ABSTRACT
The zebrafish is an emerging model for highly sophisticated medium-throughput experiments such as genetic and chemical screens. However, studies of entire protein families within this context are often hampered by poor genetic resources such as clone libraries. Here we describe a complete collection of 76 full-length open reading frame clones for the zebrafish rab protein family. While the mouse genome contains 60 rab genes and the human genome 63, we find that 18 zebrafish rab genes have 2, and in the case of rab38, 3 paralogues. In contrast, we were unable to identify zebrafish orthologues of the mammalian Rab2b, Rab17 or Rab29. We make this resource available through the Addgene repository to facilitate cell biologic approaches using this model.
KEYWORDS: in vivo, library, rab, screening, zebrafish
Introduction
The zebrafish model has undergone several transitions since its development as an experimental system in the late 1970s and early 1980s.1 Initially, the optical transparency and rapid, external development were attractive to developmental biologists. Subsequently, large clutch sizes and rapid generation times have led to its adoption for large scale mutagenesis and forward genetic screening using methodologies pioneered in invertebrates.2-5 Screens have advanced in sophistication to incorporate modifier and sensitized screens and now focus almost exclusively on questions related to human disease.6 Recent advances in microscopy, (such as light-sheet,7,8 and super-resolution microscopy9), along with a new suite of genetic tools (such as Tol2 transposition10 and CRISPR/Cas11) have left us poised for a third renaissance; the zebrafish as a model for in vivo cell biology.
Although the zebrafish is a well established model with a fully sequenced genome, genetic resources tend to be sub-optimal, and often studies rely on expression of human or mouse orthologues. Zebrafish genomes have undergone at least one ancestral genome duplication.12 Gene loss or gain followed by subfunctionalisation and/or compartmentalisation of expression is common. This has allowed an unprecedented level of genetic dissection where pleiotropic effects in other systems have been problematic, but equally has frustrated attempts to create functional nulls.13 Crucial to the momentum of in vivo cell biology in zebrafish is the creation, curation and distribution of zebrafish specific tools through repositories such as Addgene and the Zebrafish Information Network (zfin).
Here, we describe the zebrafish rab family of GTPases, which regulate virtually all membrane trafficking events in eukaryotic cells.14 While there have been studies of the canonical rabs in zebrafish,15 a comprehensive resource has not been put together as in Drosophila.16 We summarise orthology relationships between zebrafish, mouse and human rab genes, and make available a library of full-length rab open reading frame clones through the Addgene repository.
Results and Discussion
To identify all possible rabs from the zebrafish genome we used the orthology prediction algorithm built into the ensemble genome browser,17 using human and then mouse rabs as a starting point. We identified 76 individual zebrafish rab genes compared with 60 mouse genes and 63 human genes (Fig 1A). 18 genes were found to be duplicated in the zebrafish compared with human and mouse. There were only 3 genes present in mouse and human that we were unable to identify zebrafish orthologues for, rab2b, rab17 and rab29. RAB6C, RAB40A and RAB40AL were all present in human but neither mouse or zebrafish, and Rab41 was absent in mouse but present in human and zebrafish. All duplicated zebrafish paralogues clustered together in a neighbor joining tree (Fig 1B).
Figure 1.

Homology relationships between zebrafish, mouse and human rab proteins. A) Orthology between rab proteins of different species is shown with solid lines. Green shading represents paralogous genes within a species which are not duplicated within the other 2 species. Red shading represents lack of an ortholog present in the other 2 species. B) Neighbor joining tree for all zebrafish rab protein sequences. Numbers at each node represent the percentage of 10,000 bootstrap replicates. Nodes with less than 50% of bootstrap replicates are collapsed. Green shading represents paralogous genes which are not duplicated in mouse or human. All paralogues cluster together.
We next used RT-PCR and TA cloning to generate full length ORF clones of all zebrafish rab proteins. Using primers spanning the start and stop codons, we were able to amplify all sequences but one in single amplicons. We amplified Rab44 in 2 separate reactions, since the full ORF is 5.8kb, which were subsequently joined by standard directional cloning. All amplicons were cloned into pGemTeasy, fully sequenced and made available through the Addgene repository (Table 1). Finally, to demonstrate the utility of this resource we generated fluorescent protein fusion vectors for 3 canonical rab proteins by subcloning from our ORF clones. Live imaging of zebrafish periderm cells expressing the fusion proteins marked endoplasmic reticulum (rab1ab), early/recycling endosomes (rab4a) and late endosome/golgi (rab9a) (Fig 2). Details of all clones generated in this study are given in Table 1.
Table 1.
List of constructs used in this study and corresponding Addgene identifiers.
| Vector | Addgene ID | Vector | Addgene ID |
|---|---|---|---|
| Rab1aa | 86217 | Rab19 | 80504 |
| Rab1ab | 86218 | Rab20 | 80540 |
| Rab1ba | 80505 | Rab21 | 86222 |
| Rab1bb | 80506 | Rab22a | 86223 |
| Rab2a | 86219 | Rab23 | 80542 |
| Rab3aa | 80508 | Rab24 | 80543 |
| Rab3ab | 80509 | Rab25a | 80544 |
| Rab3b | 80503 | Rab25b | 80545 |
| Rab3c | 80510 | Rab26 | 86224 |
| Rab3da | 80511 | Rab27a | 80546 |
| Rab3db | 80512 | Rab27b | 80519 |
| Rab4a | 80513 | Rab28 | 80547 |
| Rab4b | 80514 | Rab30 | 80548 |
| Rab5aa | 80515 | Rab31 | 80507 |
| Rab5ab | 80516 | Rab32a | 80549 |
| Rab5b | 80517 | Rab32b | 80550 |
| Rab5c | 80518 | Rab33a | 80551 |
| Rab6a | 86220 | Rab33ba | 80552 |
| Rab6ba | 80520 | Rab33bb | 80524 |
| Rab6bb | 80521 | Rab34a | 80553 |
| Rab7a | 80522 | Rab34b | 80554 |
| Rab7b | 80523 | Rab35a | 86225 |
| Rab8a | 86221 | Rab35b | 80556 |
| Rab8b | 80525 | Rab36 | 80557 |
| Rab9a | 80526 | Rab37 | 86226 |
| Rab9b | 80527 | Rab38a | 80558 |
| Rab10 | 80528 | Rab38b | 80541 |
| Rab11a | 80529 | Rab38c | 80555 |
| Rab11al | 80530 | Rab39a | 86227 |
| Rab11ba | 80531 | Rab39ba | 80559 |
| Rab11bb | 80532 | Rab39bb | 80560 |
| Rab12 | 80533 | Rab40b | 86228 |
| Rab13 | 80534 | Rab40c | 80562 |
| Rab14 | 80535 | Rab41 | 80561 |
| Rab14l | 80536 | Rab42a | 80563 |
| Rab15 | 80537 | Rab42b | 80564 |
| Rab18a | 80538 | Rab43 | 80565 |
| Rab18b | 80539 | Rab44 | 86229 |
Figure 2.
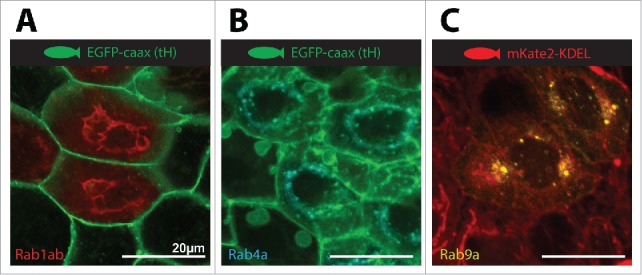
Expression of canonical zebrafish rabs in dermal cells. A) Expression of FusionRed-Rab1ab on a membrane localized EGFPcaax background marks the endoplasmic reticulum. B) Expression of TagBFP2-Rab4a on a membrane localized EGFPcaax background marks the early/recycling endosome compartment. C) Expression of mVenus-Rab9a on a endoplasmic reticulum localized mKate2-KDEL background marks the late endosome/golgi compartment.
Materials and methods
Zebrafish rab sequences were identified by orthology to annotated mouse and human sequences as described. The tree was constructed using a ClustalW alignment with the full-length protein sequences, followed by neighbor joining with 10,000 bootstrap replicates in MacVector. Primers for cDNA cloning were designed using Primer3 software, using a 40bp window surrounding the start and stop codons. RNA was extracted and cDNA generated according to established methods.18 PCR was performed using platinum taq HiFi polymerase (Invitrogen). Sequencing was performed by Macrogen. Expression vectors were generated using the Tol2 kit,19 using β actin, rcn320 and krt421 promoters. Transient expression analysis was performed as described previously.22
Animal Ethics and Biosafety
All experiments were approved by the University of Queensland Animal Ethics committee and University of Queensland Biosafety committee.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
Confocal microscopy was performed at the Australian Cancer Research Foundation (ACRF)/Institute for Molecular Bioscience Dynamic Imaging Facility for Cancer Biology, which was established with the support of the ACRF. We would also like to thank the following people for supplying initial clones and sequences which, although different to those which we make available on Addgene, were nonetheless helpful to our study. Kristen Kwan for rab3c, Michel Bagnat for Rab7b, Brian Link for rab5c, rab7a and rab11a, Uwe Strahle for Rab1ab, 6a, 7a, 5a, 12, 27a.
Funding
This work was supported by grants and a fellowship from the National Health and Medical Research Council of Australia (grant numbers APP1037320, APP1045092, APP1058565, and APP1099251 to R.G. Parton).
References
- [1].Streisinger G, Walker C, Dower N, Knauber D, Singer F. Production of clones of homozygous diploid zebra fish (Brachydanio rerio). Nature 1981; 291(5813):293-6; PMID:7248006; http://dx.doi.org/ 10.1038/291293a0 [DOI] [PubMed] [Google Scholar]
- [2].Anderson KV, Nüsslein-Volhard C. Information for the dorsal–ventral pattern of the Drosophila embryo is stored as maternal mRNA. Nature 1984; 311:223-7; PMID:6434989; http://dx.doi.org/ 10.1038/311223a0 [DOI] [PubMed] [Google Scholar]
- [3].Nüsslein-Volhard C, Frohnhofer H, Lehmann R. Determination of anteroposterior polarity in Drosophila. Science 1987; 238(4834):1675-81; PMID:3686007; http://dx.doi.org/ 10.1126/science.3686007 [DOI] [PubMed] [Google Scholar]
- [4].Haffter P, Granato M, Brand M, Mullins MC, Hammerschmidt M, Kane DA, Odenthal J, van Eeden FJ, Jiang YJ, Heisenberg CP, et al.. The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development 1996; 123:1-36; PMID:9007226 [DOI] [PubMed] [Google Scholar]
- [5].Driever W, Solnica-Krezel L, Schier AF, Neuhauss SC, Malicki J, Stemple DL, Stainier DY, Zwartkruis F, Abdelilah S, Rangini Z, et al.. A genetic screen for mutations affecting embryogenesis in zebrafish. Development 1996; 123:37-46; PMID:9007227 [DOI] [PubMed] [Google Scholar]
- [6].Lieschke GJ, Currie PD. Animal models of human disease: zebrafish swim into view. Nat Rev Genet 2007; 8:353-67; PMID:17440532; http://dx.doi.org/ 10.1038/nrg2091 [DOI] [PubMed] [Google Scholar]
- [7].Chi NC, Shaw RM, Jungblut B, Huisken J, Ferrer T, Arnaout R, Scott I, Beis D, Xiao T, Baier H, et al.. Genetic and physiologic dissection of the vertebrate cardiac conduction system. PLoS Biol 2008; 6:e109; PMID:18479184; http://dx.doi.org/ 10.1371/journal.pbio.0060109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Xiao Y, Faucherre A, Pola-Morell L, Heddleston JM, Liu T-L, Chew T-L, Sato F, Sehara-Fujisawa A, Kawakami K, López-Schier H. High-resolution live imaging reveals axon-glia interactions during peripheral nerve injury and repair in zebrafish. Disease Models Mechanisms 2015; 8(6):553-64; http://dx.doi.org/ 10.1242/dmm.018184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Middel V, Zhou L, Takamiya M, Beil T, Shahid M, Roostalu U, Grabher C, Rastegar S, Reischl M, Nienhaus GU, et al.. Dysferlin-mediated phosphatidylserine sorting engages macrophages in sarcolemma repair. Nat Comms 2016; 7:12875; http://dx.doi.org/ 10.1038/ncomms12875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kawakami K, Takeda H, Kawakami N, Kobayashi M, Matsuda N, Mishina M. A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev Cell 2004; 7:133-44; PMID:15239961; http://dx.doi.org/ 10.1016/j.devcel.2004.06.005 [DOI] [PubMed] [Google Scholar]
- [11].Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JR, Joung JK. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol 2013; 31:227-9; PMID:23360964; http://dx.doi.org/ 10.1038/nbt.2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Meyer A, Schartl M. Gene and genome duplications in vertebrates: the one-to-four (-to-eight in fish) rule and the evolution of novel gene functions. Curr Opin Cell Biol 1999; 11(6):699-704; PMID:10600714; http://dx.doi.org/ 10.1016/S0955-0674(99)00039-3 [DOI] [PubMed] [Google Scholar]
- [13].McClintock JM, Kheirbek MA, Prince VE. Knockdown of duplicated zebrafish hoxb1 genes reveals distinct roles in hindbrain patterning and a novel mechanism of duplicate gene retention. Development 2002; 129(10):2339-54. [DOI] [PubMed] [Google Scholar]
- [14].Hutagalung AH, Novick PJ. Role of Rab GTPases in membrane traffic and cell Physiology. Physiol Rev 2011; 91(1):119-49; PMID:21248164; http://dx.doi.org/ 10.1152/physrev.00059.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Clark BS, Winter M, Cohen AR, Link BA. Generation of Rab-based transgenic lines for in vivo studies of endosome biology in zebrafish. Dev Dyn 2011; 240(11):2452-65; PMID:21976318; http://dx.doi.org/ 10.1002/dvdy.22758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Zhang J, Schulze KL, Hiesinger PR, Suyama K, Wang S, Fish M, Acar M, Hoskins RA, Bellen HJ, Scott MP. Thirty-one flavors of Drosophila rab proteins. Genetics 2007; 176(2):1307-22; PMID:17409086; http://dx.doi.org/ 10.1534/genetics.106.066761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Howe K, Clark MD, Torroja CF, Torrance J, Berthelot C, Muffato M, Collins JE, Humphray S, McLaren K, Matthews L, et al.. The zebrafish reference genome sequence and its relationship to the human genome. Nature 2013; 496(7446):498-503; PMID:23594743; http://dx.doi.org/ 10.1038/nature12111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Lo HP, Nixon SJ, Hall TE, Cowling BS, Ferguson C, Morgan GP, Schieber NL, Fernandez-Rojo MA, Bastiani M, Floetenmeyer M, et al.. The caveolin-cavin system plays a conserved and critical role in mechanoprotection of skeletal muscle. J Cell Biol 2015; 210(5):833-49; PMID:26323694; http://dx.doi.org/ 10.1083/jcb.201501046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, Parant JM, Yost HJ, Kanki JP, Chien CB. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev Dyn 2007; 236(11):3088-99;PMID:17937395; http://dx.doi.org/ 10.1002/dvdy.21343 [DOI] [PubMed] [Google Scholar]
- [20].Ellis K, Bagwell J, Bagnat M. Notochord vacuoles are lysosome-related organelles that function in axis and spine morphogenesis. J Cell Biol 2013; 200(5):667-79; PMID:23460678; http://dx.doi.org/ 10.1083/jcb.201212095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Chen CF, Chu CY, Chen TH, Lee SJ, Shen CN, Hsiao CD. Establishment of a transgenic zebrafish line for superficial skin ablation and functional validation of apoptosis modulators in vivo. PLoS One 2011; 6(5):e20654; PMID:21655190; http://dx.doi.org/ 10.1371/journal.pone.0020654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Roostalu U, Strähle U. In vivo imaging of molecular interactions at damaged sarcolemma. Dev Cell 2012; 22(3):515-29; PMID:22421042; http://dx.doi.org/ 10.1016/j.devcel.2011.12.008 [DOI] [PubMed] [Google Scholar]


