Abstract
The mammalian kidney collecting ducts are critical for water, electrolyte and acid-base homeostasis and develop as a branched network of tubular structures composed of principal cells intermingled with intercalated cells. The intermingled nature of the different collecting duct cell types has made it challenging to identify unique and critical factors that mark and/or regulate the development of the different collecting duct cell lineages. Here we report that the canonical Notch signaling pathway components, RBPJ and Presinilin1 and 2, are involved in patterning the mouse collecting duct cell fates by maintaining a balance between principal cell and intercalated cell fates. The relatively reduced number of principal cells in Notch-signaling-deficient kidneys offered a unique genetic leverage to identify critical principal cell-enriched factors by transcriptional profiling. Elf5, which codes for an ETS transcription factor, is one such gene that is down-regulated in kidneys with Notch-signaling-deficient collecting ducts. Additionally, Elf5 is among the earliest genes up regulated by ectopic expression of activated Notch1 in the developing collecting ducts. In the kidney, Elf5 is first expressed early within developing collecting ducts and remains on in mature principal cells. Lineage tracing of Elf5-expressing cells revealed that they are committed to the principal cell lineage by as early as E16.5. Over-expression of ETS Class IIa transcription factors, including Elf5, Elf3 and Ehf, increase the transcriptional activity of the proximal promoters of Aqp2 and Avpr2 in cultured ureteric duct cell lines. Conditional inactivation of Elf5 in the developing collecting ducts results in a small but significant reduction in the expression levels of Aqp2 and Avpr2 genes. We have identified Elf5 as an early maker of the principal cell lineage that contributes to the expression of principal cell specific genes.
Keywords: renal development, collecting ducts, Notch, RBPJ, intercalated cell
Introduction
The mammalian kidneys are responsible for water, electrolyte and acid-base homeostasis. These functions are carried out by nephrons which are tubular structures organized into segments each consisting of unique epithelial cell types. An exception to this segregation of cell types occurs in the connecting tubule (CNT) of the distal nephron and the collecting ducts (CD) which consist of intermingled cell types [1–3]. The majority of the CD consist of vasopressin responsive principal cells that uniquely express aquaporin-2 (Aqp2) apically and arginine-vasopressin receptor-2 (Avpr2), aquaporin-3 (Aqp3) and aquaporin-4 (Aqp4) on the basolateral membranes to regulate water homeostasis [4]. Intermingled among the principal cells (PC) are the intercalated cell (IC) types that are responsible for pH homeostasis. The ICs express carbonic anhydrases (CA), vacuolar H+-ATPase pumps (H+-ATPase) and anion exchangers (AE) [5–7]. Based on the differential localization of vacuolar H+-ATPase and the type and localization of AE (apical versus basolateral) ICs are classified into α, β, and non-α and β [8, 9]. ICs belong to a type of epithelia referred to as proton-secreting cells [10] found in many mammalian organ systems, including kidneys, inner ear and epididymis. These proton-secreting cells are also present in Xenopus and zebrafish skins, where their differentiation is promoted by Foxi1-orthologs and negatively regulated by Notch signaling [11, 12].
Notch signaling regulates cell fate patterning in which neighboring cells during development adopt different fates [11, 13–16]. Notch signaling occurs between adjacent cells, with at least one of four Notch receptors being activated by ligands belonging to the Delta-like (Dll) and Jagged (Jag) family of type I transmembrane proteins [17]. Upon ligand binding, Notch receptors undergo a series of cleavage events initiated by Adam metalloproteinases and followed by γ-secretase. Presenilin1 (PS1) and Presenilin2 (PS2) are two components of the γ-secretase complex critical for releasing the Notch intracellular domain (NICD) from the membrane [18, 19]. NICD translocates to the nucleus and converts DNA-bound RBPJ-containing transcriptional repressor complexes into activators by interacting directly with RBPJ and recruiting mastermind-like to activate target genes.
The molecular regulators of collecting duct differentiation are only beginning to be identified. PCs and ICs originate from a common precursor in the developing CD. Inactivation of Foxi1, a forkhead transcription factor in mice results in CD cells expressing both PC and IC markers, possibly arresting at an intermediary developmental state [20]. Foxi1 activates expression of IC-specific genes, including Slc4A9 and genes coding for subunits of the vacuolar H+-ATPase pump [21, 22]. Foxi1 expression levels and the number of ICs are increased in Mind bomb1 (Mib1) and Adam10-deficient mouse collecting ducts. Both Mib1 and Adam10 are required for Notch receptor activation within the developing CD to ensure that a sufficient number of CD cells select the PC-fate [23, 24]. Collectively, the studies in different organisms reveal a central role for Foxi1 in specification of IC-like cells and a role for Notch signaling in repressing Foxi1 expression to allow for PC development. It remains unknown what factors directly activate the PC specific genes to turn on the PC program.
In the current study, we made use of the mouse kidneys with Notch-signaling-deficient-collecting ducts as a way of “genetically sorting” the principal and intercalated cells. We hypothesized that comparing the gene expression profiles of developing kidneys with Notch-signaling-deficient collecting ducts versus wild-type kidneys would allow for the identification of novel PC specific factors. In support of our hypothesis, we have identified Elf5 as an early PC-specific transcription factor that contributes to the regulation of mature PC genes.
Results
Inactivation of RBPJ in the developing mouse collecting ducts results in a reduction in the number of principal cells and an increase in intercalated cell number
To utilize mouse kidneys with Notch-signaling-deficient-collecting ducts as a tool to identify novel PC-specific transcription factors we inactivated RBPJf/f (RBPJ-conditional alleles) in the developing ureteric bud (UB) lineage by breeding with HoxB7->Cre mice [25]. RBPJ protein was depleted from most UB cells in HoxB7->Cre;RBPJf/− kidneys by E14.5 (Fig.1). At E14.5 the UB cells, which in HoxB7->Cre;Rosa+/EYFP kidneys are EYFP+ cells, have not differentiated into ICs or PCs as determined by the absence of Foxi1 and Aqp2 in EYFP+ cells (Fig.S1). However, EYFP−negative cells that are likely part of the CNT-segment of nephrons have begun to differentiate into PCs and ICs at E14.5 (Fig.S1). Although both CNT and CD consist of PC and IC types, the CNT is derived from the Six2+ cap mesenchyme while the CD is derived from the UB [26–28]. In HoxB7->Cre;RBPJf/− kidneys most cells of the UB lineage are deficient for RBPJ by E14.5 (Fig.1), before differentiation into ICs or PCs (Fig.S1).
Figure 1. The HoxB7->Cre transgenic line efficiently inactivates RBPJ in the ureteric bud lineage by E14.5.
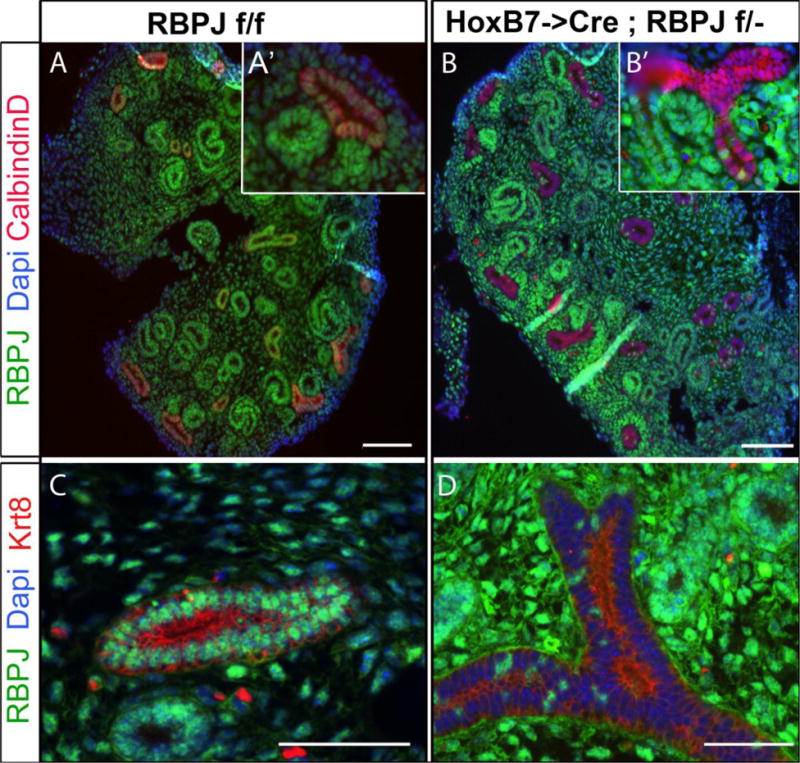
A. RBPJf/f (wild-type) E14.5 littermate kidneys express RBPJ (green) in all cells including Calbindin-D expressing (red) ureteric bud cells. A′. Higher magnification of a ureteric bud section reveals RBPJ expression in UB cells. B. Most ureteric duct cells in HoxB7->Cre;RBPJf/− (mutant) E14.5 kidneys lack RBPJ. B′. A higher magnification of a T-shaped ureteric duct reveals that most UB cells are deficient for RBPJ but not the cells of distal segment of the nascent nephron that fuses with the ureteric duct. C&D. Collecting ducts expressing cytokeratin-8 (Krt8; Red) in E14.5 RBPJf/f (C) also express RBPJ, whereas collecting ducts in HoxB7->Cre;RBPJf/− E14.5 littermate kidneys are deficient for RBPJ (D). Several kidney sections from 3 mice per genotype were analyzed at E14.5. The scale bars are 50μm.
Similar to Mib1 or Adam10-deficient CDs, inactivation of RBPJ resulted in many more ICs as evidenced by an increased number of Foxi1 (Fig.2A–D) or carbonic anhydrase II (CAII) expressing cells (Fig.2E–H), accompanied by a reduction in Aqp2 expressing cells (Fig.2A–H). The alteration in IC to PC ratio was verified by measuring the expression levels of IC-specific genes and PC-specific genes. RT-qPCR using whole kidney RNA revealed increased expression of IC-specific genes along with decreased expression of PC-specific genes in the E18.5 kidneys with RBPJ-deficient CDs when compared with that of wild-type kidneys (Fig.2I, J). There was a two-fold increase in expression level of the intercalated cell-specific gene Foxi1 and a two-fold reduction in expression levels of principal cell-specific genes, Aqp2 and Aqp4. This reduction in principal cell gene expression was accompanied by a reduced ability to concentrate urine (Fig. 2K), and occasionally hydronephrosis in one of the two kidneys resulting in loss of renal medullary tissue (Fig.S2). Similar to the Mib1-deficient and Adam10-deficient collecting ducts, PCs still develop from RBPJ-deficient-CD cells of HoxB7->Cre; RBPJf/− mice (Fig.2 and Fig.S3). Overall, Notch-mediated transcription via RBPJ is required to ensure that a sufficient number of CD cells adopt the PC fate to allow for normal CD function.
Figure 2. Notch signaling mediated patterning of collecting duct cell fates is dependent on RBPJ. A, B, E and F.
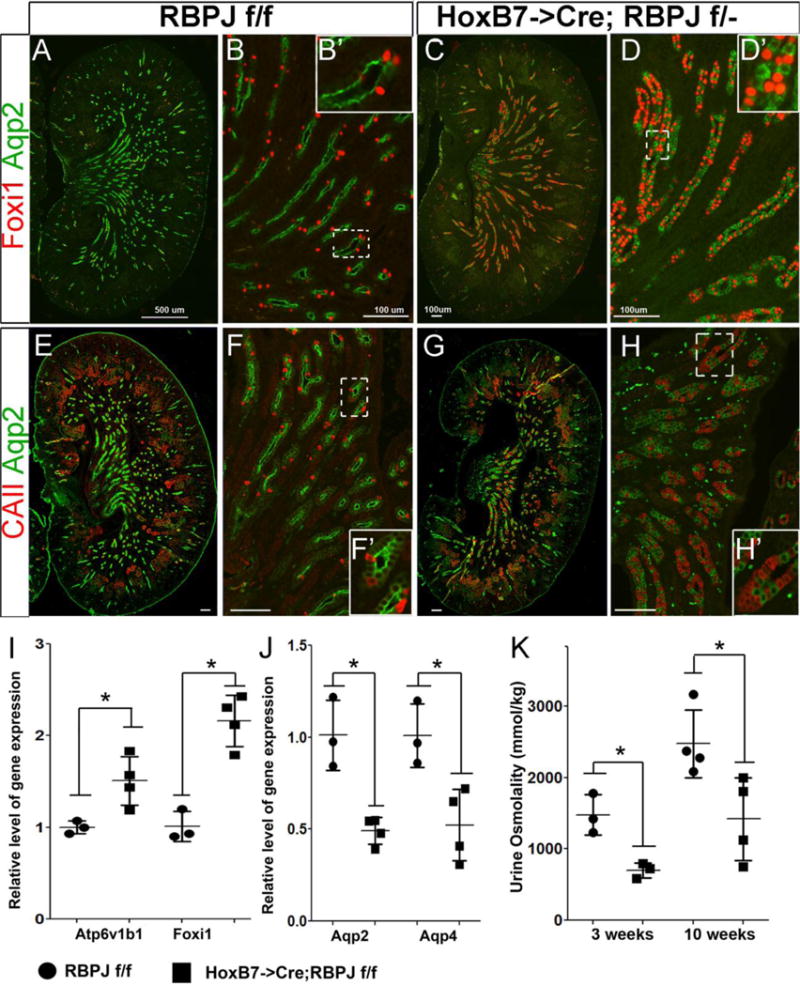
Kidney sections of wild-type post-natal day 1 RBPJf/f mice reveal many Aqp2 expressing cells intermingled with a few ICs expressing Foxi1 (A&B) or CAII (E&F) within the CDs. C, D, G and H: Kidney sections with RBPJ-deficient CDs contain many more ICs expressing Foxi1 (C&D) and CAII (G &H) and fewer Aqp2-expressing PCs. A, E, C and G are images of whole kidney sections revealing the central papillary and inner medullary regions. B, D, F and H are higher magnifications of inner medullary/papillary regions. D′ and H′ are higher magnifications of boxed areas in D and H, respectively. I &J: RT-qPCR reveals significantly increased expression of Atp6v1b1 and Foxi1, along with significantly decreased levels of Aqp2 and Aqp4 in E18.5 HoxB7->Cre;RBPJf/f kidneys (n=4) compared with RBPJf/f littermate kidneys (n=3). K. The urine concentrating ability of HoxB7->Cre;RBPJf/f mice (n=3 or 4 per time point) is significantly decreased compared with RBPJf/f wild-type littermate (n=3 or 4 per time point). A separate cohort was analyzed at each time point. * denotes p<0.05, two-tailed unpaired t-tests. Scale bars are 100μm, expect A it is 500μm.
Elf5 is down-regulated in kidneys with Notch-signaling-deficient collecting ducts and is among the earliest genes up regulated with ectopic expression of activated Notch1 in the developing collecting ducts
There is limited knowledge of the transcription factors that are specifically expressed in the PC lineage and required for PC development. We hypothesized that we could identify such factors by comparing the gene expression profiles of developing kidneys containing Notch-signaling-deficient collecting ducts versus wild-type kidneys. Since Notch-signaling-deficient collecting ducts have reduced number of PCs, genes uniquely expressed in the PCs are expected to be expressed at lower levels in the kidneys with Notch-signaling-deficient collecting ducts (Fig. 2J). To identify PC-specific transcripts we compared gene expression profiles of E18.5 HoxB7->Cre;RBPJf/−; RosaEyfp/+ (mt) to RBPJf/f; RosaEyfp/+ wild-type (wt) littermate kidneys. Profiling of whole kidney total RNA yielded approximately 80 genes down-regulated by at least 1.25-fold (p<0.05) in mt versus wt (Fig.3A). The presence of Aqp2 and Aqp4 among the down-regulated genes (arrowheads in Fig.3A) added confidence that the gene profiling experiment had technically worked and that at least a sub-set of the down-regulated genes are likely to be expressed only in the principal cells within the kidney. Among the down-regulated genes was Elf5, which codes for an E26 transformation-specific (ETS) transcription factor (arrow in Fig.3A). In further confirmation that renal Elf5 expression is dependent on Notch signaling within developing CDs, E18.5 HoxB7Cre; PS1−/f; PS2−/− kidneys have a reduction in Elf5, Aqp2 and Aqp4, along with an increase in Foxi1 and Atp6v1b1 expression levels, when compared to PS1f/f; PS2−/− littermates (Fig.3B). Based on the rationale that the first transcription factors to be up regulated by Notch signaling within the collecting ducts are likely to be critical early PC-specific transcription factors we tested whether Elf5 is among the earliest genes activated by ectopic expression of Notch1 intracellular domain (N1-ICD) in the developing collecting ducts. We generated HoxB7->Cre; Rosa+/NICD mice, in which N1-ICD is expressed in the ureteric duct cell lineage from a time prior to normal PC and IC differentiation. Mouse kidneys with ectopic N1-ICD expression in the collecting ducts at E13.5 had a two-fold increase in Elf5 (n=3, p<0.05), a 2.5-fold variable increase in Aqp4 (n =3; p=0.052) and a 4-fold variable increase in Aqp2 (n=3, p= 0.1) gene expression (Fig.3C). Hes1, a known target of Notch signaling, is also activated while Foxi1 expression is unchanged (Fig.3C). We decided to follow up on Elf5 as it is among the earliest genes up regulated by Notch signaling and the function and expression of Elf5 in the kidney has not been characterized. Additionally Elf5 in other tissues, such as the mammary gland and trophoblast, regulates cell fate selection/cell differentiation [29–31].
Figure 3. Elf5 expression is down-regulated in mouse kidneys with Notch-signaling- deficient-collecting ducts.
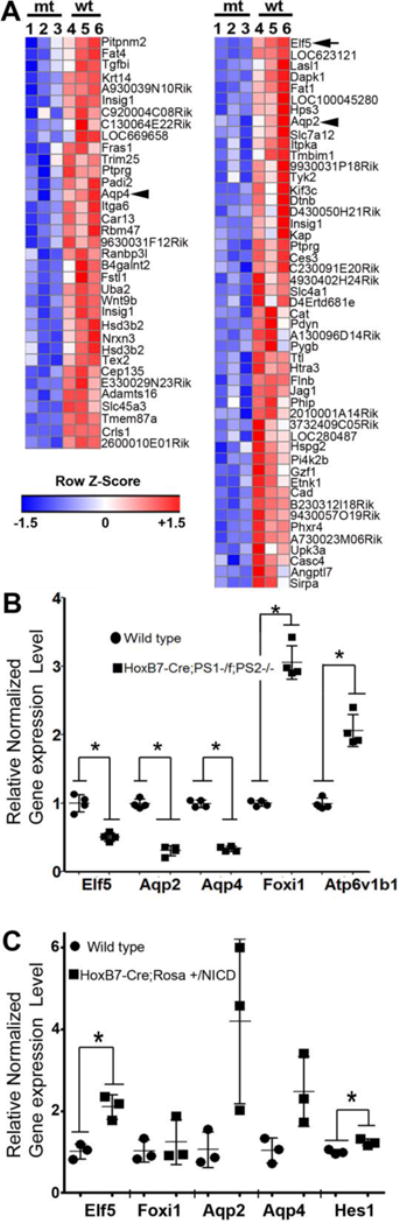
A. Heatmap of genes with reduced expression in the kidneys of three mutant (mt) HoxB7->Cre; RBPJ−/f;Rosa+/EYFP E18.5 embryos compared with that of three wild type (wt) RBPJf/f ;Rosa+/EYFP E18.5 embryos. The genes listed were down-regulated in the mt kidneys by at least 1.25 fold, n=3 per genotype, p<0.05. The values in each row were normalized by subtracting the row mean and dividing by row standard deviation to calculate the z-scores. The heatmap color scale is −1.5 to 1.5 (z-score), with intensity of red correlating with increased level of expression and intensity of blue correlating with lower level of expression compared with mean value within each row, which is represented as white. The arrowheads point at known PC genes Aqp2 and Aqp4, while the arrow points at Elf5. B. Elf5, Aqp2 and Aqp4 expression levels are reduced, while that of Foxi1 and Atp6v1b1 are increased in E18.5 kidneys of HoxB7Cre; PS1−/f; PS2−/− (inactivation of presenilin1 (PS1) and presenilin2 (PS2) when compared with PS1f/f;PS2−/− littermates, n=4 per genotype and asterisks denote p<0.05, two-tailed unpaired t-test.
Elf5 expression is activated early in the developing collecting ducts and remains on in mature principal cells
To determine the pattern of Elf5 expression we used Elf5+/LacZ mice in which β-galactosidase reports Elf5 expression [32], and the Elf5->GFP transgenic mice which harbor a 120kb mouse Elf5 genomic region with GFP inserted at the start of Elf5 coding sequence [33]. Examination of GFP expression at post-natal day 0 revealed Elf5 expression only in a subset of cells within the cytokeratin8+ collecting ducts of mouse kidneys (Fig. 4A, D, G). Within the nephrogenic zone and cortex, GFP was detected consistently within the stalk of the terminal collecting duct branches (arrow in Fig. 4A). In very rare instances we observed a luminal cell within the duct tip that was GFP+ in the Bac Elf5->GFP kidneys (arrowhead in Fig.4B). As we move further away from the duct tips and towards the medulla, more and more cells within the cortical collecting duct stalks express Elf5 (Fig. 4D–F), and the majority of cells within the medullary collecting ducts express Elf5 (Fig.4G–I). The strongest Elf5 expression occurs in medullary CD cells (Fig.4G), is detectable by E16.5 (Supplementary Fig.4) and remains on in Aqp2+ PCs at postnatal day 0 (Fig.4B,E,H and Supplementary Fig.4) and even at post-natal day 21 (Supplementary Fig.4). Elf5 expression was not detected in Foxi1+ cells (Fig.4C, F, I, Supplementary Fig.4F–J). Foxi1+ cells (arrows in Fig.4C, F, I) are always found adjacent to Elf5-expressing cells in the cortical and medullary collecting ducts. Interestingly, Elf5 expression occurs in more cortical collecting duct cells closer to the duct tips (arrowheads in C and F) that are not adjacent to Foxi1+ cells (arrows in C and F), and do not express Aqp2 (arrows in B and E). Aqp2 expression begins to occur in Elf5-expressing cortical collecting duct cells that are located further away from the duct tips. In addition, Aqp2 is also expressed in cells that do not express Elf5 (arrowheads in Fig.4H), revealing heterogeneity among Aqp2+ cells.
Figure 4. Elf5 expression overlaps with Aqp2 expression and occurs in a subset of the branching collecting duct cells.
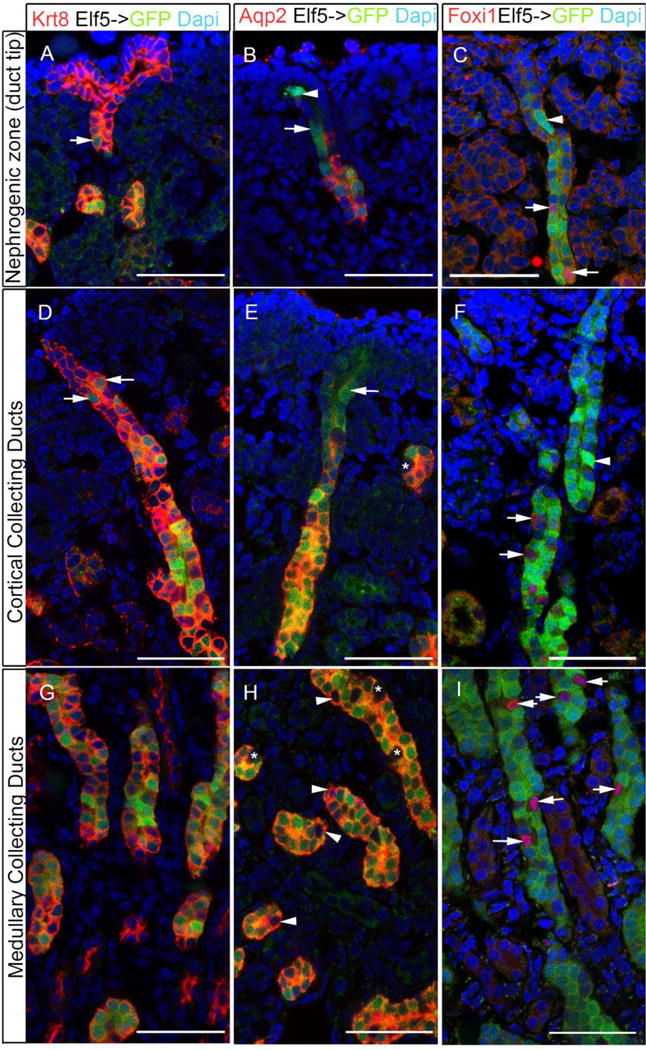
Analysis of Elf5->GFP transgenic post-natal day 0 mouse kidneys revealed GFP expression in a subset of cytokeratin 8 (krt8) positive collecting duct cells (A,D,G). Using GFP as an indicator of Elf5 expression, we observe Elf5 expression in Aqp2+ cells within the collecting ducts present in the nephrogenic zone (B), cortex (E) and medulla (H). Elf5 expression starts in more cortical collecting duct cells closer to the duct tips (arrowheads in C and F) that do not express Aqp2 (arrows in B and E). The asterisks in E and H mark Elf5− and Aqp2− cells, while the arrowheads in H mark Aqp2+ cells that do not express Elf5. Foxi1+ cells (arrows in C, F and I) are adjacent to Elf5 expressing cells. All scale bars are 50μm.
Elf5 activates principal cell specific gene promoters and ensures normal level of Aqp2 and Avpr2 expression in developing kidneys
Considering that Elf5 expression appears to precede Aqp2 at least in a subset of collecting duct cells, we tested whether ectopic expression of Elf5 can activate proximal promoters of Aqp2 and Avpr2 in immature CD cell lines, isolated from E12.5 HoxB7->Cre;ROSAEYFP/+;Immorto+/TG mouse kidneys (Fig.5A–C). Three cell lines, M1, M3 and M5, were established from E12.5 branching ureteric duct that expressed GFP. Analysis of genes expressed in these cells revealed that they do not express the mature principal (Aqp2, Aqp4) or intercalated (Foxi1) cell specific genes (Fig.5D). The M1 and M3 cells do not express Elf5, while the M5 cells express a low level of Elf5, possibly indicating that the M5 cells are at an early stage of becoming principal cells (Fig.5D). Interestingly, GFRα1 expression is lower in M5 cells than in M1 and M3 cells, however c-Ret levels appear similar (Fig.5D). Ectopic expression of Elf5 in these immature CD cell lines activated a proximal 1kb Avpr2 promoter significantly in all three cell lines, with the highest activation of 2.0-fold in the M5 cells (Fig.5E). Additionally, Elf5 significantly activated a proximal 1kb Aqp2 promoter in M3 and M5 cells, with the highest activation of 1.6-fold in the M5 cells (Fig.5E).
Figure 5. Elf5 modestly activates the proximal promoters of Aqp2 and Avpr2 in immature collecting duct cell lines derived from the developing kidney of mouse embryos.
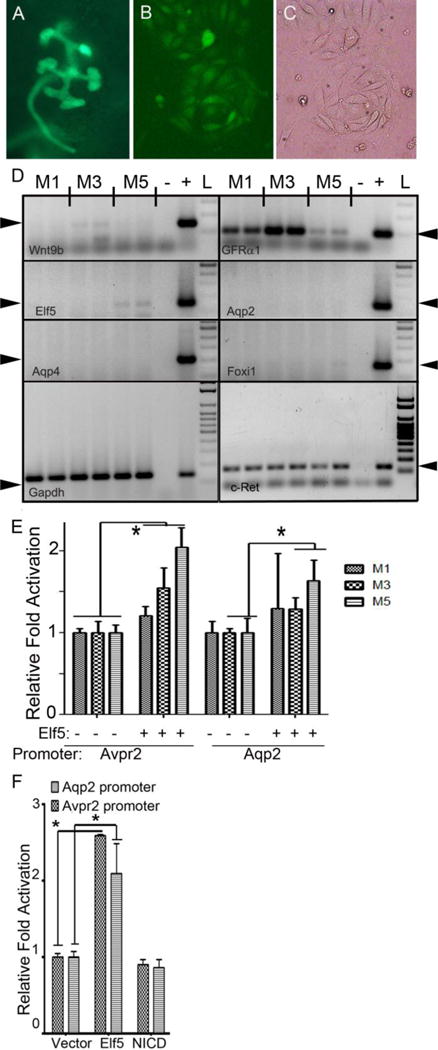
A Image of kidney with EYFP expression in the ureteric duct isolated from E12.5 Immorto; HoxB7->Cre; RosaEYFP/+ mouse. B & C. GFP and bright-field images of an isolated EYFP+ colony derived from the ureteric duct. D. RT-PCR of RNA extracted from M1, M3 and M5 cell lines isolated from ureteric ducts reveals variable GFRα1 and c-Ret expression in all cells, low levels of Wnt9b in some cell lines and no expression of mature markers of collecting duct cells such as Aqp2, Aqp4 and Foxi1. The M5 cells express a low level of Elf5, suggestive that they may be further along in the differentiation process compared with M1 and M3. +: RT-PCR of embryonic mouse total kidney mRNA, −: water control, L: DNA ladder. Arrowheads point to the expected sizes of RT-PCR products. E. The mouse Avpr2 proximal promoter is activated by Elf5 by as much as 2-fold in the immature collecting duct cell lines. Elf5 activated the proximal Aqp2 promoter in only one of the three immature duct cell lines by 1.5-fold. N=6 per condition per cell line. F. Elf5 increases the transcriptional activity of mouse 1Kb Avpr2 and 1Kb Aqp2 proximal promoters by at least two fold in MDCK cells, whereas ectopic expression of NICD is unable to increase the transcriptional activity. N=6 per condition, * denotes p<0.05, unpaired t-tests.
We next tested whether Elf5 can activate principal cell specific gene promoters in MDCK cells which have properties of mature PCs and are of CD origin [34]. Ectopic expression of Elf5 in MDCK cells can activate Aqp2 and Avpr2 proximal promoters by 2.5-fold and 2-fold, respectively (Fig.5F). In further confirmation that Elf5 can activate principal cell genes we observed that transient expression of Elf5 in the mature principal cell line mpkCCDc14, derived from mouse kidneys [35], increased the transcript levels of Aqp2 and Avpr2 by about 1.5-fold on average (n=6 per condition, p<0.05) as measured by RT-qPCR (Fig. 6A). To determine the contribution of Elf5 in the regulation of PC-specific genes we inactivated floxed alleles of Elf5f/f [31] using Cdh16->Cre transgene which drives Cre expression in distal tubules and CDs [36]. Neonatal mouse kidneys deficient for Elf5 expression (Cdh16->Cre; Elf5*/f, where *= floxed or null allele) express 25% less Aqp2 and 25% less Avpr2 compared with control littermates (Fig.6B). This decrease in Aqp2 and Avpr2 expression was not due to a reduction in number of PCs in any region of the kidney (Fig. 6C–I). The percentage of CAII+ collecting duct cells, as well as the percentage of Aqp2+ collecting duct cells were not significantly different in Elf5f/f (wild-type) versus Cdh16->Cre;Elf5*/f (Elf5-deficient) mouse kidneys (Fig. 6C). Consistent with no change in the principal to intercalated cell ratio, Foxi1 expression levels were the same in Elf5-deficient kidneys and control littermate kidneys (Fig. 6B). Additionally, not all principal cell specific genes, for example Aqp4, were significantly down-regulated in the absence of Elf5 (Fig. 6B).
Figure 6. Elf5 regulates the level of Avpr2 and Aqp2 expression in mouse kidneys.
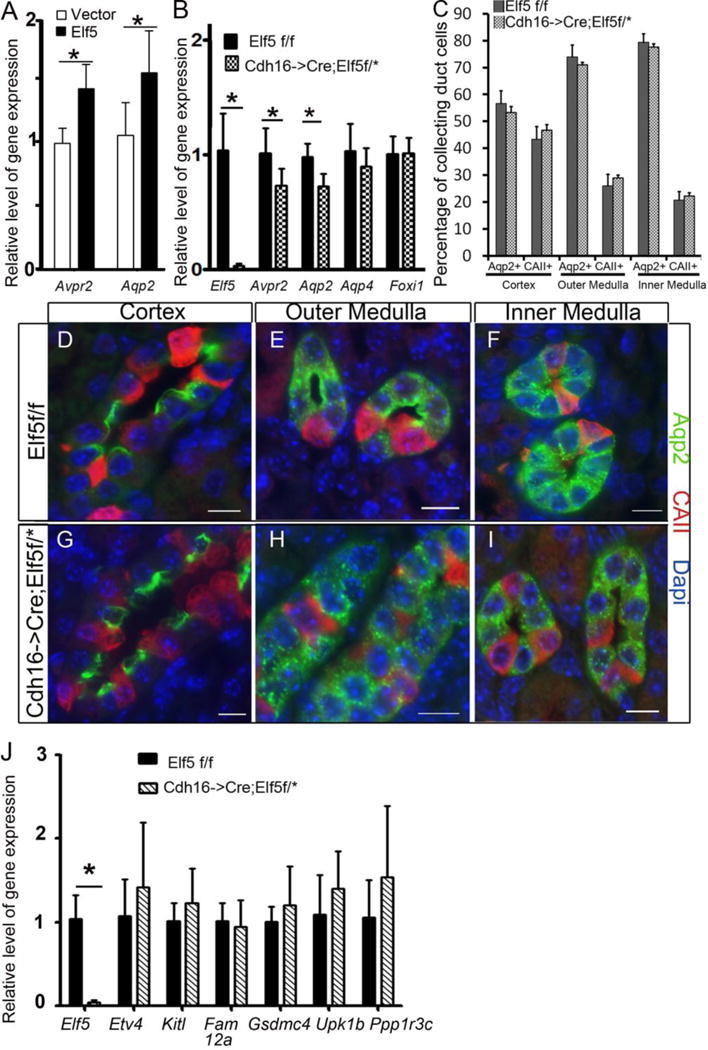
A Transient transfection of Elf5 increases the endogenous Avpr2 and Aqp2 transcript levels in mpkCCDC14 cells as determined by RT-qPCR. B. RT-qPCR reveals that Elf5 expression is essentially absent in Cdh16->Cre; Elf5f/* mouse neonatal kidneys compared with wild type littermates, where * denotes floxed (f) or null allele. In the absence of Elf5 expression, Avpr2 and Aqp2 expression are reduced to 75% of wild type levels on average, p<0.05, n= 6 wild type and 8 Cdh16-Cre;Elf5f/*. C. The percentage of collecting duct cells that express Aqp2 is not significantly different in the cortex, outer medulla and inner medulla of Elf5-deficient collecting ducts compared with similar regions in the kidneys of wild-type littermates at post-natal day one (P1), n>6 40X images per region of kidney for each mouse and 3 mice per genotype. Images of P1 kidney sections from Elf5f/f (D–F) and Cdh16->Cre;Elf5f/* (G–I) stained for Aqp2 and CAII to determine the percentage of principal and intercalated present in the collecting ducts. Scale bars are 10μm. (J) Collecting duct segment specific genes are not expressed at significantly different levels in the Cdh16->Cre;Elf5f/* mouse neonatal kidneys compared with wild type littermate kidneys as determined by RT-qPCR, n= 5 per group.
Considering that Elf5 is expressed in a subset of Aqp2+ principal cells, with the highest expression in medullary collecting ducts, we next tested if Elf5 contributes more to the differentiation of cells within a specific segment of the collecting ducts. For this we analyzed the expression levels of genes previously characterized to have restricted expression within certain segments of the developing collecting ducts [37, 38]. Etv4, a duct tip specific gene [38] and Kitl which is expressed in the tip and cortical collecting duct [37] were not significantly differentially expressed in the P0 wild-type versus Elf5-deficient kidneys (Fig.6J). Additionally, Fam129a and Gsdmc4 which are restricted to the medullary collecting ducts [37] were not expressed at significantly different levels in the wild-type versus Elf5-deficient kidneys (Fig.6J). We also observed no significant difference in the expression levels of Upk1b and Ppp1r3c, which are enriched in the medullary collecting ducts [37], between wild-type versus the Elf5-deficient kidneys (Fig.6J).
Elf3 and Ehf are also capable of regulating principal genes
Elf5 belongs to the class IIa subfamily of ETS transcription factors [39]. This classification is based on similar DNA binding properties mediated by the winged helix-turn-helix DNA binding domain found in all ETS factors. Among the class IIa subfamily of ETS transcription factors Elf3 and Ehf are reported to be expressed in the mouse developing collecting duct system [38]. Considering the modest contribution of Elf5 to the activation of Aqp2 and Avpr2 genes, we wondered whether Elf3 and Ehf can perform the same functions as Elf5 in the collecting duct cells. We observed that both Elf3 and Ehf are capable of activating the Aqp2 and Avpr2 proximal promoters in immature collecting duct cell lines. Ehf activated Aqp2 and Avpr2 promoters by 2.4-fold and 4-fold, respectively, in M5 cells (Fig.7A). Elf3 activated Aqp2 and Avpr2 promoters by 21.5-fold and 28-fold, respectively, in M5 cells (Fig.7A). Interestingly, Elf3 but not Ehf activated both Aqp2 and Avpr2 principal cell gene promoters in the M1 and M3 cells (Fig.7A). These observations indicate that Elf3 and Ehf could potentially compensate for loss of Elf5 in the developing collecting ducts.
Figure 7. Elf3 and Ehf can also activate principal cell genes.
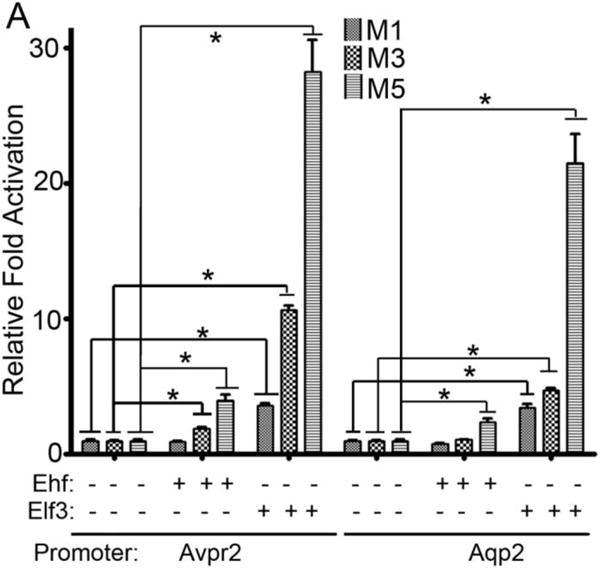
A. Elf3 and Ehf increase the transcriptional activities of mouse 1Kb Avpr2 and 1Kb Aqp2 proximal promoters in immature collecting duct cell lines. Elf3 significantly activates both principal cell gene promoter in all three immature collecting duct cell lines, with a maximum activation of the Avpr2 promoter by 28-fold and Aqp2 promoter by 21-fold in M5 cells. Ehf significantly activates both gene promoters in M5 cells and only the Avpr2 promoter in M3 cells. The asterisks denote p<0.05, n=6 per condition per cell type, unpaired t-tests.
Elf5-expressing cells are committed to the principal cell lineage by E16.5
Although Elf5 has only a modest role in activating Aqp2 and Avpr2 genes in the collecting ducts we next wanted to test if Elf5 expression could serve as a marker of the principal cell lineage. Considering that Elf5 is expressed prior to Aqp2 in some duct cells at E16.5 and its expression is restricted to Aqp2+ cells by E18.5, it is seems likely that the early Elf5 expression occurs in collecting duct cells that are committed to becoming principal cells but have not yet turned on Aqp2 expression. Alternatively, it is possible that the Elf5 expression at E16.5 occurs in duct cells that still have not committed to becoming principal cells, and that Elf5 expression becomes restricted to the principal cell lineage only towards the end of collecting duct development. To distinguish between these possibilities we performed lineage tracing experiments using Elf5->rtTA-IRES-GFP mice [40] which should express the reverse tetracycline transactivator (rtTA) and GFP in Elf5 expressing cells. We confirmed that GFP expression within the developing kidneys of Elf5->rtTA-IRES-GFP mice is similar (data not shown) to that of β-galactosidase in the Elf5+/LacZ mice and to the GFP expression in the Elf5->GFP mice (Fig. 4). To genetically label the early Elf5-expressing cells we provided doxycycline in the drinking water of pregnant Rosatdtomato/tdtomato [41] females from embryonic day 16.5 to 17.5, that had been mated with Tet-O-Cre [42]; Elf5->rtTA-IRES-GFP males. In experiment#1 (Fig.8A) we analyzed embryonic kidneys at E17.5, immediately after the doxycycline pulse, and observed tdtomato expression (red fluorescence) within epithelial structures only in embryos inheriting Elf5->rtTA-IRES-GFP along with Tet-O-Cre; Rosa+/tdtomato (arrowheads in Fig. 8B and C). Most (29 out of 30) tdtomato expressing epithelial cells also expressed Aqp4, a specific marker of principal cells (arrowhead in Fig. 8B). None of the epithelia marked with tdtomato were positive for Atp6v1b1, which is expressed only in the intercalated cells within the developing kidneys. All embryos inheriting Tet-O-Cre; Rosa+/tdtomato with or without Elf5-rtTA-IRES-GFP contain tdtomato expressing interstitial cells within the kidneys (arrows in Fig. 8D and E, Fig.9B, C and D). The interstitial expression of tdtomato occurs independently of inheritance of Elf5-rtTA-IRES-GFP and is likely due to the expression of Cre in a subpopulation of renal interstitial cells that is not dependent on the tet-response-elements being activated by rtTA in combination with doxycycline. In theory this could occur if the Tet-O-Cre transgene inserted into a locus that is near a renal-interstitial-cell-specific promoter that drives gene expression only in a subset of renal interstitial cells. More importantly the doxycycline administered between E16.5 and E17.5 only labeled the principal cells in the epithelial structures of the Elf5->rtTA->GFP; Tet-O-Cre; Rosa+/tdtomato embryos and not in Tet-O-Cre; Rosa+/tdtomato embryos. All the tdtomato+ epithelial cells also expressed GFP at E17.5 which supports that the genetic labeling within epithelial cells is restricted to Elf5 expressing cells.
Figure 8. Elf5->rtTA-IRES-GFP mice can be used to genetically pulse label Aqp4+ collecting duct epithelial cells between E16.5 and E17.5.
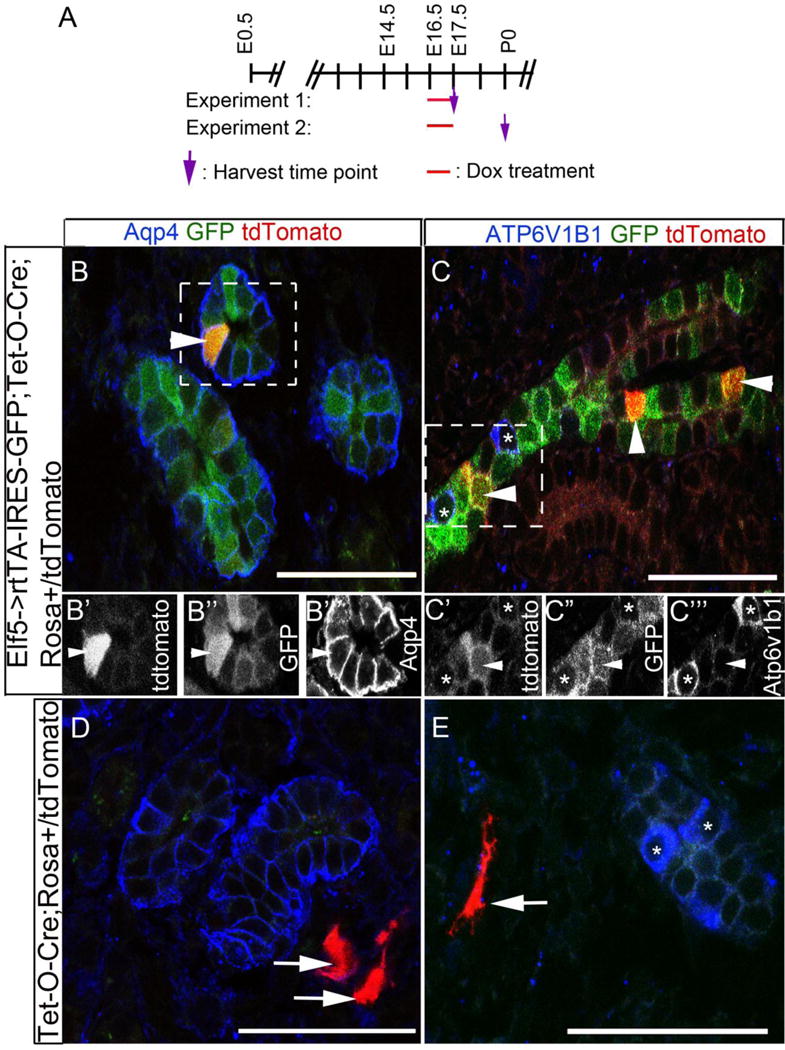
A. Schematic of lineage tracing experiments 1 and 2 are depicted, in which 1mg/ml of doxycycline was provided to pregnant females in their drinking water between E16.5 and E17.5. Images of sections through E17.5 kidneys are shown in panels B–E. B and C are sections from Elf5->rtTA-IRES-GFP; Tet-O-Cre; Rosa+/tdtomato kidneys, while D and E are sections from Tet-O-Cre; Rosa+/tdtomato kidneys. The arrowheads are pointing at tdtomato+ epithelial cells, and the arrows are pointing at tdtomato+ interstitial cells. B′, B″ and B″′ are black and white images of individual channels of the boxed area in B. C′, C″ and C″′ are black and white images of individual channels of the boxed area in C. The tdtomato+ genetic labeling occurred in epithelial cells only in Elf5->rtTA-IRES-GFP; Tet-O-Cre; Rosa+/tdtomato kidney sections (B and C) and never in Tet-O-Cre; Rosa+/tdtomato kidney sections (D and E). The tdtomato+ epithelial cells are frequently also Aqp4+ (B to B”’) and are never Atp6v1b1+ (C to C”’). Asterisks mark the Atp6v1b1+ epithelial cells. Scale bars are 50μm.
Figure 9. Elf5-expressing ureteric duct cells commit to the principal cell fate as early as E16.5.
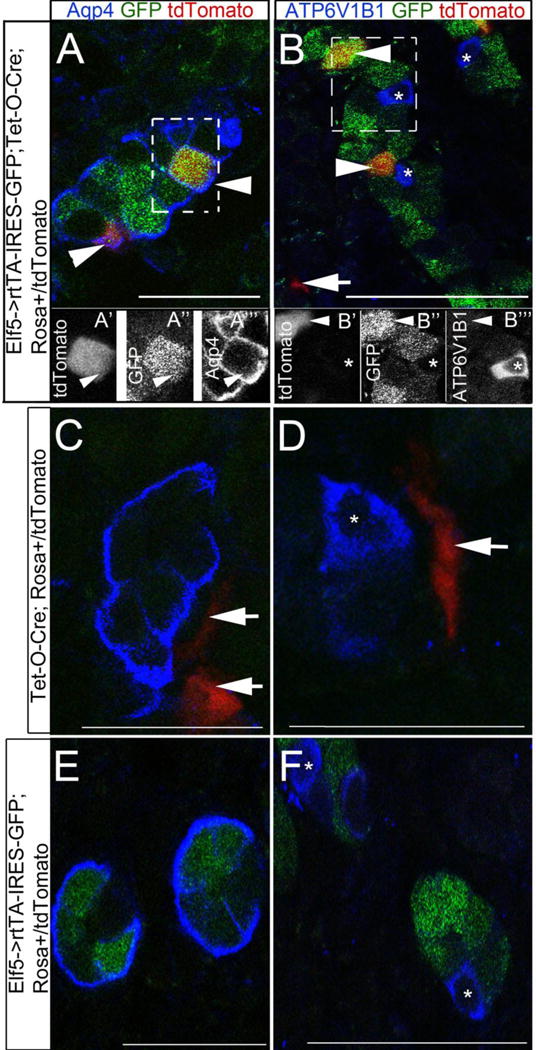
In the second lineage tracing experiment 1mg/ml of doxycycline was provided to pregnant females in their drinking water between E16.5 and E17.5 and kidney sections of the offspring were analyzed at post-natal day 0 and representative images are shown A–F. The arrowheads are pointing at tdtomato+ epithelial cells, and the arrows are pointing at tdtomato+ interstitial cells. A’, A” and A”’ are black and white images of individual channels of the boxed area in A. B’, B” and B”’ are black and white images of individual channels of the boxed area in B. The tdtomato+ genetic labeling occurred in epithelial cells only in Elf5->rtTA-IRES-GFP; Tet-O-Cre; Rosa+/tdtomato kidney sections (A & B) and never in Tet-O-Cre; Rosa+/tdtomato kidney sections (C & D) or in Elf5->rtTA-IRES-GFP; Rosa+/tdtomato kidney sections (E & F). The tdtomato+ epithelial cells are also Aqp4+ (A to B”’) and are never Atp6v1b1+ (B to B”’). Asterisks mark the Atp6v1b1+ epithelial cells. Scale bars are 25μm, except in B and F they are 50μm.
Since not all collecting duct cells have differentiated by E17.5, we genetically pulse labeled a subset of Elf5-expressing cells between E16.5 and E17.5 and analyzed the kidneys at post-natal day 0 (P0; Fig.9). Epithelial cells expressing tdtomato were present only in Elf5->rtTA-IRES-GFP; Tet-O-Cre; Rosa+/tdtomato P0 kidneys (Fig.9A and B), and not present in Tet-O-Cre; Rosa+/tdtomato P0 kidneys (Fig. 9C and D) or in Elf5->rtTA-IRES-GFP; Rosa+/tdtomato P0 kidneys (Fig.9E and F). Once again consistent with the Tet-O-Cre driving Cre expression independent of rtTA in a subset of non-epithelial renal cells we observed tdtomato+ interstitial cells in both Elf5->rtTA-IRES-GFP; Tet-O-Cre; Rosa+/tdtomato and in Tet-O-Cre; Rosa+/tdtomato P0 kidneys (arrowheads in Fig.9B, C and D) but not in Elf5->rtTA-IRES-GFP; Rosa+/tdtomato P0 kidneys (Fig.9E and F). All epithelial cells expressing tdtomato (n=45) also expressed Aqp4 (Fig. 9A) and none expressed Atp6v1b1 (Fig. 9B). Hence, the early Elf5 expression at E16.5 occurs in cells that are committed to the principal cell fate.
Discussion
Elf5 is a novel collecting duct specific transcription factor that functions down-stream of Notch/RBPJ-mediated signaling and contributes to the regulation of some PC-specific genes
Our observations extend previous studies of Mib1 and Adam10 deficient mice [23, 24] by uncovering a role for RBPJ and Presenilin1 and Presenilin2 in development of an appropriate number of principal cells within the collecting ducts. Together with the previous studies we confirm a role for the canonical Notch signaling pathway in ensuring the development of sufficient number of principal cells. The role of Notch signaling in patterning of the mammalian collecting ducts into principal cells intermingled with intercalated cells is reminiscent of Notch’s role in the “salt and pepper” patterning of multiciliated cells versus the transporter cells in the zebrafish distal pronephros [16, 43, 44], the patterning of the lungs with clara cells intermingled with multiciliated cells [45], and the patterning of the Xenopus and zebrafish skins with proton-secreting ionocytes (intercalated-like cells) intermingled with transport cells [11, 12]. In all these systems Notch signaling is considered to only repress the minority cell fate, for example by preventing the expression of Foxi1 orthologs that promote the intercalated-like cell fate [12] or Multicillin/Mcidas orthologs that promote the multiciliated cell fate [46, 47]. Here we have utilized the kidneys with Notch-signaling-deficient collecting ducts as a tool to identify genes that may be specifically expressed in the principal cells. Among these we verified that Elf5, one of six ETS Class-IIa subfamily members, is a novel Notch signaling dependent transcription factor that is expressed in the developing collecting ducts. Additionally, we determined that Elf5 is among the earliest genes up regulated with ectopic activation of Notch signaling in the developing collecting ducts. However, it remains to be determined whether or not the relationship between Notch signaling and activation of Elf5 expression is direct. Elf5 expression prior to Aqp2 in a subset of developing collecting duct cells at E16.5, along with the ability of Elf5 to modestly activate Aqp2 and Avpr2 proximal promoters and the requirement of Elf5 for normal level of Aqp2 and Avpr2 expression in the mouse kidneys allows us to conclude that Elf5 contributes to principal cell-specific gene expression. Elf5 however is not essential for principal cell differentiation. It remains possible that Elf5’s role in the principal cells may become critical under physiologic stress which we have not evaluated. Considering the redundancy built into the molecular mechanisms driving kidney development [48], it is possible that other ETS Class-IIa subfamily members may function redundantly alongside Elf5 within the developing collecting ducts. The continued expression of Elf5 in mature principal cells, combined with the report of increased nuclear abundance of Elf3 in principal cells in response to the vasopressin analog dDAVP [49] further support the idea that multiple ETS Class-IIa subfamily members could be involved in mediating mature principal cell functions.
We speculate that the modest contribution of Elf5 to Aqp2 and Avpr2 expression is likely because Elf5 mediates only part of the function of Notch signaling to promote the principal cell program in a subset of PCs. The Notch independent pathway that promotes PC differentiation may involve the other ETS Class IIa subfamily members and/or other transcription factors such as Gata2 which contribute to normal levels of Aqp2 expression in the adult kidney [50]. This speculation is based on the following observations: (i) Elf5 is not expressed in all Aqp2+ (principal) cells, (ii) there is a Notch independent pathway that regulates PC differentiation since PCs do develop in the absence of Notch signaling, (iii) conditional inactivation of RBPJ in the developing collecting ducts reduced Aqp2 levels to 50% of normal whereas conditional inactivation of Elf5 reduced Aqp2 levels to 75% of normal, and (iv) Elf3 and Ehf, which are expressed in the collecting ducts, are able to activate Aqp2 and Avpr2 proximal promoters.
The Elf5-expressing cells are committed to becoming mature principal cells
Elf5 expression turned on consistently in the branching collecting duct system from E16.5 onwards and resembled the pattern observed at post-natal day 0 (Fig.4). The very rare cells within the lumen of the collecting duct tips that were GFP+ in the Elf5->GFP kidneys may have been undergoing mitosis considering that luminal mitosis occurs in the developing collecting duct tips [51]. Further studies are required to determine if Elf5 is turned on occasionally in one or both of the daughter cells following luminal mitosis. Elf5 expression is more pervasive among the collecting duct cells in the medulla compared to the cortex, which is consistent with a higher ratio of principal to intercalated cells in the medullary collecting ducts compared to the cortical collecting ducts. The absence of Elf5 expression in a subset of Aqp2+ cells indicates that not all Aqp2+ cells are equal and we still do not know the full extent of the heterogeneity among non-intercalated collecting duct cells. Previous studies indicate that the terminal regions of the inner medullary collecting ducts contain morphologically and functionally distinct Aqp2+ cells compared to the Aqp2+ collecting duct cells in the cortex and outer medulla [52, 53]. Future studies examining the transcriptional profiles of individual Elf5-expressing cells may reveal the existence of different subpopulations of Elf5-expressing collecting duct cells and identify novel genes uniquely expressed in the morphologically distinct subsections of the collecting ducts.
The lineage tracing experiments using the Elf5->rtTA-IRES-GFP mice revealed that the expression of Elf5 at E16.5 is restricted to cells that have already selected to become principal cells. At E16.5 Elf5 is a marker of the principal cell lineage since during normal mouse kidney collecting duct development the cells marked by Elf5 expression do not become intercalated cell types. Various observations reported over the years suggest that the principal and intercalated cell fates are not fixed following collecting duct development. For example, lithium treatment [54] results in increased number of intercalated cells along with a decrease in principal cell number which may be due to principal cell trans-differentiation into intercalated cells. More recently the inactivation of dot1l, a histone H3-K79-methyltransferase, in Aqp2-expressing cells resulted in increased intercalated cell numbers and reduced principal cells numbers [55]. In other instances such as culturing isolated intercalated cells or during the recovery after cessation of lithium treatment the intercalated cells possibly convert into principal cells [56, 57]. However, definitive proof that trans-differentiation of principal cells occurs is still lacking. In this regard the Elf5->rtTA-IRES-GFP mice, which allow for the genetic labeling of principal cells, are likely to be useful in testing whether or not principal to intercalated cell trans-differentiation really occurs in the mature kidney collecting ducts.
Material and Methods
Mice
All experiments involving mice were approved by the Sanford Research IACUC. Details of mouse lines used are listed in Supplementary Table1. Mice used in this study were maintained on mixed backgrounds and genotyped following a universal PCR genotyping protocol [58]. For the lineage tracing experiments pregnant female mice were given doxycycline in the drinking water (1mg/ml; Sigma). Timed-mating of mice were observed daily and noon on the day a plug was observed was considered E0.5. Primer sequences are available upon request.
Histology and immunohistochemistry
Analysis of GFP, β-galactosidase, and RBPJ expression patterns were done as previously described [23, 59]. For other immunohistochemistry kidneys were fixed in Bouin’s fixative overnight at 4°C, rinsed in 70% ethanol prior to paraffin embedding and sectioning at 8 to 12 μm thickness. Staining was done as previously described [60]. Primary antibody details are listed in Supplementary Table 2.
Microarray analysis
Embryonic kidneys from three E18.5 HoxB7->Cre;RBPJ−/f;RosaEyfp/+ and three RBPJf/f;RosaEyfp/+ littermates were harvested. The total embryonic kidney RNA was extracted using RNeasy Mini-Kit (Qiagen) and the quality of RNA confirmed using BioAnalyzer (Agilent Tech.). Samples were analyzed using Illumina Mouse 6.0 Array platform by the Sanford-Burnham Microarray Core. The results of the microarray reported in this publication have been deposited in NCBI’s Gene Expression Omnibus [61] and are accessible through GEO series accession number GSE68389 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE68389).
Quantitative PCR
RNA was reverse transcribed using random hexamers or oligo dT with reverse transcription kit (Promega). Quantitative PCR was performed using Power SYBR Green (Life Technologies), gene specific primers with the forward and reverse primers designed from different exons, and an ABI 7500 instrument (Applied Biosystems). Standard curves were generated analyzing serially diluted cDNA reverse transcribed from mouse kidneys to determine the efficiency of each primer pair. Each sample was measured either in duplicate or triplicate and relative gene expression levels were normalized to that of GAPDH or beta-2 microglobulin. The relative expression levels were determined using the ΔΔCt method [62]. Primer sequences are available upon request.
Cell lines and plasmids
We utilized MDCK cells (from ATCC, CCL-34) which we confirmed during the course of these experiment have principal cell properties by gene expression profiling and IHC. We also used mpkCCDc14 cells from Dr. Vandewalle [35], which were grown on trans-well filters. We isolated ureteric duct cells from E12.5 HoxB7->Cre;ROSAEYFP/+;Immorto+/TG mouse kidneys and established three individual clones M1, M3 and M5 following a method previously described [63]. The proximal promoters used here were amplified from mouse genomic DNA and cloned into pGL3-Basic vector.
Dual luciferase assays
For dual luciferase reporter assays, ~80000 cells plated in each well of 24-well plates and transfected with 200ng reporter plasmid, 20ng normalization plasmid (pTK-Renilla; Promega) and 200ng expression plasmid. Dual luciferase reporter assays (Promega) were performed 48h after transfection according to manufacturer’s instructions. For each well, firefly (reporter) luciferase activity was divided by renilla (normalizer) luciferase activity to determine the normalized level of promoter activation. Each condition in the reporter assays had six biologic replicates.
Statistical Analysis
Our initial studies of four Notch-signaling-deficient mouse kidneys and three wild-type littermate kidneys revealed these numbers were sufficient to detect significant differences between the two groups by two-tailed t-test, with α=0.05 and Power=0.8, resulting in an effect size of 2.4 or higher. We therefore continued the studies with n=3 to 8 per group with the exact numbers depending on the size of and genotype within the litters. In the graphs, the height of each bar represents the mean and the error bars represent one standard deviation. In the scatter plots the large horizontal line through a group of data points represents the mean and the smaller horizontal lines flanking the mean in each group represent one standard deviation. Excel was used to perform two-tailed unpaired t-tests to compare two groups of mice or cells, after verification that samples had a normal distribution and testing for equal variance between groups using the F-test. The resulting p values are stated in the text and figure legends.
Supplementary Material
Highlights.
RBPJ mediates Notch signaling dependent patterning of collecting duct cell types
Lineage tracing reveals Elf5 to be an early principal cell lineage specific factor
Elf5 contributes to principal cell gene expression in the kidney
Acknowledgments
We thank Dr. Raphael Kopan for his guidance and for allowing for this project to be initiated in his laboratory. We thank Dr. Indra Chandrasekar, Dr. Karla Otterpohl, and Ryan Hart for critically reviewing this manuscript. We thank Sarah Garcia for technical assistance.
Funding
Research reported in this publication was supported by NIH grant P20GM103620 and by NIDDK of NIH under award number R01DK106135. We thank the Molecular Pathology Core for assisting with tissue processing and Imaging Core for maintenance of microscopes used in this study. These Cores at Sanford Research are supported by NIH grants P20GM10358 and P20GM103620.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
J.G., M.M., C.H., J.D., E.F., and M.B. performed experiments. S.S. reviewed manuscript, contributed intellectually to Elf5 related experiments and provided Elf5 related reagents. J.V. reviewed manuscript and provided the Elf5->rtTA-IRES-GFP mice for lineage tracing experiments. H.Z. contributed reagents and reviewed early versions of this manuscript. K.S. designed and performed experiments, and wrote the manuscript.
Data availability
Microarray data: (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE68389).
References
- 1.Dressler GR. Advances in early kidney specification, development and patterning. Development. 2009;136(23):3863–74. doi: 10.1242/dev.034876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Costantini F, Kopan R. Patterning a complex organ: branching morphogenesis and nephron segmentation in kidney development. Dev Cell. 2010;18(5):698–712. doi: 10.1016/j.devcel.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaissling B. Ultrastructural characterization of the connecting tubule and the different segments of the collecting duct system in the rabbit kidney. Curr Probl Clin Biochem. 1977;8:435–46. [PubMed] [Google Scholar]
- 4.Brown D, Weyer P, Orci L. Vasopressin stimulates endocytosis in kidney collecting duct principal cells. Eur J Cell Biol. 1988;46(2):336–41. [PubMed] [Google Scholar]
- 5.Brown D, Kumpulainen T. Immunocytochemical localization of carbonic anhydrase on ultrathin frozen sections with protein A-gold. Histochemistry. 1985;83(2):153–8. doi: 10.1007/BF00495146. [DOI] [PubMed] [Google Scholar]
- 6.Bastani B. Immunocytochemical localization of the vacuolar H(+)-ATPase pump in the kidney. Histol Histopathol. 1997;12(3):769–79. [PubMed] [Google Scholar]
- 7.Kim J, Tisher CC, Madsen KM. Differentiation of intercalated cells in developing rat kidney: an immunohistochemical study. Am J Physiol. 1994;266(6 Pt 2):F977–90. doi: 10.1152/ajprenal.1994.266.6.F977. [DOI] [PubMed] [Google Scholar]
- 8.Emmons C, Kurtz I. Functional characterization of three intercalated cell subtypes in the rabbit outer cortical collecting duct. J Clin Invest. 1994;93(1):417–23. doi: 10.1172/JCI116976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fejes-Toth G, et al. Differential expression of AE1 in renal HCO3-secreting and -reabsorbing intercalated cells. J Biol Chem. 1994;269(43):26717–21. [PubMed] [Google Scholar]
- 10.Brown D, Breton S. Mitochondria-rich, proton-secreting epithelial cells. J Exp Biol. 1996;199(Pt 11):2345–58. doi: 10.1242/jeb.199.11.2345. [DOI] [PubMed] [Google Scholar]
- 11.Quigley IK, Stubbs JL, Kintner C. Specification of ion transport cells in the Xenopus larval skin. Development. 2011;138(4):705–14. doi: 10.1242/dev.055699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Janicke M, Carney TJ, Hammerschmidt M. Foxi3 transcription factors and Notch signaling control the formation of skin ionocytes from epidermal precursors of the zebrafish embryo. Dev Biol. 2007;307(2):258–71. doi: 10.1016/j.ydbio.2007.04.044. [DOI] [PubMed] [Google Scholar]
- 13.Kageyama R, et al. Dynamic Notch signaling in neural progenitor cells and a revised view of lateral inhibition. Nat Neurosci. 2008;11(11):1247–51. doi: 10.1038/nn.2208. [DOI] [PubMed] [Google Scholar]
- 14.Heitzler P, Simpson P. The choice of cell fate in the epidermis of Drosophila. Cell. 1991;64(6):1083–92. doi: 10.1016/0092-8674(91)90263-x. [DOI] [PubMed] [Google Scholar]
- 15.Bray S. Notch signalling in Drosophila: three ways to use a pathway. Semin Cell Dev Biol. 1998;9(6):591–7. doi: 10.1006/scdb.1998.0262. [DOI] [PubMed] [Google Scholar]
- 16.Liu Y, et al. Notch signaling controls the differentiation of transporting epithelia and multiciliated cells in the zebrafish pronephros. Development. 2007;134(6):1111–22. doi: 10.1242/dev.02806. [DOI] [PubMed] [Google Scholar]
- 17.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137(2):216–33. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Strooper B, et al. A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature. 1999;398(6727):518–22. doi: 10.1038/19083. [DOI] [PubMed] [Google Scholar]
- 19.Steiner H, et al. A loss of function mutation of presenilin-2 interferes with amyloid beta-peptide production and notch signaling. J Biol Chem. 1999;274(40):28669–73. doi: 10.1074/jbc.274.40.28669. [DOI] [PubMed] [Google Scholar]
- 20.Blomqvist SR, et al. Distal renal tubular acidosis in mice that lack the forkhead transcription factor Foxi1. J Clin Invest. 2004;113(11):1560–70. doi: 10.1172/JCI20665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vidarsson H, et al. The forkhead transcription factor Foxi1 is a master regulator of vacuolar H-ATPase proton pump subunits in the inner ear, kidney and epididymis. PLoS One. 2009;4(2):e4471. doi: 10.1371/journal.pone.0004471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurth I, et al. The forkhead transcription factor Foxi1 directly activates the AE4 promoter. Biochem J. 2006;393(Pt 1):277–83. doi: 10.1042/BJ20051094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo Q, et al. Adam10 Mediates the Choice between Principal Cells and Intercalated Cells in the Kidney. J Am Soc Nephrol. 2015;26(1):149–59. doi: 10.1681/ASN.2013070764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jeong HW, et al. Inactivation of Notch signaling in the renal collecting duct causes nephrogenic diabetes insipidus in mice. J Clin Invest. 2009;119(11):3290–300. doi: 10.1172/JCI38416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu J, Carroll TJ, McMahon AP. Sonic hedgehog regulates proliferation and differentiation of mesenchymal cells in the mouse metanephric kidney. Development. 2002;129(22):5301–12. doi: 10.1242/dev.129.22.5301. [DOI] [PubMed] [Google Scholar]
- 26.Oxburgh L, et al. TGFbeta superfamily signals are required for morphogenesis of the kidney mesenchyme progenitor population. Development. 2004;131(18):4593–605. doi: 10.1242/dev.01324. [DOI] [PubMed] [Google Scholar]
- 27.Kobayashi A, et al. Six2 defines and regulates a multipotent self-renewing nephron progenitor population throughout mammalian kidney development. Cell Stem Cell. 2008;3(2):169–81. doi: 10.1016/j.stem.2008.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Georgas K, et al. Analysis of early nephron patterning reveals a role for distal RV proliferation in fusion to the ureteric tip via a cap mesenchyme-derived connecting segment. Dev Biol. 2009;332(2):273–86. doi: 10.1016/j.ydbio.2009.05.578. [DOI] [PubMed] [Google Scholar]
- 29.Pearton DJ, et al. Elf5 counteracts precocious trophoblast differentiation by maintaining Sox2 and 3 and inhibiting Hand1 expression. Dev Biol. 2014;392(2):344–57. doi: 10.1016/j.ydbio.2014.05.012. [DOI] [PubMed] [Google Scholar]
- 30.Chakrabarti R, et al. Elf5 regulates mammary gland stem/progenitor cell fate by influencing notch signaling. Stem Cells. 2012;30(7):1496–508. doi: 10.1002/stem.1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choi YS, et al. Elf5 conditional knockout mice reveal its role as a master regulator in mammary alveolar development: failure of Stat5 activation and functional differentiation in the absence of Elf5. Dev Biol. 2009;329(2):227–41. doi: 10.1016/j.ydbio.2009.02.032. [DOI] [PubMed] [Google Scholar]
- 32.Choi YS, et al. Generation and analysis of Elf5-LacZ mouse: unique and dynamic expression of Elf5 (ESE-2) in the inner root sheath of cycling hair follicles. Histochem Cell Biol. 2008;129(1):85–94. doi: 10.1007/s00418-007-0347-x. [DOI] [PubMed] [Google Scholar]
- 33.Pearton DJ, et al. Elf5 regulation in the trophectoderm. Dev Biol. 2011;360(2):343–50. doi: 10.1016/j.ydbio.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 34.Gekle M, et al. Characterization of two MDCK-cell subtypes as a model system to study principal cell and intercalated cell properties. Pflugers Arch. 1994;428(2):157–62. doi: 10.1007/BF00374853. [DOI] [PubMed] [Google Scholar]
- 35.Bens M, et al. Corticosteroid-dependent sodium transport in a novel immortalized mouse collecting duct principal cell line. J Am Soc Nephrol. 1999;10(5):923–34. doi: 10.1681/ASN.V105923. [DOI] [PubMed] [Google Scholar]
- 36.Shao X, Somlo S, Igarashi P. Epithelial-specific Cre/lox recombination in the developing kidney and genitourinary tract. J Am Soc Nephrol. 2002;13(7):1837–46. doi: 10.1097/01.asn.0000016444.90348.50. [DOI] [PubMed] [Google Scholar]
- 37.Thiagarajan RD, et al. Identification of anchor genes during kidney development defines ontological relationships, molecular subcompartments and regulatory pathways. PLoS One. 2011;6(2):e17286. doi: 10.1371/journal.pone.0017286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu J, et al. Identification of molecular compartments and genetic circuitry in the developing mammalian kidney. Development. 2012;139(10):1863–73. doi: 10.1242/dev.074005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wei GH, et al. Genome-wide analysis of ETS-family DNA-binding in vitro and in vivo. EMBO J. 2010;29(13):2147–60. doi: 10.1038/emboj.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rios AC, et al. In situ identification of bipotent stem cells in the mammary gland. Nature. 2014;506(7488):322–7. doi: 10.1038/nature12948. [DOI] [PubMed] [Google Scholar]
- 41.Madisen L, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13(1):133–40. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perl AK, et al. Early restriction of peripheral and proximal cell lineages during formation of the lung. Proc Natl Acad Sci U S A. 2002;99(16):10482–7. doi: 10.1073/pnas.152238499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma M, Jiang YJ. Jagged2a-notch signaling mediates cell fate choice in the zebrafish pronephric duct. PLoS Genet. 2007;3(1):e18. doi: 10.1371/journal.pgen.0030018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Y, et al. Zebrafish nephrogenesis is regulated by interactions between retinoic acid, mecom, and Notch signaling. Dev Biol. 2014;386(1):111–22. doi: 10.1016/j.ydbio.2013.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morimoto M, et al. Canonical Notch signaling in the developing lung is required for determination of arterial smooth muscle cells and selection of Clara versus ciliated cell fate. J Cell Sci. 2010;123(Pt 2):213–24. doi: 10.1242/jcs.058669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kyrousi C, et al. Mcidas and GemC1 are key regulators for the generation of multiciliated ependymal cells in the adult neurogenic niche. Development. 2015;142(21):3661–74. doi: 10.1242/dev.126342. [DOI] [PubMed] [Google Scholar]
- 47.Stubbs JL, et al. Multicilin promotes centriole assembly and ciliogenesis during multiciliate cell differentiation. Nat Cell Biol. 2012;14(2):140–7. doi: 10.1038/ncb2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ye X, et al. Genetic mosaic analysis reveals a major role for frizzled 4 and frizzled 8 in controlling ureteric growth in the developing kidney. Development. 2011;138(6):1161–72. doi: 10.1242/dev.057620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schenk LK, et al. Quantitative proteomics identifies vasopressin-responsive nuclear proteins in collecting duct cells. J Am Soc Nephrol. 2012;23(6):1008–18. doi: 10.1681/ASN.2011070738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu L, et al. GATA2 regulates body water homeostasis through maintaining aquaporin 2 expression in renal collecting ducts. Mol Cell Biol. 2014;34(11):1929–41. doi: 10.1128/MCB.01659-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Packard A, et al. Luminal mitosis drives epithelial cell dispersal within the branching ureteric bud. Dev Cell. 2013;27(3):319–30. doi: 10.1016/j.devcel.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Madsen KM, Clapp WL, Verlander JW. Structure and function of the inner medullary collecting duct. Kidney Int. 1988;34(4):441–54. doi: 10.1038/ki.1988.201. [DOI] [PubMed] [Google Scholar]
- 53.Clapp WL, et al. Morphologic heterogeneity along the rat inner medullary collecting duct. Lab Invest. 1989;60(2):219–30. [PubMed] [Google Scholar]
- 54.Christensen BM, et al. Changes in cellular composition of kidney collecting duct cells in rats with lithium-induced NDI. Am J Physiol Cell Physiol. 2004;286(4):C952–64. doi: 10.1152/ajpcell.00266.2003. [DOI] [PubMed] [Google Scholar]
- 55.Xiao Z, et al. Dot1l deficiency leads to increased intercalated cells and upregulation of V-ATPase B1 in mice. Exp Cell Res. 2016;344(2):167–75. doi: 10.1016/j.yexcr.2015.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fejes-Toth G, Naray-Fejes-Toth A. Differentiation of renal beta-intercalated cells to alpha-intercalated and principal cells in culture. Proc Natl Acad Sci U S A. 1992;89(12):5487–91. doi: 10.1073/pnas.89.12.5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Trepiccione F, et al. Evaluation of cellular plasticity in the collecting duct during recovery from lithium-induced nephrogenic diabetes insipidus. Am J Physiol Renal Physiol. 2013;305(6):F919–29. doi: 10.1152/ajprenal.00152.2012. [DOI] [PubMed] [Google Scholar]
- 58.Stratman JL, Barnes WM, Simon TC. Universal PCR genotyping assay that achieves single copy sensitivity with any primer pair. Transgenic Res. 2003;12(4):521–2. doi: 10.1023/a:1024225408961. [DOI] [PubMed] [Google Scholar]
- 59.Surendran K, et al. The contribution of Notch1 to nephron segmentation in the developing kidney is revealed in a sensitized Notch2 background and can be augmented by reducing Mint dosage. Dev Biol. 2010;337(2):386–95. doi: 10.1016/j.ydbio.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Surendran K, et al. Reduced Notch signaling leads to renal cysts and papillary microadenomas. J Am Soc Nephrol. 2010;21(5):819–32. doi: 10.1681/ASN.2009090925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–10. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29(9):e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tai G, Hohenstein P, Davies JA. Making immortalized cell lines from embryonic mouse kidney. Methods Mol Biol. 2012;886:165–71. doi: 10.1007/978-1-61779-851-1_15. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


