Abstract
Companion animals are considered as one of the reservoirs of antimicrobial-resistant (AR) bacteria that can be cross-transmitted to humans. However, limited information is available on the possibility of AR bacteria originating from companion animals being transmitted secondarily from owners to non-owners sharing the same space. To address this issue, the present study investigated clonal relatedness among AR E. coli isolated from dog owners and non-owners in the same college classroom or household. Anal samples (n=48) were obtained from 14 owners and 34 non-owners; 31 E. coli isolates were collected (nine from owners and 22 from non-owners). Of 31 E. coli, 20 isolates (64.5%) were resistant to at least one antimicrobial, and 16 isolates (51.6%) were determined as multi-drug resistant E. coli. Six isolates (19.4%) harbored integrase genes (five harbored class I integrase gene and one harbored class 2 integrase gene, respectively). Pulsed-field gel electrophoretic analysis identified three different E. coli clonal sets among isolates, indicating that cross-transmission of AR E. coli can easily occur between owners and non-owners. The findings emphasize a potential risk of spread of AR bacteria originating from pets within human communities, once they are transferred to humans. Further studies are needed to evaluate the exact risk and identify the risk factors of secondarily transmission by investigating larger numbers of isolates from pets, their owners and non-owners in a community.
Keywords: antimicrobial resistance, companion animals, secondary bacterial transmission
The number of people living with companion animals has been increasing annually worldwide. According to the 2013 to 2014 American Pet Products Association survey, about 70% of U.S. households include companion animals [20]. In the Korean pet industry, the market size associated with companion animals is rapidly increasing and estimated to be $5.4 billion by 2020 [27]. Additionally, most owners consider their pets as family members and go to great lengths for their medical treatment [32]. As such, the use of antimicrobials in pets is increasing, which has resulted in the emergence and spread of antimicrobial-resistant (AR) bacteria. Companion animals are often considered as one of the reservoirs of AR bacteria that could be transferrable to their owners through direct or indirect contact [2, 25]. Direct contact includes a bite, lick or scratch and handling of animal feces, whereas indirect contact can occur by sharing the bed or toilet environment or being bitten by arthropods originating from pets [24].
AR bacteria in companion animals can be cross-transmitted to humans [2, 25]. Bacterial transmission among humans frequently occurs in confined environments, such as schools and households [6]; indeed, the spread of hemolytic uremic syndrome and bloody diarrhea caused by infection with a same clone of Shiga toxin-producing Escherichia coli has been reported in these environments [14, 18, 21]. Likewise, owners of companion animals could spread AR bacteria originating from their pets to other persons via close contact. However, there have been no studies investigating this possibility. We addressed this in the present study by comparing the genetic similarity of AR E. coli isolates from owners of dogs and non-owners sharing a classroom or household to determine the risk of secondary transmission of AR bacteria between humans.
MATERIALS AND METHODS
Sampling
All study participants provided written, informed consent for their participation. All protocols and procedures were approved by the institutional review board at the Seoul National University (IRB No. 1208/001-004). A total of 48 anal samples were collected from owners of dogs and non-owners at a college classroom and households located in Seoul, Korea, from April in 2010 to November in 2012. We used the sampling method described in previous studies [8, 15]. Owner samples (n=14) were collected from 11 undergraduate students as well as three of their family members; non-owner samples (n=34) were collected from 28 undergraduate students sharing the classroom with 11 owner students as well as six of their family members. Samples were placed in individual collection tubes containing Amies transport medium (Yu-Han Lab Tech, Seoul, Korea) and transported to our laboratory on ice within 6 hr of collection.
E. coli isolation and identification
For non-selective enrichment of microorganisms in samples, the swabs were mixed by vortexing in 10 ml buffered peptone water (BD Biosciences, Franklin Lakes, NJ, U.S.A.) and incubated at 37°C for 24 hr [31]. One milliliter aliquot of culture was inoculated in 9 mlE. coli broth and incubated at 37°C for 24 hr. The cultures were streaked on MacConkey agar plates and incubated at 37°C for 24 hr to isolate coliform bacteria, including E. coli [12]. Pink colonies suspected as E. coli were selected according to a standard protocol previously established in our laboratory [3]. Strain-specific PCR targeting 16S ribosomal RNA was carried out to confirm the bacterial species as E. coli [29]. E. coli ATCC 25922 (American Type Culture Collection, Manassas, VA, U.S.A.) was used as a positive control strain.
Antimicrobial susceptibility tests
Antimicrobial susceptibility was tested by the standard disk diffusion method according to Clinical and Laboratory Standard Institute guidelines [35]. The antimicrobial disks (BD Biosciences) used in this study were as follows: ampicillin (AM, 10 μg), amoxicillin/clavulanic acid (AMC, 20/10 μg), aztreonam (ATM, 30 μg), ceftazidime (CAZ, 30 μg), cefotaxime (CTX, 30 μg), cefotetan (CTT, 30 μg), ceftriaxone (CRO, 30 μg), chloramphenicol (30 μg), ciprofloxacin (CIP, 5 μg), imipenem (IMP, 10 μg), gentamicin (GM, 10 μg), nalidixic acid (NA, 30 μg), sulfamethoxazole/trimethoprim (SXT, 1.25/23.75 μg) and tetracycline (TE, 30 μg). Resistance, intermediate resistance and susceptibility to antimicrobials were established as described by Clinical and Laboratory Standards Institute guidelines [35]. E. coli ATCC 25922 was used as a reference strain. Multidrug resistance (MDR) was defined as resistance to three or more different classes of antimicrobial [13].
Detection of integrase genes in E. coli isolates
To determine the association between MDR and the presence of mobile genetic elements, integrase genes responsible for horizontal gene transfer, were detected in all E. coli isolates. Briefly, the integrase genes were amplified by PCR using the common integrase primer set, hep35 (5’-TGCGGGTYAARGATBTKGATTT-3’) and hep36 (5’-CARCACATGCGTRTARAT-3’). For positive isolates, PCR fragments were first digested with HinfI restriction enzyme (New England Biolabs, Ipswich, MA, U.S.A.) and analyzed by gel-electrophoresis. The class of integron was determined based on the number and size of DNA bands as previously described [34].
Molecular fingerprinting
To investigate cross-transmission of AR E. coli between owners and non-owners, the genetic relatedness of AR E. coli isolates was evaluated by standard pulsed-field gel electrophoresis (PFGE) using CHEF MAPPER (Bio-Rad, Hercules, CA, U.S.A.) [10]. Briefly, isolates cultured overnight in tryptic soy broth (BD Biosciences) were streaked on tryptic soy agar (BD Biosciences) plates and incubated at 37°C for 14–18 hr. The turbidity of bacterial suspensions was adjusted to 4.0 McFarland, and cells were embedded in 1.0% agarose plugs that were lysed with proteinase K prepared as a 20 mg/ml stock solution (Sigma-Aldrich, St. Louis, MO, U.S.A.), followed by digestion for 2 hr with 50 U XbaI (New England Biolabs) at 37°C. Digested plugs were then placed on 1.0% SeaKem Gold agarose (Lonza, Allendale, NJ, U.S.A.), and PFGE was carried out at 6.0 V for 19 hr with a ramped pulse time of 6.76–35.38 sec in 0.5× Tris-Borate-EDTA buffer at 14°C. BioNumerics software (Applied Maths, Sint-Martens-Latem, Belgium) was used to analyze DNA restriction patterns using the dice coefficient (0.5% optimization and 1.0% tolerance) and the unweighted pair group method. E. coli ATCC 25922 was used as a reference strain.
RESULTS
Isolation of E. coli from swab samples
A total of 31 E. coli isolates were obtained from 48 swab samples (64.6%), with 9/14 (64.3%) and 22/34 (64.7%) collected from owners and non-owners, respectively (Table 1).
Table 1. Antibiogram of 31 E. coli isolates from owners and non-owners.
| Antimicrobial | Non-owner | Owner | Total |
|---|---|---|---|
| (n=22) | (n=9) | (n=31) | |
| AM | 13 (59.1)a) | 3 (33.3) | 16 (51.6) |
| AMC | 11 (50.0) | 2 (22.2) | 13 (41.9) |
| GM | 2 (9.1) | 1 (11.1) | 3 (9.7) |
| TE | 11 (50.0) | 2 (22.2) | 13 (41.9) |
| CIP | 4 (18.2) | 1 (11.1) | 5 (16.1) |
| NA | 11 (50.0) | 4 (44.4) | 15 (48.4) |
| SXT | 7 (31.8) | 2 (22.2) | 9 (29.0) |
| C | 0 | 1 (11.1) | 1 (3.2) |
| CRO | 1 (4.5) | 0 | 1 (3.2) |
| CTX | 1 (4.5) | 0 | 1 (3.2) |
| CAZ | 0 | 0 | 0 |
| CTT | 0 | 0 | 0 |
| IMP | 0 | 0 | 0 |
| ATM | 0 | 0 | 0 |
| MDR | 13 (59.1) | 3 (33.3) | 16 (51.6) |
| AR b) | 15 (68.2) | 5 (55.6) | 20 (64.5) |
a) Number of E. coli isolates showing resistance to each indicated antimicrobial is presented with the antimicrobial resistant rate in parenthesis. b) Antimicrobial resistant. This row shows the number of E. coli isolates showing resistance to at least one antimicrobial tested. AM, ampicillin; AMC, amoxicillin/clavulanic acid; ATM, aztreonam; C, chloramphenicol; CAZ, ceftazidime; CIP, ciprofloxacin; CRO, ceftriaxone; CTT, cefotetan; CTX, cefotaxime; GM, gentamicin; IMP, imipenem; MDR, multidrug resistance; NA, nalidixic acid; SXT, sulfamethoxazole/trimethoprim; TE, tetracycline.
Antibiogram of 31 E. coli isolates
The number of E. coli isolates showing resistance to each antimicrobial is shown in Table 1. A total of 20/31 isolates (64.5%) from non-owners (n=15) and owners (n=5) were resistant to at least one antimicrobial (Table 1). All isolates were susceptible to CAZ, CTT, IMP and ATM. The antibiogram analysis revealed frequencies of AR E. coli isolates of 51.6% (n=16) for AM followed by 48.4% (n=15) for NA, 41.9% (n=13 each) for AMC and TE, 29.0% (n=9) for SXT and 16.1% (n=5) for CIP. In addition, nine isolates showed resistance to each of GM, CRO and CTX. A total of 16 isolates (51.6%) were identified as harboring MDR E. coli.
Detection of integrase genes in E. coli isolates
Six of the 31 E. coli isolates (19.4%) harbored integrase genes (Table 2). Of these, four originated from non-owners, and two were from owners. The four isolates from non-owners harbored only the class 1 integrase gene intI1, whereas both intI1 and the class 2 integrase gene intI2 were detected in the two isolates from owners. All six isolates were defined as having MDR, since they harbored at least three different antimicrobial resistance genes.
Table 2. Characterization of six AR E. coli isolates harboring integrase genes.
| Isolate no. | Integron class | Sample group | Antibiogram | Integron-associated genesa) |
|---|---|---|---|---|
| P99 | Class 1/intI1 | Non-owner (S)b) | AM, AMC, TE, CIP, NA, SXT | OXA, tetB, dfrA17 |
| P130-2 | Class 1/intI1 | Non-owner (S) | AM, TE, SXT | OXA, tetA, dfrA7 |
| P154-1 | Class 1/intI1 | Non-owner (S) | AM, AMC, TE, CIP, NA, SXT | OXA, tetB, dfrA17 |
| P150 | Class 1/intI1 | Non-owner (S) | TE, NA, SXT | tetA, tetB, sul1, dfrA12/13 |
| P143-1 | Class 1/intI1 | Owner (S) | GM, TE, NA, SXT | tetB, sul1, dfrA17 |
| P117-1 | Class 2/intI2 | Owner (F) | AM, AMC, TE, SXT, C | OXA, tetB, sul1, dfrA1, cat1 |
a) cat1, chloramphenicol resistance gene; dfrA1/A7/A17/A12/A13, trimethoprim resistance genes; OXA, ampicillin resistance gene; sul1, sulfamethoxazole resistance gene; tetA/B, tetracycline resistance genes. b) S and F indicate college student and family member, respectively. AM, ampicillin; AMC, amoxicillin/clavulanic acid; C, chloramphenicol; CIP, ciprofloxacin; GM, gentamicin; NA, nalidixic acid; SXT sulfamethoxazole/trimethoprim; TE, tetracycline.
Genetic relatedness of E. coli isolates from owners and non-owners
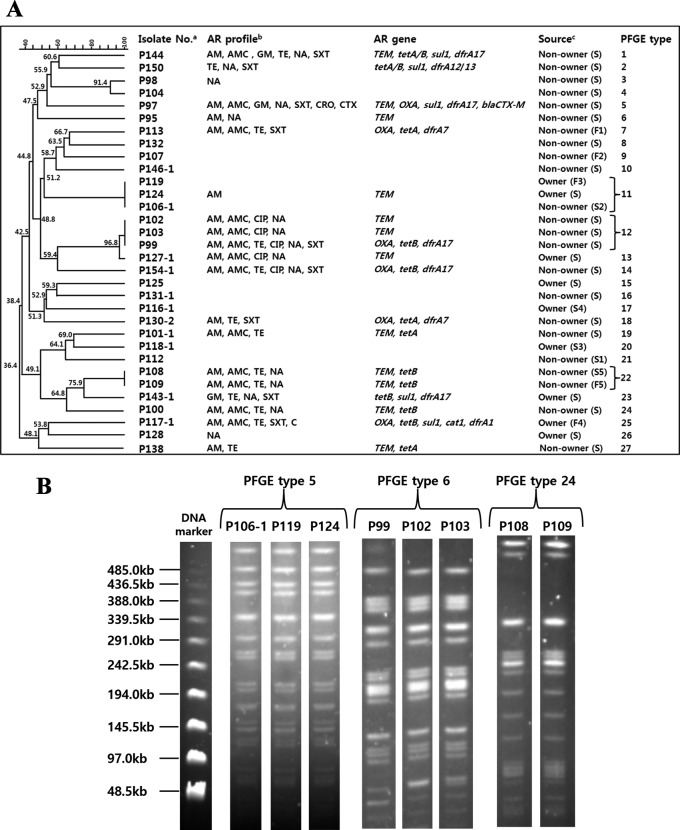
To determine the risk of cross-transmission between owners and non-owners, we analyzed the genetic relatedness of the 31 E. coli isolates by PFGE. Three clonal sets (PFGE types 5, 6 and 24) were identified (Fig. 1A and 1B). For type 5, two E. coli isolates (nos. P106-1 and P124) were obtained from two students (one non-owner and one owner) who shared a classroom, whereas the other isolate (no. P119) was from a family member of an owner student. For type 6, the three isolates (nos. P102, P103 and P99) were obtained from three non-owner students, but they had slightly different antibiogram profiles. For type 24, the two isolates (nos. P108 and P109) were from two non-owners living in the same household who showed identical antibiogram profiles.
Fig. 1.
PFGE analysis of 31 E. coli isolates. (A), Dendrogram of all PFGE patterns; (B), PFGE results of types 5, 6 and 24. Levels of similarity were determined using the Dice coefficient (0.5% optimization and 1.0% tolerance) and the unweighted pair-group method. Individual PFGE patterns are summarized with their antimicrobial resistance profiles and genes and sample sources (A). PFGE results of each isolate belonging to types 5, 6 and 24 are presented (B). AM, ampicillin; AMC, amoxicillin/clavulanic acid; C, chloramphenicol; CIP, ciprofloxacin; CRO, ceftriaxone; CTX, cefotaxime; GM, gentamicin; NA, nalidixic acid; SXT, sulfamethoxazole/trimethoprim; TE, tetracycline. a) Identification number of each E. coli isolate from pet owners and non-owners. b) Antimicrobial resistance profiles. c) Sample sources: college students (S) and family members (F). Students and family members living in the same house are represented by a combination of a letter and the same number, as follows: S1 (college student) and F1 (family member who lives in the same household as S1).
DISCUSSION
AR bacteria can be cross-transmitted between humans and animals [7, 17]. Transmission is usually determined by detecting the same clonal isolates from different hosts [2]. Livestock (e.g., horse, goat and cattle) and wild animals (e.g., free-roaming elk) are sources of enteric pathogens and AR bacteria that can be transmitted to humans via direct or indirect contact [9]. Companion animals are also considered as sources of AR bacteria and infectious human pathogens [4, 7]. However, there is no direct evidence that AR bacteria originating from companion animals are cross-transmitted between owners and non-owners living in a confined community. Common enteric microorganisms, such as E. coli, are easily and inadvertently transferred between individuals via hand-to-hand contact [16]. In addition, E. coli isolated from feces is considered as a good indicator for antimicrobial resistance in a population [1, 5]. The current study was carried out in order to establish the risk of cross-transmission of AR bacteria originating from companion animals within a confined human community.
Of 31 E. coli isolates, 64.5% were identified as AR bacteria showing resistance to at least one antimicrobial. This rate is similar to that in healthy humans (67.1%), but much lower than that in human clinical specimens (98.5%) reported by a previous study from Korea [11]. Integrons are known to play an important role in the horizontal transfer of antimicrobial resistance genes by conjugative plasmids and transposons, and closely associated with the development of MDR in enterobacteria [22, 23]. Class I is the predominant class of integron detected in many countries including Korea [11, 19, 26]. Likewise, all integron positive E. coli isolates in this study were found to be multi-drug resistant, and the prevalent type was class I. PFGE analysis revealed that all isolates carrying integrons had distinct PFGE types (Fig. 1A). This suggests that the dissemination of integrons was not due to clonal spread, but to horizontal gene transfer of plasmids or transposons, emphasizing the important role of integrons in the spread of antimicrobial resistance genes. The prevalence of integrons (19.4%) in E. coli isolates from healthy humans was higher than that reported in an earlier study from Korea [11], but lower than that in other countries [19, 30].
Three clonal sets were identified among 31 E. coli isolates, providing evidence of clonal expansion of resistant strains within the study population (Fig. 1A and 1B). PFGE types 6 and 24 indicated the spreading of AR E. coli within a classroom and family, respectively, whereas PFGE type 5 included three isolates from a pet owner student, a non-owner student and an owner family member. Although we did not analyze E. coli isolates from the dogs in this study, our results indirectly demonstrate the possibility of cross-transmission of AR bacteria from companion animals to non-owners.
In most countries, the overall amount of antimicrobials used for companion animals is not reliably measured. However, antimicrobials used in human and veterinary hospitals are almost identical [7]; as such, resistance patterns in bacteria originating from animals and humans are very similar [28, 33]. There is an increasing concern that AR bacteria from pet animals can spread among humans. Although this study only investigated the clonal expansion of AR E. coli in a confined human community, the results indicate that once these bacteria are transmitted from pets to their owners, they can spread to other humans through social activities. Further investigations are required to provide more direct evidence and identify the risk factors of secondary transmission by studying larger numbers of bacterial isolates from pets, their owners and non-owners in a community.
Acknowledgments
This study was supported by grants from the Animal and Plant Quarantine Agency, Ministry of Food, Agriculture, Forestry, and Fisheries, Republic of Korea (Z-AD13-2011-11-06 and Z-1543081-2016-17-01). Additional support was obtained from the BK 21 Plus Program for Veterinary Science Research.
REFERENCES
- 1.Alekshun M. N., Levy S. B.2006. Commensals upon us. Biochem. Pharmacol. 71: 893–900. doi: 10.1016/j.bcp.2005.12.040 [DOI] [PubMed] [Google Scholar]
- 2.Chung Y. S., Kwon K. H., Shin S., Kim J. H., Park Y. H., Yoon J. W.2014. Characterization of veterinary hospital-associated isolates of Enterococcus species in Korea. J. Microbiol. Biotechnol. 24: 386–393. doi: 10.4014/jmb.1310.10088 [DOI] [PubMed] [Google Scholar]
- 3.Chung Y. S., Song J. W., Kim D. H., Shin S., Park Y. K., Yang S. J., Lim S. K., Park K. T., Park Y. H.2016. Isolation and characterization of antimicrobial-resistant Escherichia coli from national horse racetracks and private horse-riding courses in Korea. J. Vet. Sci. 17: 199–206. doi: 10.4142/jvs.2016.17.2.199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doyle M. P., Ruoff K. L., Pierson M., Weinberg W., Soule B., Michaels B. S.2000. Reducing Transmission of Infectious Agents in the Home-Part I: Sources of Infection. Dairy Food Environ. Sanit. 20: 330–337. [Google Scholar]
- 5.EFSA2012. The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2010. EFSA J. 10: 2598. doi: 10.2903/j.efsa.2012.2598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greig J. D., Todd E. C., Bartleson C. A., Michaels B. S.2007. Outbreaks where food workers have been implicated in the spread of foodborne disease. Part 1. Description of the problem, methods, and agents involved. J. Food Prot. 70: 1752–1761. doi: 10.4315/0362-028X-70.7.1752 [DOI] [PubMed] [Google Scholar]
- 7.Guardabassi L., Schwarz S., Lloyd D. H.2004. Pet animals as reservoirs of antimicrobial-resistant bacteria. J. Antimicrob. Chemother. 54: 321–332. doi: 10.1093/jac/dkh332 [DOI] [PubMed] [Google Scholar]
- 8.Gustavsson L., Westin J., Andersson L. M., Lindh M.2011. Rectal swabs can be used for diagnosis of viral gastroenteritis with a multiple real-time PCR assay. J. Clin. Virol. 51: 279–282. doi: 10.1016/j.jcv.2011.05.025 [DOI] [PubMed] [Google Scholar]
- 9.Hale C. R., Scallan E., Cronquist A. B., Dunn J., Smith K., Robinson T., Lathrop S., Tobin-D’Angelo M., Clogher P.2012. Estimates of enteric illness attributable to contact with animals and their environments in the United States. Clin. Infect. Dis. 54Suppl 5: S472–S479. doi: 10.1093/cid/cis051 [DOI] [PubMed] [Google Scholar]
- 10.Jaros P., Dufour M., Gilpin B., Freeman M. M., Ribot E. M.2015. PFGE for Shiga toxin-producing Escherichia coli O157:H7 (STEC O157) and non-O157 STEC. Methods Mol. Biol. 1301: 171–189. doi: 10.1007/978-1-4939-2599-5_15 [DOI] [PubMed] [Google Scholar]
- 11.Kang H. Y., Jeong Y. S., Oh J. Y., Tae S. H., Choi C. H., Moon D. C., Lee W. K., Lee Y. C., Seol S. Y., Cho D. T., Lee J. C.2005. Characterization of antimicrobial resistance and class 1 integrons found in Escherichia coli isolates from humans and animals in Korea. J. Antimicrob. Chemother. 55: 639–644. doi: 10.1093/jac/dki076 [DOI] [PubMed] [Google Scholar]
- 12.Kiskó G., Roller S.2005. Carvacrol and p-cymene inactivate Escherichia coli O157:H7 in apple juice. BMC Microbiol. 5: 36. doi: 10.1186/1471-2180-5-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kos V. N., Desjardins C. A., Griggs A., Cerqueira G., Van Tonder A., Holden M. T., Godfrey P., Palmer K. L., Bodi K., Mongodin E. F., Wortman J., Feldgarden M., Lawley T., Gill S. R., Haas B. J., Birren B., Gilmore M. S.2012. Comparative genomics of vancomycin-resistant Staphylococcus aureus strains and their positions within the clade most commonly associated with Methicillin-resistant S. aureus hospital-acquired infection in the United States. MBio 3: e00112–e00112. doi: 10.1128/mBio.00112-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuijper E. J., Soonawala D., Vermont C., van Dissel J. T.2011. Household transmission of haemolytic uraemic syndrome associated with Escherichia coli O104:H4 in the Netherlands, May 2011. Euro Surveill. 16: 19897. [PubMed] [Google Scholar]
- 15.Laarhoven L. M., de Heus P., van Luijn J., Duim B., Wagenaar J. A., van Duijkeren E.2011. Longitudinal study on methicillin-resistant Staphylococcus pseudintermedius in households. PLoS ONE 6: e27788. doi: 10.1371/journal.pone.0027788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee H., Choi S.2009. Hand washing awareness among students in Seoul and antibiotic resistance of staphylococcus aureus isolated on their hands. J. Environ. Health Sci. 35: 278–286. [Google Scholar]
- 17.Lee J. H.2003. Methicillin (Oxacillin)-resistant Staphylococcus aureus strains isolated from major food animals and their potential transmission to humans. Appl. Environ. Microbiol. 69: 6489–6494. doi: 10.1128/AEM.69.11.6489-6494.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee M. B., Greig J. D.2010. A review of gastrointestinal outbreaks in schools: effective infection control interventions. J. Sch. Health 80: 588–598. doi: 10.1111/j.1746-1561.2010.00546.x [DOI] [PubMed] [Google Scholar]
- 19.Li B., Zhao Z. C., Wang M. H., Huang X. H., Pan Y. H., Cao Y. P.2014. Antimicrobial resistance and integrons of commensal Escherichia coli strains from healthy humans in China. J. Chemother. 26: 190–192. doi: 10.1179/1973947813Y.0000000113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindstrom N. M., Moore D. M., Zimmerman K., Smith S. A.2015. Hematologic assessment in pet rats, mice, hamsters, and gerbils: Blood sample collection and blood cell identification. Clin. Lab. Med. 35: 629–640. doi: 10.1016/j.cll.2015.05.011 [DOI] [PubMed] [Google Scholar]
- 21.Locking M. E., Pollock K. G., Allison L. J., Rae L., Hanson M. F., Cowden J. M.2011. Escherichia coli O157 infection and secondary spread, Scotland, 1999−2008. Emerg. Infect. Dis. 17: 524–527. doi: 10.3201/eid1703.100167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martinez-Freijo P., Fluit A. C., Schmitz F. J., Grek V. S., Verhoef J., Jones M. E.1998. Class I integrons in Gram-negative isolates from different European hospitals and association with decreased susceptibility to multiple antibiotic compounds. J. Antimicrob. Chemother. 42: 689–696. doi: 10.1093/jac/42.6.689 [DOI] [PubMed] [Google Scholar]
- 23.Recchia G. D., Hall R. M.1995. Gene cassettes: a new class of mobile element. Microbiology 141: 3015–3027. doi: 10.1099/13500872-141-12-3015 [DOI] [PubMed] [Google Scholar]
- 24.Robinson R. A., Pugh R. N.2002. Dogs, zoonoses and immunosuppression. J. R. Soc. Promot. Health 122: 95–98. doi: 10.1177/146642400212200210 [DOI] [PubMed] [Google Scholar]
- 25.So J. H., Kim J., Bae I. K., Jeong S. H., Kim S. H., Lim S. K., Park Y. H., Lee K.2012. Dissemination of multidrug-resistant Escherichia coli in Korean veterinary hospitals. Diagn. Microbiol. Infect. Dis. 73: 195–199. doi: 10.1016/j.diagmicrobio.2012.03.010 [DOI] [PubMed] [Google Scholar]
- 26.Solberg O. D., Ajiboye R. M., Riley L. W.2006. Origin of class 1 and 2 integrons and gene cassettes in a population-based sample of uropathogenic Escherichia coli. J. Clin. Microbiol. 44: 1347–1351. doi: 10.1128/JCM.44.4.1347-1351.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song H. G., Lim S. U.2015. Assessing pet industry in Korea using service quality improvement gap model. Int. J. Technology 15: 2–20. [Google Scholar]
- 28.Strommenger B., Kehrenberg C., Kettlitz C., Cuny C., Verspohl J., Witte W., Schwarz S.2006. Molecular characterization of methicillin-resistant Staphylococcus aureus strains from pet animals and their relationship to human isolates. J. Antimicrob. Chemother. 57: 461–465. doi: 10.1093/jac/dki471 [DOI] [PubMed] [Google Scholar]
- 29.Tsen H. Y., Lin C. K., Chi W. R.1998. Development and use of 16S rRNA gene targeted PCR primers for the identification of Escherichia coli cells in water. J. Appl. Microbiol. 85: 554–560. doi: 10.1046/j.1365-2672.1998.853535.x [DOI] [PubMed] [Google Scholar]
- 30.Vinué L., Sáenz Y., Somalo S., Escudero E., Moreno M. Á., Ruiz-Larrea F., Torres C.2008. Prevalence and diversity of integrons and associated resistance genes in faecal Escherichia coli isolates of healthy humans in Spain. J. Antimicrob. Chemother. 62: 934–937. doi: 10.1093/jac/dkn331 [DOI] [PubMed] [Google Scholar]
- 31.Wales A. D., Pearson G. R., Skuse A. M., Roe J. M., Hayes C. M., Cookson A. L., Woodward M. J.2001. Attaching and effacing lesions caused by Escherichia coli O157:H7 in experimentally inoculated neonatal lambs. J. Med. Microbiol. 50: 752–758. doi: 10.1099/0022-1317-50-9-752 [DOI] [PubMed] [Google Scholar]
- 32.Walsh F.2009. Human-animal bonds I: the relational significance of companion animals. Fam. Process 48: 462–480. doi: 10.1111/j.1545-5300.2009.01296.x [DOI] [PubMed] [Google Scholar]
- 33.Weese J. S., Dick H., Willey B. M., McGeer A., Kreiswirth B. N., Innis B., Low D. E.2006. Suspected transmission of methicillin-resistant Staphylococcus aureus between domestic pets and humans in veterinary clinics and in the household. Vet. Microbiol. 115: 148–155. doi: 10.1016/j.vetmic.2006.01.004 [DOI] [PubMed] [Google Scholar]
- 34.White P. A., McIver C. J., Deng Y., Rawlinson W. D.2000. Characterisation of two new gene cassettes, aadA5 and dfrA17. FEMS Microbiol. Lett. 182: 265–269. doi: 10.1111/j.1574-6968.2000.tb08906.x [DOI] [PubMed] [Google Scholar]
- 35.Wikler M. A.2009. Performance Standards for Antimicrobial Susceptibility Testing: Nineteenth Informational Supplement. Clinical and Laboratory Standards Institute. [Google Scholar]