ABSTRACT
Intrinsically disordered proteins (IDPs) do not have a well-defined and stable 3-dimensional fold. Some IDPs can function as either transient or permanent binders of other proteins and may interact with an array of ligands by adopting different conformations. A novel outer membrane lipoprotein, bacterial interleukin receptor I (BilRI) of the opportunistic oral pathogen Aggregatibacter actinomycetemcomitans binds a key gatekeeper proinflammatory cytokine interleukin (IL)-1β. Because the amino acid sequence of the novel lipoprotein resembles that of fibrinogen binder A of Haemophilus ducreyi, BilRI could have the potential to bind other proteins, such as host matrix proteins. However, from the tested host matrix proteins, BilRI interacted with neither collagen nor fibrinogen. Instead, the recombinant non-lipidated BilRI, which was intrinsically disordered, bound various pro/anti-inflammatory cytokines, such as IL-8, tumor necrosis factor (TNF)-α, interferon (IFN)-γ and IL-10. Moreover, BilRI played a role in the in vitro sensing of IL-1β and IL-8 because low concentrations of cytokines did not decrease the amount of extracellular DNA in the matrix of bilRI− mutant biofilm as they did in the matrix of wild-type biofilm when the biofilms were exposed to recombinant cytokines for 22 hours. BilRI played a role in the internalization of IL-1β in the gingival model system but did not affect either IL-8 or IL-6 uptake. However, bilRI deletion did not entirely prevent IL-1β internalization, and the binding of cytokines to BilRI was relatively weak. Thus, BilRI might sequester cytokines on the surface of A. actinomycetemcomitans to facilitate the internalization process in low local cytokine concentrations.
KEYWORDS: Aggregatibacter actinomycetemcomitans, bacterial cytokine receptor, biofilm matrix composition, intrinsically disordered protein, outer membrane lipoprotein
Introduction
Intrinsically disordered proteins (IDPs) go against the structure-defines-function paradigm given that they lack a well-defined 3-dimensional fold; yet, they are elementary components in a myriad of cellular processes.1 The proportion of IDP increases when moving from simple microorganisms to more complex eukaryotes, suggesting an evolutionary advantage of having flexible proteins that may possess several functions. For instance, the proteome of Escherichia coli has been predicted to contain approximately 15% proteins having more than 30 amino acid disordered segments, whereas in Saccharomyces cerevisiae, the ratio is approximately 50-60%.2 In eukaryotes, many IDPs have roles in signal transduction, where they may bind to multiple ligands with variable affinities.3
The oral opportunistic pathogen Aggregatibacter actinomycetemcomitans can be found from multispecies biofilms in diseased periodontal pockets of patients suffering from aggressive or chronic forms of periodontitis.4-6 Among the diverse changes in the host response to multispecies biofilms, periodontal diseases are characterized by alterations in the levels of various inflammatory cytokines, such as interleukin (IL)-1β, IL-6, and IL-8, and the anti-inflammatory cytokine IL-10.7 The highly leucotoxic JP2 genotype of A. actinomycetemcomitans has been suggested to be an important etiological agent in disease initiation,8 where the inflammatory reaction is caused by inflammophilic dysbiotic multispecies bacterial biofilm whose existence may be favored by the micromilieu in inflammation.9 This biofilm grows attached to the tooth surface and invades between the tooth and gingival tissue toward the junctional epithelium.10 The host tissue, including alveolar bone, is mainly destroyed by the host response to the pathogenic biofilm. A. actinomycetemcomitans may have systemic effects on host health because it has been linked to the etiology of cardiovascular diseases,11,12 endocarditis,13 and brain abscesses.14 Thus, its pathogenic properties may have broader significance to human health than merely oral health.
Human pathogens have several strategies to disturb and evade the host innate immune defense systems. Bacterial cells may grow as protective communities known as biofilms, in which the extracellular matrix provides protection from antibodies, antibiotics and cellular immune defense cells, such as macrophages.15 Adhesive type IV Flp-pili, poly-N-acetylglucosamine (PGA) and extracellular DNA (eDNA) are the main biofilm matrix components of A. actinomycetemcomitans.16,17 Of these, long bundled Flp-pili protein fiber plays the most important role in autoaggregation, nonspecific adherence, biofilm formation and virulence in a rat model.17-20
Various pathogens possess receptors that bind host inflammatory cytokines.21-24 The binding of cytokines to bacteria may change the properties of the bacteria, such as their biofilm formation25,26 and virulence gene expression,21,23,27 and may also manipulate complex host inflammatory reactions, leading to debilitated host defense against colonizing or invading pathogens. We have shown that A. actinomycetemcomitans is able to bind the central proinflammatory cytokine IL-1β 26 and to internalize IL-1β 26,28 and that intracellular IL-1β binds to at least 2 bacterial proteins.26,28 In addition, IL-1β decreases the metabolic activity of A. actinomycetemcomitans biofilms.26 In our recent study, we have identified an outer membrane lipoprotein of A. actinomycetemcomitans, bacterial interleukin receptor I (BilRI),24 which is most likely one of the first-line binders of IL-1β on the extracellular side of the bacterium. Whether this novel outer membrane protein is involved merely in the response of A. actinomycetemcomitans biofilm to IL-1β or whether it could bind host proteins and cytokines other than IL-1β was not known. Thus, the aims of the present study was to resolve the 3-dimensional structure of BilRI, to investigate the host protein- and cytokine-binding capacity of the Pasteurellaceae-specific BilRI and to study the phenotype and response to cytokines of a single-gene-deletion mutant of bilRI.
Our results indicate that BilRI is not a specific receptor of IL-1β in vitro and binds to other inflammatory cytokines, such as IL-8 and IL-10. We also found that BilRI is an IDP, which most likely explains the existence of several ligands. bilRI deletion did not completely prevent cytokine internalization, but it significantly decreased IL-1β uptake and impeded the response of biofilm to low concentrations of IL-1β and IL-8. Because the binding of cytokines to the BilRI was relatively weak, BilRI might function as a non-specific cytokine concentrator on the surface of A. actinomycetemcomitans that facilitates the internalization process, especially in low concentrations of cytokines.
Results
BilRI is an intrinsically disordered protein
The proton (1H) spectrum of BilRI measured at 600 MHz exhibits features typical of a disordered protein, including a collapsed chemical shift dispersion in the amide proton region (8.2 ± 0.3 1H ppm) and the lack of shielded methyl protons, i.e., clustering of methyl protons to so-called random coil shift, 0.7 ppm (Fig. 1A). To confirm these observations, we also performed a 2-dimensional 1H, 15N heteronuclear single-quantum coherence (15N HSQC) experiment at the 800-MHz 1H frequency of BilRI (Fig. 1B). To slow down the chemical exchange of labile amide protons with solvent protons, we measured the 15N HSQC spectrum of BilRI under mildly acidic conditions (pH 5). This spectrum more clearly highlights the same features already visible in the corresponding 1H spectrum, i.e., poor dispersion of amide proton chemical shifts, indicating that BilRI remains disordered in solution and under slightly acidic conditions.29 The amino acid sequence analysis supported this finding, showing high numbers of charged and polar residues and a low number of hydrophobic bulky amino acids (Fig. 1C). Moreover, the BilRI sequence had a low complexity, i.e., biased amino acid composition: it did not have any aromatic amino acids, such as phenylalanine, tyrosine and tryptophan, and 48% of the sequence is made up of 3 residues: alanine, lysine and aspartate (Fig. 1C). All of the above-mentioned amino acid sequence features are typical for IDPs.
Figure 1.
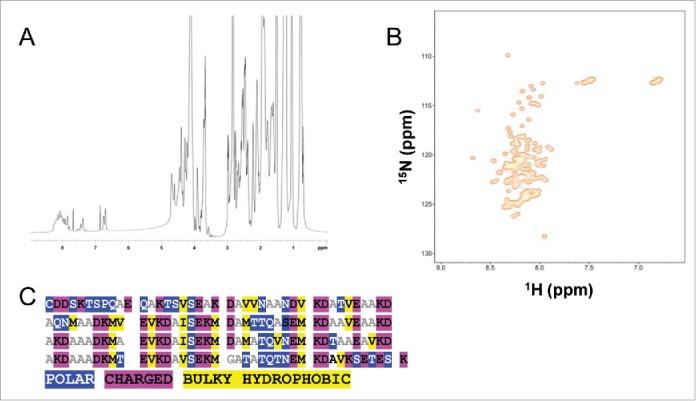
BilRI is an IDP lacking a specific fold without a binding ligand. A) 1H spectrum of BilRI (pH 7.0, 25°C) at 600 MHz. The lack of signal dispersion in the 1H methyl (< 1 ppm) and amide proton (1HN, 7-8 ppm) regions is indicative of the disordered nature of BilRI in solution. B) 1H-15N correlation spectrum (15N-HSQC) of BilRI (pH 5.0, 25°C) at 800 MHz. A two-dimensional 1H-15N correlation map highlights the poorly dispersed 1HN region, confirming observations of the intrinsic disorder of BilRI, based on a 1H spectrum at 600 MHz at pH 7. C) Amino acid sequence analysis confirmed the IDP nature of BilRI. The high proportions of either polar (blue) or charged (magenta) and the low numbers of bulky hydrophobic (yellow) amino acids are typical for IDPs.
Recombinant BilRI binds to various cytokines but not to the host matrix proteins collagen and fibrinogen
A microplate assay showed that recombinant BilRI bound to various cytokines, of which the binding to IL-8 was high compared with the binding of BilRI to the negative control protein bovine serum albumin (BSA; p = 0.008; paired-samples T-test; Fig. 2A). However, the binding to IL-6-coated wells was weak and almost as inefficient as the binding to BSA, which was used as a blocking agent in the assay (Fig. 2A). We decided to use C-tagged recombinant BilRI in our binding assays because binding to IL-1β was originally shown with a similar protein.24 However, we also tested an N-tagged variant of BilRI, which did not show increased binding to IL-1β, IL-8, or IL-6 compared with the C-tagged protein (data not shown). BilRI did not bind to fibrinogen- (Fig. 2B) or to collagen (Fig. 2C)-coated wells. Moreover, BilRI binding to IL-8 was weaker than the fibrinogen-binding of positive control protein clumping factor A (ClfA) of Staphylococcus aureus and the collagen-binding of positive control protein YadA of Yersinia enterocolitica (Fig. 2C).
Figure 2.
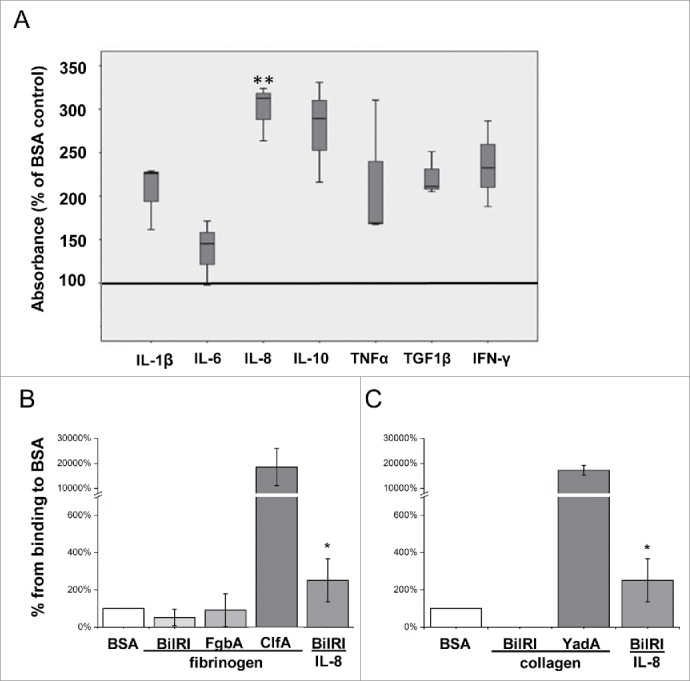
BilRI bound to various human inflammatory cytokines but not to fibrinogen or collagen. A) Recombinant BilRI containing an 8-histidine-long C-terminal tag bound to various recombinant human cytokines in a microplate assay. BSA served as a negative control and was used as a blocking agent in the assays. The bound BilRI was detected with HRP-labeled HisProbe™. The BilRI binding to IL-8 was high compared to the binding to the control protein BSA (**:p = 0.008, paired-samples T-test). B) Recombinant BilRI containing an 8-histidine-long C-terminal tag did not bind to fibrinogen-coated wells in a microplate assay when detected with europium-labeled antibody against the histidine tag. Although the positive control recombinant ClfA of S. aureus bound fibrinogen efficiently, the recombinant FgbA, which has been reported to bind fibrinogen,42 did not show positive binding in the assay. Only BilRI binding to IL-8 showed a statistically significant positive difference compared with the BSA control (p = 0.028, Mann-Whitney U-test) from the test experiments. C) Recombinant BilRI containing an 8-histidine-long C-terminal tag did not bind to collagen-coated wells in the microplate assays when detected with europium-labeled antibody against the histidine tag. The positive control recombinant YadA of Y. enterocolitica bound collagen efficiently. Only BilRI binding IL-8 showed a statistically significant positive difference compared with control BSA (p = 0.028, Mann-Whitney U-test) from the test experiments.
Viable biofilm of wild-type A. actinomycetemcomitans bound IL-8 and IL-6
When wild-type A. actinomycetemcomitans biofilm was co-cultured together with an organotypic gingival mucosa in the absence of antibiotics, the biofilm sequestered both IL-8 and IL-6 (Fig. 3A). However, when the co-culture was performed in the presence of antibiotics, which decreased the viability of the biofilm,28 the immunohistological staining of the biofilm with anti-IL-8 and anti-IL-6 was faint (Fig. 3A). However, the epithelium contained more cytokines in the presence than in the absence of antibiotics (Fig. 3A). In addition, the growth medium contained slightly elevated amounts of IL-6 and IL-8 (Fig. 3B) when antibiotics were used in the biofilm-gingival tissue co-culture, suggesting that the cytokines leaked from the system when not sequestered by the viable biofilm. However, due to inter-sample variance, the difference was not statistically significant. In similar organotypic gingival tissue – biofilm co-cultures with a slightly thinner keratinocyte layer28 A. actinomycetemcomitans cells efficiently internalized IL-1β (Fig. 3C).
Figure 3.
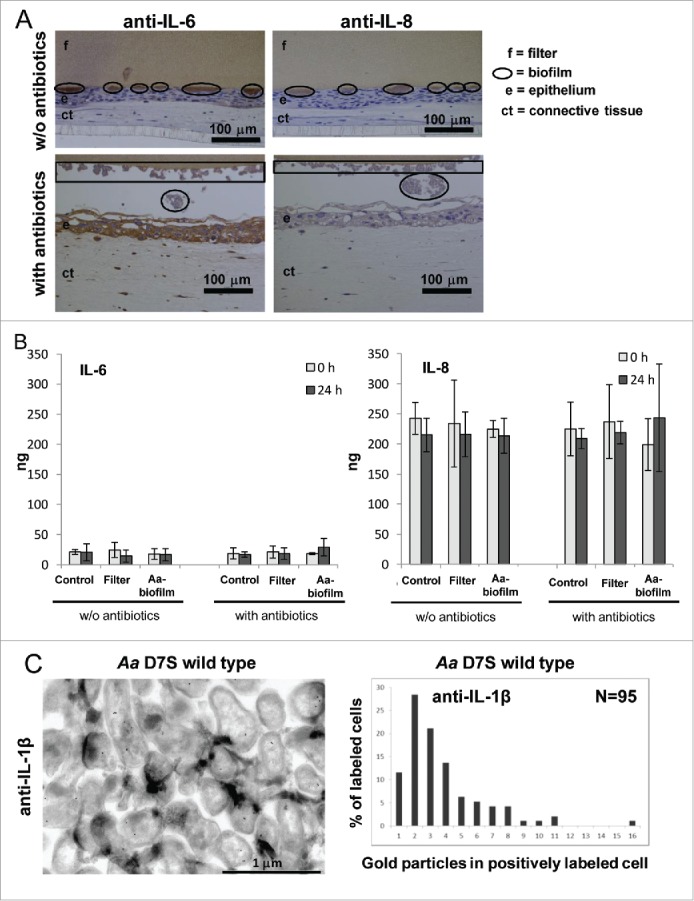
Viable wild-type A. actinomycetemcomitans biofilm bound IL-8 and IL-6, and internalized IL-1β when co-cultured with organotypic gingival mucosa. A) A. actinomycetemcomitans wild-type biofilm bound both IL-8 and IL-6 when co-cultured with organotypic gingival mucosa in the absence of the antibiotics penicillin and streptomycin. In the presence of these antibiotics, the biofilm bound less IL-8 and IL-6 whereas the epithelium contained elevated amounts of IL-8 and IL-6. B) The amount of IL-8 produced was approximately 10 times the amount of IL-6 in the organotypic gingival mucosa tissue culture model. The co-culture system released slightly more IL-8 and IL-6 to the culture medium when stimulated with A. actinomycetemcomitans biofilm in the presence of antibiotics than in the absence of antibiotics. Due to the standard deviation between the samples, the difference was not statistically significant. N = 3. C) In the organotypic gingival tissue culture model, which produced approximately 200 pg of IL-1β to the culture medium during a 24-h incubation with viable A. actinomycetemcomitans wild-type biofilm,28 the uptake efficiency of IL-1β was estimated by counting the number of gold particles in anti-IL-1β-stained immuno-EM samples.
Deletion of stand-alone gene bilRI altered the biofilm matrix composition in rich medium
The prokaryotic operon database (ProOpDB, http://operons.ibt.unam.mx/OperonPredictor)30 predicted that bilRI is a stand-alone gene. When cultured on blood agar plates, the single-gene-deletion mutant of bilRI produced typical colonies with a rough colony morphology (Fig. 4A). Although the bilRI−mutant colonies were slightly more adherent to the agar than the wild-type colonies, cell suspensions31 could be produced similarly from both strains (Fig. 4B). However, BilRI overexpression resulted in a tiny colony size, and only small amounts of bacteria could be harvested from the plates. Nonetheless, an even cell suspension could be attained (Fig. 4B). The bilRI− mutant formed as much biofilm as the wild-type strain in rich medium, whereas the overexpression of BilRI in A. actinomycetemcomitans almost completely disappeared the cell's capacity to form biofilm (p = 0.0003, paired-samples T-test with Bonferroni corrections) (Fig. 4C). In biofilm, the cell morphology of bilRI− mutants did not differ from the morphology of the wild-type strain (Fig. 4D). BilRI overexpression appeared to cause outer membrane lysis (Fig. 4D), explaining the tiny colonies (Fig. 4A) and small cell size (Fig. 4D). In rich medium, the young biofilm, i.e., the biofilm that had not started to detach by releasing cells into the medium,32 of the bilRI− mutant strain contained more total protein in proportion to the biofilm mass than the wild-type A. actinomycetemcomitans strain (p = 0.009; Mann-Whitney U-test) (Fig. 5A). In contrast, the bilRI− mutant biofilm contained less eDNA than the wild-type strain (p = 0.021; Mann-Whitney U-test) (Fig. 5B). Because some outer membrane proteins of A. actinomycetemcomitans or close relative species bind to host proteins, such as collagen and fibrinogen, we studied the binding of the bilRI− mutant to these host proteins. bilRI deletion did not decrease the binding of A. actinomycetemcomitans to either fibrinogen- or collagen-coated wells (Fig. 5C). In contrast, the bilRI− mutant bound collagen slightly more efficiently than the corresponding wild-type strain, but the difference was not statistically significant (p = 0.275; Mann-Whitney U-test, Fig. 5C).
Figure 4.
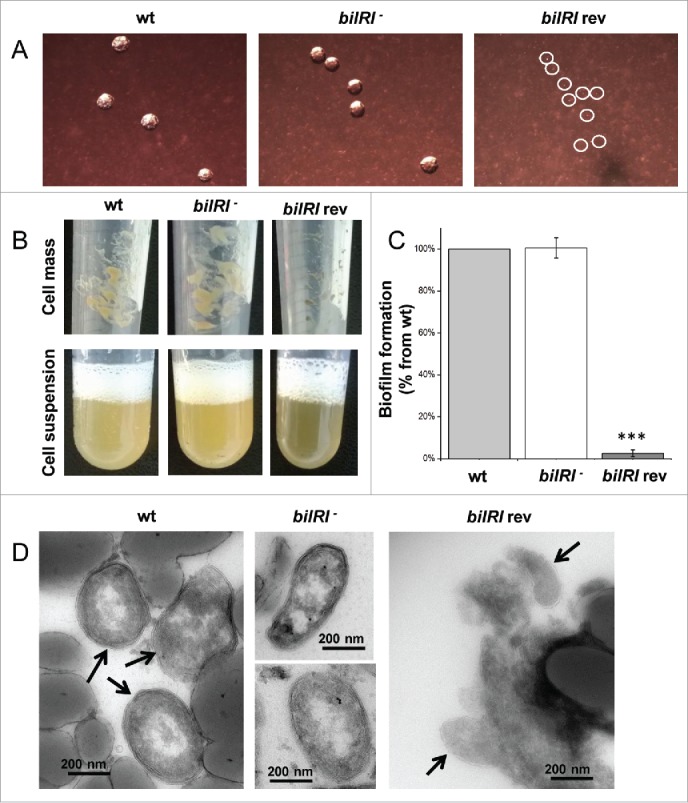
The outer membrane lipoprotein BilRI was not essential for the formation of typical A. actinomycetemcomitans rough-type colonies, biofilm or cell size and shape. BilRI overexpression-induced lysis of the outer membrane resulted in tiny colonies and significantly reduced biofilm amounts. A) On blood agar plates, the bilRI− mutant formed typical rough-type colonies, whereas the BilRI-overexpressing strain (bilRI rev) formed very tiny colonies (circled in white). B) Uniform cell suspensions could be produced similarly with the wild-type and bilRI− mutant strains following the method described by Karched et al.31 Because the BilRI-overexpressing strain bilRI rev grew slowly on agar plates, it was difficult to harvest a sufficient cell mass to obtain a dense cell suspension. C) The bilRI− mutant formed as much biofilm as the wild-type strain after 20-24 hours, as estimated through Crystal violet staining.32 The overexpression of BilRI almost completely eliminated the capacity of the strain (bilRI rev) to form biofilm (***:p = 0.0003, paired-samples T-test with Bonferroni corrections). D) Transmission electron microscopy revealed that the shape and size of the bilRI− mutant cells resembled those of wild-type cells. The overexpression of BilRI (bilRI rev) lysed the bacterial outer membrane, resulting in a smaller cell size. Arrows indicate the A. actinomycetemcomitans cells in images in which other structures, such as the filter disc, are visible.
Figure 5.
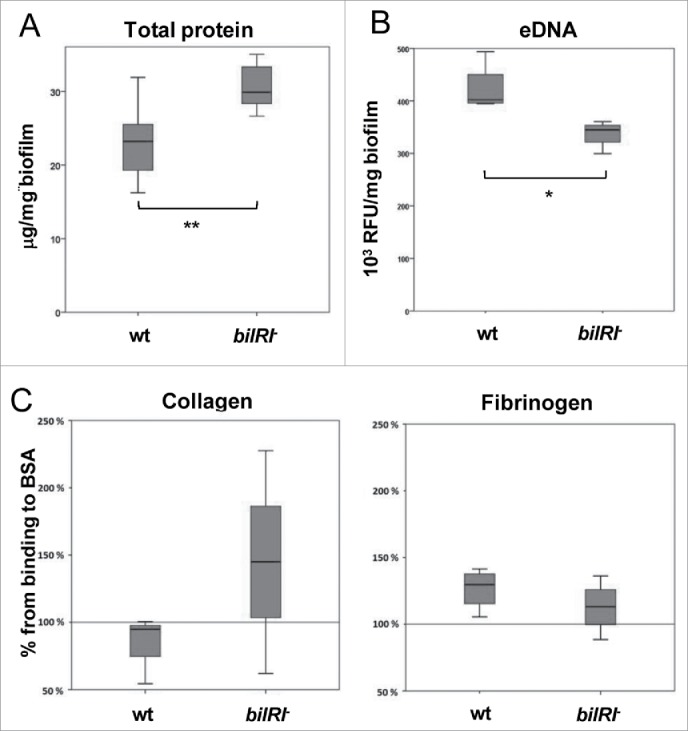
A. actinomycetemcomitans bilRI− mutant cells differed from wild-type cells in the composition of the biofilm and their capacity to bind collagen and fibrinogen. A) The bilRI− mutant biofilm contained more total protein than the corresponding A. actinomycetemcomitans wild-type strain. N = 7, ** p = 0.009 (Mann-Whitney U-test). B) The bilRI− mutant biofilm contained less eDNA than the corresponding A. actinomycetemcomitans wild-type strain. N = 4, * p = 0.021 (Mann-Whitney U-test). C) A. actinomycetemcomitans bilRI− mutant cells did not differ significantly from the wild-type cells in terms of binding to collagen-coated or fibrinogen-coated wells.
BilRI played a role in IL-1β internalization
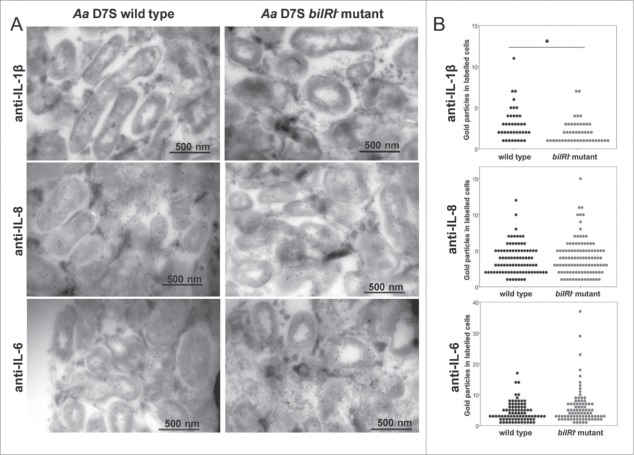
Because the viable biofilm of wild-type A. actinomycetemcomitans bound both IL-8 and IL-6, the uptake of these cytokines and the role of BilRI in their uptake were studied by incubating A. actinomycetemcomitans wild-type and bilRI− biofilms with gingival keratinocyte monolayers. Previously reported IL-1β uptake28 was used as a positive control. The wild-type A. actinomycetemcomitans biofilm cells internalized IL-8, IL-6, and IL-1β (Fig. 6A) in these conditions. When bilRI was deleted from the A. actinomycetemcomitans genome, the amount of IL-1β inside and attached to the biofilm cells, which were co-cultured with human gingival epithelial cells, was significantly lower than for corresponding wild-type cells (p = 0.007; Mann-Whitney U-test, Fig. 6A and 6B). However, the bilRI− mutant cells did not differ from the wild-type cells in their IL-8 and IL-6 uptake efficiencies (p = 0.649 and p = 0.128, respectively; Mann-Whitney U-test, Fig. 6A and B).
Figure 6.
A. actinomycetemcomitans wild-type and bilRI− mutant strains internalized all tested inflammatory cytokines: IL-1β, IL-8 and IL-6. The outer membrane lipoprotein BilRI had a role in the uptake of only IL-1β in the test system. A) Both A. actinomycetemcomitans wild-type and bilRI− mutant biofilm cells internalized IL-1β, IL-8 and IL-6 when incubated for 24 h with human gingival keratinocyte monolayers. Cytokine uptake was studied with anti-cytokine IgG antibodies combined with protein A-gold labeling and transmission electron microscopy. B) Deletion of the bilRI gene decreased only IL-1β uptake (p = 0.007, Mann-Whitney U-test), while IL-8 and IL-6 uptake levels were not affected. The uptake efficiencies were estimated by counting the amounts of gold labeling in the positively stained cells.
Deletion of bilRI abolished biofilm response to IL-1β and IL-8
When exposed to low concentrations of IL-1β and IL-8, the matrix composition of the wild-type biofilm changed, i.e., the amount of eDNA decreased, whereas the amount of PGA, total protein and total biofilm mass remained unchanged (Table 1). The deletion of bilRI rendered the biofilm unresponsive to IL-1β and IL-8, as determined by measuring the composition of the biofilm (Table 1).
Table 1.
Effect of the cytokines IL-1β and IL-8 (22 h incubation) on the amount and composition of pre-formed A. actinomycetemcomitans D7S wild-type (wt) and bilRI− mutant biofilms. The data are shown as the means ± SD from 4 independent experiments. The statistically significant differences (p ≤ 0.05, Mann-Whitney U-test with Bonferroni corrections) between the cytokine-treated and cytokine-untreated biofilms are given in parenthesis.
| Percentage of the control(without cytokines) |
|||||
|---|---|---|---|---|---|
| Strain | Cytokine | Biofilm mass | eDNA | PGA | Total protein |
| D7S wt | IL-1β | 94 ± 7 | 56 ± 16 (0.018) | 89 ± 8 | 86 ± 17 |
| D7S wt | IL-8 | 96 ± 12 | 63 ± 23 (0.028) | 88 ± 15 | 88 ± 19 |
| D7S bilRI− | IL-1β | 92 ± 5 | 106 ± 26 | 94 ± 7 | 89 ± 9 |
| D7S bilRI− | IL-8 | 104 ± 11 | 103 ± 22 | 89 ± 9 | 94 ± 32 |
Discussion
Although IL-1β was the cytokine that was originally used in the identification of BilRI, it was only moderately bound by this bacterial protein compared with the other tested cytokines. Our novel finding that BilRI is an IDP could explain the existence of multiple ligands. The results of the nuclear magnetic resonance (NMR) studies, which indicated the absence of a specific fold, were supported by the amino acid analysis showing high numbers of charged and polar residues and a low number of hydrophobic bulky amino acids, a composition that is typical for IDPs.1 In addition, the BilRI sequence had low complexity: it lacks all aromatic amino acids, such as phenylalanine, tyrosine and tryptophan, and 48% of the sequence is made up of 3 residues: alanine, lysine and aspartate. The unadorned peptides of IDPs are often involved in molecular interactions, in which they may bind the ligand with variable affinities. Thus, an IDP can function as a scavenger/effector protein if it has a strong affinity or as a chaperone/recognition motif if it has a weak affinity for its ligands.33 Our results confirmed that BilRI had relatively weak affinity for the cytokines, suggesting that it might function as a cytokine concentrator in the outer membrane of A. actinomycetemcomitans binding cytokines only temporarily before sending them forward to the next binding motif in the internalization chain. This hypothesis was supported by the observation that the deletion of bilRI did not completely inhibit the internalization of IL-1β but significantly decreased the uptake efficacy.
Various molecules released by the A. actinomycetemcomitans biofilm are known to induce IL-8 and IL-6 production in human whole blood.34 Because IL-8 showed the highest affinity to BilRI, it was selected for further studies to investigate whether it affects the composition of A. actinomycetemcomitans biofilm and is internalized by the biofilm cells in a BilRI-dependent or BilRI-independent manner. A. actinomycetemcomitans responded to BilRI in a manner dependent on low concentrations (10 ng/ml) of IL-1β and IL-8 by decreasing the eDNA amount in biofilm. This was the only observed change in the biofilm composition because the cytokines did not alter either the PGA or total protein amounts. Moreover, the total biofilm mass did not change in response to the cytokines. In general, eDNA is suggested to play an important role in the early stages of biofilm development, enhancing adhesion to the surface and stabilizing the young biofilm (for a review, see ref.35). Although eDNA protects at least young biofilms from antimicrobial agents,36 host defense factors,37 and mechanical stress,38 it may also compromise the bacterial viability by acting as a pathogen-associated molecular pattern (PAMP)39 and boosting the innate immune defense. The observed decrease in the amount of eDNA in response to IL-1β and IL-8 could impede immune defense by reducing the amount of potential PAMPs.
In a gingival epithelial cell co-culture model, the IL-8 and IL-6 uptake efficiencies were not affected by bilRI deletion. This observation was expected in the case of IL-6, which did not bind to BilRI in vitro. The BilRI-independent uptake of IL-8 might be explained by the high concentration of IL-8 in the system. For example, our organotypic gingival tissue culture system produces approximately 200 ng IL-8 in 24 h compared with 200 pg of IL-1β28 during the same time period. The A. actinomycetemcomitans biofilm virtually bathes in IL-8, which may allow efficient IL-8 uptake without a cell surface concentrator. In our test systems, the IL-8 concentration always exceeded that of IL-6. In the in vivo environment of periodontitis-associated biofilm, a similar surplus of IL-8 is observed with approximately one hundred times more IL-8 than IL-6 in gingival crevicular fluid.40
The deletion of bilRI exerted only minor effects on the phenotype of A. actinomycetemcomitans, which were mainly observed as a change in the composition of the biofilm matrix. However, the overexpression of BilRI caused lysis of the outer membrane. In addition, our previous study showed that E. coli cells are more prone to cell lysis when expressing BilRI under a strong promoter.24 Due to the vulnerability of the outer membrane, the expression of outer membrane proteins of Gram-negative bacteria needs to be precisely regulated.41 We decided to use a constitutively expressed strong ltxP promoter instead of the endogenous bilRI promoter, which may be more strictly regulated, to ascertain efficient complementation. Moreover, we were interested in investigating how the overproduction of BilRI affects the phenotype. BilRI was not involved in binding collagen and fibrinogen, although the wild-type A. actinomycetemcomitans cells clearly bound the proteins. Both experiments with the bilRI− mutant and purified BilRI showed similar results. Our findings are partly contradictory to those obtained in previous study conducted by Bauer and co-workers,42 which showed that a similar protein of Haemophilus ducreyi, which was named fibrinogen-binding protein A (FgbA), interacts with human fibrinogen. The incapability of C-tagged BilRI to interact with fibrinogen cannot be explained by the location of the histidine tag because N-tagged BilRI showed similar results (data not shown) and the control protein FgbA, which was N-tagged, could not bind fibrinogen. More recent studies have confirmed that another protein, i.e., ducreyi serum resistance A (DsrA), a trimeric autotransporter, is, in fact, the main binder of fibrinogen in H. ducreyi and that FgbA does not play a central role in fibrinogen binding.43 Our results are in line with those obtained in the more recent later study because we also found that the slightly truncated form of FgbA, which can be found in some strains of H. ducreyi, does not bind to fibrinogen. However, FgbA undoubtedly promotes H. ducreyi virulence; thus, the major functions of FgbA and similar proteins, such as BilRI, are worth studying.
The differential affinity and capacity to uptake various cytokines may provide the pathogen with the means to modulate the host inflammatory response and the cytokine balance. In healthy periodontal tissue, IL-8 forms a concentration gradient with higher concentrations in the coronal parts of the junctional epithelium, near the bacterial biofilm.7 During acute inflammation, neutrophils are the first innate immune cells to enter the site. However, their activity, i.e., the release of reactive oxygen species and proteases, causes severe tissue damage if not limited by regulative actions. IL-6 signaling is known to suppress chemokines, such as IL-8, which attract neutrophils and directly causes neutrophil apoptosis.44 The immune system redirects from innate to acquired immunity by replacing the neutrophils with monocytes and T cells. IL-6 is involved in this process by inducing the production of chemokines that attract monocytes (for a review, see ref. 45), augmenting monocyte differentiation into macrophages,46 recruiting T cells 47 and impeding their apoptosis.48 Periodontitis is characterized by progressive bone loss in tooth supportive tissues, which is associated with a high receptor activator of nuclear factor κ-B (RANK) ligand (RANKL) / osteoprotegerin (OPG) ratio, i.e., RANKL causes bone destruction by binding RANK, which leads to the induction of osteoclast production.49 However, OPG can inhibit osteoclastogenesis by sequestering RANKL.50 Various cytokines, such as IL-1β, IL-6, IL-11, IL-17 and TNF-α, increase the expression of RANKL over OPG (for a review, see ref. 49). IL-6 activates osteoclastogenesis together with soluble IL-6 receptor (sIL-6R) 51 by provoking RANKL expression.52 Thus, high IL-6 concentration in the inflammatory milieu resolves the acute inflammation reaction that is detrimental to the host tissue by enhancing the clearance of neutrophils and moves the balance to acquired immunity by increasing the recruitment of monocytes and T cells.44-48 However, bone homeostasis is skewed in the direction of osteoclastogenesis and bone degradation due to the increased RANKL/OPG ratio.52 By decreasing local IL-6 amounts in inflammation, A. actinomycetemcomitans could decelerate the clearance of acute inflammation and could extend the time of the neutrophil-skewed immune reaction.
In conclusion, the role of intrinsically disordered BilRI is most likely to concentrate small proteins, such as different host cytokines, on the surface of A. actinomycetemcomitans, which facilitates the efficient uptake of cytokines using as yet unknown machinery. The affinity of BilRI to the cytokines is relatively weak when compared with, for example, the binding of Y. enterocolitica YadA to collagen. The weak affinity is most likely needed for the proficient transfer of the cytokine to the next binding protein in the chain of internalization. Because periodontitis is an inflammatory disease caused by multispecies biofilm, cytokine binding and uptake might not benefit only A. actinomycetemcomitans. By binding and internalizing cytokines, A. actinomycetemcomitans could help other species in a periodontal biofilm to persist in an inflammatory environment.53 The uptake of cytokines by opportunistic pathogens may disturb the balance of cytokines with low local concentrations, whereas the effect on cytokines with high local concentrations, such as IL-8, might be only marginal. Moreover, in low cytokine concentrations, the role of BilRI, a potential cytokine concentrator, might be emphasized in facilitating the uptake of cytokines at the surface of A. actinomycetemcomitans.
Materials and methods
Cloning and expression of recombinant proteins: BilRI, FgbA, ClfA, IL-8, YadA
To study the interaction of BilRI with various cytokines, the bilRI gene was cloned into the pET36b expression vector, which inserts an 8-histidine long tag into the C-terminal end (Novagen, Darmstadt, Germany) using the forward primer 5′-ATT CATATG GATGACAGCAAAACTTCACC-3′ and the reverse primer 5′- ATA CTCGAG TTTGCTTTCAGTTTCGC-3′ during PCR. The underlined sequences are the NdeI and XhoI restriction sites, respectively. The bilRI gene was amplified from a previously produced expression vector,24 which contained the gene from D7S. The plasmid was transformed into bacterial cells from the BL21 CodonPlus (DE3)-RIL E. coli protein expression strain (Stratagene, San Diego, CA, USA).
The recombinant BilRI containing amino acids 21-181 was expressed in Terrific broth medium (12 g/L tryptone, 24 g/L yeast extract, 0.4% glycerol, 17 mM KH2PO4, 72 mM K2HPO4) containing 30 µg/mL kanamycin. Protein expression was induced with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) when the OD600nm was 1.2. Cells were grown for 3 h under induction, after which they were harvested by centrifugation (6 400×g, 10 min, 4°C), and cell pellets were stored at −20°C.
To purify the intracellular recombinant protein, 8-10 g of cells were defrosted and suspended to 30 mL in binding buffer (20 mM NaH2PO4/Na2HPO4, 800 mM NaCl, 20 mM imidazole, pH 7.5) including DNAse I (Roche, Mannheim, Germany ), 5 mM MgCl2 and 0.2 mM phenylmethylsulfonyl fluoride (PMSF) protease inhibitor. Cells were sonicated 5×15 s with a 100 Watt MSE ultrasonic disintegrator. Cell debris was removed by centrifugation (48 000×g, 25 min, 4°C), and the clarified supernatant containing the recombinant BilRI protein was loaded in a balanced 5-mL HisTrap HP (GE Healthcare, Uppsala, Sweden) column. The unbound material was washed out with 5% elution buffer (20 mM NaH2PO4/Na2HPO4, 800 mM NaCl, 500 mM Imidazole, pH 7.5), and the His-tagged BilRI was eluted with 50% elution buffer. The eluate was loaded into a size-exclusion chromatography Superdex 200 26/60 (GE Healthcare) column in PBS1 (2.7 mM KCl, 1.8 mM KH2PO4, 140 mM NaCl, and 10 mM Na2HPO4, pH 7.4). The recombinant BilRI does not include any tryptophans or any other aromatic residues; therefore, it is nonvisible at 280 nm. However, the protein could be detected in fractions based on both A220nm readings and Bio-Safe™ Coomassie (Bio-Rad, Hercules, CA, USA)-stained SDS-PAGE (Thermo Fisher Scientific Precise™ 4-20% Tris-Glycine Gels). According to the analysis, BilRI-containing protein fractions were concentrated using an Amicon Ultra-15 Centrifugal Filter Unit with an Ultracel-10 membrane (Millipore, Billerica, MA, USA), and the protein amount in the final concentrate was determined using the method by Lowry et al.54 Protein purity was verified with SDS-PAGE, and the homogeneity was determined by native PAGE (PhastGel Gradient, 8-25, GE Healthcare).
Synthetic DNA with optimized codon usage for E. coli expression was ordered from Eurofins Genomics for FgbA, including the gene for residues 20-105 from H. ducreyi HMC112, the fibrinogen-binding segment of S. aureus strain NCTC 8325 ClfA, the gene fragment for residues 230-542 and the cDNA of the coding residues 23-99 of human IL-8. N-terminal NdeI and C-terminal XhoI restriction sites were added for all 3 synthetic genes. DNA fragments were ligated into the pET15b plasmid (Novagen, Darmstadt, Germany), and DNA sequences were verified with sequencing.
FgbA was expressed, purified and identified by the same method as BilRI because it lacks all aromatic residues. ClfA was expressed and purified similarly to FgbA, except the binding and elution buffers contained 300 mM NaCl. The ClfA concentration was measured on a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA) using an A2800.1% of 0.999.
IL-8 was purified as a mature protein. The N-terminal His-tag was cut by digesting in a HisTrap column with 200 NIH units of thrombin (MP Biomedicals, Santa Ana, CA, USA) at RT overnight. Digested IL-8 was eluted with binding buffer, and the protein was purified from other proteins by size-exclusion chromatography. After concentration with an Amicon Ultra-15 Centrifugal Filter Unit with an Ultracel-10 membrane (Millipore), the protein concentration was determined with A280 nm using an A2800.1%of 0.863. The IL-8 molcular mass was verified with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI TOF MS) (Bruker). Analysis yielded a mass of 8381.99 Da (+H form) (+/-1 Da) for IL-8, whereas the theoretical mass with 2 cysteines was 8381.67 Da, indicating that the recombinant IL-8 had 72 amino acids, suggesting that the endogenous thrombin site of IL-8 was exposed.
The plasmid pHN-1 and the E. coli M15(pREP4) strain (Qiagen, Hilden, Germany) for the collagen-binding fragment of Y. enterocolitica adhesin (YadA) expression were kind gifts from Professor Mikael Skurnik (University of Helsinki, Finland). YadA expression and purification were performed as published by Nummelin et al.55 except that the size-exclusion chromatography buffer was PBS1.
All proteins were deep-frozen with liquid nitrogen and stored at −85°C. All recombinant protein preparations has high purity, as observed in the Coomassie-stained 4-20% Tris-glycine SDS-PAGE gel (Fig. 7).
Figure 7.
The produced recombinant proteins YadA (23 kDa), ClfA (36 kDa), FgbA (12 kDa), IL-8 (9 kDa) and BilRI (18 kDa) were pure, as observed in a Coomassie-stained SDS-PAGE gel. A total amount of 1 μg of each protein was run in the gel.
NMR spectroscopy studies of BilRI structure
NMR spectra were collected at 298 K using either Varian INOVA 600 MHz or INOVA 800 MHz NMR spectrometers (Agilent, Santa Clara, CA, USA), both equipped with cryogenically cooled 1H, 13C, 15N triple-resonance probeheads with z-gradient coils. For 1H NMR spectra, measured at 600 MHz, the recombinant BilRI was diluted in 95%/5% H2O/D2O, 50 mM NaCl, pH 7 buffer in a Shigemi microcell (250 μL). The final BilRI concentration was 4.6 mM. The 1H spectrum was sampled with 20,438 complex points using 64 transients per free induction decay (FID), resulting in an acquisition time of 500 ms in the 1H dimension. The two-dimensional 1H-15N HSQC spectrum of BilRI at pH 5 was measured at the 800 MHz 1H frequency using 128 and 852 complex points in 15N and 1H dimensions, corresponding to acquisition times of 49 ms and 85.2 ms, respectively. A total of 256 transients per FID were used to assure sufficient signal accumulation. The total experimental time was 18 h. Spectra were processed with VnmrJ (Agilent, Santa Clara, CA, USA) and analyzed with Sparky (T. D. Goddard and D. G. Kneller, University of California, San Francisco, CA, USA) software packages.
Cytokine-binding assay for recombinant BilRI
Because BilRI produced unwanted spontaneous dimers when the cysteine at position 20 was included in the recombinant protein, the construct that was used in the cytokine-binding assays contains neither the signal sequence (the first 19 amino acids) nor the C20. Moreover, the recombinant BilRI contained an 8-histidine long tail in the C-terminus to allow detection with His-Probe (Thermo Fisher Scientific).
A total of 100 ng of each cytokine (IL-1β/IL-6/IL-8/IL-10/tumor necrosis factor [TNF]-α/interferon [INF]-γ/transforming growth factor [TGF]-β1) diluted in PBSN buffer (0.05% sodium azide in PBS1,) was incubated in a Nunc MaxiSorp 96-well plate (Affymetrix, Santa Clara, CA, USA) at RT overnight. Wells were washed 3 times with ion-exchanged water, after which the wells were blocked with blocking buffer (0.25% BSA, 0.02% sodium azide in PBS-T) at 37°C for 3 h. The wells were again washed as above, and 400 ng of C-His-tagged BilRI21-181 was added to the wells and incubated at 4°C overnight. The wells were washed 4 times with PBS-T using Delfia Platewash (Perkin Elmer, Turku, Finland). His-Probe-HRP™ (Thermo Fisher Scientific) was diluted to 1:5000, and 50 µl of the dilution was incubated in the wells at RT for 15 min. The wells were washed again 4 times with PBS-T as described above, and detection was performed with 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) diammonium salt (Sigma-Aldrich) in citrate buffer (10 mM sodium citrate and 0.03% H2O2, pH 4.2). The A414nm value was read using a Multiscan Go plate reader (Thermo Fisher Scientific).
Collagen- and fibrinogen-binding assay for recombinant BilRI (EuLISA)
The binding of BilRI to type V collagen and fibrinogen was determined in a microplate assay modified from the method described by Yu et al.56 Type V collagen from human plasma (Sigma-Aldrich) was dissolved in 0.5 M acetic acid, and fibrinogen from human placenta (Sigma-Aldrich) was dissolved in 0.85% NaCl at 37°C with gentle mixing for 5 h and filtered through a 0.2-μm syringe filter. Collagen and fibrinogen were diluted in PBS1, and a total of 1 μg of each was incubated in the wells of a Nunc MaxiSorp 96-well plate (Affymetrix) at 4°C overnight. Equal amounts of BSA (Sigma-Aldrich) and IL-8 (production described above) were used as negative and positive controls, respectively.
Unbound proteins were removed by washing once with PBS1 using a Delfia Platewash (Perkin Elmer). The wells were blocked with 200 μg of BSA in PBS1 at RT for 1-2 h and washed as above. A total of 1 μg of C-His-tagged BilRI21-181 was diluted in Delfia Assay Buffer (Perkin Elmer) and incubated in wells at RT for 1 h. YadA (0.8 μg), FgbA (1.6 μg) and ClfA (0.5 μg) were used as positive collagen or fibrinogen binders. The production of these proteins is described above. The wells were washed 3 times with PBS1 using Delfia Platewash (Perkin Elmer). Then, 25 ng of DELFIA® Eu-N1 Anti-6xHis antibody (Perkin Elmer) in 50 μL of Delfia Assay Buffer was incubated in wells at RT for 1 h. The wells were washed as in the previous step. Detection was performed measuring time-resolved fluorescence using a Victor3 multilabel plate reader (Perkin Elmer) after a 5-min incubation in DELFIA® Enhancement Solution (Perkin Elmer).
Binding of IL-8 and IL-6 by the viable A. actinomycetemcomitans biofilm
A. actinomycetemcomitans biofilms were co-cultured in a gingival mucosa model as described by Paino et al.28 In brief, in the model, human gingival fibroblasts (HGFs)57 and spontaneously immortalized human gingival keratinocytes (HGKs)58 were cultured at an air-liquid interphase to obtain the 3-dimensional tissue organization. First, HGFs (passages 13-18) in Dulbecco's modified Eagle's medium (DMEM; Gibco, Life Technologies, Paisley, UK) were suspended in collagen solution (PureCol®, Advance Biomatrix, AZ, USA), and an aliquot containing 1.5 × 105 fibroblasts was transferred to cell culture inserts and grown for 1 day submerged in Green's medium.59 To obtain the epithelial layer in the top layer of the connective tissue, 4 × 105 HGKs (passage 18-22) were added on top of the fibroblast-collagen layer. The epithelial cells were cultured submerged for 1 day, and the tissue model was then lifted in the air-liquid interface and allowed to mature for 5 days, after which the separately grown A. actinomycetemcomitans biofilm was added on top of the tissue culture model. The biofilms were co-cultured with the tissue models for 24 h. Culture medium was collected before and after the 24 h co-culture and stored at −80°C. In half of the cultures, penicillin (63.4 IU/ml) and streptomycin (63.4 μg/ml) were used in culture media to decrease biofilm viability. The co-cultures were fixed with 10% formalin solution overnight, and the sectioning of paraffin-embedded samples was performed using standard histological techniques.
Before staining with anti-IL-8 and anti-IL-6 antibodies, the specimens were mechanically deparaffinized, and heat-mediated antigen retrieval was conducted in 10 mM citrate buffer (pH 6.0) with microwaving. The staining was performed with Dako TechMate™ 500 Plus Autostainer (Dako, Glostrup, Denmark) using 20 μg/mL of primary polyclonal rabbit IgG against IL-8 (NBP2-16958; Novus Biologicals, Cambridge, UK) and IL-6 (NBP2-16957; Novus Biologicals) and a Dako REAL™ Detection System, Peroxidase/DAB+, Rabbit/Mouse (Code K5001; Dako) as instructed by the producer. The histological samples were imagined with Leica DM RXA light microscope using Leica HC PL APO 20x / 0.70 objective.
The culture media samples collected prior to the co-culture indicating the basal level of cytokine expression, along with the samples collected after 24 h co-culture, were analyzed with IL-8- and IL-6-specific enzyme-linked immunosorbent assay (ELISA) kits (SABiosciences, Qiagen, Germantown, MD, USA). Because the volume of the medium varied slightly between different experiments, the amount of cytokine that was excreted into the medium was calculated as a total amount (ng) leaked into the culture medium in 24 h.
Prediction of the size of the mRNA expressing BilRI
The Prokaryotic Operon Database (ProOpDB, http://operons.ibt.unam.mx/OperonPredictor)30 was used to predict whether bilRI is a stand-alone gene or belongs to an operon. Because A. actinomycetemcomitans strain D7S was not deposited into the database, A. actinomycetemcomitans strain D11S was used. The hypothetical protein D11S_0933 of A. actinomycetemcomitans D11S-1 (GenBank accession number CP001733.1) has an identical amino acid sequence to the BilRI protein of A. actinomycetemcomitans D7S. In addition, the genes surrounding the gene encoding BilRI are similar in both strains. Downstream of bilRI is a gene encoding septum site-determining protein MinC, whereas genes encoding the SixA phosphohistidine phosphatase, phosphoglucosamine mutase and dihydropteroate synthase are found upstream of bilRI.
Markerless bilRI-deletion mutant
A single-gene-deletion mutant of bilRI was produced from A. actinomycetemcomitans strain D7S.60 The strain was recovered from −80°C frozen storage cultures by culturing on modified tryptone soy agar (TSA) plates consisting of 3 % tryptone soy broth (TSB, Lab-M, Lancashire, UK), 0.3 % yeast extract (Lab-M), 1.5 % agar and 5 % heat-inactivated horse serum (HyClone, SH30074.03, Thermo Fisher Scientific) in a candle jar at 37°C for 2.5 d. In addition, 2 types of TSB media were used. TSB1 contained 3 % TSB and 0.6 % yeast extract. TSB2 was additionally supplemented with 0.8% separately autoclaved glucose. Whenever necessary, the cultures were supplemented with the appropriate antibiotics: either 50 µg/mL spectinomycin or 6 µg/mL tetracycline.
The plasmids used for mutant generation were generous gifts from Professor Casey Chen (University of Southern California, Los Angeles, CA, USA). The pLox2-Spe plasmid contained a spe cassette flanked by loxP sites.61 The pAT-Cre plasmid contained the cre recombinase and tet(O) genes.62,63 The plasmids were amplified in Escherichia coli strain TOP10 (Invitrogen).
The gene encoding BilRI (NC_017846.1 AaD7S_02241) was deleted using the Cre-loxP mediated recombination method optimized for A. actinomycetemcomitans.61,62 The primer sequences used for PCR product generation in the target-gene-deletion mutant are listed in Table 2. First, a 2960-bp PCR product containing the bilRI gene was amplified from the genome of A. actinomycetemcomitans D7S using bilRI_nest primers. The PCR product was then used to generate 2 PCR fragments flanking the bilRI gene in both the downstream and upstream directions. The primer pair ycgL_FD/bilRI_RD_BamHI was used to amplify the downstream region, and primer pair phoGlu-R/sixA_FD_SalI was used for the upstream region. The PCR fragments and pLox2-spe-plasmid were digested with BamHI and/or SalI restriction enzymes (FastDigest restriction enzymes, Thermo Fisher Scientific). After fragment isolation, the ligation was completed by incubating the amplicons and the spe-cassette fragment (130 ng each) in the presence of T4 DNA ligase (Thermo Fisher Scientific).
Table 2.
The nucleotide sequences of primers that were used in producing the markerless bilRI deletion mutant of A. actinomycetemcomitans D7S.
| Primer name | Sequence |
|---|---|
| bilRI_nest-F | 5′- GTATGGTGCCTGACTTTCGG-3 |
| bilRI_nest-R | 5′-TTATGGTGGATCACCTTGGT-3′ |
| ycgL-FD | 5′CCAAGGCTGGAAAGCGATATT-3′ |
| bilRI_RD_BamHI | 5′-CTAGGATCCTGAAAGCAAATAAAAAAGCAGTCTA-3′ |
| phoGlu-R | 5′-GCGACCAAGCCTTATTTA −3′ |
| sixA_FD_SalI | 5′- CGT GTC GAC TTA ATA TAG GTC AAA ATT TAT CT −3′ |
The natural transformation was performed according to a previously described method.60,64 In brief, suspensions of plate-grown A. actinomycetemcomitans cells were prepared in TSB1, and the bacterial cell number was estimated according to the method described by Karched et al.31 Then, 2×107 colony-forming units (CFUs) were plated on TSA-plates, and the cells were grown in a candle jar at 37°C for 2 h after mixing the recipient cells with the ligation mix (250 ng of DNA). After culturing for 5 h, the cells were scraped off the plate, resuspended in 150 µL of TSB1 and plated on a spectinomycin-supplemented TSA plate. Colony PCR was used to confirm the presence of a deletion in the bilRI gene site in the A. actinomycetemcomitans D7S genome. Using this method, a loopful of bacteria was suspended in lysis buffer (20 µg/mL proteinase K, 2.5% glycerol in 10 mM Tris-HCl, pH 8.0). Twenty microliters of the resulting suspension were added to a PCR reaction using bilRI_nest-primers, and the correct 3550-bp PCR product was detected. The pAT-Cre plasmid was then transformed into electrocompetent primary bilRI-deletion mutant cells by electroporation (5 ms, 1250 V) using a BTV ECM399 electroporation apparatus (BTX Instrument Division, Harvard Apparatus, Inc., Holliston, MA, USA) to remove the spe-cassette. After culturing the cells in TSB2 for 2 h, the cells were plated on TSA-plates supplemented with tetracycline and grown for a few days until visible colonies were formed. The selected colonies were further plated onto TSA-plates with no antibiotics and with tetracycline and spectinomycin and then grown for 4 d. Colonies sensitive to both antibiotics were considered potential markerless bilRI mutants. Colony PCR using minc_F_1 (5′-CGCGCTATCAACCGACTAAA-3′) and SixA_R_2 primers (5′-TTTATCTCGGTGATGAGC-GC-3′) was used to select products of the correct size (2100 bp), and the products were further verified by sequencing the flanking regions of the bilRI gene in both directions by Eurofins Genomics (Ebersberg, Germany). Moreover, the absence of bilRI in the bilRI− mutant was verified by PCR using genomic DNA as the template and primers 24 that amplify the whole bilRI gene, including the signal sequence.
Restoration of BilRI expression in the bilRI-deletion mutant
Because we did not succeed in restoring the bilRI gene to the markerless bilRI deletion mutant despite many attempts, we decided to restore BilRI expression using an A. actinomycetemcomitans/E. coli shuttle plasmid under a constitutively expressed leucotoxin promoter (ltxP). The bilRI gene was amplified from A. actinomycetemcomitans strain D7S by PCR using the 5′-ATACTCGAGTTTAGGAGTAACGATG-3′ forward primer and the 5′-TTTCTGCAGTTAT-TTGCTTTCAGTT-3′ reverse primer, which contained XhoI and PstI restriction sites, respectively. The bilRI gene was inserted into ltxP from the pVT1296 plasmid65 by ligating the bilRI PCR product into XhoI- and PstI-digested pVT1296. The final ltxP-bilRI construct was moved to the pPK1-based 66 pVT1503 plasmid67 by cutting the pVT1-296-based construct with PstI, blunting the ends with Klenow, cutting ltxP-bilRI from the plasmid with KpnI, and ligating ltxP-bilRI to KpnI- and EcoRV-digested pVT1503 (KanR). The correct insert size was confirmed through KpnI-EcoRI double digestion of the final expression plasmid pVT1503-ltxP-bilRI. The bilRI-containing product of the KpnI-EcoRI-digested expression plasmid pVT1503-ltxP-bilRI was ligated into the KpnI-EcoRI-digested pUC19 plasmid (New England Biolabs, Ipswich, MA, USA) and sequenced (Eurofins Genomics). The expression plasmid pVT1503-ltxP-bilRI was then transformed into a markerless bilRI-deletion mutant through natural transformation as described above. This time, 300 ng of DNA was mixed with cells and supplemented with 1 mM CaCl2 to improve the transformation efficiency.68 After incubation, the transformants were screened on TSA plates supplemented with 30 μg/mL kanamycin to select a BilRI-overexpressing variant containing the pVT1503-based expression plasmid.
Effect of BilRI on biofilm formation
A. actinomycetemcomitans D7S wild-type, bilRI− mutant and BilRI-overexpressing strains were compared to determine the effect of BilRI on biofilm formation, which was measured through crystal violet staining.32 Briefly, A. actinomycetemcomitans D7S wild-type and bilRI− strains were grown on TSA-blood-plates (37 g/L TSA [Lab-M], 3 g/L agar, 5% defibrinated sheep blood) in a candle jar at 37°C for 3 d. A uniform cell suspension was prepared from plate-grown bacteria in TSB2 medium, and the cell density was determined by measuring the optical density.31 The cell suspension was added to the wells of a 48-well microtitre plate such that each well contained 2.5×107 CFUs in a total volume of 0.5 mL. Seven replicates of each strain were prepared. The plate was incubated in a candle jar at 37°C overnight. The medium was removed with suction, 200 μL of Gram-staining reagent (20 mg/mL crystal violet, 8 mg/mL ammonium oxalate, and 20% ethanol) was added to each well, and the samples were incubated at RT for 10 min. The Gram stain was removed with suction, and the wells were washed 7 times with ultrapure water. After 200 μL of 95% ethanol was added to the wells, the plates were incubated at RT for 10 min. The amount of released stain was measured by transferring 100 μL of liquid from each well to a 96-well microtitre plate, and the A620nm value was measured using a Multiscan Go plate reader (Thermo Fisher Scientific).
Effect of BilRI on biofilm composition
Because the bilRI− mutant formed similar amounts of biofilm as the wild-type A. actinomycetemcomitans strain, we further analyzed the biofilm composition of the wild-type and bilRI− mutant strains. However, because the bilRI− mutant in which BilRI expression was restored with a plasmid loses its viability when expressing elevated amounts of the outer membrane protein BilRI (Fig. 4A–4D), its biofilm composition could not be studied. Biofilm cultures were generated as described above with the exception that the biofilms were grown in 50-mL cell culture bottles (Cellstar #690160, Greiner Bio-One, Frickenhausen, Germany) by adding 2.5 × 108 CFUs in a total volume of 5 mL of TSB2 medium. The biofilms were grown in a candle jar at 37°C overnight. Samples were collected from the culture medium and cultured on blood agar plates to ensure that the biofilms were not contaminated. The TSB2 medium was removed, the biofilms were washed 3 times with 10 mL of PBS1, and the biofilm was scraped into 3 mL of PBS1 with an inoculation loop. The samples were divided into 3 1-mL aliquots, the biofilm mass of centrifuged (12,000×g, 15 min) pellets from each sample was weighed, and the amounts of total protein and eDNA were estimated using the methods described below.
For the total protein measurement, the pre-weighed biofilm pellets were suspended in 200 μL of ultrapure water with mild sonication (2×5 s, 5-μm amplitude, 100-Watt MSE ultrasonic disintegrator), and the volume was then doubled by adding sodium dodecyl sulfate (SDS) to a final concentration of 2% in 0.5× PBS1. The samples were boiled for 10 min, the insoluble fraction was separated through a short centrifugation, and the total protein amount in the supernatant was measured using the method described by Lowry et al.54
For eDNA extraction,69 the pre-weighed biofilm pellet was suspended in 0.9% NaCl to obtain 9 mg/mL, and the suspension was homogenized using mild sonication, as described above. The suspension was supplemented with 1× Glyko Buffer 2 (New England Biolabs) and 250 units of PNGase F (New England Biolabs). After the mixture was incubated at 37°C for 30 min, proteinase K (Thermo Fisher Scientific) was added to a final concentration of 5 μg/mL, the samples were incubated at 37°C for 30 min. The samples were filtered through a 0.2-μm polyethersulfone (PES) membrane (VWR) before the amount of eDNA was determined with propidium iodide, as described by Rose et al.70 Briefly, 25 μL of biofilm extract was mixed with an equal volume of 6 μM propidium iodide in a white 96-well plate (Thermo Fisher Scientific). The plate was incubated in the dark at RT for 15 min before the fluorescence was read using a Hidex Sense Microplate reader (Hidex, Turku, Finland) with a 535-nm excitation filter and a 620-nm emission filter.
Binding of bilRI− mutant cells on collagen and fibrinogen
The binding of A. actinomycetemcomitans to type V collagen and fibrinogen was determined using a microplate assay modified from the methods described by Yu et al 56 and Tang and Mintz.71 Collagen and fibrinogen solutions were prepared as in the collagen- and fibrinogen-binding assay for recombinant BilRI. A total of 1 μg of collagen in sodium bicarbonate buffer (16 mM sodium carbonate, 34 mM sodium bicarbonate, and 0.02% sodium azide, pH 9.6) or 25 ng of fibrinogen in PBSN1 (0.05% sodium azide in PBS1) was added to the wells of a Nunc MaxiSorp 96-well plate (Affymetrix). The plate was coated at 4°C overnight. Liquid was removed from the wells by decanting, and the wells were washed 4 times with ion-exchanged water. The wells were blocked with 1 mg of BSA in PBS1 at RT for 2 h, and the wells were then washed as described above. Wild-type A. actinomycetemcomitans and the bilRI− mutant were collected from TSA-blood plates, a uniform bacterial suspension in PBS1 was prepared, and the number of bacterial cells was estimated as described above. One hundred microliters of bacterial suspension (1.25×106, 2.5×106, 5×106 and 1×107 CFUs) were added, and the mixture was incubated in a candle jar at 37°C for 1 h. After the liquid was removed from the wells by suction, the plate was washed 3 times with 200 μL of PBS-T (PBS1 with 0.05% Tween-20). A volume of 100 μL of anti-serotype A antibody 72 (1/1000, diluted in PBS-T supplemented with 0.25% BSA) was added to each well, and the plate was incubated at RT for 1 h. The wells were washed 4 times with PBS-T using a Delfia Platewash (Perkin Elmer). After 100 μL of anti-rabbit IgG–horseradish peroxidase (HRP) antibody (Promega, 1/9000, diluted into PBS-T) was added to each well, the plate was incubated at RT for 1 h. The wells were washed as in the previous step, and detection was conducted as in the investigation of BilRI binding to cytokines but measured at A405nm.
Role of BilRI in the binding and internalization of IL-8 and IL-6 by the biofilm cells
A clinical wild-type A. actinomycetemcomitans strain D7S, the bilRI− mutant strain and the BilRI-overexpressing strain were revived from frozen milk stocks or TSB2 containing 20% glycerol through growth on TSA-blood plates for 4 d. Bacterial suspensions were prepared in TSB2 medium, and the number of bacterial cells was estimated as described above. Then, 2.5 mL of suspension (5×108 CFUs) was added to sterile hydrophilic PES membranes (Supor®-200; diameter of 25 mm; 0.2-μm pore size; Pall Corporation, Ann Arbour, MI, USA) in a 6-well culture plate followed by incubation in a candle jar at 37°C for 24 h. To remove non-adherent bacteria, the membranes were briefly washed twice with 0.85% NaCl prior to a 24-h incubation in RPMI-1640 medium (Sigma-Aldrich) supplemented with 0.6 g/L L-glutamine (Sigma-Aldrich).
In parallel to biofilm formation, spontaneously immortalized HGKs58 were maintained in keratinocyte SFM growth medium (#17005-075, Gibco®, Thermo Fisher Scientific, Paisley, UK) containing the supplement provided by the manufacturer. Briefly, the HGKs (passages 12-16) were grown to confluence in 175-cm2 cell culture flasks with a medium change every 4–5 d. The same day on which the biofilms were incubated with RPMI-1640 medium, the confluent epithelial cells in flasks were reseeded into 6-well plates (4×105) and grown for 24 h. Prior to co-culturing, the biofilms were gently washed with PBS2(10 mM Na2HPO4 and 150 mM NaCl, pH 7.4). Then, the biofilms were placed on top of HGKs, and the co-cultures were incubated at 37°C in 5% CO2 for 24 h.
After the co-culture with gingival epithelial cells, the biofilms were fixed initially in 4 % paraformaldehyde with 2.5 % sucrose in 0.1 M phosphate buffer pH 7.4 at RT for 6 h. Then, the biofilms were moved to 4°C, and an extra 1-h fixation was applied in the same fixative. After the fixation was completed, the co-cultures were stored in 2.3 M sucrose in PBS2 at 4°C. For immuno-electron microscopy (immuno-EM) detection, small spherical samples (with a diameter of 2 mm) were taken from the co-cultures using a biopsy punch (Miltex, Lake Success, NY, USA).
The immuno-EM detection of IL-1β, IL-8 and IL-6 in the spherical biofilm samples was performed as described previously.28 Briefly, the samples were stored in 2.3 M PBS2 at 4°C before freezing in liquid nitrogen and cryosectioning. The sections were incubated in 0.2% gelatin-PBS2 followed by 0.1% glycine-PBS2. The primary antibodies, rabbit anti-IL-1β (NB600-633; Novus Biologicals), rabbit anti-IL-8 (NBP2-16958; Novus Biologicals), and rabbit anti-IL-6 (NBP2-16957), were diluted in 1% BSA-PBS2 and incubated with the samples for 60 min. After washing with 1% BSA-PBS2, the bound antibodies were detected by incubating with protein A-gold complex (10 nm) diluted in 0.1% BSA-PBS2.73 Negative controls were prepared similarly, except primary antibodies were omitted from the protocol. The labeled sections were embedded in methylcellulose and examined with a Philips CM100 transmission electron microscope (FEI Company, Eindhoven, The Netherlands). Two independent repetitions of the experiments were performed, of which the amounts of gold labels in 39-104 labeled cells were counted from 9-18 representative pictures.
Effect of IL-1β and IL-8 on biofilm composition of A. actinomycetemcomitans
Because it was impossible to control the amounts of cytokines in the organotypic mucosa co-culture model, we studied the effects of IL-1β and IL-8 on the composition of the biofilm matrix by exposing A. actinomycetemcomitans biofilms to similar amounts (10 ng/mL) of recombinant cytokines in 50-mL tissue culture bottles or 48-well standard tissue culture-treated plates.
A. actinomycetemcomitans D7S wild-type and bilRI− mutant strains were grown on TSA-blood plates for 4 d. An even cell suspension was prepared in TSB2 medium, and the number of bacterial cells was estimated as described above. The cell suspension was added to 50-mL cell culture bottles (Cellstar #690160, Greiner Bio-One, Frickenhausen, Germany) to obtain a cell density of 1×109 CFUs in 5 mL of TSB2. After a 5-h incubation in a candle jar at 37°C, the medium was discarded, and the attached biofilms were washed with 9 mL of RPMI-1640 medium (Sigma-Aldrich) supplemented with 0.6 g/L L-glutamine (Sigma-Aldrich). Five milliliters of the same medium was added to the culture bottles and supplemented with 10 ng/mL IL-1β or IL-8. One bottle per strain was prepared without cytokines as a control. The biofilms were grown in a candle jar overnight at 37°C. The following morning, the medium was replaced by fresh medium supplemented with 10 ng/mL cytokines when needed. The biofilms were grown for an additional 5 h and then collected as described above. The eDNA and protein amounts in the biofilms were determined from pre-weight cell pellets as described above.
To determine the effect of cytokines on the PGA amount and total biofilm formation, biofilms were prepared using a protocol similar to that described above with the exception that they were grown on a 48-well microtiter plate instead of culture bottles and 3.8×107 CFUs were added to each well. Each sample was prepared in triplicate. After the biofilms were grown for 5 h in TSB2 and for 22 h in RPMI-1640 (supplemented with L-glutamine and cytokines as described above), the biofilms were washed with ultrapure water and stained with Congo red to determine the PGA amount in the biofilms using the method described by Izano et al.16 with some modifications. Briefly, the biofilms were stained with 200 μL of 1% Congo red dye (Sigma-Aldrich) diluted in ultrapure water. The stain was incubated for 2 min, and the wells were washed twice with ultrapure water. The bound dye was solubilized with 200 μL of 50% DMSO (Sigma-Aldrich) at RT for 1 h. The absorbance was measured using a Multiscan GO plate reader (Thermo Fisher Scientific) at 405 nm. To determine the overall biofilm formation, identically prepared biofilms were alternatively stained with Crystal violet stain as described above.
Statistics
The binding of recombinant BilRI to various cytokines was analyzed through related-samples Friedman's 2-way analysis of variance on ranks, and this was followed by an analysis of BilRI binding to IL-8 through a paired-samples T-test (IBM SPSS Statistics 22). Due to the small sample size, which was always less than 10, the differences in the biofilm composition, binding capacities and uptake efficiencies of various cytokines of wild-type and bilRI−mutant strains were analyzed using the nonparametric Mann-Whitney U-test (IBM SPSS Statistics 22). The effects of cytokines on the biofilm amount and composition were analyzed using a Kruskall-Wallis test followed by paired Mann-Whitney U-tests with Bonferroni corrections (IBM SPSS Statistics 22) when needed. Differences were regarded as statistically significant at p < 0.05.
Abbreviations
- BilRI
bacterial interleukin receptor I
- BSA
bovine serum albumin
- CFUs
colony forming units
- ClfA
clumping factor A
- DsrA
Ducreyi serum resistance A
- eDNA
extracellular DNA
- ELISA
enzyme-linked immunosorbent assay
- FgbA
fibrinogen binder A
- FID
free induction decay
- HGF
human gingival fibroblast
- HGK
human gingival keratinocyte
- HSQC
heteronuclear single quantum coherence spectroscopy
- IDP
intrinsically disordered protein
- IL
interleukin
- IFN
interferon
- IPTG
isopropyl β-D-1-thiogalactopyranoside
- ltxP
leucotoxin promoter
- MALDI TOF MS
matrix-assisted laser desorption/ionization time-of-flight mass spectrometry
- NMR
nuclear magnetic resonance
- OPG
osteoprotegerin
- PAMP
pathogen-associated molecular pattern
- PGA
poly-N-acetylglucosamine
- PMSF
phenylmethylsulfonyl fluoride
- RANK
receptor activator of nuclear factor κ-B
- RANKL
RANK ligand
- RT
room temperature
- sIL-6R
soluble IL-6 receptor
- TGF
transforming growth factor
- TNF
tumor necrosis factor
- TSA
tryptone soy agar
- TSB
tryptone soy broth
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
Keith P. Mintz is acknowledged for kindly providing pVT1296 and pVT1503 plasmids. Mrs Katja Sampalahti, Mrs Mariia Valkama and Mrs Marja-Riitta Uola are thanked for their skilful technical assistance in tissue culture and immunohistological staining. MSc Kalle Sipilä is thanked for help in the EuLISA analysis and Ms Nelli Vahvelainen for assistance in analyzing the biofilm composition. The immuno-EM studies were performed at the EM laboratory of Biocenter Oulu, University of Oulu, Finland. The light microscopic imaging was performed at the Cell Imaging Core (Turku Center for Biotechnology, University of Turku and Åbo Akademi University).
Funding
This work was supported by the Academy of Finland grants 265609 and 272960 to RI and 288235 to PP and the Central Foundation of Finnish Cultural Foundation to TA.
References
- [1].Uversky VN. Intrinsically disordered proteins from A to Z. Int J Biochem Cell Biol 2011; 43:1090-103; PMID:21501695; http://dx.doi.org/ 10.1016/j.biocel.2011.04.001 [DOI] [PubMed] [Google Scholar]
- [2].Tompa P, Dosztanyi Z, Simon I. Prevalent structural disorder in E. coli and S. cerevisiae proteomes. J Proteome Res 2006; 5:1996-2000; PMID:16889422; http://dx.doi.org/ 10.1021/pr0600881 [DOI] [PubMed] [Google Scholar]
- [3].Wright PE, Dyson HJ. Intrinsically disordered proteins in cellular signalling and regulation. Nat Rev Mol Cell Biol 2015; 16:18-29; PMID:25531225; http://dx.doi.org/ 10.1038/nrm3920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Zambon JJ. Actinobacillus actinomycetemcomitans in human periodontal disease. J Clin Periodontol 1985; 12:1-20; PMID:3882766; http://dx.doi.org/ 10.1111/j.1600-051X.1985.tb01348.x [DOI] [PubMed] [Google Scholar]
- [5].Haffajee AD, Socransky SS. Microbial etiological agents of destructive periodontal diseases. Periodontol 2000 1994; 5:78-111; PMID:9673164; http://dx.doi.org/ 10.1111/j.1600-0757.1994.tb00020.x [DOI] [PubMed] [Google Scholar]
- [6].Teles RP, Gursky LC, Faveri M, Rosa EA, Teles FR, Feres M, Socransky SS, Haffajee AD. Relationships between subgingival microbiota and GCF biomarkers in generalized aggressive periodontitis. J Clin Periodontol 2010; 37:313-23; PMID:20447254; http://dx.doi.org/ 10.1111/j.1600-051X.2010.01534.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Darveau RP. Periodontitis: A polymicrobial disruption of host homeostasis. Nat Rev Microbiol 2010; 8:481-90; PMID:20514045; http://dx.doi.org/ 10.1038/nrmicro2337 [DOI] [PubMed] [Google Scholar]
- [8].Haubek D, Johansson A. Pathogenicity of the highly leukotoxic JP2 clone of Aggregatibacter actinomycetemcomitans and its geographic dissemination and role in aggressive periodontitis. J Oral Microbiol 2014; 6:23980; http://dx.doi.org/ 10.3402/jom.v6.23980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hajishengallis G. The inflammophilic character of the periodontitis-associated microbiota. Mol Oral Microbiol 2014; 29:248-57; PMID:24976068; http://dx.doi.org/ 10.1111/omi.12065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Bosshardt DD, Lang NP. The junctional epithelium: From health to disease. J Dent Res 2005; 84:9-20; PMID:15615869; http://dx.doi.org/ 10.1177/154405910508400102 [DOI] [PubMed] [Google Scholar]
- [11].Hyvärinen K, Mäntylä P, Buhlin K, Paju S, Nieminen MS, Sinisalo J, Pussinen PJ. A common periodontal pathogen has an adverse association with both acute and stable coronary artery disease. Atherosclerosis 2012; 223:478-84; http://dx.doi.org/ 10.1016/j.atherosclerosis.2012.05.021 [DOI] [PubMed] [Google Scholar]
- [12].Kozarov EV, Dorn BR, Shelburne CE, Dunn WA Jr, Progulske-Fox A. Human atherosclerotic plaque contains viable invasive Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis. Arterioscler Thromb Vasc Biol 2005; 25:e17-8; PMID:15662025; http://dx.doi.org/ 10.1161/01.ATV.0000155018.67835.1a [DOI] [PubMed] [Google Scholar]
- [13].Das M, Badley AD, Cockerill FR, Steckelberg JM, Wilson WR. Infective endocarditis caused by HACEK microorganisms. Annu Rev Med 1997; 48:25-33; PMID:9046942; http://dx.doi.org/ 10.1146/annurev.med.48.1.25 [DOI] [PubMed] [Google Scholar]
- [14].Rahamat-Langendoen JC, van Vonderen MG, Engström LJ, Manson WL, van Winkelhoff AJ, Mooi-Kokenberg EA. Brain abscess associated with Aggregatibacter actinomycetemcomitans: Case report and review of literature. J Clin Periodontol 2011; 38:702-6; PMID:21539594; http://dx.doi.org/ 10.1111/j.1600-051X.2011.01737.x [DOI] [PubMed] [Google Scholar]
- [15].Donlan RM, Costerton JW. Biofilms: Survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev 2002; 15:167-93; PMID:11932229; http://dx.doi.org/ 10.1128/CMR.15.2.167-193.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Izano EA, Sadovskaya I, Wang H, Vinogradov E, Ragunath C, Ramasubbu N, Jabbouri S, Perry MB, Kaplan JB. Poly-N-acetylglucosamine mediates biofilm formation and detergent resistance in Aggregatibacter actinomycetemcomitans. Microb Pathog 2008; 44:52-60; PMID:17851029; http://dx.doi.org/ 10.1016/j.micpath.2007.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Inoue T, Shingaki R, Sogawa N, Sogawa CA, Asaumi J, Kokeguchi S, Fukui K. Biofilm formation by a fimbriae-deficient mutant of Actinobacillus actinomycetemcomitans. Microbiol Immunol 2003; 47:877-81; PMID:14638999; http://dx.doi.org/ 10.1111/j.1348-0421.2003.tb03454.x [DOI] [PubMed] [Google Scholar]
- [18].Kachlany SC, Planet PJ, Desalle R, Fine DH, Figurski DH, Kaplan JB. Flp-1, the first representative of a new pilin gene subfamily, is required for non-specific adherence of Actinobacillus actinomycetemcomitans. Mol Microbiol 2001; 40:542-54; PMID:11359562; http://dx.doi.org/ 10.1046/j.1365-2958.2001.02422.x [DOI] [PubMed] [Google Scholar]
- [19].Schreiner HC, Sinatra K, Kaplan JB, Furgang D, Kachlany SC, Planet PJ, Perez BA, Figurski DH, Fine DH. Tight-adherence genes of Actinobacillus actinomycetemcomitans are required for virulence in a rat model. Proc Natl Acad Sci U S A 2003; 100:7295-300; PMID:12756291; http://dx.doi.org/ 10.1073/pnas.1237223100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Wang Y, Chen C. Mutation analysis of the flp operon in Actinobacillus actinomycetemcomitans. Gene 2005; 351:61-71; PMID:15837433; http://dx.doi.org/ 10.1016/j.gene.2005.02.010 [DOI] [PubMed] [Google Scholar]
- [21].Wu L, Estrada O, Zaborina O, Bains M, Shen L, Kohler JE, Patel N, Musch MW, Chang EB, Fu YX, et al.. Recognition of host immune activation by Pseudomonas aeruginosa. Science 2005; 309:774-7; PMID:16051797; http://dx.doi.org/ 10.1126/science.1112422 [DOI] [PubMed] [Google Scholar]
- [22].Zav'yalov VP, Chernovskaya TV, Navolotskaya EV, Karlyshev AV, MacIntyre S, Vasiliev AM, Abramov VM. Specific high affinity binding of human interleukin 1 β by Caf1A usher protein of Yersinia pestis. FEBS Lett 1995; 371:65-8; PMID:7664886; http://dx.doi.org/ 10.1016/0014-5793(95)00878-D [DOI] [PubMed] [Google Scholar]
- [23].Mahdavi J, Royer PJ, Sjölinder HS, Azimi S, Self T, Stoof J, Wheldon LM, Brännström K, Wilson R, Moreton J, et al.. Pro-inflammatory cytokines can act as intracellular modulators of commensal bacterial virulence. Open Biol 2013; 3:130048; PMID:24107297; http://dx.doi.org/ 10.1098/rsob.130048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Paino A, Ahlstrand T, Nuutila J, Navickaite I, Lahti M, Tuominen H, Välimaa H, Lamminmäki U, Pöllänen MT, Ihalin R. Identification of a novel bacterial outer membrane interleukin-1Beta-binding protein from Aggregatibacter actinomycetemcomitans. PLoS One 2013; 8:e70509; PMID:23936223; http://dx.doi.org/ 10.1371/journal.pone.0070509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].McLaughlin RA, Hoogewerf AJ. Interleukin-1beta-induced growth enhancement of Staphylococcus aureus occurs in biofilm but not planktonic cultures. Microb Pathog 2006; 41:67-79; PMID:16769197; http://dx.doi.org/ 10.1016/j.micpath.2006.04.005 [DOI] [PubMed] [Google Scholar]
- [26].Paino A, Tuominen H, Jääskeläinen M, Alanko J, Nuutila J, Asikainen SE, Pelliniemi LJ, Pöllänen MT, Chen C, Ihalin R. Trimeric form of intracellular ATP synthase subunit β of Aggregatibacter actinomycetemcomitans binds human interleukin-1beta. PLoS One 2011; 6:e18929; PMID:21533109; http://dx.doi.org/ 10.1371/journal.pone.0018929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kanangat S, Postlethwaite A, Cholera S, Williams L, Schaberg D. Modulation of virulence gene expression in Staphylococcus aureus by interleukin-1beta: Novel implications in bacterial pathogenesis. Microbes Infect 2007; 9:408-15; PMID:17307379; http://dx.doi.org/ 10.1016/j.micinf.2006.12.018 [DOI] [PubMed] [Google Scholar]
- [28].Paino A, Lohermaa E, Sormunen R, Tuominen H, Korhonen J, Pöllänen MT, Ihalin R. Interleukin-1beta is internalised by viable Aggregatibacter actinomycetemcomitans biofilm and locates to the outer edges of nucleoids. Cytokine 2012; 60:565-74; PMID:22898394; http://dx.doi.org/ 10.1016/j.cyto.2012.07.024 [DOI] [PubMed] [Google Scholar]
- [29].Hellman M, Tossavainen H, Rappu P, Heino J, Permi P. Characterization of intrinsically disordered prostate associated gene (PAGE5) at single residue resolution by NMR spectroscopy. PLoS One 2011; 6:e26633; PMID:22073178; http://dx.doi.org/ 10.1371/journal.pone.0026633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Taboada B, Ciria R, Martinez-Guerrero CE, Merino E. ProOpDB: Prokaryotic operon DataBase. Nucleic Acids Res 2012; 40:D627-31; PMID:22096236; http://dx.doi.org/ 10.1093/nar/gkr1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Karched M, Paul-Satyaseela M, Asikainen S. A simple viability-maintaining method produces homogenic cell suspensions of autoaggregating wild-type Actinobacillus actinomycetemcomitans. J Microbiol Methods 2007; 68:46-51; PMID:16904783; http://dx.doi.org/ 10.1016/j.mimet.2006.06.004 [DOI] [PubMed] [Google Scholar]
- [32].Kaplan JB, Meyenhofer MF, Fine DH. Biofilm growth and detachment of Actinobacillus actinomycetemcomitans. J Bacteriol 2003; 185:1399-404; PMID:12562811; http://dx.doi.org/ 10.1128/JB.185.4.1399-1404.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Tompa P, Schad E, Tantos A, Kalmar L. Intrinsically disordered proteins: Emerging interaction specialists. Curr Opin Struct Biol 2015; 35:49-59; PMID:26402567; http://dx.doi.org/ 10.1016/j.sbi.2015.08.009 [DOI] [PubMed] [Google Scholar]
- [34].Oscarsson J, Karched M, Thay B, Chen C, Asikainen S. Proinflammatory effect in whole blood by free soluble bacterial components released from planktonic and biofilm cells. BMC Microbiol 2008; 8:206; PMID:19038023; http://dx.doi.org/ 10.1186/1471-2180-8-206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Okshevsky M, Meyer RL. The role of extracellular DNA in the establishment, maintenance and perpetuation of bacterial biofilms. Crit Rev Microbiol 2015; 41:341-52; PMID:24303798; http://dx.doi.org/ 10.3109/1040841X.2013.841639 [DOI] [PubMed] [Google Scholar]
- [36].Chiang WC, Nilsson M, Jensen PO, Hoiby N, Nielsen TE, Givskov M, Tolker-Nielsen T. Extracellular DNA shields against aminoglycosides in pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother 2013; 57:2352-61; PMID:23478967; http://dx.doi.org/ 10.1128/AAC.000-01-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Jones EA, McGillivary G, Bakaletz LO. Extracellular DNA within a nontypeable Haemophilus influenzae-induced biofilm binds human β defensin-3 and reduces its antimicrobial activity. J Innate Immun 2013; 5:24-38; PMID:22922323; http://dx.doi.org/ 10.1159/000339961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Peterson BW, van der Mei HC, Sjollema J, Busscher HJ, Sharma PK. A distinguishable role of eDNA in the viscoelastic relaxation of biofilms. MBio 2013; 4:e00497-13; PMID:24129256; http://dx.doi.org/ 10.1128/mBio.00497-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Bauer S, Kirschning CJ, Hacker H, Redecke V, Hausmann S, Akira S, Wagner H, Lipford GB. Human TLR9 confers responsiveness to bacterial DNA via species-specific CpG motif recognition. Proc Natl Acad Sci U S A 2001; 98:9237-42; PMID:11470918; http://dx.doi.org/ 10.1073/pnas.161.293498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Khalaf H, Lonn J, Bengtsson T. Cytokines and chemokines are differentially expressed in patients with periodontitis: Possible role for TGF-beta1 as a marker for disease progression. Cytokine 2014; 67:29-35; PMID:24680479; http://dx.doi.org/ 10.1016/j.cyto.2014.02.007 [DOI] [PubMed] [Google Scholar]
- [41].Lee SY, Choi JH, Xu Z. Microbial cell-surface display. Trends Biotechnol 2003; 21:45-52; PMID:12480350; http://dx.doi.org/ 10.1016/S0167-7799(02)00006-9 [DOI] [PubMed] [Google Scholar]
- [42].Bauer ME, Townsend CA, Doster RS, Fortney KR, Zwickl BW, Katz BP, Spinola SM, Janowicz DM. A fibrinogen-binding lipoprotein contributes to the virulence of Haemophilus ducreyi in humans. J Infect Dis 2009; 199:684-92; PMID:19199547; http://dx.doi.org/ 10.1086/596656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Fusco WG, Elkins C, Leduc I. Trimeric autotransporter DsrA is a major mediator of fibrinogen binding in Haemophilus ducreyi. Infect Immun 2013; 81:4443-52; PMID:24042118; http://dx.doi.org/ 10.1128/IAI.00743-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Kaplanski G, Marin V, Montero-Julian F, Mantovani A, Farnarier C. IL-6: A regulator of the transition from neutrophil to monocyte recruitment during inflammation. Trends Immunol 2003; 24:25-9; PMID:12495721; http://dx.doi.org/ 10.1016/S1471-4906(02)00013-3 [DOI] [PubMed] [Google Scholar]
- [45].Scheller J, Chalaris A, Schmidt-Arras D, Rose-John S. The pro- and anti-inflammatory properties of the cytokine interleukin-6. Biochim Biophys Acta 2011; 1813:878-88; PMID:21296109; http://dx.doi.org/ 10.1016/j.bbamcr.2011.01.034 [DOI] [PubMed] [Google Scholar]
- [46].Chomarat P, Banchereau J, Davoust J, Palucka AK. IL-6 switches the differentiation of monocytes from dendritic cells to macrophages. Nat Immunol 2000; 1:510-4; PMID:11101873; http://dx.doi.org/ 10.1038/82763 [DOI] [PubMed] [Google Scholar]
- [47].McLoughlin RM, Jenkins BJ, Grail D, Williams AS, Fielding CA, Parker CR, Ernst M, Topley N, Jones SA. IL-6 trans-signaling via STAT3 directs T cell infiltration in acute inflammation. Proc Natl Acad Sci U S A 2005; 102:9589-94; PMID:15976028; http://dx.doi.org/ 10.1073/pnas.0501794102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Curnow SJ, Scheel-Toellner D, Jenkinson W, Raza K, Durrani OM, Faint JM, Rauz S, Wloka K, Pilling D, Rose-John S, et al.. Inhibition of T cell apoptosis in the aqueous humor of patients with uveitis by IL-6/soluble IL-6 receptor trans-signaling. J Immunol 2004; 173:5290-7; PMID:15470075; http://dx.doi.org/ 10.4049/jimmunol.173.8.5290 [DOI] [PubMed] [Google Scholar]
- [49].Cochran DL. Inflammation and bone loss in periodontal disease. J Periodontol 2008; 79:1569-76; PMID:18673012; http://dx.doi.org/ 10.1902/jop.2008.080233 [DOI] [PubMed] [Google Scholar]
- [50].Yasuda H, Shima N, Nakagawa N, Yamaguchi K, Kinosaki M, Mochizuki S, Tomoyasu A, Yano K, Goto M, Murakami A, et al.. Osteoclast differentiation factor is a ligand for osteoprotegerin/osteoclastogenesis-inhibitory factor and is identical to TRANCE/RANKL. Proc Natl Acad Sci U S A 1998; 95:3597-602; PMID:9520411; http://dx.doi.org/ 10.1073/pnas.95.7.3597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Tamura T, Udagawa N, Takahashi N, Miyaura C, Tanaka S, Yamada Y, Koishihara Y, Ohsugi Y, Kumaki K, Taga T. Soluble interleukin-6 receptor triggers osteoclast formation by interleukin 6. Proc Natl Acad Sci U S A 1993; 90:11924-8; PMID:8265649; http://dx.doi.org/ 10.1073/pnas.90.24.11924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Hashizume M, Hayakawa N, Mihara M. IL-6 trans-signalling directly induces RANKL on fibroblast-like synovial cells and is involved in RANKL induction by TNF-α and IL-17. Rheumatology (Oxford) 2008; 47:1635-40; PMID:18786965; http://dx.doi.org/ 10.1093/rheumatology/ken363 [DOI] [PubMed] [Google Scholar]
- [53].Hajishengallis G. Periodontitis: From microbial immune subversion to systemic inflammation. Nat Rev Immunol 2015; 15:30-44; PMID:25534621; http://dx.doi.org/ 10.1038/nri3785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the folin phenol reagent. J Biol Chem 1951; 193:265-75; PMID:14907713 [PubMed] [Google Scholar]
- [55].Nummelin H, El Tahir Y, Ollikka P, Skurnik M, Goldman A. Expression, purification and crystallization of a collagen-binding fragment of yersinia adhesin YadA. Acta Crystallogr D Biol Crystallogr 2002; 58:1042-4; PMID:12037311; http://dx.doi.org/ 10.1107/S0907444902005231 [DOI] [PubMed] [Google Scholar]
- [56].Yu C, Ruiz T, Lenox C, Mintz KP. Functional mapping of an oligomeric autotransporter adhesin of Aggregatibacter actinomycetemcomitans. J Bacteriol 2008; 190:3098-109; PMID:18310342; http://dx.doi.org/ 10.1128/JB.01709-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Oksanen J, Hormia M. An organotypic in vitro model that mimics the dento-epithelial junction. J Periodontol 2002; 73:86-93; PMID:11846204; http://dx.doi.org/ 10.1902/jop.2002.73.1.86 [DOI] [PubMed] [Google Scholar]
- [58].Mäkelä M, Salo T, Larjava H. MMP-9 from TNF α-stimulated keratinocytes binds to cell membranes and type I collagen: A cause for extended matrix degradation in inflammation? Biochem Biophys Res Commun 1998; 253:325-35; http://dx.doi.org/ 10.1006/bbrc.1998.9641 [DOI] [PubMed] [Google Scholar]
- [59].Green H, Fuchs E, Watt F. Differentiated structural components of the keratinocyte. Cold Spring Harb Symp Quant Biol 1982; 46 Pt 1:293-301; PMID:6179694; http://dx.doi.org/ 10.1101/SQB.1982.046.01.031 [DOI] [PubMed] [Google Scholar]
- [60].Wang Y, Goodman SD, Redfield RJ, Chen C. Natural transformation and DNA uptake signal sequences in Actinobacillus actinomycetemcomitans. J Bacteriol 2002; 184:3442-9; PMID:12057937; http://dx.doi.org/ 10.1128/JB.184.13.3442-3449.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Cheng YA, Jee J, Hsu G, Huang Y, Chen C, Lin CP. A markerless protocol for genetic analysis of Aggregatibacter actinomycetemcomitans. J Formos Med Assoc 2014; 113:114-23; PMID:24530245; http://dx.doi.org/ 10.1016/j.jfma.2012.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Fujise O, Wang Y, Chen W, Chen C. Adherence of Aggregatibacter actinomycetemcomitans via serotype-specific polysaccharide antigens in lipopolysaccharides. Oral Microbiol Immunol 2008; 23:226-33; PMID:18402609; http://dx.doi.org/ 10.1111/j.1399-302X.2007.00416.x [DOI] [PubMed] [Google Scholar]
- [63].Sternberg N, Sauer B, Hoess R, Abremski K. Bacteriophage P1 cre gene and its regulatory region. evidence for multiple promoters and for regulation by DNA methylation. J Mol Biol 1986; 187:197-212; PMID:3486297; http://dx.doi.org/ 10.1016/0022-2836(86)90228-7 [DOI] [PubMed] [Google Scholar]
- [64].Wang Y, Shi W, Chen W, Chen C. Type IV pilus gene homologs pilABCD are required for natural transformation in Actinobacillus actinomycetemcomitans. Gene 2003; 312:249-55; PMID:12909361 http://dx.doi.org/ 10.1016/S0378-1119(03)00620-6 [DOI] [PubMed] [Google Scholar]
- [65].Lippmann JE, Froeliger EH, Fives-Taylor PM. Use of the Actinobacillus actinomycetemcomitans leukotoxin promoter to drive expression of the green fluorescent protein in an oral pathogen. Oral Microbiol Immunol 1999; 14:321-5; PMID:10551160; http://dx.doi.org/ 10.1034/j.1399-302X.1999.140509.x [DOI] [PubMed] [Google Scholar]
- [66].Sreenivasan PK, Fives-Taylor P. Isolation and characterization of deletion derivatives of pDL282, an Actinobacillus actinomycetemcomitans/Escherichia coli shuttle plasmid. Plasmid 1994; 31:207-14; PMID:8029328; http://dx.doi.org/ 10.1006/plas.1994.1022 [DOI] [PubMed] [Google Scholar]
- [67].Mintz KP, Moskovitz J, Wu H, Fives-Taylor PM. Peptide methionine sulfoxide reductase (MsrA) is not a major virulence determinant for the oral pathogen Actinobacillus actinomycetemcomitans. Microbiol 2002; 148:3695-703; PMID:12427959; http://dx.doi.org/ 10.1099/00221287-148-11-3695 [DOI] [PubMed] [Google Scholar]
- [68].Hisano K, Fujise O, Miura M, Hamachi T, Matsuzaki E, Nishimura F. The pga gene cluster in Aggregatibacter actinomycetemcomitans is necessary for the development of natural competence in Ca(2+) -promoted biofilms. Mol Oral Microbiol 2014; 29:79-89; PMID:24450419; http://dx.doi.org/ 10.1111/omi.12046 [DOI] [PubMed] [Google Scholar]
- [69].Wu J, Xi C. Evaluation of different methods for extracting extracellular DNA from the biofilm matrix. Appl Environ Microbiol 2009; 75:5390-5; PMID:19561191; http://dx.doi.org/ 10.1128/AEM.00400-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Rose SJ, Babrak LM, Bermudez LE. Mycobacterium avium possesses extracellular DNA that contributes to biofilm formation, structural integrity, and tolerance to antibiotics. PLoS One 2015; 10:e0128772; PMID:26010725; http://dx.doi.org/ 10.1371/journal.pone.0128772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Tang G, Mintz KP. Glycosylation of the collagen adhesin EmaA of Aggregatibacter actinomycetemcomitans is dependent upon the lipopolysaccharide biosynthetic pathway. J Bacteriol 2010; 192:1395-404; PMID:20061477; http://dx.doi.org/ 10.1128/JB.01453-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Saarela M, Asikainen S, Alaluusua S, Pyhälä L, Lai CH, Jousimies-Somer H. Frequency and stability of mono- or poly-infection by Actinobacillus actinomycetemcomitans serotypes a, b, c, d or e. Oral Microbiol Immunol 1992; 7:277-9; PMID:1494451; http://dx.doi.org/ 10.1111/j.1399-302X.1992.tb00588.x [DOI] [PubMed] [Google Scholar]
- [73].Slot JW, Geuze HJ. A new method of preparing gold probes for multiple-labeling cytochemistry. Eur J Cell Biol 1985; 38:87-93; PMID:4029177 [PubMed] [Google Scholar]