Abstract
Magnetic resonance imaging (MRI) of pediatric brain provides invaluable information for early normal and abnormal brain development. Longitudinal neuroimaging has spanned various research works on examining infant brain development patterns. However, studies on predicting postnatal brain image evolution remain scarce, which is very challenging due to the dynamic tissue contrast change and even inversion in postnatal brains. In this paper, we unprecedentedly propose a dual image intensity and anatomical structure (label) prediction framework that nicely links the geodesic image metamorphosis model with sparse patch-based image representation, thereby defining spatiotemporal metamorphic patches encoding both image photometric and geometric deformation. In the training stage, we learn the 4D metamorphosis trajectories for each training subject. In the prediction stage, we define various strategies to sparsely represent each patch in the testing image using the training metamorphosis patches; as we progressively increment the richness of the patch (from appearance-based to multimodal kinetic patches). We used the proposed framework to predict 6, 9 and 12-month brain MR image intensity and structure (white and gray matter maps) from 3 months in 10 infants. Our seminal work showed promising preliminary prediction results for the spatiotemporally complex, drastically changing brain images.
1 Introduction
Studying early brain development and neurodevelopmental disorders has been at the heart of various recent neuroimaging studies that aim to improve our limited understanding of early brain workings as well as the dynamics of brain abnormalities [1]. Part of the ongoing research in this exploding field aims to not only understand how infant brains grow, but also to learn how to predict their evolution in space and time. In other words, given a single MR image acquired at a specific timepoint, can we build a learning framework that can robustly output how the brain looks like at late timepoints (in both image appearance and anatomical structure)? This will greatly help understand the dynamic, nonlinear early brain development and provide fundamental insights into neurodevelomental disorders.
To the best of our knowledge, the problem of jointly predicting both image appearance (intensity) and structure (e.g., brain tissue labels: white and gray matters) during the first postnatal year from a single acquired image has not been explored yet. In the sate-of-the art, a few methods [2, 3] have dealt with extrapolating image appearance at earlier or late timepoints (backward or forward prediction) using geodesic shape regression models to estimate diffeomeorphic (i.e. smooth and invertible) deformation trajectories; however, they required at least two images for prediction or extrapolation. This confined scope of publications for predicting postnatal early brain development can be partly explained by the drastic change of tissue contrast and anatomical structures in developing brains, which renders prediction more challenging (Fig. 1).
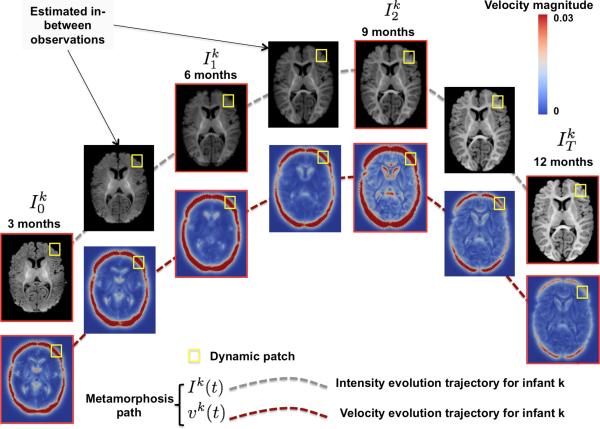
Fig. 1.
Spatiotemporal image metamorphosis. For each infant k in the training dataset, we estimate a dynamic photometric and kinetic paths (vector field is displayed); which can be more locally traced within a dynamic patch.
In this paper, we propose the first dual image and label prediction framework that nicely links the geodesic image metamorphosis model [4] (training stage) with sparse patch-based image representation (prediction stage). The image-to-image metamorphosis model elegantly solves the problem of contrast change between di erent MR brain images (see 3-month vs. 6-month T1-w images in Fig. 1) as it morphs a baseline image I0 into a target image IT by jointly estimating optimal spatiotemporal velocity deformation vector field vt (i.e. dynamic kinetic path) and intensity scalar field It (dynamic photometric path), with t ∈ [0, T]. In the learning stage, we first propose an extension of the image-to-image metamorphosis into a spatiotemporal metamorphosis that exactly fits the baseline image to subsequent observations in an ordered set of images.
The training stage comprises the estimation of a spatiotemporal metamorphosis for each training subject using a patch-based image encoding. A wide spectrum of patch-based methods has been for infant images labeling and registration [5, 6]. The richness and locality of a patch made it a good candidate for examining individual and population-based image variation. Besides, the in-vogue sparse patch-based image representation methods take into account the neighborhood of corresponding patches and simultaneously select the best candidate patches for representation, unlike patch-based non-local mean or majority voting methods. In the prediction stage, for each voxel x in the testing baseline image I0, we sparsely encode each patch p(x) centered at x using a dynamic dictionary composed of patches from the training subjects. For prediction, the sparse representation will be propagated to late timepoints as each patch is dynamic and has a metamorphosis path p(x, t) estimated during the training stage. The propagation is applied to both dynamic label map dictionaries and intensity dictionaries; thereby allowing a simultaneous image and label prediction. We test the proposed framework through progressively upgrading the richness of the dynamic patch from an appearance-based patch to an appearance-and-structure-based patch up to a multimodal kinetic patch. Finally, we predict for each of these baseline patches their intensity and structure evolution paths up to the last timepoint in the training dataset.
2 Spatiotemporal image metamorphosis framework (training stage)
As introduced by [4], the metamorphosis image theory presents a solid mathematical ground for tracking both local intensity and shape changes in images; which is fundamental to learning early brain dynamic growth. The idea of metamorphosis stems from integrating the infinitesimal variation of the image appearance (i.e. residual) with the diffeormophic metric used to estimate image deformation trajectories; therefore accounting for intensity change and producing the metamorphic metric. Basically, a baseline image I0 morphs under the action of a velocity vector field vt that advects the scalar intensity It (i.e. time-evolving image intensity) [4]. Solving the advection equation with a residual allows to estimate both image intensity evolution and the velocity at which it evolves (Fig. 1). In other words, this model registers one source image I0 to a target image IT while estimating two optimal evolution paths linking these images: (1) a geometric path {vt}[0,T] encoding the smooth velocity deforming one image into another, and (2) a photometric path {It}[0,T] representing the change in image intensity. Both paths characterize the dynamics of the image metamorphosis from the source into the target image in small discrete time and space intervals.
The definition of the metamorphic metric requires the use of appropriate spaces to which the images, the force acting on the images, and the velocity driving the evolution of the baseline image towards the target image belong. An image I is an element of the square integrable set of functions L2 considered as a Riemannian manifold denoted M (the object space). A curve (t → It, t ∈ [0, T]) on M is the path of evolution of a baseline image I0 (Fig. 1). M is equipped with the usual metric on L2. We denote by Ω the space where an image I is defined. The action (force) ϕ is a diffeomorphic transformation (a mapping) that belongs to a Lie group G endowed with a Lie algebra . In classical diffeomorphic deformation theory, we associate to the action ϕ a velocity v that satisfies the flow equation:
A curve (t → ϕt, t ∈ [0, T]) on G acting on an image I ∈ M describes a deformation path morphing I over the time interval [0, T ]. The velocity field vt, for all t ∈ [0, T], belongs to the vector space V, which is the tangent space to the action group G. We adopt similar construction as in [7] for V with an inner product < ., . >V defined through a differential Cauchy-Navier type operator L (with adjoint L†) given by: < f, g >V =< Lf, Lg >L2 =< L†Lf, g >L2) where < ., . >2 is the standard L2 L2 inner-product for square integrable vector fields on M and L = (−α∇2+γ)Id (Id is the identity matrix and α and γ two deformation parameters).
We estimate the optimal metamorphosis path (It, vt) from I0 to IT by minimizing the following cost functional U using a standard alternating steepest gradient descent algorithm [8]:
σ weighs the trade-off between the deformation smoothness (first term) and fidelity-to-data (second term). The term ∇It.vt represents the spatial variation of the moving image It in the direction vt. Furthermore, the moving intensity scalar field It is defined under the action of the diffeomorphism ϕt on a baseline image I0 : It = ϕt.I0.
We extend the image-to-image metamorphosis to a spatiotemporal metamorphosis passing through the images by adding to the boundary conditions for the intensity path It beginning at I(t = 0) = I0 and reaching I(t = T ) = IN the intermediate conditions, where we force the evolving metamorphic path It to successively fit a set of observations {I1, . . . , IT−1}.
3 Sparse patch-based image intensity and label prediction from baseline (testing stage)
Weighted sparse representation of a new image using estimated metamorphosis path
The idea that similar brain images in appearance and structure will deform in a similar way was recently used for image registration prediction in [9]. In the context of prediction from a single image, we also adopt the same idea within a patch-based image representation framework. Indeed, the local anatomical structure can be better captured within the richness of local patches. Besides, the patch size can explore different scales of the image and can be easily adjusted to upgrade the method performance. For a new testing image, we first sample cubic overlapping patches at each voxel x ∈ Ω. Then we trace forth the photometric evolution path for each patch in every training subject k (Fig.1-gray curve), thereby defining a set of dynamic patches centered at x: pk(x, t)[0,T],s=1...,S, where S denotes the number of the training subjects. We use these training patches in a column-wise manner to define a dynamic dictionary at each voxel x: D(x, t)[0,T]. To overcome the possible registration error, we expand D(x, t) by adding all patches centered at y, s.t. y ∈ Wx, where Wx is a search window centered at x. Next, for a new testing baseline appearance patch p(x, 0), we select the most similar patches in D(x, 0) using the squared absolute difference as a metric, thus generating a ‘filtered’ dictionary Dselect(x, 0). Last, we sparsely represent p(x, 0) by the coefficient vector α and Dselect(x, 0) through minimizing the following functional:
where Dselect ∈ Rd×SW, p(x, 0) ∈ Rd, α ∈ RSW and η > 0. d is the patch dimension and SW is the total number of patches in the dictionary, which depends on both the number of training subjects S and the number of patches inside the window W. The first term measures the discrepancy between the baseline patch p(x, 0) and the reconstructed baseline patch Dselect(x, 0)α, and the second term is L1 LASSO regularization on the coefficient vector α.
Patch-wise prediction of both photometric and label evolution paths for a testing subject
To predict the image intensity Î(x, t) located at x, at a specific timepoint t, we use the sparse coefficient vector α to select and weigh patches from the built dictionary at t as follows: p̂(x, t) = Dselect(x, t)α; where for t > 0 Dselect(., t) contains only appearance patches. Then we take the center value of the appearance patch as a predicted value Î(x, t).
Assuming that we have Y possible labels {l1, . . . , lY} in the training images, we predict the label L̂(x, t) associated with Î(x, t) by weighted majority voting as follows: where lp denotes the label of the patch p center in Dselect(x, t) and the Dirac function δ(lp, lm) = 1 when lp = lm and 0 otherwise.
Multiple strategies for baseline dictionary construction
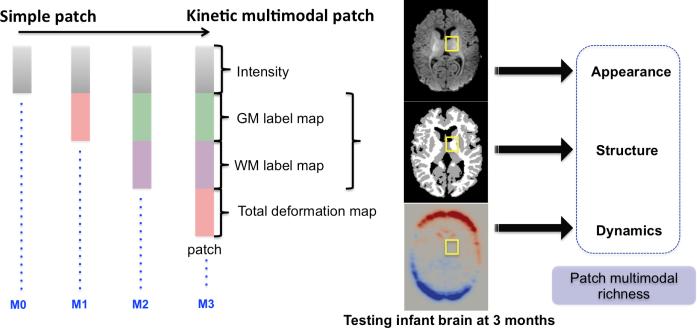
To investigate the role of the baseline dictionary D(., 0) building and its richness in sparse prediction performance, we define three methods to build Dselect(x, t) for t ∈ [0, T] in addition to the default method M0 which only considers appearance (intensity) patches (Fig. 3):
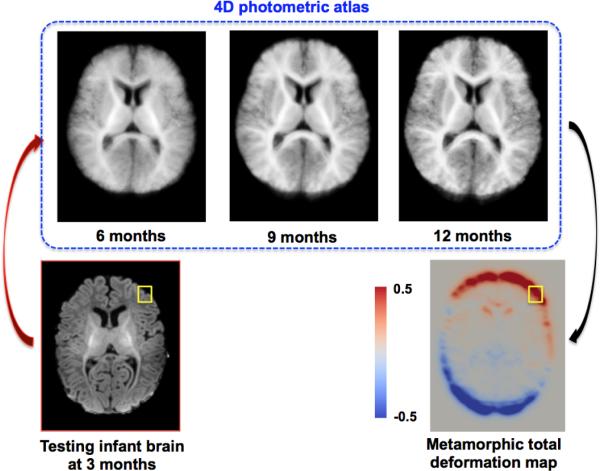
Fig. 3.
Spatiotemporal testing image metamorphosis into a 4D photometric atlas to generate total deformation map.
٭ Appearance-and-deformation-based dictionary (Method1 or M1)
To incorporate the kinetic information provided by the metamorphosis model, which may be of value, we estimated the total deformation map, which is integrated over the dynamic kinetic path (red curve in Fig. 1) of a testing image when morphed into a 4D photometric atlas (Fig. 3). This provides a mean photometric evolution of a baseline testing image –that may help guide the prediction of how it locally changes. Thus, we stack up the total deformation patch behind the appearance patch (Fig. 2–M1).
Fig. 2.
Multiple strategies for baseline dictionary construction.
٭ Appearance-and-structure-based dictionary (Method2 or M2)
We build the dictionary using column patches, each composed of three sub-patches: the top patch encodes the image appearance, the middle patch represents the GM (gray matter) label map and the bottom patch represents the WM (white matter) label map (Fig. 2–M2). This enables to upgrade the richness of the column patch by incorporating guiding structural information.
٭ Multimodal kinetic dictionary (Method3 or M3)
We build multimodal patches which include appearance, structure, and kinetic local information to construct the baseline dictionary (Fig. 2–M3).
4 Results and Discussion
Data and parameters setting
We evaluated the proposed framework on T1-w images in 10 infants, each with MRI scans acquired at 3, 6, 9 and 12 months of age. We fixed the metamorphosis parameters at the same values for all infants: α = 0.01, α = 4 and γ = 0.05. For patch-based representation, the window size W is of the patch size; η = 0.01. Patch-size was set to 3 × 3 × 3. We also used the patch size 5 × 5 × 5 for comparison.
Leave-one-out cross validation and evaluation metrics
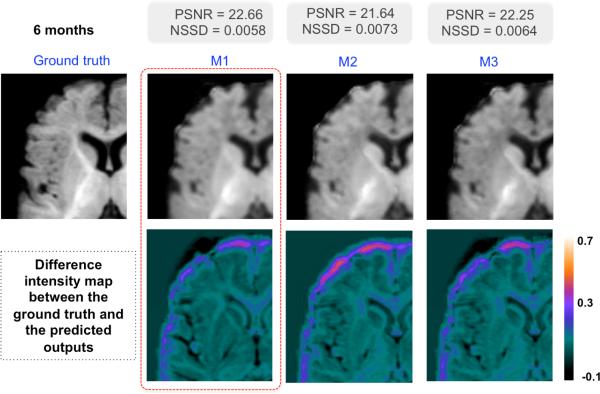
We use Dice index to evaluate label prediction accuracy (Table 1) and normalized sum of squared differences (NSSD) and peak signal-to-noise ratio (PSNR) to evaluate intensity prediction accuracy (Table 2). Interestingly, the multimodal kinetic patch (Method 3) showed the best performance in GM prediction accuracy, while the appearance-and-label based patch (Method 2) led to a better prediction for WM. The former results can be explained by the high dynamics of the GM well captured in the total deformation map (i.e. cortical growth) (Fig. 2), unlike the WM deformation map which is less dynamic and lacks details, eventually due to the fuzziness of the 4D photometric atlas. One could further improve WM prediction results when using the multimodal kinetic patch through estimating a sharper 4D atlas that preserves the anatomical details. For the intensity prediction, method 1 displays the best performance (appearance-and-deformation patch). As the image domain differs from the label domain, one could easily expect different performances. One way to solve this inconsistency in performance in both domains is to bridge the gap between both domains through the estimation of a mapping that separately evolves the appearance dictionary towards the structure or deformation dictionaries.
Table 1.
Mean Dice index across 10 infants (patch size 3 × 3 × 3)
| Method | Methodl | Method2 | Method3 | |
|---|---|---|---|---|
| 6 months | GM | 0.7530 ±0.0636 | 0.7660 ±0.0564 | 0.7690 ±0.0572 |
| WM | 0.6320 ±0.0636 | 0.7080 ±0.0432 | 0.6970 ±0.0481 | |
| 9 months | GM | 0.7640 ±0.0519 | 0.7510 ±0.0428 | 0.7740 ±0.0504 |
| WM | 0.6340 ±0.0640 | 0.7080 ±0.0329 | 0.6740 ±0.0430 | |
| 12 months | GM | 0.7500 ±0.0503 | 0.7210 ±0.0423 | 0.7600 ±0.0523 |
| WM | 0.6590 ±0.0647 | 0.7350 ±0.0334 | 0.6890 ±0.0482 | |
Table 2.
Average intensity prediction error across 10 infants (patch size 3×3×3).
| Month | Method1 | Method2 | Method3 | |||
|---|---|---|---|---|---|---|
| PSNR | NSSD | PSNR | NSSD | PSNR | NSSD | |
| 6 months | 31.23 | 0.0239 | 30.39 | 0.0282 | 30.15 | 0.0306 |
| 9 months | 28.28 | 0.0438 | 28.12 | 0.0455 | 27.59 | 0.0508 |
| 12 months | 28.57 | 0.0389 | 28.61 | 0.0413 | 28.2 | 0.0419 |
Besides, when we used a larger patch (5×5×5), we noticed that the prediction accuracy for Method 3 (appearance-and-label patch) remarkably increased up to (GM = 0.84 ± 0.043, WM = 0.71 ± 0.063) at 6 months, (GM = 0.811 ± 0.038, WM = 0.676 ± 0.042) at 9 months and (GM = 0.778 ± 0.039, WM = 0.70 ± 0.043) at 12 months; however, the intensity prediction became more blurry. Our prediction results are promising with regard to the dynamics and complexity of brain development and the contrast drastic shifts (Fig.1 3 months vs. 12 months).
5 Conclusion
We proposed the first sparse metamorphosis patch-based prediction model for joint image intensity and structure evolution during early brain development. The results are quite promising and can be further improved through including more shape information as well as sharpening the 4D photometric atlas. Furthermore, in our future work, the selection of the best patches from the dynamic baseline dictionary can be further refined by including multimodal features (e.g., shape or anatomical structure) –instead of only relying on the 3 month image intensity.
Fig. 4.
Prediction results in a representative infant at 6 months for the 3 tested methods: M1 (appearance-and-deformation), M2 (appearance-and-labels), and M3 (appearance-labels-and-deformation).
References
- 1.Knickmeyer R, Gouttard S, Kang C, Evans D, Wilber K, Smith J, Hamer R, Lin W, Gerig G, Gilmore J. A structural mri study of human brain development from birth to 2 years. J Neurosci. 2008;28:12176–12182. doi: 10.1523/JNEUROSCI.3479-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fletcher P. Geodesic regression and the theory of least squares on riemannian manifolds. International journal of computer vision. 2013;105:171–185. [Google Scholar]
- 3.Niethammer M, Huang Y, Vialard FX. Geodesic regression for image time-series. MICCAI. 2011;6892:655–662. doi: 10.1007/978-3-642-23629-7_80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Trouvé A, Younes L. Metamorphoses through lie group action. Foundations of Computational Mathematics. 2005;5:173–198. [Google Scholar]
- 5.Wu G, Kim M, Sanroma G, Wang Q, Munsell B, Shen D. Hierarchical multi-atlas label fusion with multi-scale feature representation and label-specific patch partition. Neuroimage. 2015;106:34–46. doi: 10.1016/j.neuroimage.2014.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi F, Wang L, Wu G, Li G, Gilmore J, Lin W, Shen D. Neonatal atlas construction using sparse representation. Hum Brain Mapp. 2014;35:4663–4677. doi: 10.1002/hbm.22502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beg M, Miller M, Trouvé A, Younes L. Computing large deformation metric mappings via geodesic flows of di eomorphisms. International Journal of Computer Vision. 2005;61:139–157. [Google Scholar]
- 8.Garcin L, Younes L. Geodesic image matching: a wavelet based energy minimization scheme. 2005:349–364. [Google Scholar]
- 9.Wang Q, Kim M, Shi Y, Wu G, Shen D. Predict brain MR image registration via sparse learning of appearance and transformation. Med Image Anal. 2015;20:61–75. doi: 10.1016/j.media.2014.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]