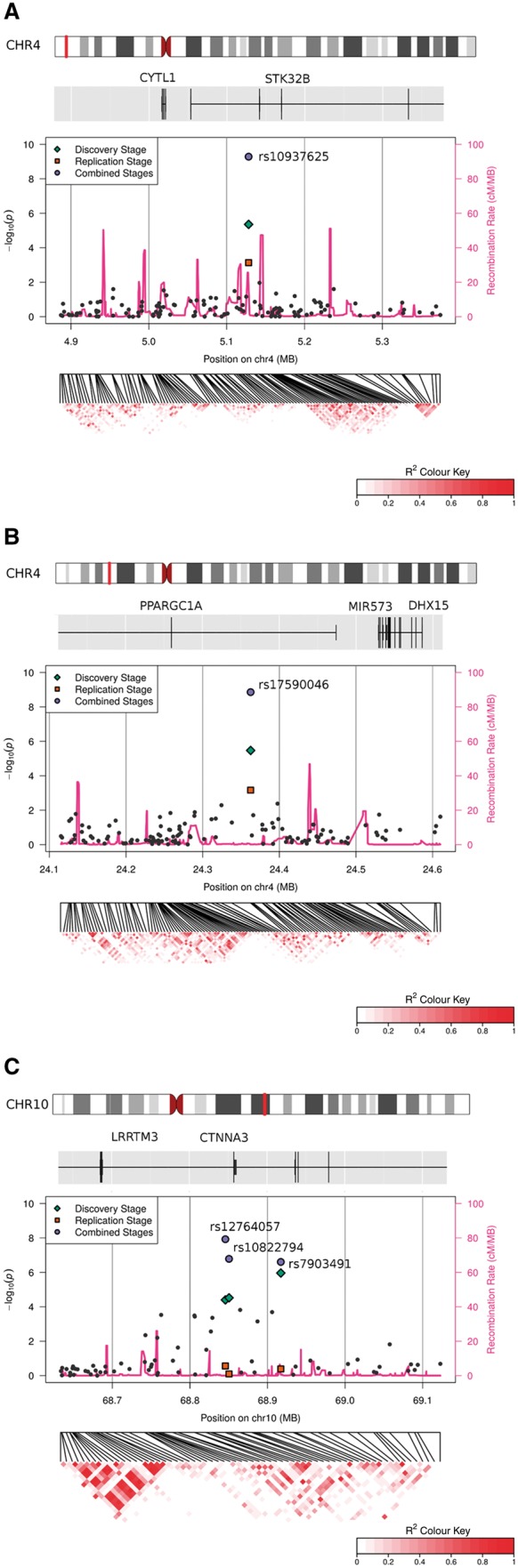
Fine mapping of 500 kb windows centred around each of the three essential tremor-associated loci.Top to bottom: Chromosome ideogram followed by genes (horizontal line) and their coding regions (vertical bars), negative logarithmic P-values of all markers for the discovery stage (left y-axis) against chromosomal position (x-axis) and recombination rates for the genomic positions (right y-axis) in pink. Bottom: Heatmap indicating LD. Saturated red indicating high LD, white indicating no LD. (A) STK32B; (B) PPARGC1A; and (C) CTNNA3.
No replication of previously reported associations
We genotyped SNPs rs3794087 in the SLC1A2 gene and rs9652490 in LINGO1, which were associated with essential tremor in previous GWAS in all available samples (2847 cases/1977 controls) (Supplementary material). Controls provided by Kora and PopGen could not be included because we could not obtain DNA for these samples. Neither of the two SNPs nor neighbouring SNPs showed a significant association with essential tremor (Supplementary Table 7, Supplementary Figs 13 and 14).
The STK32B SNP rs10937625 is located in a regulatory region
Variant annotation using HaploReg3 revealed that rs10937625 is located in a DNase hypersensitive region but did not provide further information for the other essential tremor-associated SNPs. A BioGRID search did not find any evidence for interactions or shared GO Processes between the genes STK32B, PPARGC1A and CTNNA3.
Increased expression of STK32B in brains of patients with essential tremor
We examined the expression of STK32B, PPARGC1A and CTNNA3 in lateral cerebellar cortex of 19 patients with essential tremor and 17 control subjects by reverse transcription (RT)-qPCR. Age at death, post-mortem intervals, and sex did not significantly differ between patients and controls. The expression of STK32B was significantly increased in essential tremor brains compared to control brains (P = 0.026) (Supplementary Table 8 and Supplementary Fig. 15). The expression difference was not associated with genotype at rs10937625 (Supplementary Table 9 and Supplementary Fig. 16). A linear regression model identified disease status as the only determinant of STK32B expression (Supplementary Fig. 15). We found no genotype or phenotype related differences in the expression of PPARGC1A or CTNNA3.
STK32B marker rs10937625 associated with eQTL in public database
In the Braineac eQTL database, the expression of probeset 2716778 (Affymetrix Probeset ID, chr4:5532972-5533090) located in the STK32B coding region is highly associated with the genotype of SNP rs1093762 (P < 0.0008) in cerebellar cortex but not in other brain regions (Trabzuni et al., 2011) (Supplementary Fig. 18). We observed an additive, negative effect of the minor, protective allele on expression of the STK32B probe set. Expression of PPARGC1A and CTNNA3 were not associated with the genotypes of the corresponding essential tremor-associated SNPs in the Braineac data.
Discussion
Standard analysis of our discovery stage data using PLINK suggested a high level of population stratification. GEMMA’s relatedness matrix is known to be particularly suited for the correction of population stratification. The very low mean χ2 metric of 1.0048 in the GEMMA analysis made us suspect significant levels of overcorrection and loss of power. To minimize type 1 errors (PLINK) as well as type 2 errors (GEMMA) we selected the top SNPs from all analysis types for replication. Only the third and fourth ranked SNPs in the GEMMA analysis of the discovery stage (rs10937625, rs17590046) were significantly associated with essential tremor in the replication stage. We choose to report three additional SNPs in the CTNNA3 gene, which were among the top variants in the discovery sample in both the PLINK as well as the GEMMA analysis. However, it has to be noted that these SNPs failed replication and carry an increased risk of a type 1 error.
The biological function of the serine/threonine kinase STK32B is unknown. The gene is located in a 520 kb deletion causing the rare skeletal dysplasia Ellis-Van-Creveld syndrome (Temtamy et al., 2008). The association of rs10937625 in the STK32B gene with essential tremor, the increased expression in the cerebellar cortex of patients with essential tremor and the association of the protective minor allele with reduced STK32B expression in cerebellar cortex in the Braineac eQTL database support a role of STK32B in the pathogenesis of essential tremor. We found no association between the rs10937625 genotype and STK32B expression in our samples which might be a related to the small sample size.
The other essential tremor-associated SNP, rs17590046 is located in the PPARGC1A gene encoding the peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α). PGC-1α is a transcriptional coactivator of genes involved in energy metabolism and mitochondrial function (Wu et al., 1999). Genetic and functional studies suggest a role in the pathogenesis of various neurodegenerative disorders (Weydt et al., 2006). We found no evidence that rs17590045 is related to the expression of PPARGC1A. The CTNNA3 gene, harbouring the SNPs rs7903491, rs12764057, rs10822974 codes for catenin alpha 3, a cell–cell adhesion molecule genetically linked to Alzheimer’s disease (Miyashita et al., 2007).
We are fully aware that the most severe limitations of our study are the lack of genome-wide significance of the replicated markers in the discovery stage and incomplete harmonization of clinical data throughout our cases. The lack of genome-wide significance in the GEMMA analysis of the first stage might be due to overcorrection in the GEMMA based analysis. Therefore, we would like to strongly encourage replication to confirm or refute the hypotheses provided by our study. Nevertheless, we would also like to emphasize that this is by far the largest GWAS in essential tremor and we are confident that it will be the driving force for the next generation of genetics and molecular studies to come.
Supplementary Material
Acknowledgements
We want to thank the patients and healthy individuals that participated in this study. We are grateful to the following persons for their help in recruiting cases and controls: Jay van Gerpen, Ryan J. Uitti, Audrey J Strongosky, Maria A. Pastor and Virginia Norris.
Funding
This study was made possible by funding from the CIHR, Canada Research Chair in genetics of the nervous system and from the DFG (KU1194/9-1) and from the National Institutes of Health (NIH R01 NS073872). S.L.G. is grateful to FRQS for financial support. G.A.R. is grateful to CIHR and to the Canada Research Chair in genetics of the nervous system. G.K., G.D. and F.H. are grateful to the DFG for financial support (KU1194/9-1). E.D.L. and L.N.C. were supported by NIH R01 NS073872. KORA research was supported within the Munich Center of Health Sciences (MC-Health), Ludwig-Maximilians-Universität, as part of LMUinnovativ. The Mayo Clinic Florida is a Morris K. Udall Parkinson’s Disease Research Center of Excellence (NINDS P50 #NS072187), and this work was supported by the Mayo Clinic Florida Research Committee. This research was supported by a donation of the Rhein-Taunus-Krematorium. C.M.T. is grateful to the Emory ADRC and National Institute of Aging (P50 AG025688).
Web resources
The URLs for data presented herein are as follows:
HaploReg V3: https://www.broadinstitute.org/mammals/haploreg/haploreg_v3.php
BioGRID: http://thebiogrid.org/
BRAINEAC: http://www.braineac.org/
Supplementary material
Supplementary material is available at Brain online.
Glossary
Abbreviations
- GWAS
genome-wide association study
- LD
linkage disequilibrium
- RT-qPCR
reverse transcription-quantitative polymerase chain reaction
- SNP
single nucleotide polymorphism
References
- Anderson CA, Pettersson FH, Clarke GM, Cardon LR, Morris AP, Zondervan KT. Data quality control in genetic case-control association studies. Nat Protoc 2010; 5: 1564–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuschl G, Bain P, Brin M. Consensus statement of the Movement Disorder Society on Tremor. Ad Hoc Scientific Committee. Mov Disord 1998; 13 (Suppl 3): 2–23. [DOI] [PubMed] [Google Scholar]
- Krawczak M, Nikolaus S, von Eberstein H, Croucher PJ, El Mokhtari NE, Schreiber S. PopGen: population-based recruitment of patients and controls for the analysis of complex genotype-phenotype relationships. Community Genet 2006; 9: 55–61. [DOI] [PubMed] [Google Scholar]
- Kuhlenbäumer G, Hopfner F, Deuschl G. Genetics of essential tremor: meta-analysis and review. Neurology 2014; 82: 1000–7. [DOI] [PubMed] [Google Scholar]
- Lemieux Perreault LP, Provost S, Legault MA, Barhdadi A, Dube MP. pyGenClean: efficient tool for genetic data clean up before association testing. Bioinformatics 2013; 29: 1704–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz D, Poremba C, Papengut F, Schreiber S, Deuschl G. The psychosocial burden of essential tremor in an outpatient- and a community-based cohort. Eur J Neurol 2011; 18: 972–9. [DOI] [PubMed] [Google Scholar]
- Louis ED, Agnew A, Gillman A, Gerbin M, Viner AS. Estimating annual rate of decline: prospective, longitudinal data on arm tremor severity in two groups of essential tremor cases. J Neurol Neurosurg Psychiatry 2011; 82: 761–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis ED, Ferreira JJ. How common is the most common adult movement disorder? Update on the worldwide prevalence of essential tremor. Mov Disord 2010; 25: 534–41. [DOI] [PubMed] [Google Scholar]
- Miyashita A, Arai H, Asada T, Imagawa M, Matsubara E, Shoji M, et al. Genetic association of CTNNA3 with late-onset Alzheimer's disease in females. Hum Mol Genet 2007; 16: 2854–69. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81: 559–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajput A, Robinson CA, Rajput AH. Essential tremor course and disability: a clinicopathologic study of 20 cases. Neurology 2004; 62: 932–6. [DOI] [PubMed] [Google Scholar]
- Stefansson H, Steinberg S, Petursson H, Gustafsson O, Gudjonsdottir IH, Jonsdottir GA, et al. Variant in the sequence of the LINGO1 gene confers risk of essential tremor. Nat Genet 2009; 41: 277–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temtamy SA, Aglan MS, Valencia M, Cocchi G, Pacheco M, Ashour AM, et al. Long interspersed nuclear element-1 (LINE1)-mediated deletion of EVC, EVC2, C4orf6, and STK32B in Ellis-van Creveld syndrome with borderline intelligence. Hum Mutat 2008; 29: 931–8. [DOI] [PubMed] [Google Scholar]
- Thier S, Lorenz D, Nothnagel M, Poremba C, Papengut F, Appenzeller S, et al. Polymorphisms in the glial glutamate transporter SLC1A2 are associated with essential tremor. Neurology 2012; 79: 243–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabzuni D, Ryten M, Walker R, Smith C, Imran S, Ramasamy A, et al. Quality control parameters on a large dataset of regionally dissected human control brains for whole genome expression studies. J Neurochem 2011; 119: 275–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weydt P, Pineda VV, Torrence AE, Libby RT, Satterfield TF, Lazarowski ER, et al. Thermoregulatory and metabolic defects in Huntington's disease transgenic mice implicate PGC-1alpha in Huntington's disease neurodegeneration. Cell Metab 2006; 4: 349–62. [DOI] [PubMed] [Google Scholar]
- Wichmann HE, Gieger C, Illig T, Group MKS. KORA-gen–resource for population genetics, controls and a broad spectrum of disease phenotypes. Gesundheitswesen 2005; 67 (Suppl 1): S26–30. [DOI] [PubMed] [Google Scholar]
- Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell 1999; 98: 115–24. [DOI] [PubMed] [Google Scholar]
- Zhou X, Stephens M. Genome-wide efficient mixed-model analysis for association studies. Nat Genet 2012; 44: 821–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


