ABSTRACT
MARK2/Par1b/EMK1, a serine/threonine kinase, is required for correct apical/basolateral membrane polarization in epithelial cells. However, the specific substrates mediating MARK2 action are less well understood. We have now found that MARK2 phosphorylates Rab11-FIP1B/C at serine 234 in a consensus site similar to that previously identified in Rab11-FIP2. In MDCK cells undergoing repolarization after a calcium switch, antibodies specific for pS234-Rab11-FIP1 or pS227-Rab11-FIP2 demonstrate that the spatial and temporal activation of Rab11-FIP1 phosphorylation is distinct from that for Rab11-FIP2. Phosphorylation of Rab11-FIP1 persists through calcium switch and remains high after polarity has been reestablished whereas FIP2 phosphorylation is highest early in reestablishment of polarity but significantly reduced once polarity has been re-established. MARK2 colocalized with FIP1B/C/D and p(S234)-FIP1 in vivo. Overexpression of GFP-Rab11-FIP1C wildtype or non-phosphorylatable GFP-Rab11-FIP1C(S234A) induced two significant phenotypes following calcium switch. Overexpression of FIP1C wildtype and FIP1C(S234A) caused a psuedo-stratification of cells in early time points following calcium switch. At later time points most prominently observed in cells expressing FIP1C(S234A) a significant lateral lumen phenotype was observed, where F-actin-rich lateral lumens appeared demarcated by a ring of ZO1 and also containing ezrin, syntaxin 3 and podocalyxin. In contrast, p120 and E-Cadherin were excluded from the new apical surface at the lateral lumens and now localized to the new lateral surface oriented toward the media. GFP-FIP1C(S234A) localized to membranes deep to the lateral lumens, and immunostaining demonstrated the reorientation of the centrosome and the Golgi apparatus toward the lateral lumen. These results suggest that both Rab11-FIP1B/C and Rab11-FIP2 serve as critical substrates mediating aspects of MARK2 regulation of epithelial polarity.
KEYWORDS: epithelial cell polarity, lateral lumen, MARK2, Rab11-FIP1
Introduction
PAR-1 is a serine/threonine kinase that has a conserved role in cell polarity in Caenorhabditis elegans (Par-1),1 Yeast (Kin1),2 Drosophila3 and mammals (MARK2/EMK1).4,5 Originally identified in C. elegans,6 PAR-1, or MARK2 in humans, is one of 6 partitioning genes involved in asymmetric cell division, potentially through the influence of Par-1b on mitotic spindle orientation.7 In Drosophila oocytes, Par-1 is required for anterior-posterior polarity and axis formation.3,8 Furthermore, PAR-1 functions to regulate the plus ends of microtubules by capping them at the basal cortex in flies.9
In mammals, Par1b or MAP/Microtubule Affinity-Regulating Kinase 2 (MARK2), localizes to cell membranes and controls microtubule stability through phosphorylation of microtubule-associated proteins.4,10 Studies in the brain have shown that MARK2 is required for neurite outgrowth, cortical neuronal migration, directed neuroblast migration to the olfactory bulb and axon formation and transport.11,12,13–17 In Madin Darby Canine Kidney cells (MDCK), the Müsch laboratory demonstrated that MARK2 (EMK1) regulated the polarity axis leading to lateral lumen formation and apical targeting pathways independent of the polarity axis.5 Knockdown of MARK2 in Madin Darby Canine Kidney (MDCK) cells elicited a reduction in apical surface proteins and prevented lateral lumen formation in a collagen overlay assay.5 Overexpression of MARK2 caused a reorganization of microtubules and reorientation of MDCK cell's apical surface to the lateral membrane in collagen overlay assays.5,18
MARK2 phosphorylates at least one apical trafficking protein, Rab11 family interacting protein 2 (Rab11-FIP2).19 Previous investigations have identified Rab11-FIP2 as an interacting partner for both Rab11 family members and myosin Vb (MYO5B).20,21 Rab11a is associated with subapical recycling vesicles in epithelial cells.22,23 Rab11-FIP2 interacting with Rab11 and Myosin Vb is important for plasma membrane recycling, regulating trafficking at multiple steps during transcytosis and apical recycling.20,24,25 Phosphorylation of Rab11-FIP2 by MARK2 at serine 227 is upregulated early in junction re-assembly following readdition of extracellular calcium after an overnight incubation in low calcium medium (calcium switch), and overexpression of a phospho-mimetic (S227E) form of Rab11-FIP2 in MDCK cells causes a multi-lumen phenotype in 3-dimenstional cyst culture.26 Interestingly, pS227-Rab11-FIP2 does not associate with Rab11 or Myosin Vb.26 Thus, Rab11-FIP2, when phosphorylated by MARK2, is involved in cellular polarization and lumen formation independent of its Rab11-dependent trafficking roles.
The Rab11-FIP1 (FIP1) gene undergoes alternative splicing to form at least 3 distinct protein products: FIP1A, FIP1B, and FIP1C.27 FIP1B contains all 6 exons from the FIP1 gene. FIP1C lacks exon 4, the largest exon, making FIP1C roughly half the size of FIP1B. Rab11-FIP1A lacks an amino-terminal C2-domain and 2 other domains that are present in FIP1B and FIP1C. FIP1B is expressed ubiquitously, however, FIP1A and FIP1C have differential tissue expression.27 While little is known about FIP1A and FIP1B function, FIP1C (also known as Rab Coupling Protein (RCP)) plays an important role in membrane recycling trafficking.28 FIP1C has primarily been identified as a membrane trafficking protein.27,29 However, it has been implicated in regulation of many different pathways including endocytic sorting,29–31 integrin trafficking,32–35 mitochondrial remodeling36 and actin assembly36 and can also function as either an oncogene or a tumor suppressor.37,38 Importantly, studies in non-polarized cells have demonstrated that FIP1A, FIP1B and FIP1C define distinct domains within the dynamic tubular recycling system.39,40
Through sequence analysis, we have now identified a MARK2 consensus sequence, similar to that found in Rab11-FIP2, in Rab11-FIP1B and Rab11-FIP1C.19 In order to analyze the role of MARK2 phosphorylation on FIP1B/C, we developed a phosphorylation-specific antibody against phospho-serine-S234-FIP1 (pS234-FIP1). Here we show that phosphorylated Rab11-FIP1 is differentially regulated both spatially and temporally in comparison with MARK2 phosphorylated pS227-Rab11-FIP2. Levels for phosphorylated Rab11-FIP1 remained high throughout calcium switch and did not cycle up and down, as seen with Rab11-FIP2.26 Overexpression of a non-phosphorylatable mutant of Rab11-FIP1C, Rab11-FIP1C(S234A), promoted the reorganization of cell polarity following calcium switch with formation of lateral lumen structures showing apical membrane markers and the reorganization of intracellular organelles orienting toward this neo-apical surface. Thus, overexpression of the non-phosphorylatable mutant FIP1C(S234A) promotes a reorganization of the polarity axis in MDCK cells, similar to that seen with overexpression of MARK2.5,41 These results suggest that coordinated phosphorylation of both Rab11-FIP1B/C and Rab11-FIP2 by MARK2 regulates the establishment of epithelial polarity.
Results
In previous studies we documented a consensus sequence for MARK2 phosphorylation in FIP2 of SMSXL.25 Through sequence analysis of the Rab11-FIPs, FIP1B and FIP1C were identified to contain the same MARK2 phosphorylation consensus sequence as FIP2 (Fig. 1A). Using in vitro phosphorylation assays with purified recombinant human FIP1C protein and radiolabeled [γ−32P]-ATP, MARK2 did phosphorylate recombinant FIP1C, and mutation of serine 234 to alanine abolished phosphorylation (Fig. 1B).
Figure 1.
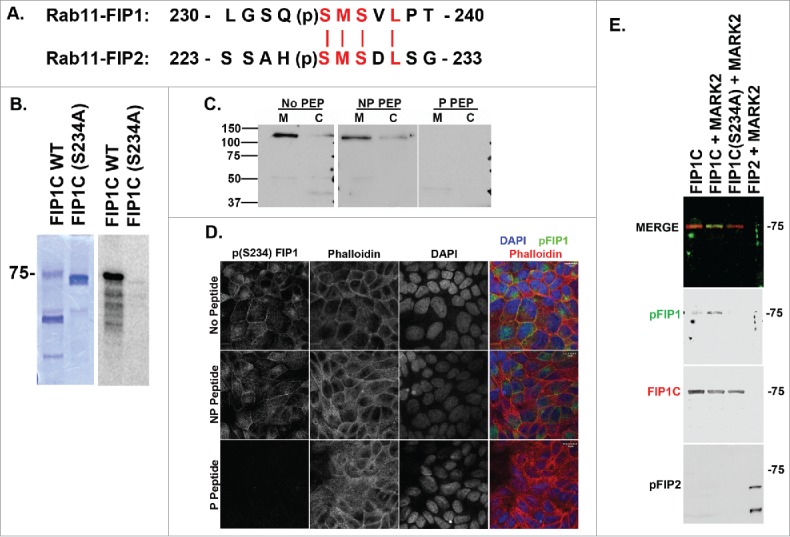
Rab11-FIP1C is phosphorylated by MARK2. A. Protein alignment of Rab11-FIP1 and Rab11-FIP2 around the predicted MARK2 phosphorylation sequence. FIP1 is predicted to be phosphorylated on S234 and FIP2 is phosphorylated on S227. B. In vitro phosphorylation of FIP1C. Coomassie blue staining of recombinant Rab11-FIP1C and Rab11-FIP1C(S234A) preparations are shown at right with autoradiograph of [32P]-incorporation at left. Purified FIP1C, but not FIP1C(S234A), was phosphorylated by MARK2. C. A phospho-specific antibody against pS234-FIP1 identified a 130kDa band on western blot in MDCK (M) and Caco (C) cells consistent with the molecular size of FIP1B. Labeling was blocked by phospho-peptide (P PEP), but not non-phosphorylated peptide (NP PEP). Molecular size markers (kDa) are indicated at the left. D. Immunostaining for pS234-FIP1C showed staining at junctions and in the subapical regions of MDCK cells. Staining for pS234-FIP1 was abolished by incubations with phosphorylated peptide, but not with the non-phosphorylated peptide. E. In vitro phosphorylation assay using purified recombinant FIP1C, FIP1C(S234A) and Rab11-FIP2. Western blot analysis utilizing the pS234-FIP1 antibody detected Rab11-FIP1C phosphorylated by MARK2 (blots were co-stained with antibody against FIP1C to demonstrate equal loading), but no phosphorylation was detected in S234A FIP1C mutant or in MARK2 phosphorylated Rab11-FIP2. In a duplicate blot, the pS227A-Rab11-FIP2 antibody detected MARK2 phosphorylated Rab11-FIP2, but not FIP1 phosphorylated by MARK2. Note that two bands are observed for Rab11-FIP2 because of breakdown of the recombinant FIP2 to a smaller fragment that also is phosphorylated. Position of the 75 kDa molecular weight marker is indicated at the right.
To analyze further the phosphorylation of FIP1 by MARK2, a phosphorylation-specific antibody was generated against (pS234)-FIP1B/C and analyzed for specificity using both T23-MDCK cells as well as CaCo2-BBE cells (Fig. 1C). Based on molecular size, the major endogenous phosphorylated species in MDCK cells was FIP1B. However, no endogenous FIP1C phosphorylation was detected (predicted size of FIP1C is 75kD). Fig. 1D shows complete peptide blocking of the (pS234) antibody with the phosphorylated antigen peptide, but not a non-phosphorylated peptide in lysates of polarized MDCK cells grown on filters. The (pS234) antibody was specific for phosphorylated FIP1 and did not detect (pS227)-FIP2 (Fig. 1E). Similarly, our previously characterized (pS227)-FIP2 antibody did not detect (pS234)-FIP1 (Fig. 1E). These two antibodies therefore showed specificity in vivo and in vitro.
We have previously demonstrated that FIP2 is phosphorylated on serine 227 during re-establishment of polarity in MDCK cells after calcium switch.26 To compare (pS227)-FIP2 to (pS234)-FIP1 during epithelial cell polarization, a calcium switch assay was used to assess either de novo junction formation or junction reformation during cell polarization.42 Polarized MDCK cells cultured in low calcium overnight undergo junction disassembly, but not complete degradation of junctional components, and upon calcium re-addition, junctions reform within 2 to 4 hours.43 Cells were fixed in methanol because the anti-phospho-S227-Rab11-FIP2 antibody performs best for staining MDCK cells after alcohol fixation.26 Following re-addition of calcium to the media, the phosphorylated Rab11-FIP proteins showed very different patterns of localization and abundance during repolarization (Fig. 2). As previously reported, FIP2 phosphorylation was upregulated 0–30 minutes after calcium re-addition and persisted until 2 hours after calcium addition (Fig. 2B), whereas phospho-S234-FIP1 staining was elevated after overnight incubation in low calcium and were maintained following re-addition of calcium and remained high 4–8 hours after calcium switch (Fig. 2A). Western blot demonstrated an increase in FIP1B phosphorylation after overnight low calcium incubation with maintenance of phosphorylation after re-addition of calcium (Fig. S1). The amount of phosphorylated FIP1 in non-switched stable monolayer cells was also higher, when compared with phosphorylated FIP2. These results suggest that phosphorylation of FIP1 and FIP2 by MARK2 occurs along differentiable spatial and temporal pathways.
Figure 2.
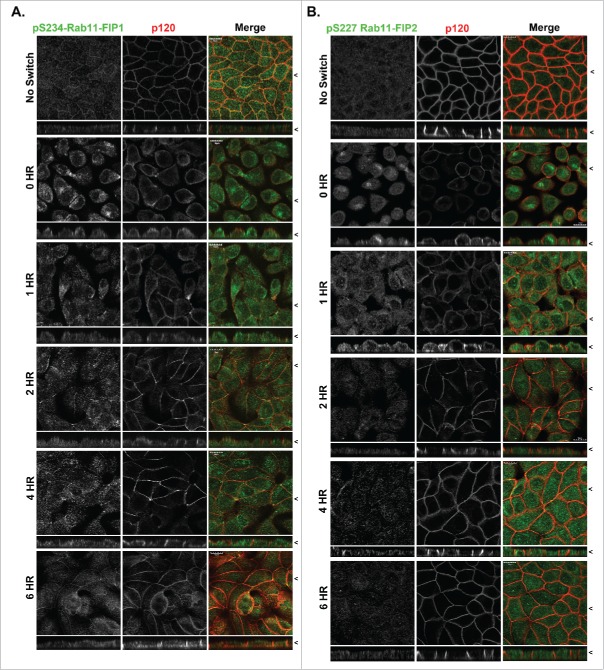
Localization of p(S234)-Rab11-FIP1 is distinct from p(S227)-Rab11-FIP2 during re-polarization of MDCK cells. T23 MDCK cells labeled with either p(S234)-Rab11-FIP1 antibodies (A) or p(S227)-Rab11-FIP2 antibodies (B) along with antibodies against p120 in non-switched cells (NS) and then 0, 1, 2, 4, or 6 hours after re-addition of calcium. Cells were fixed with methanol. Merged dual color images are shown at the right. Localization of p(S234)-Rab11-FIP1 was punctate/vesicular as well as junctional with the greatest density in the apical region and was maintained throughout the calcium switch. Phosphorylation of p(S227)-Rab11-FIP2 was upregulated in calcium switch, however, p(S227)-Rab11-FIP2 labeling peaked at 2 hours after calcium switch and then declined. X/Z images are shown below X/Y images, < indicates where images were taken for X/Y and X/Z planes. All scale bars = 10 µm.
To analyze the subcellular localization of MARK2 in MDCK cells, we utilized a commercially available antibody. MARK2 distributed throughout polarized MDCK cells, with a significant concentration subapically. Addition of the antigen peptide blocked all staining in MDCK cells, verifying specificity (Fig. S2). MARK2 localized throughout 4% paraformaldehyde-fixed MDCK cells during repolarization with a significant accumulation in the subapical domain (Fig. 3A). Interestingly, MARK2 colocalized with a FIP1B/C specific antibody and the (pS234)-FIP1 antibody (Fig. 3A). Colocalization was calculated and Pearson's coefficients were compared. MARK2 colocalized with pFIP1 and FIP1B/C throughout calcium switch, however, there was a small but statistically significant increase in colocalization at the later 6 hour time point (Fig. 3B, p < 0.05).
Figure 3.
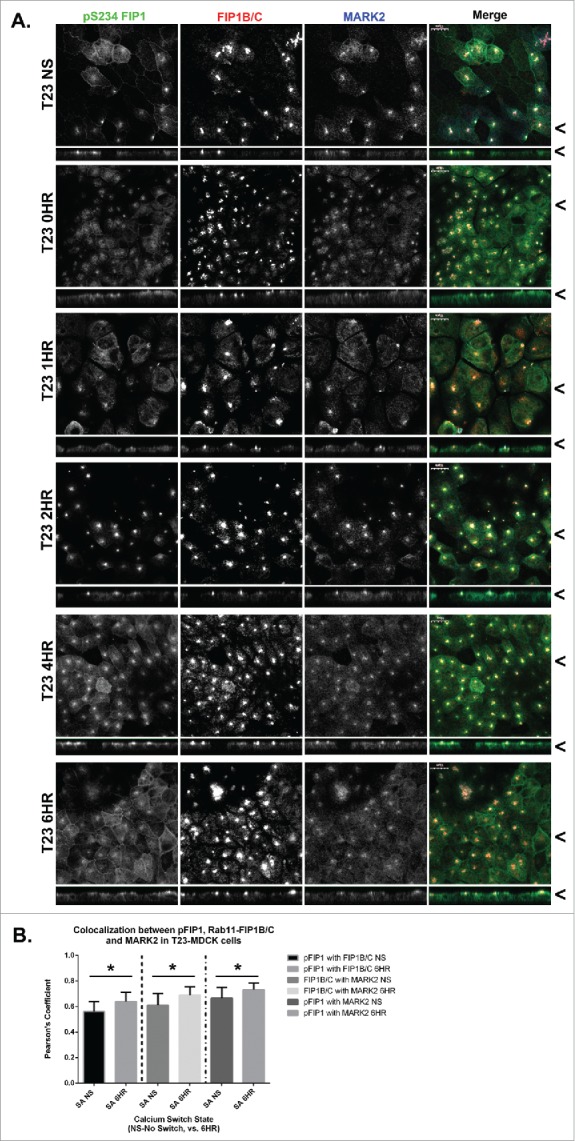
MARK2 colocalizes with Rab11-FIP1B/C and p(S234)-Rab11-FIP1 throughout calcium switch. A. Non-switched T23 MDCK cells (NS) and cells at 0, 1, 2, 4, and 6 hours after calcium re-addition were fixed with 4% paraformaldehyde and immunostained using the p(S234)-FIP1 antibody, a Rab11-FIP1B/C antibody, and a MARK2 antibody. The MARK2 colocalized with both the p(S234)-FIP1 and FIP1B/C staining. Colocalization was observed between p(S234)-FIP1 and MARK2 at the lateral membrane 4 or more hours after calcium switch. X/Z images are shown below X/Y images, < indicates where images were taken for X/Y and X/Z planes. All scale bars = 10 µm. B. Colocalization analysis was conducted comparing p(S234)-FIP1 (pFIP1) with Rab11-FIP1B/C, pFIP1 with MARK2, and MARK2 with Rab11-FIP1B/C. All proteins colocalized together, and in each case, colocalization increased significantly through calcium switch (pFIP1 and FIP1B/C p = 0.024, FIP1B/C and MARK2 p = 0.026, pFIP1 and MARK2 p = 0.031, graphs plot mean Pearson's Coefficient, error bars represent standard deviation).
To understand how FIP1 protein phosphorylation might affect MDCK cell polarity, stable expression lines were generated using the pTRE2 TET-OFF system for T23 MDCK cells stably expressing GFP-FIP1C wildtype, GFP-FIP1C(S234A), or GFP-FIP1C(S234D). These stable lines were evaluated under no doxycycline conditions to induce expression and cells were subjected to calcium switch and assessed using fluorescence microscopy. Cells expressing wildtype FIP1C start to pseudo-stratify as early as one hour after calcium re-addition (Fig. 4A). Four hours following calcium addition MDCK cells started forming F-actin enriched lateral lumens in all overexpression lines (Fig. 4A, 4B, Fig. S3). The FIP1C(S234A)-expressing line, however, displayed significantly more lateral lumens per field than other lines (see Figs. 4B-8HR). Quantitation of lateral lumen abundance revealed lateral lumens formed in approximately 1% of WT and FIP1C(S234D)-expressing cells. However, 25% of FIP1C(S234A)-expressing cells contained lateral lumens 4–8 hours after calcium addition (Fig. 4C). Podocalyxin appeared to relocate to the surface of the lateral lumen, however, no colocalization was seen between GFP-FIP1C and Podocalyxin (Fig. 4D, E).
Figure 4.
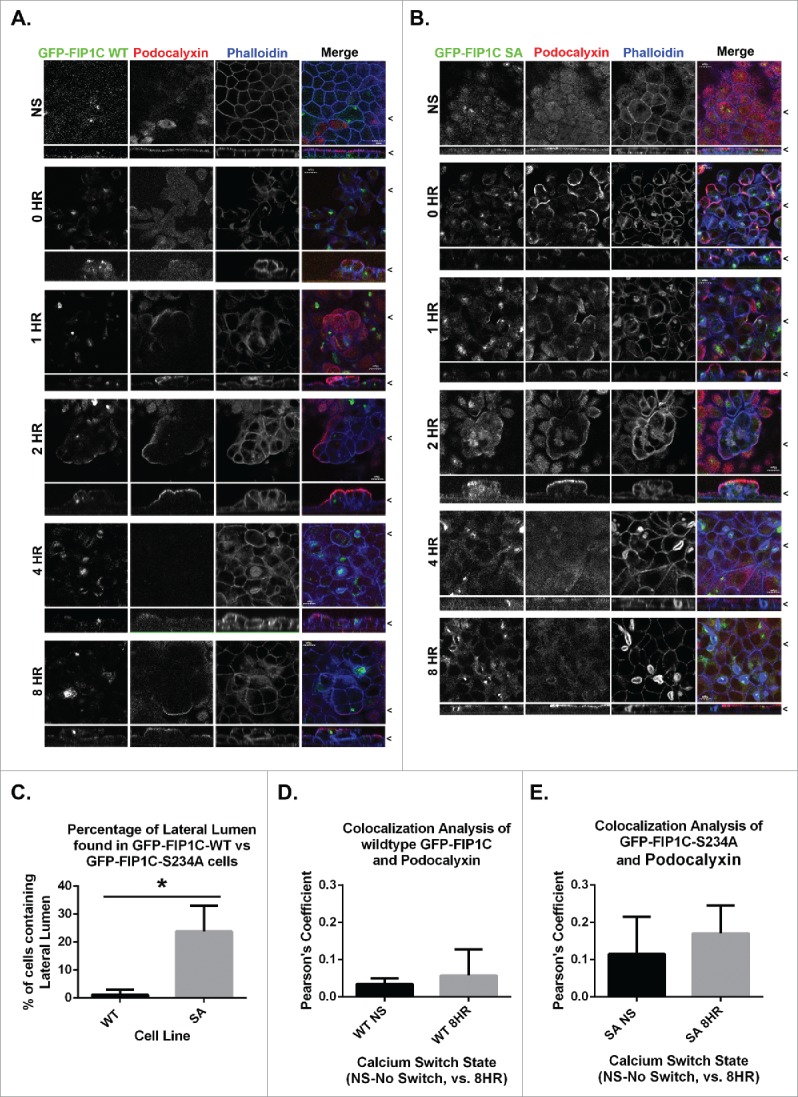
Overexpression of GFP-Rab11-FIP1C wildtype and GFP-Rab11-FIP1C(S234A) causes two distinct phenotypes during calcium switch: pseudo-stratification and lateral lumen formation. MDCK cell lines stably expressing either GFP-Rab11-FIP1C wildtype (A) or GFP-Rab11-FIP1C(S234A) (B) were examined in the non-switched (NS) condition and 0,1, 2, 4, and 8 hours after calcium re-addition with immunostaining with antibody against Podocalyxin along with fluorescent-Phalloidin to visualize F-actin. Overexpression of GFP-Rab11-FIP1C wildtype protein caused a dramatic pseudo-stratification 2 hours into a calcium switch assay. Podocalyxin localized only to the most apical surface of these pseudo-stratified layers. The expression of the GFP-Rab11-FIP1C(S234A) mutant caused some cell stratification, but also induced F-actin accumulation at the lateral membrane greater than 4 hours after calcium switch in lateral lumens. Podocalyxin also lined the lateral lumens. The lateral lumens are seen in WT overexpression lines (see A. 4 HR), however, to a much lesser extent. X/Z images are shown below X/Y images, < indicates where images were taken for X/Y and X/Z planes. All scale bars = 10µm. C. Quantitation of lateral lumen formation. Wildtype GFP-FIP1C overexpression caused lateral lumens to form in ∼1% of cells, whereas, overexpression of FIP1C(S234A) causes lateral lumen to form in ∼25% of cells following calcium switch (*p = 0.00006, error bars represent standard deviation). Colocalization analysis of wildtype GFP-FIP1C (D) and GFP-FIP1C(S234A) with Podocalyxin colocalization demonstrated little colocalization. Graphs plot mean Pearson's Coefficient, error bars represent standard deviation, WT p = 0.17, SA p = 0.19.
To analyze lateral lumen formation further, using the GFP-FIP1C(S234A) line and the parent T23-MDCK line as a control, cells were either not switched to low calcium media or calcium switched and fixed 4–8 hours after calcium addition. Cells were immunostained for subcellular markers. The most striking change observed was the redistribution of F-actin from the apical membrane to the lateral lumens (Fig. 4A,B). GFP-FIP1C(S234A) was localized on membranes in the cytoplasm located adjacent to the lateral lumens. We utilized this redistribution of F-actin to identify lateral lumens. Phosphorylated FIP1 localization appeared to be altered in cells expressing GFP-FIP1C constructs (Fig. 5). In both the wildtype and S234A expressing cells, the non-calcium switched cells showed pFIP1 localization increased at the lateral membrane compared with the T23 parental cell line (Fig. 5). Theoretically, the GFP-FIP1C wildtype protein (Fig. 5B) should be phosphorylated by MARK2, however, this was not readily observed. We did observe some colocalization between pFIP1 and GFP, but it was limited.
Figure 5.
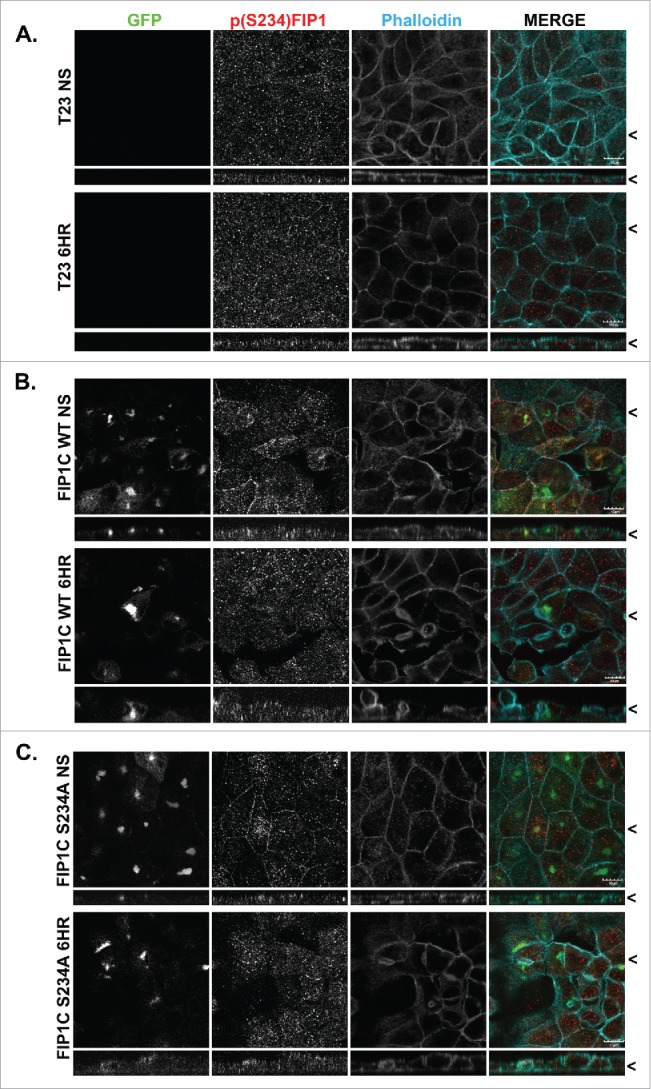
Effects of overexpression of GFP-Rab11-FIP1C wildtype and GFP-Rab11-FIP1C(S234A) on p(S234)-FIP1 abundance and localization. Parental T32 MDCK cells (A), and T23 MDCK cells expressing GFP-Rab11-FIP1C wildtype (B) or GFP-FIP1C(S234A) (C) were examined in non-switched cells (NS) and 6 hours after re-addition of calcium for immunostaining for p(S234)-Rab11-FIP1C and phalloidin staining. Overexpression of wildtype GFP-FIP1C and GFP-FIP1C(S234A) increased the lateral localization of pFIP1 staining in non-switched cells. Overexpression of GFP constructs did not inhibit phosphorylation of FIP1 by MARK2. X/Z images are shown below X/Y images, < indicates where images were taken for X/Y and X/Z planes. All scale bars = 10 µm.
To investigate the alteration in pFIP1 localization further, a complete calcium switch was performed in GFP-FIP1C(S234A) cells (No switch, 0 HR, 1 HR, 2 HR, 4 HR, 6 HR) (Fig. 6A). Phosphorylated FIP1 did not appear to be reduced in FIP1C(S234A) expressing cells and accumulation of pFIP1 was increased at the lateral membrane earlier in calcium switch (Fig. 6A, 1HR, 2HR). Strong colocalization was seen between GFP-FIP1C(S234A) and endogenous FIP1A/B, with an increase in calcium switched cells (Fig. 6C). FIPs are known to hetero- and homo-dimerize with other family members.39,44 Thus, some co-localization may accrue from GFP-FIP1C(S234A) hetero-dimerization with endogenous FIP1A and/or FIP1B. There was also a sub-population of pFIP1 protein that was not colocalized with FIP1A/B (Fig. 6A-1HR, lateral membrane staining).
Figure 6.
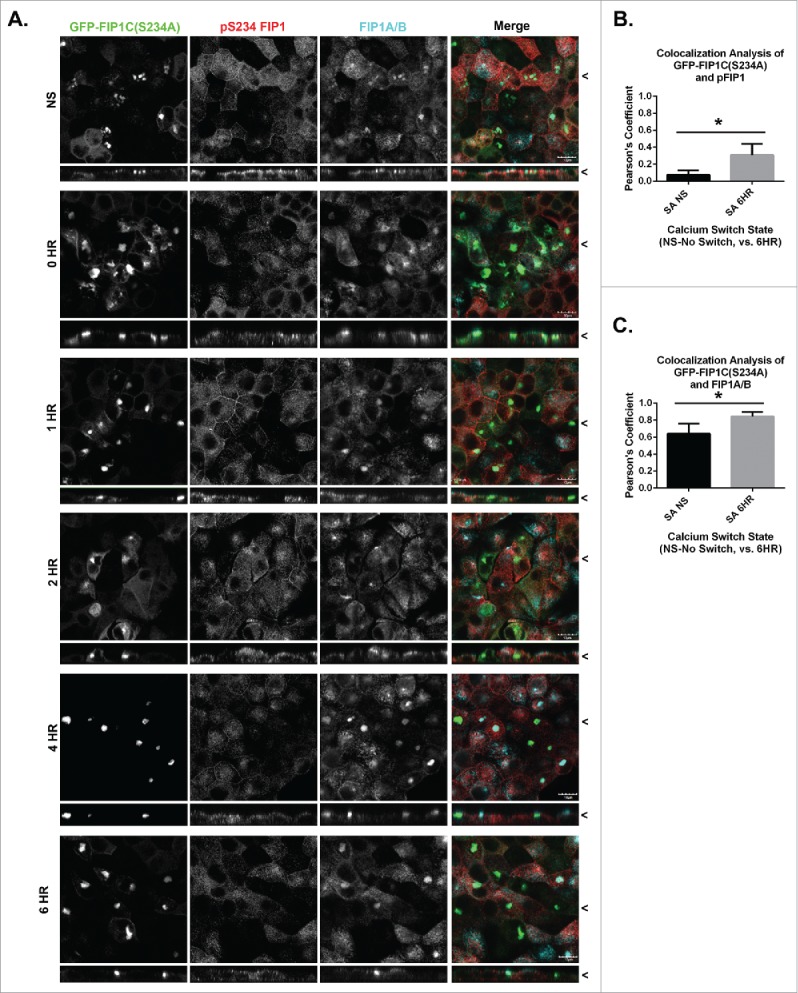
Endogenous FIP1A/B and MARK2 colocalize with GFP-FIP1C(S234) through calcium switch. A. Stable GFP-FIP1C(S234A) expressing MDCK cells were calcium switched and fixed at 0, 1, 2, 4 or 6 hours following calcium re-addition. Cells were then stained for p(S234)-FIP1 and FIP1A/B. FIP1A/B and pFIP1 colocalized with the GFP-FIP1C(S234A) near the apical membrane. X/Z images are shown below X/Y images, < indicates where images were taken for X/Y and X/Z planes. All scale bars = 10 µm. B. Colocalization analysis confirmed a statistically significant increase in colocalization of GFP-FIP1C(S234A) with pFIP1 after calcium switch (p = 0.000024). C. Strong colocalization was seen between GFP-FIP1C(S234A) and FIP1A/B both at non-switched and switched time points. However, there was a significant increase in colocalization following calcium switch (*p = 0.0001). (B and C.) Graphs plot mean Pearson's Coefficient and error bars represent standard deviation.
Subcellular markers were used in no switch and 4–8 HR calcium switched cells to understand in greater detail the extent of the lateral lumen phenotype. Endogenous Rab11a and Rab11b were redistributed and colocalized with GFP-FIP1C(S234A) adjacent to the lateral lumen in 8HR calcium switched cells (Fig. 7A, 7D). Interestingly, colocalization analysis showed an increase in colocalization between GFP-FIP1C(S234A) and Rab11a in 8HR calcium switched cells as compared with the no switch control (Fig. 7C). The opposite was observed with Rab11b, which showed greater colocalization with GFP-FIP1C(S234A) in the no switch cells (Fig. 7D, 7F). Three dimensional reconstructions rendered using Imaris software, demonstrate the relationship of Rab11a and Rab11b with GFP-FIP1C(S234A) (Fig. 7B, 7E, Movie S1-Rab11a).
Figure 7.
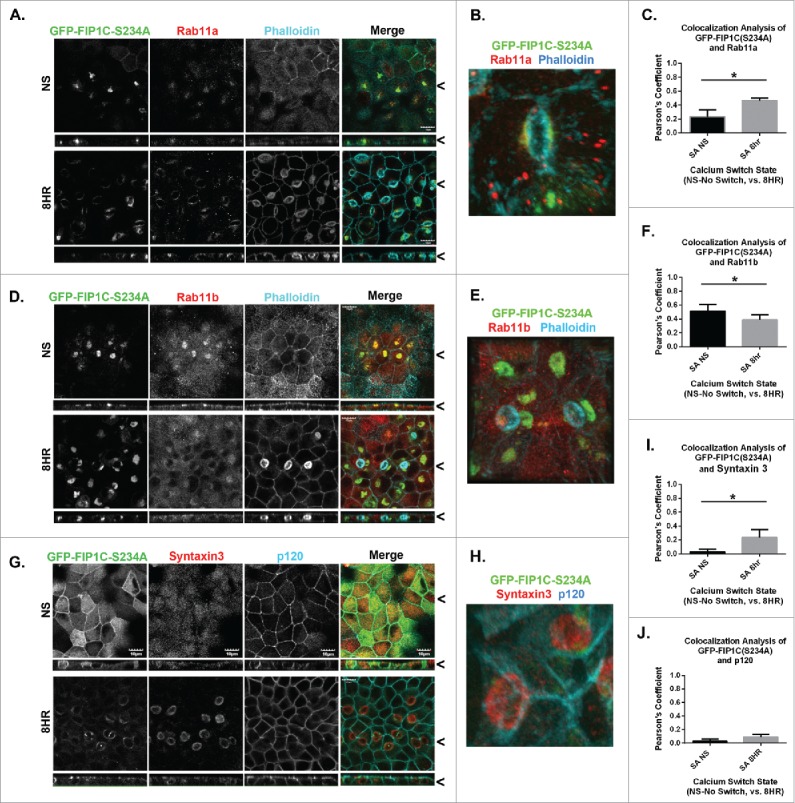
Overexpression of GFP-Rab11-FIP1C(S234A) causes reorganization of recycling system markers after 8-hour calcium switch. GFP-FIP1C(S234A) expressing cells, either non-switched (NS) or fixed at 8 hours following calcium re-addition were examined for localization of Rab11a (A-C) or Rab11b (D-F) along with phalloidin staining or for Syntaxin 3 with p120 staining (G-I). A. Endogenous Rab11a colocalized with GFP-Rab11-FIP1C(S234A) in non-switched cells and at 8 hours after calcium switch. A still image taken from a 3-dimensional reconstruction shows Rab11a co-localizing with GFP-FIP1C(S234A) beneath the surface of a lateral lumen (B, see Movie S1). C. Colocalization analysis confirmed an increase in colocalization between Rab11a and GFP-FIP1C(S234A) following calcium switch (*p = 0.0001). D. Endogenous staining of Rab11b also colocalized with GFP-Rab11-FIP1C (S234A), however, to a lesser extent than Rab11a. The colocalization decreased significantly in cells after calcium switch (see F; *p = 0.003). E. A still image taken from a 3-dimensional reconstruction of Rab11b co-localizing with GFP-FIP1C(S234A) beneath a lateral lumen (Movie not shown). G. While Syntaxin 3 localized to the apical membrane region in non-switched cells, Syntaxin 3 relocalized following calcium switch in GFP-FIP1C(S234A) cells to the lateral lumen. P120 was used to label lateral membranes and was excluded from the lateral lumen surface. H. A still image taken from a 3-dimensional reconstruction of Syntaxin 3 at the surface of a lateral lumen (see Movie S2). P120 is absent from the surface of the lateral lumen (see Movie S3). I. Colocalization analysis confirms an increase in colocalization between GFP-FIP1C(S234A) and Syntaxin3 (*p = 0.000005). J. Little colocalization was present between GFP-FIP1C(S234A) and p120. X/Z images are shown below X/Y images, < indicates where images were taken for X/Y and X/Z planes. All scale bars = 10 µm. (C. F. I. J.) Graphs plot mean Pearson's Coefficient and error bars represent standard deviation.
Apical membrane proteins, such as Syntaxin 3, were also redistributed to the lateral lumen (Fig. 7G, Movie S2). Colocalization analysis showed very little colocalization of Syntaxin 3 with GFP-FIP1C(S234A) at the no switch time point, but colocalization was increased through calcium switch (Fig. 7I). P120 was excluded from the lateral lumen and redistributed to the apical membrane (Fig. 7G, 7H, Movie S3). No colocalization was seen between GFP-FIP1C(S234A) and p120 (Fig. 7J).
To analyze further the boundaries of lateral lumens induced by GFP-FIP1C(S234A), no switch and 6HR calcium switched cells were stained for ZO-1, E-Cadherin and pERM (Fig. 8). ZO-1 was redistributed around the lateral lumen (ZO-1, Fig. 7A, 7B) forming tight junctions in a circular domain around the lateral lumen surface (Fig. 7B, Movie S4). Colocalization with GFP-FIP1C(S234A) was low with ZO-1 (Fig. 8C). ZO-1 localization was also significantly distinct from Ezrin in calcium switched cells (Fig. 8D). Lateral membrane markers, p120 and E-Cadherin, were excluded from the surface of the lateral lumen and redistributed to the apical membrane (p120- Fig. 7G and H, E-cadherin- Fig. 8E and F, Movie S5). No colocalization with GFP-FIP1C(S234A) was seen for E-Cadherin (Fig. 8G). P-ERM was redistributed to the lateral lumen in calcium switched cells (Fig. 8H and I, Movie S6). P-ERM was not lost in cells with lower levels of GFP-FIP1C(S234A) expression (Fig. 8I, Movie S6). No change in colocalization was detected (Fig. 8J). These results suggested that the lateral lumens represented redistribution of apical and junctional components to the lateral surface of polarized cells.
Figure 8.
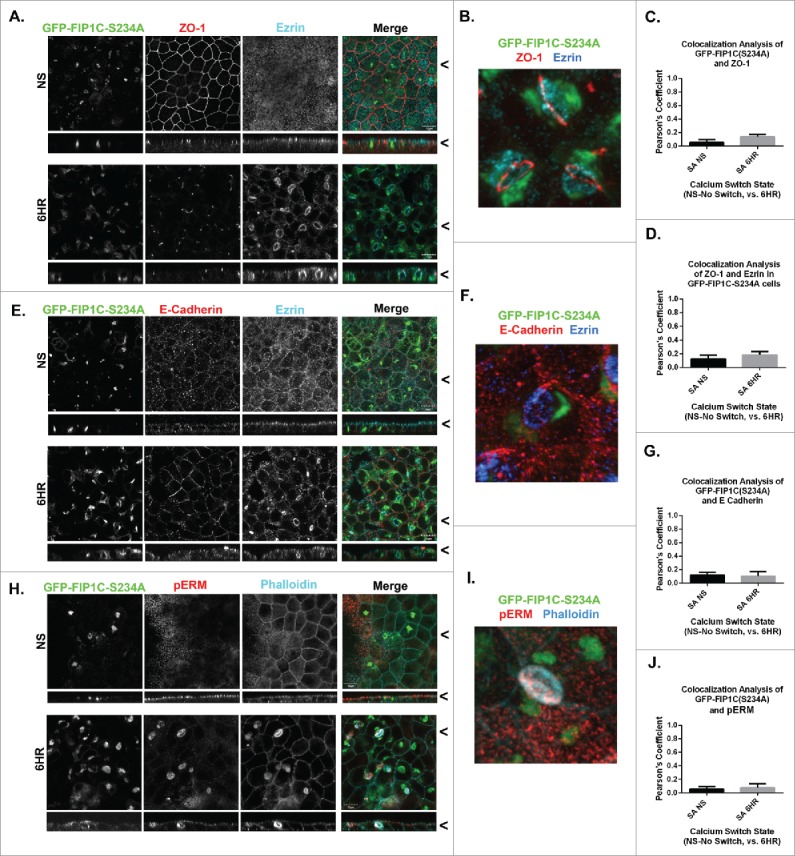
Overexpression of GFP-Rab11-FIP1C-(S234A) causes relocalization of apical markers to lateral lumens after 6-hour calcium switch. GFP-FIP1C(S234A) expressing cells, either non-switched (NS) or fixed at 6 hours following calcium re-addition, were examined for localization of ZO-1 (A-C) or E-cadherin (D-G) along with ezrin immunostaining or for phospho-ERM (pERM) with phalloidin staining (H-J). A. Endogenous staining of ZO-1 6 hours after calcium switch in GFP-FIP1C(S234A) line showed redistribution of the tight junction protein to the border of lateral lumens. B. A still image taken from a 3-dimensional reconstruction showed ZO-1 completely encircling a lateral lumen. (see Movie S4). Colocalization analysis showed little colocalization between GFP-FIP1C(S234A) and ZO-1 (C) or between ZO-1 and Ezrin (D). E. E-Cadherin was excluded from the lateral lumen membrane following calcium switch. E-cadherin staining also relocated to the new luminal lateral membrane in these cells. F. In a still image from a 3-dimensional reconstruction, E-Cadherin was absent from the new apical membrane, marked with Ezrin in blue (see Movie S5). G. No significant colocalization of GFP-FIP1C(S234A) and E-Cadherin following calcium switch was observed. H. Endogenous staining for pERM in calcium switched, GFP-FIP1C(S234A) line showed pERM relocation to the lateral lumen surface. I. A still image taken from 3-dimensional reconstruction shows pERM co-labeling with phalloidin on the membrane of a lateral lumen (see Movie S6). J. No colocalization was observed for pERM with GFP-FIP1C(S234A) following calcium switch. X/Z images are shown below X/Y images, < indicates where images were taken for X/Y and X/Z planes. All scale bars = 10 µm. In C. D. G. and J, graphs plot mean Pearson's Coefficient and error bars represent standard deviation.
The pattern of trafficking is strongly influenced by the orientation of the microtubule cytoskeleton and is reflected in the positioning of the centrosome and the Golgi apparatus. We therefore evaluated the positions of the centrosome and Golgi apparatus in MDCK cells expressing GFP-FIP1C(S234A). In non-switched cells, both the centrosome and the Golgi apparatus occupied positions below the apical membrane. Interestingly, in calcium switched cells expressing GFP-FIP1C(S234A), the Golgi apparatus and centrosome reoriented toward the lateral membrane (Pericentrin-Fig. 9A, Movie S7, Giantin-Fig. 9D, Movie S8), further confirming the realignment of subcellular components from a vertical axis of polarity to a horizontal axis of polarity. Colocalization between Pericentrin and GFP-FIP1C(S234A) was not detected (Fig. 9B), nevertheless, higher resolution images show close proximity between GFP-FIP1C(S234A) and pericentrin adjacent to a lateral lumen (Fig. 9B). No co-localization of Giantin and GFP-FIP1C(S234A) was observed in calcium switched cells (Fig. 9E, 9F).
Figure 9.
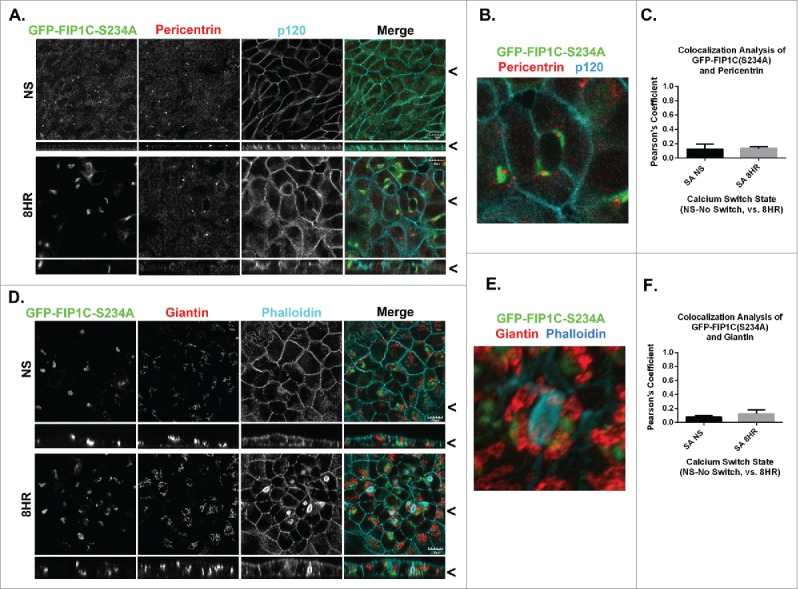
Overexpression of GFP-Rab11-FIP1C(S234A) causes reorientation of the centrosome-based axis after 8-hour calcium switch. GFP-FIP1C(S234A) expressing cells, either non-switched (NS) or fixed at 6 hours following calcium re-addition were examined for localization of the centrosome marker Pericentrin (A-C) along with p120 staining or for the Golgi apparatus marker Giantin with phalloidin staining (G-I). A. Endogenous pericentrin in non-switched cells localized to a nidus below the apical membrane. However, 8 hours after calcium switch pericentrin relocalized to a point adjacent to lateral lumens in close proximity to GFP-FIP1C(S234A). B. In a still image taken from 3-dimensional reconstruction, the Pericentrin-staining centrosome is located directly adjacent to GFP-FIP1C(S234A) in association with a lateral lumen (Movie S7). C. No colocalization was seen between pericentrin and GFP-FIP1C(S234A). D. In non-switched cells, Giantin was located deep to the apical membrane. However, 8 hours after calcium switch, Giantin, labeling the Golgi apparatus, reoriented to face the lateral lumen surface. E. In a still image taken from 3-dimensional reconstruction, Giantin stain localizes adjacent to GFP-FIP1C(S234A) however, very little colocalization (F) is seen (Movie S8). X/Z images are shown below X/Y images, < indicates where images were taken for X/Y and X/Z planes. All scale bars = 10 µm. In C and F, graphs plot mean Pearson's Coefficient and error bars represent standard deviation.
To understand the effects of GFP-FIP1C(S234A) expression, cells were calcium switched for four hours, a point before significant lateral lumen formation, and analyzed for MARK2 localization. MARK2 did not colocalize with wildtype GFP-FIP1C (Fig. 10A) and there was no change in the colocalization between non-switched and calcium switched cells (Fig. 10B). Remarkably, however, MARK2 localization was altered in GFP-FIP1C(S234A) cells (Fig. 10C). Even though the FIP1C(S234A) mutant cannot be phosphorylated, MARK2 accumulated with GFP-FIP1C(S234A) throughout the calcium switch (Fig. 10D). MARK2 remained apical in GFP-FIP1C wildtype-expressing cells regardless of calcium switch state. However, in cells expressing GFP-FIP1C(S234A) MARK2 was no longer only in the apical region, but rather was relocalized into membranes containing GFP-FIP1C(S234A) (Fig. 10C). Overall, colocalization between MARK2 and GFP-FIP1C(S234A) was low, however, the increase in colocalization was statistically significant in calcium switched cells (Fig. 10D, p = 0.006). Thus, a portion of MARK2 is sequestered on internal membranes by GFP-FIP1C(S234A).
Figure 10.
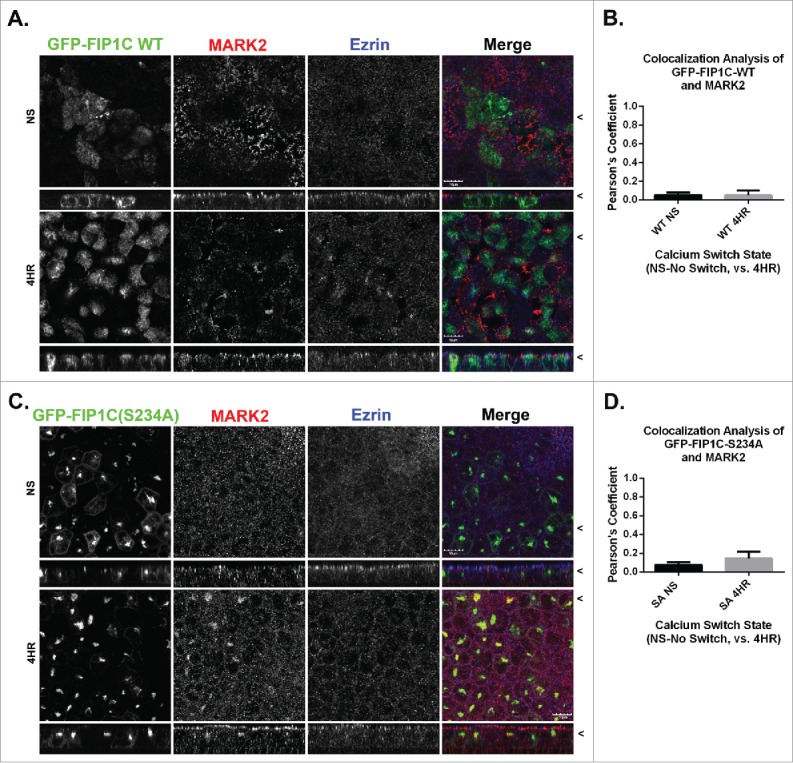
MARK2 localization is altered in FIP1C(S234A) overexpression lines. Non-switched GFP-FIP1C wildtype and GFP-FIP1C(S234A)-expressing MDCK cells or cells at 4 hours following calcium re-addition were fixed and immunostained for MARK2 and Ezrin. A and B. GFP-FIP1C wildtype did not colocalize with MARK2 in either non-switched cells or 4 hours after calcium switch. C. In non-switched GFP-FIP1C(S234A)-expressing cells, MARK2 did not localize with GFP-FIP1C(S234A). However, in switched cells at 4 hours after re-addition of calcium, a point before lateral lumens are formed, MARK2 colocalization with GFP-FIP1C(S234A) was observed. D. Colocalization of MARK2 and GFP-FIP1C(S234A) overall was weak. However, there was a statistically significant increase from NS to 4 HR. X/Z images are shown below X/Y images, < indicates where images were taken for X/Y and X/Z planes. All scale bars = 10 µm. In B and D, graphs plot mean Pearson's Coefficient and error bars represent standard deviation.
Discussion
The kinase MARK2/Par1b is a crucial regulator of the establishment and maintenance of polarity. While initial studies focused on a role in the regulation of the microtubule cytoskeleton, recent investigations now suggest that major targets for MARK2 phosphorylation may be modifiers of vesicle trafficking. Previous studies established a role for MARK2 phosphorylation of Rab11-FIP2 in regulating polarity.19,26 Our present studies demonstrate that Rab11-FIP1B/C also represent substrates for MARK2 phosphorylation. Importantly, the spatial and temporal aspects of endogenous Rab11-FIP1 and Rab11-FIP2 phosphorylation during the process of re-polarization after calcium switch are remarkably different. While Rab11-FIP2 is phosphorylated early and transiently,26 Rab11-FIP1 phosphorylation is more sustained. These findings indicate that an orderly progression of phosphorylation events mediated by MARK2 within cells during polarization may be required for successful and appropriate segregation of apical and basolateral domains.
In our previous studies, expression of the non-phosphorylatable S227A mutant of Rab11-FIP2 in MDCK cells caused delay of repolarization following calcium switch.19 Expression of GFP-Rab11-FIP2(S227A) in MDCK cells also blocked lumen formation in cells grown in three-dimensional cultures.26 In contrast, expression of the phospho-mimetic, GFP-Rab11-FIP2(S227E), caused multiple lumen formation when MDCK cells were grown in a matrigel 3-dimensional matrix.26 In the present studies, the phospho-mimetic mutant of Rab11-FIP1, Rab11-FIP1(S234D), did not induce any appreciable effect in cell behavior following calcium switch. Nevertheless, expression of Rab11-FIP1(S234A) caused a marked alteration of cell polarity after calcium switch with reorientation of the centrosome and microtubule array toward the lateral membranes and formation of lateral lumens bounded by tight junctions and containing apical surface proteins and specializations. Such changes in cell polarity have been seen before with manipulations of MARK2 activity, where overexpression of MARK2 leads to the re-orientation of the microtubule array and formation of lateral lumens.18,41,45 Interestingly, in the present studies, expression of a non-phosphorylatable FIP1C protein leads to a similar lateral lumen phenotype. Our present studies show that MARK2 tends to localize with Rab11-FIP1C and that expression of Rab11-FIP1C(S234A) caused accumulation of MARK2 on a collapsed membrane compartment lying deep to lateral lumens formed after calcium switch. The relocation of MARK2 may therefore lead to a sequestration of MARK2 away from appropriate sites of phosphorylation, thus altering the spatial and temporal coordination of MARK2 phosphorylation of its critical substrates. Given the regulation of microtubule orientation by MARK2,5 the sequestration of MARK2 by GFP-Rab11-FIP1C(S234A) expression may account for the formation of lateral lumens after calcium switch. It is important to note that the lateral lumen phenotype is lost 24 hours after calcium switch and is not present in non-switched cells. Thus it appears that such a sequestration would more likely be affecting the dynamic resolution of the ongoing membrane sorting process during the progression of repolarization.
Some of the lateral lumen phenotype may also represent effects of sequestration of endogenous Rab11-FIP1 substrates. Multiple splice variants of Rab11-FIP1 exist.27 These splice variants appear to mark differential regions of the recycling system.39 Western blot analysis suggests that the major substrate for MARK2 phosphorylation is the largest splice variant, Rab11-FIP1B. The Rab11-FIP1 proteins are able to dimerize with different Rab11-FIP1 family members.44 MARK2 strongly colocalized with pS234-FIP1 and FIP1B/C in normal T23-MDCK cells (see Fig. 3). Thus, the non-phosphorylatable GFP-FIP1C(S234A) could dimerize with endogenous Rab11-FIP1B and sequester it away from its normal localization and also inhibit its phosphorylation by MARK2 at appropriate times, thereby altering efficient repolarization.
In summary, we have identified the phosphorylation by MARK2 of a second Rab11-FIP protein group, Rab11-FIP1B/C. Phosphorylation on Rab11-FIP1B/C occurs in a distinct spatial and temporal time frame compared with MARK2 phosphorylation of Rab11-FIP2. It is of note that while phosphorylated Rab11-FIP2 is not associated with Rab11a,26 phosphorylated Rab11-FIP1 does seem to be present on Rab11a-containing elements. Additionally, MARK2 is more closely associated with Rab11-FIP1B/C. Thus, MARK2 may maintain a more constant association with elements of the recycling system containing Rab11-FIP1B/C proteins. Further investigation will be necessary to delineate what sub-compartments of the apical recycling system are specifying MARK2 distribution in polarized epithelial cells.
Materials and methods
Protein purification
Human Rab11-FIP1C was cloned into pET30a using normal cloning techniques. Full length as well as truncations of FIP1C were constructed containing the S234A, S234D, and S234E mutations using dual-primer site directed mutagenesis (QuikChange protocol). Vectors were transfected into BL-21 GOLD RIL E. coli (Agilent Technologies, Inc.) and grown overnight in LB media. Log phase bacteria were incubated overnight at 25°C in autoinduction media.46 Cells were pelleted and the pellet was either stored at −80°C or sonicated and protein extracted using a standard His-tag and nickel bead protocol. Proteins were assessed for purity by Coomassie blue staining in SDS-PAGE gels.
Phosphorylation assays
Phosphorylation assays were performed using catalytically active MARK2 purchased from Life Technologies (PV3878). Reactions were assembled using kinase buffer (20mM Tris HCl pH 7.4, 10 mM MgCl2, 2 mM EDTA), 20 ng MARK2, 5 µg of substrate protein, and 20 µM ATP, where the ATP was added immediately before a 5-minute incubation at 30°C. Radioactive [γ−32P]- ATP was used in initial experiments prior to validation of phospho-specific antibody. Western blots were analyzed by measuring band intensity using LI-COR Image Studio software.
Antibodies and immunofluorescence
Sources of antibodies used can be found in Table 1 along with concentrations used for both western blotting (where applicable) and immunofluorescence, as well as the fixative protocol used. An affinity-purified phospho-specific antibody was generated to Rab11-FIP1(pS234) by EZBioLab against peptide sequence- PLSQ(pS)MSVLPTSK-C. The antibody detected mouse, human and dog Rab11-FIP1(pS234) protein. We validated the antibody in HeLa, Caco-BBE and MDCK cells by western blot and immunofluorescence analysis. Several different fixation protocols were used based on different antibody requirements. Methanol fix performed using −20°C methanol for 5 minutes at −20°C. Also, a 4% PFA fixative was used for 20 minutes at room temperature. All cells were permeabilized and blocked with 0.03% Triton-X and 10% normal donkey serum in 1X PBS before staining with specified antibodies. F-actin was visualized by incubation with Alexa-647-conjugated-Phalloidin (Invitrogen). Secondary antibodies were purchased from Jackson Immunochemicals.
Table 1.
Antibody reagents used for immunostaining.
| Antibody | Species | Concentration | Fixative | Producer | Catalog # or reference |
|---|---|---|---|---|---|
| Rab11-FIP1B/C | chicken | 1:500 | PFA | Sigma | GW21574A |
| p(S234)-FIP1 | rabbit | 1:100 | PFA | Goldenring Lab | Current paper |
| p(S227)-FIP2 | rabbit | 1:300 | Methanol | Goldenring Lab | Lapierre, et al.26 |
| Rab11a | rabbit | 1:200 | PFA | Goldenring Lab | Ducharme, et al.19 |
| Rab11b | rabbit | 1:100 | PFA | Goldenring Lab | Lapierre, et al.48 |
| ZO-1 | rat | 1:400 | Methanol | DSHB | R26.4C |
| Ezrin | rabbit | 1:200 | PFA/Methanol | Cell Signaling | 3145s |
| p-ERM | rabbit | 1:200 | PFA | Cell Signaling | 3149s |
| Syntaxin-3 | rabbit | 1:200 | PFA | Abcam | ab4113 |
| Giantin | goat | 1:500 | PFA | Santa Cruz | sc-46993 |
| Pericentrin | mouse | 1:500 | PFA | Abcam | ab4448 |
| p120 | mouse | 1:200 | PFA/Methanol | BD Biosciences | 610133 |
| E-Cadherin | rat | 1:200 | Methanol | Abcam | ab11512 |
| MARK-2 | goat | 1:200 | PFA | Abcam | ab77641 |
Western blot analysis
Cells were grown at confluence for at least 5 d to insure polarization and then harvested off either transwell filters or plastic by scraping cells into a 1X PBS solution. Cells were pelleted using a 2000 xg 10 min spin in a tabletop centrifuge. Then cells were lysed using RIPA buffer (150 mM NaCl, 10 mM Tris pH 7.4, 0.1% SDS, 1% Triton X-100, 1% Deoxycholate, 5 mM EDTA) containing mammalian protease inhibitors, phosphatase inhibitor cocktail 2 and phosphatase inhibitor cocktail 3 (Sigma). Peptide blocking was used to assess antibody specificity. Both phospho-S234-FIP1 and non-phosphorylated FIP1 peptides (obtained from EZBioLabs) as well as the MARK2 peptide (QNGKDSTAPQR- synthesized by GenScript USA Inc.) were resuspended at 1 mg/ml in water and used at 1:1000.
Cell lines
Stable T23-MDCK 47 cell lines were made by expressing pTRE2-GFP-FIP1C (wildtype, S234A, S234D, S234E) vectors. These lines were made by transfection, hygromycin selection and clonal isolation as previously described.19 Several different clones were selected for each vector and analyzed using immunofluorescence and western blot analysis to assess levels of overexpression.
Imaging
All images were taken sequentially on an Olympus FV1000 using a 60X objective with a 3x digital zoom. Z-stacks were imaged every 0.50 micron and phalloidin staining of F-actin (Ezrin or p120 in Methanol fixed cells) was used to determine cell boundaries. Images were processed using Olympus software and all figures were assembled in Photoshop. All correlative images were taken using the same settings as predetermined using the T23-parental MDCK cell line.
Calcium switch
Cells were plated at confluence in tetracycline-free media and allowed to grow for 7 d on transwell filters. By day 5, the GFP expression of each protein was visible. The cells were then switched into calcium free media overnight and calcium was added back 16–24 hours later. Cells were fixed at specific time points after calcium re-addition; common time points were: no switch (NS), 0 min, 30 min, 1 hour, 2 hours, 4 hours and 6–8 hours, up to 48 hours. Normal MDCK cells recover fully functional junctions between 2 and 4 hours after calcium re-addition.
3-Dimensional reconstructions
Reconstructions were done using Imaris software and sequential Z-stacks taken with the Olympus FV1000 microscope. The Imaris software suite automatically generates a 3-Dimensional reconstruction anytime an image file is opened containing z-stacks. Videos were recorded using the Imaris software and manually rotating the 3-dimensional reconstruction. Videos can then be sped up and slowed down using video editing software and compressed to accommodate size requirements.
Colocalization analysis
Colocalization was analyzed using the ImageJ JACoP plugin. Merged images were split by channel and then Pearson's coefficients were taken comparing channels of interest. Each field of cells contained between 20 and 50 cells and at least 10 fields of view were analyzed totaling between 200 and 500 cells compared. Statistical significance was measured between time points using a students unpaired, two-tailed, t test. Data collected from ImageJ was analyzed using Excel and GraphPad Prism (version 6.01).
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the National Institute of Health (NIH) grants R01 DK48370 and R01 DK70856 to J.R.G. Confocal microscopy was performed through the VUMC Cell Imaging Shared Resource and the Vanderbilt Mass Spectrometry Resource, respectively, supported by NIH grants CA68485, DK20593, DK58404 and HD15052.
References
- [1].Guo S, Kemphues KJ. par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell 1995; 81:611-20. [DOI] [PubMed] [Google Scholar]
- [2].Elbert M, Rossi G, Brennwald P. The yeast par-1 homologs kin1 and kin2 show genetic and physical interactions with components of the exocytic machinery. Mol Biol Cell 2005; 16:532-49; PMID:15563607; http://dx.doi.org/ 10.1091/mbc.E04-07-0549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Shulman JM, Benton R, St Johnston D. The Drosophila homolog of C. elegans PAR-1 organizes the oocyte cytoskeleton and directs oskar mRNA localization to the posterior pole. Cell 2000; 101:377-88. [DOI] [PubMed] [Google Scholar]
- [4].Drewes G, Ebneth A, Preuss U, Mandelkow EM, Mandelkow E. MARK, a novel family of protein kinases that phosphorylate microtubule-associated proteins and trigger microtubule disruption. Cell 1997; 89:297-308; PMID:9108484; http://dx.doi.org/ 10.1016/S0092-8674(00)80208-1 [DOI] [PubMed] [Google Scholar]
- [5].Cohen D, Brennwald PJ, Rodriguez-Boulan E, Musch A. Mammalian PAR-1 determines epithelial lumen polarity by organizing the microtubule cytoskeleton. J Cell Biol 2004; 164:717-27; PMID:14981097; http://dx.doi.org/ 10.1083/jcb.200308104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Kemphues KJ, Priess JR, Morton DG, Cheng NS. Identification of genes required for cytoplasmic localization in early C. elegans embryos. Cell 1988; 52:311-20; PMID:3345562; http://dx.doi.org/ 10.1016/S0092-8674(88)80024-2 [DOI] [PubMed] [Google Scholar]
- [7].Slim CL, Lazaro-Dieguez F, Bijlard M, Toussaint MJ, de Bruin A, Du Q, Müsch A, van Ijzendoorn SC. Par1b induces asymmetric inheritance of plasma membrane domains via LGN-dependent mitotic spindle orientation in proliferating hepatocytes. PLoS Biol 2013; 11:e1001739; PMID:24358023; http://dx.doi.org/ 10.1371/journal.pbio.1001739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Cox DN, Lu B, Sun TQ, Williams LT, Jan YN. Drosophila par-1 is required for oocyte differentiation and microtubule organization. Curr Biol 2001; 11:75-87; PMID:11231123; http://dx.doi.org/ 10.1016/S0960-9822(01)00027-6 [DOI] [PubMed] [Google Scholar]
- [9].Doerflinger H, Benton R, Shulman JM, St Johnston D. The role of PAR-1 in regulating the polarised microtubule cytoskeleton in the Drosophila follicular epithelium. Development 2003; 130:3965-75; PMID:12874119; http://dx.doi.org/ 10.1242/dev.00616 [DOI] [PubMed] [Google Scholar]
- [10].Sato Y, Akitsu M, Amano Y, Yamashita K, Ide M, Shimada K, Yamashita A, Hirano H, Arakawa N, Maki T, et al.. The novel PAR-1-binding protein MTCL1 has crucial roles in organizing microtubules in polarizing epithelial cells. J Cell Sci 2013; 126:4671-83; PMID:23902687; http://dx.doi.org/ 10.1242/jcs.127845 [DOI] [PubMed] [Google Scholar]
- [11].Mandelkow EM, Thies E, Trinczek B, Biernat J, Mandelkow E. MARK/PAR1 kinase is a regulator of microtubule-dependent transport in axons. J Cell Biol 2004; 167:99-110; PMID:15466480; http://dx.doi.org/ 10.1083/jcb.200401085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Yoshimura Y, Terabayashi T, Miki H. Par1b/MARK2 phosphorylates kinesin-like motor protein GAKIN/KIF13B to regulate axon formation. Mol Cell Biol 2010; 30:2206-19; PMID:20194617; http://dx.doi.org/ 10.1128/MCB.01181-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Sapir T, Sapoznik S, Levy T, Finkelshtein D, Shmueli A, Timm T, Mandelkow EM, Reiner O. Accurate balance of the polarity kinase MARK2/Par-1 is required for proper cortical neuronal migration. J Neurosci 2008; 28:5710-20; PMID:18509032; http://dx.doi.org/ 10.1523/JNEUROSCI.0911-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Mejia-Gervacio S, Murray K, Sapir T, Belvindrah R, Reiner O, Lledo PM. MARK2/Par-1 guides the directionality of neuroblasts migrating to the olfactory bulb. Mol Cell Neurosci 2012; 49:97-103; PMID:22061967; http://dx.doi.org/ 10.1016/j.mcn.2011.10.006 [DOI] [PubMed] [Google Scholar]
- [15].Uboha NV, Flajolet M, Nairn AC, Picciotto MR. A calcium- and calmodulin-dependent kinase Ialpha/microtubule affinity regulating kinase 2 signaling cascade mediates calcium-dependent neurite outgrowth. J Neurosci 2007; 27:4413-23; PMID:17442826; http://dx.doi.org/ 10.1523/JNEUROSCI.0725-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Thies E, Mandelkow EM. Missorting of tau in neurons causes degeneration of synapses that can be rescued by the kinase MARK2/Par-1. J Neurosci 2007; 27:2896-907; PMID:17360912; http://dx.doi.org/ 10.1523/JNEUROSCI.4674-06.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Sapir T, Shmueli A, Levy T, Timm T, Elbaum M, Mandelkow EM, Reiner O. Antagonistic effects of doublecortin and MARK2/Par-1 in the developing cerebral cortex. J Neurosci 2008; 28:13008-13; PMID:19036994; http://dx.doi.org/ 10.1523/JNEUROSCI.2363-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Cohen D, Rodriguez-Boulan E, Musch A. Par-1 promotes a hepatic mode of apical protein trafficking in MDCK cells. Proc Natl Acad Sci U S A 2004; 101:13792-7; PMID:15365179; http://dx.doi.org/ 10.1073/pnas.0403684101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Ducharme NA, Hales CM, Lapierre LA, Ham AJ, Oztan A, Apodaca G, Goldenring JR. MARK2/EMK1/Par-1Balpha phosphorylation of Rab11-family interacting protein 2 is necessary for the timely establishment of polarity in Madin-Darby canine kidney cells. Mol Biol Cell 2006; 17:3625-37; PMID:16775013; http://dx.doi.org/ 10.1091/mbc.E05-08-0736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Hales CM, Vaerman JP, Goldenring JR. Rab11 family interacting protein 2 associates with Myosin Vb and regulates plasma membrane recycling. J Biol Chem 2002; 277:50415-21; PMID:12393859; http://dx.doi.org/ 10.1074/jbc.M209270200 [DOI] [PubMed] [Google Scholar]
- [21].Lapierre LA, Kumar R, Hales CM, Navarre J, Bhartur SG, Burnette JO, Jr Provance DW, Mercer JA, Bähler M, Goldenring JR. Myosin vb is associated with plasma membrane recycling systems. Mol Biol Cell 2001; 12:1843-57; PMID:11408590; http://dx.doi.org/ 10.1091/mbc.12.6.1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Casanova JE, Wang X, Kumar R, Bhartur SG, Navarre J, Woodrum JE, Altschuler Y, Ray GS, Goldenring JR. Association of Rab25 and Rab11a with the apical recycling system of polarized Madin-Darby canine kidney cells. Mol Biol Cell 1999; 10:47-61; PMID:9880326; http://dx.doi.org/ 10.1091/mbc.10.1.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Goldenring JR, Smith J, Vaughan HD, Cameron P, Hawkins W, Navarre J. Rab11 is an apically located small GTP-binding protein in epithelial tissues. AmJPhysiol 1996; 270(33):G515-G25. [DOI] [PubMed] [Google Scholar]
- [24].Ducharme NA, Williams JA, Oztan A, Apodaca G, Lapierre LA, Goldenring JR. Rab11-FIP2 regulates differentiable steps in transcytosis. Am J Physiol Cell Physiol 2007; 293:C1059-72; PMID:17626244; http://dx.doi.org/ 10.1152/ajpcell.00078.2007 [DOI] [PubMed] [Google Scholar]
- [25].Ducharme NA, Ham AJ, Lapierre LA, Goldenring JR. Rab11-FIP2 influences multiple components of the endosomal system in polarized MDCK cells. Cell Log 2011; 1:57-68; PMID:21686255; http://dx.doi.org/ 10.4161/cl.1.2.15289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Lapierre LA, Avant KM, Caldwell CM, Oztan A, Apodaca G, Knowles BC, Roland JT, Ducharme NA, Goldenring JR. Phosphorylation of Rab11-FIP2 regulates polarity in MDCK cells. Mol Biol Cell 2012; 23:2302-18; PMID:22553350; http://dx.doi.org/ 10.1091/mbc.E11-08-0681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Jin M, Goldenring JR. The Rab11-FIP1/RCP gene codes for multiple protein transcripts related to the plasma membrane recycling system. Biochim Biophys Acta 2006; 1759:281-95; PMID:16920206; http://dx.doi.org/ 10.1016/j.bbaexp.2006.06.001 [DOI] [PubMed] [Google Scholar]
- [28].Lindsay AJ, Hendrick AG, Cantalupo G, Senic-Matuglia F, Goud B, Bucci C, McCaffrey MW. Rab coupling protein (RCP), a novel Rab4 and Rab11 effector protein. J Biol Chem 2002; 277:12190-9; PMID:11786538; http://dx.doi.org/ 10.1074/jbc.M108665200 [DOI] [PubMed] [Google Scholar]
- [29].Lindsay AJ, McCaffrey MW. The C2 domains of the class I Rab11 family of interacting proteins target recycling vesicles to the plasma membrane. J Cell Sci 2004; 117:4365-75; PMID:15304524; http://dx.doi.org/ 10.1242/jcs.01280 [DOI] [PubMed] [Google Scholar]
- [30].Peden AA, Schonteich E, Chun J, Junutula JR, Scheller RH, Prekeris R. The RCP-Rab11 complex regulates endocytic protein sorting. Mol Biol Cell 2004; 15:3530-41; PMID:15181150; http://dx.doi.org/ 10.1091/mbc.E03-12-0918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Qi M, Williams JA, Chu H, Chen X, Wang JJ, Ding L, Akhirome E, Wen X, Lapierre LA, Goldenring JR, et al.. Rab11-FIP1C and Rab14 direct plasma membrane sorting and particle incorporation of the HIV-1 envelope glycoprotein complex. PLoS Pathog 2013; 9:e1003278; PMID:23592992; http://dx.doi.org/ 10.1371/journal.ppat.1003278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Rainero E, Caswell PT, Muller PA, Grindlay J, McCaffrey MW, Zhang Q, Wakelam MJ, Vousden KH, Graziani A, Norman JC. Diacylglycerol kinase alpha controls RCP-dependent integrin trafficking to promote invasive migration. J Cell Biol 2012; 196:277-95; PMID:22270919; http://dx.doi.org/ 10.1083/jcb.201109112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Caswell PT, Chan M, Lindsay AJ, McCaffrey MW, Boettiger D, Norman JC. Rab-coupling protein coordinates recycling of alpha5beta1 integrin and EGFR1 to promote cell migration in 3D microenvironments. J Cell Biol 2008; 183:143-55; PMID:18838556; http://dx.doi.org/ 10.1083/jcb.200804140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Eva R, Dassie E, Caswell PT, Dick G, ffrench-Constant C, Norman JC, Fawcett JW. Rab11 and its effector Rab coupling protein contribute to the trafficking of beta 1 integrins during axon growth in adult dorsal root ganglion neurons and PC12 cells. J Neurosci 2010; 30:11654-69; PMID:20810886; http://dx.doi.org/ 10.1523/JNEUROSCI.2425-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Muller PA, Caswell PT, Doyle B, Iwanicki MP, Tan EH, Karim S, Lukashchuk N, Gillespie DA, Ludwig RL, Gosselin P, et al.. Mutant p53 drives invasion by promoting integrin recycling. Cell 2009; 139:1327-41; PMID:20064378; http://dx.doi.org/ 10.1016/j.cell.2009.11.026 [DOI] [PubMed] [Google Scholar]
- [36].Landry MC, Champagne C, Boulanger MC, Jette A, Fuchs M, Dziengelewski C, Lavoie JN. A functional interplay between the small GTPase Rab11a and mitochondria-shaping proteins regulates mitochondrial positioning and polarization of the actin cytoskeleton downstream of Src family kinases. J Biol Chem 2014; 289:2230-49; PMID:24302731; http://dx.doi.org/ 10.1074/jbc.M113.516351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Boulay PL, Mitchell L, Turpin J, Huot-Marchand JE, Lavoie C, Sanguin-Gendreau V, Jones L, Mitra S, Livingstone JM, Campbell S, et al.. Rab11-FIP1C Is a Critical Negative Regulator in ErbB2-Mediated Mammary Tumor Progression. Cancer Res 2016; 76:2662-74; PMID:26933086; http://dx.doi.org/ 10.1158/0008-5472.CAN-15-2782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Zhang J, Liu X, Datta A, Govindarajan K, Tam WL, Han J, George J, Wong C, Ramnarayanan K, Phua TY, et al.. RCP is a human breast cancer-promoting gene with Ras-activating function. J Clin Invest 2009; 119:2171-83; PMID:19620787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Baetz NW, Goldenring JR. Rab11-Family Interacting Proteins define spatially and temporally distinct regions within the dynamic Rab11a-dependent recycling system. Molec Biol Cell 2013; 24:643-58; PMID:23283983; http://dx.doi.org/ 10.1091/mbc.E12-09-0659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Baetz NW, Goldenring JR. Distinct patterns of phosphatidylserine localization within the Rab11a-containing recycling system. Cell Log 2014; 4:e28680; PMID:25210648; http://dx.doi.org/ 10.4161/cl.28680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Cohen D, Musch A. Apical surface formation in MDCK cells: regulation by the serine/threonine kinase EMK1. Methods 2003; 30:269-76; PMID:12798141; http://dx.doi.org/ 10.1016/S1046-2023(03)00033-1 [DOI] [PubMed] [Google Scholar]
- [42].Ivanov AI, McCall IC, Babbin B, Samarin SN, Nusrat A, Parkos CA. Microtubules regulate disassembly of epithelial apical junctions. BMC Cell Biol 2006; 7:12; PMID:16509970; http://dx.doi.org/ 10.1186/1471-2121-7-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Gonzalez-Mariscal L, Contreras RG, Bolivar JJ, Ponce A, Chavez De Ramirez B, Cereijido M. Role of calcium in tight junction formation between epithelial cells. Am J Physiol 1990; 259:C978-86; PMID:2124417 [DOI] [PubMed] [Google Scholar]
- [44].Wallace DM, Lindsay AJ, Hendrick AG, McCaffrey MW. The novel Rab11-FIP/Rip/RCP family of proteins displays extensive homo- and hetero-interacting abilities. Biochem Biophys Res Commun 2002; 292:909-15; PMID:11944901; http://dx.doi.org/ 10.1006/bbrc.2002.6736 [DOI] [PubMed] [Google Scholar]
- [45].Cohen D, Tian Y, Musch A. Par1b promotes hepatic-type lumen polarity in Madin Darby canine kidney cells via myosin II- and E-cadherin-dependent signaling. Mol Biol Cell 2007; 18:2203-15; PMID:17409351; http://dx.doi.org/ 10.1091/mbc.E07-02-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif 2005; 41:207-34; PMID:15915565; http://dx.doi.org/ 10.1016/j.pep.2005.01.016 [DOI] [PubMed] [Google Scholar]
- [47].Barth AI, Pollack AL, Altschuler Y, Mostov KE, Nelson WJ. NH2-terminal deletion of beta-catenin results in stable colocaization of mutant beta-catenin with adenomatous polyposis coli protein and altered MDCK adhesion. JCell Biol 1997; 136:693-706; http://dx.doi.org/ 10.1083/jcb.136.3.693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Lapierre LA, Dorn MC, Zimmerman CF, Navarre J, Burnette JO, Goldenring JR. Rab11b resides in a vesicular compartment distinct from Rab11a in parietal cells and other epithelial cells. Exp Cell Res 2003; 290:322-31; PMID:14567990; http://dx.doi.org/ 10.1016/S0014-4827(03)00340-9 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.