Abstract
Background
Our aim was to assess the influence of age, gender and lifestyle factors on the effect of the obesity-promoting alleles of FTO and MCR4.
Methods
The HUNT study comprises health information on the population of Nord-Trøndelag county, Norway. Extreme phenotype participants (gender-wise lower and upper quartiles of waist-hip-ratio and BMI ≥ 35 kg/m2) in the third survey, HUNT3 (2006–08), were genotyped for the single-nucleotide polymorphisms rs9939609 (FTO) and rs17782313 (MC4R); 25686 participants were successfully genotyped. Extreme sampling was chosen to increase power to detect genetic and gene-environment effects on waist-hip-ratio and BMI. Statistical inference was based on linear regression models and a missing-covariate likelihood approach for the extreme phenotype sampling design. Environmental factors were physical activity, diet (artificially sweetened beverages) and smoking. Longitudinal analysis was performed using material from HUNT2 (1995–97).
Results
Cross-sectional and longitudinal genetic effects indicated stronger genetic associations with obesity in young than in old, as well as differences between women and men. We observed larger genetic effects among physically inactive compared to active individuals. This interaction was age-dependent and seen mainly in 20–40 year olds. We observed a greater FTO effect among men with a regular intake of artificially sweetened beverages, compared to non-drinkers. Interaction analysis of smoking was mainly inconclusive.
Conclusions
In a large all-adult and area-based population survey the effects of obesity-promoting minor-alleles of FTO and MCR4, and interactions with life style factors are age- and gender-related. These findings appear relevant when designing individualized treatment for and prophylaxis against obesity.
Introduction
It is well recognized that both genetic and non-genetic factors are operative in the development and persistence of obesity. Polymorphisms of the FTO gene are shown to be the strongest genetic determinant of obesity [1] compared with the > 40 other genes which affect body weight [2, 3]. Given the primacy of FTO, the minor-allele exerts only modest effects in most studies. It is possible that interactions with other factors may under some conditions lead to an increase (or decrease) in effects. However, whether the impact of FTO on obesity is modified by lifestyle factors, such as diet, physical activity and smoking has only been partly elucidated. Also gene-environment interactions should be tested in relation to age and gender, something that has so far only been rarely done.
Interactions with physical activity have been demonstrated when results from a large number of different populations have been pooled together [4, 5]. However, such meta-analyses have also revealed differences between populations [5], something which is also evident in individual studies, some demonstrating an interaction [6–8] others not [9]. Such differences reduce the possibilities to characterize an interaction in detail. A large individual study could then be preferable for detailed characterization. This also applies to diet where results are discordant between investigations as to interactions with FTO [10–15], and non-conclusive as to smoking [16]. A study encompassing different ages and both genders would also allow the assessment of whether age and gender modify interactions with the lifestyle factors mentioned. The potential importance of age and gender for FTO effects was previously recognised [17, 18] but the effects have so far been insufficiently characterized.
The HUNT all-population survey provides data that appear to be representative of Norway and Scandinavia [19]. Here we designed a study for the associations between FTO (rs9939609) and obesity, including gene-environment interactions, based on data from HUNT3 (2006–2008). Results on FTO were compared with results on MCR4 (rs17782313), which is also an influential gene [20], albeit not with the same impact as FTO in relation to obesity [21]. We previously analysed waist-hip-ratio (WHR) with respect to a range of dietary variables in the HUNT material [22]. Here, we obtained genotypes for approximately half of the HUNT3 participants by selecting individuals according to an extreme phenotype sampling design (extremes being lowest and highest quartiles of WHR, and additionally all individuals with BMI ≥ 35 kg/m2), rather than drawing a random sample. Genotyping the phenotypic extremes is considered to increase the statistical power to detect associations between genetic variants and continuous phenotypes [23]. In addition to large sample sizes, choosing a powerful design can be particularly important for analysis of gene-environment interactions (GEIs), which one would expect to have low power to detect [24–26]. By genotyping individuals with extreme WHR and BMI values, and by carefully selecting and constructing environmental variables from the HUNT material we should have good power to detect GEIs. Statistical inference methods that correctly account for extreme sampling have been proposed in the statistical literature [23], but to our knowledge not applied to the extent that we have done here. By adapting these methods, we have assessed over-all impact as well as gene-environment interactions with respect to WHR and BMI. Analysis was performed gender-wise in three age groups. Although most of the data analysed were cross-sectional, we also assessed longitudinal effects, then using data from HUNT2 (1995–97).
Materials and methods
The HUNT study [19] comprises a database of health and medical information on the population of Nord-Trøndelag county, Norway, collected in three surveys; HUNT1 (1984–86), HUNT2 (1995–97) and HUNT3 (2006–08). In HUNT3 the participation rate out of the 93860 invited was 54.1%. Trained nurses performed measurements of waist circumference, hip circumference, weight and height. From these measurements we calculated WHR and BMI. We carefully constructed three environmental lifestyle variables based on Questionnaire 1 in HUNT3 to represent physical activity, diet and smoking. We summarized four questions on physical activity into a 9-leveled index. The index then captured frequency, intensity and duration of exercise, in addition to overall daily physical inactivity. Four questions on cigarette smoking were summarized in the variable of pack years, defined as the average number of packs of 20 cigarettes smoked per year during the period of daily smoking. Mostad et al has previously analysed the relationship between WHR and dietary variables in HUNT3 [22]. Here we selected artificially sweetened beverages (measured in glasses drunk per week) as a dietary explanatory variable because artificially sweetened beverages was found to be strongly associated with WHR (57% higher intake among individuals with central obesity compared to individuals without central obesity [22]). For details on the construction of environmental lifestyle variables from Questionnaire 1, see S1 Appendix.
Extreme phenotype participants in HUNT3 were genotyped for FTO (rs9939609) and MC4R (rs17782313). Extreme phenotypes were defined as the gender-wise lower and upper quartiles of WHR (WHR < 0.817 and WHR > 0.917 for women, WHR < 0.895 and WHR > 0.981 for men) and additionally BMI ≥ 35 kg/m2. Of the 25981 extreme phenotype individuals, 25686 were successfully genotyped for both SNPs (98.9%). See S1 Appendix for details on the genotyping procedure.
The Regional Committees for Medical and Health Research Ethics for central Norway (REC Central) approved this research project.
Statistical methods
We assumed that in the underlying population, WHR and BMI could be modelled with linear regression models with environmental covariates (age, physical activity (PA), artificially sweetened beverages (ASB) and smoking (PCYR)) and genetic covariates (FTO, MC4R). Age was coded into 5-year intervals and treated as a categorical variable, denoted xage. The physical activity, diet and smoking variables were treated as continuous explanatory variables, xPA, xASB and xPCYR. We analysed data from men and women separately. Genetic covariates were genotypes of the FTO and MC4R SNPs, xFTO and xMC4R and coded as 0, 1 or 2 according to the number of copies of the minor-allele (FTO: 0 = TT, 1 = TA, 2 = AA, MC4R: 0 = TT, 1 = TC, 2 = CC). Our basic regression model for over-all genetic effects was
| (1) |
where Y is a trait (WHR or BMI), ε is normally distributed with mean 0 and variance σ2, and
For longitudinal analysis, we modelled the difference in traits ΔY between HUNT3 and HUNT2 by
| (2) |
where η is as in Model (1). All lifestyle and genetic covariates were taken from HUNT3 while changes in WHR and BMI were calculated based on HUNT2 and HUNT3 data. Longitudinal analysis was then based on individuals participating in both HUNT2 and HUNT3. For gene-environment interactions we included a statistical interaction term between the relevant environmental covariate and SNPs and thereby assessed departures from additive effects. For example for interactions with artificially sweetened beverages the regression model was
| (3) |
where η is as in Model (1), and similarly for physical activity and smoking. In the following, the term “estimated effect of FTO” refers to estimates of the parameter βFTO (over-all or longitudinal effect) while the term “estimated GEI effect” refers to estimates of βFTO*ENV (where ENV is any of the three environmental variables), and similarly for MC4R.
By the extreme sampling design the variables y, xage, xPA, xASB and xPCYR were observed for the full sample, while the genetic variables xFTO and xMC4R were only observed for the extreme phenotype individuals. The genetic covariates were then missing at random in the full sample, and we used likelihood methods for missing covariate data (S2 Appendix). In such missing covariate likelihoods, the distribution of the missing covariates (here FTO and MC4R) must be estimated. We assumed that SNP genotypes could be modelled by multinomial distributions that were independent of all non-genetic covariates. We verified this assumption by testing for independence between genotypes and environmental covariates in the extreme genotyped samples, and by sensitivity analyses (S2 Appendix). Maximum likelihood estimates of parameters in the three regression models were found by numerical optimization. We implemented the score test for extreme sampling data to test H0: βFTO = 0 against H1: βFTO ≠ 0, and H0: βMC4R = 0 against H1: βMC4R ≠ 0 in the over-all effects Model (1) and the longitudinal Model (2). We implemented the likelihood ratio test for extreme sampling data to test GEIs such as H0: βFTO*PA = 0 against H1: βFTO*PA ≠ 0 in Model (3). All statistical methods were implemented and executed in R [27], see S2 Appendix for further details.
An important assumption for linear regression models is independence. The HUNT3 population is a stable population; hence many individuals should be related. Due to privacy regulations any familial ties between study participants were unknown to us. We divided the sample into age groups; 20–40 years (20 ≤ age < 40), 40–60 years (40 ≤ age < 60), 60–80 years (60 ≤ age < 80) that we analysed separately. This should eliminate to a large extent the unknown but assumed co-presence of parents and children in each sample. Importantly, such stratified analysis also allowed us to detect age- and gender-related influences on the genetic impact of FTO and MCR4.
A large number of tests were performed in our analysis because we analysed six age and gender groups separately for two outcomes (WHR and BMI). For each of these, we considered over-all effects, longitudinal effects and three different GEIs; in total 120 tests. To account for the large number of tests, we have chosen to control the false discovery rate (FDR) at the 5% level using the Benjamini-Hochberg step-up procedure [28].
Power-simulation study
We performed a power simulation study to verify that the extreme sampling design could in fact be more powerful than a random sampling design for our analysis (S3 Appendix). We used all non-genetic covariates from the 20–40 year female stratum, and then simulated a genetic covariate. We used the actual parameter estimates from this stratum and simulated a new response. Then the estimated statistical power to detect a non-zero GEI effect at significance level 0.05 was 84% in the full (simulated) data set. We then considered genotyping only the extremes (upper and lower quartile), compared to genotyping a random sample. We found that the extreme sampling design had 80% power, while a random sampling design had 56% power. Thus, to detect GEIs in our data when a fixed number of individuals (approximately half of all HUNT3 participants) could be genotyped, it seems crucial to select those with extreme phenotypes, rather than a random sample.
Results
Relevant sample sizes for our analysis are presented in Table 1. The age and gender groups varied in size between 3000 and 8000 participants, the largest being the 40–60 years age group. Estimates of βFTO and βMC4R in the over-all effects Model (1) and longitudinal effects Model (2) are presented Figs 1 and 2 and in S1 and S2 Tables. In S1 and S2 Tables we also report FDR-adjusted p-values for the hypothesis tests H0: βFTO = 0 against H1: βFTO ≠ 0, and H0: βMC4R = 0 against H1: βMC4R ≠ 0. Estimates of GEIs in Model (3), and adjusted p-values for the hypothesis tests H0: βFTO*ENV = 0 and H0: βMC4R*ENV = 0 are presented in Table 2 (physical activity), Table 3 (artificially sweetened beverages) and Table 4 (pack years of smoking). Out of the 120 tests for association that we performed, 24 tests had FDR-adjusted p-values below 0.05. Thus, by controlling the false discovery rate at a 0.05 significance level we have 24 findings. Below we summarize main findings based on both estimated effect sizes and significance of test results.
Table 1. Sample sizes.
| 20–40 years | 40–60 years | 60–80 years | ||||
|---|---|---|---|---|---|---|
| Men | Women | Men | Women | Men | Women | |
| Complete sample | 3237 | 4817 | 6694 | 7944 | 4084 | 3948 |
| Genotyped sample | 2029 | 2740 | 3081 | 3794 | 1925 | 2037 |
| HUNT2/HUNT3 sample | 993* | 1505* | 5216 | 6397 | 3693 | 3660 |
Sample sizes gender-wise for each age group, 20–40 years (*30–40 years in HUNT2-HUNT3 analysis because individuals younger than 30 years in HUNT3 did not participate in HUNT2), 40–60 years and 60–80 years. Genotyped sample refers to individuals who were genotyped based on the extreme phenotype criteria.
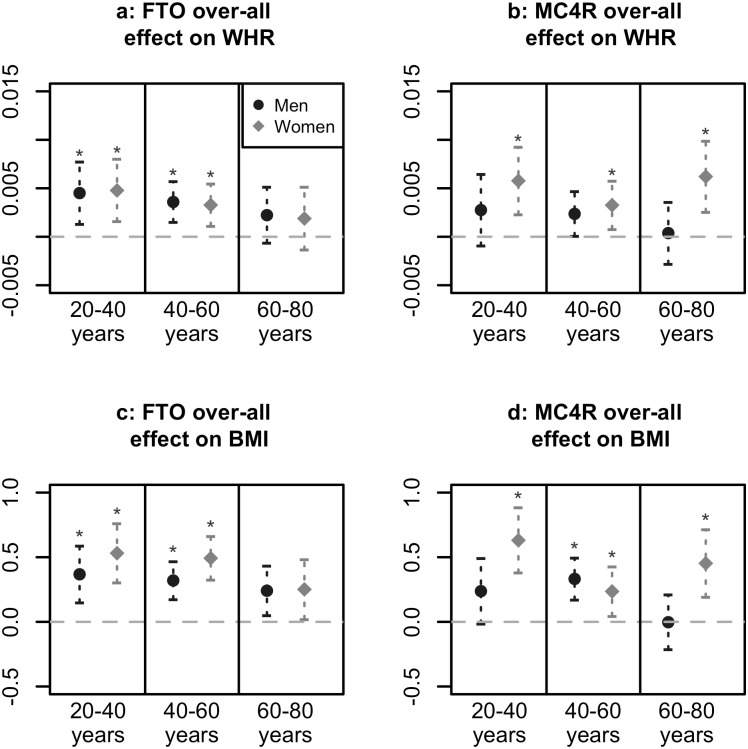
Fig 1. Over-all genetic effects.
Over-all effects of FTO and MC4R with 95% confidence intervals, for men and women in three age groups. Asterisk indicates FDR-adjusted p-value below 0.05 for testing H0: βSNP = 0 against H1: βSNP ≠ 0.
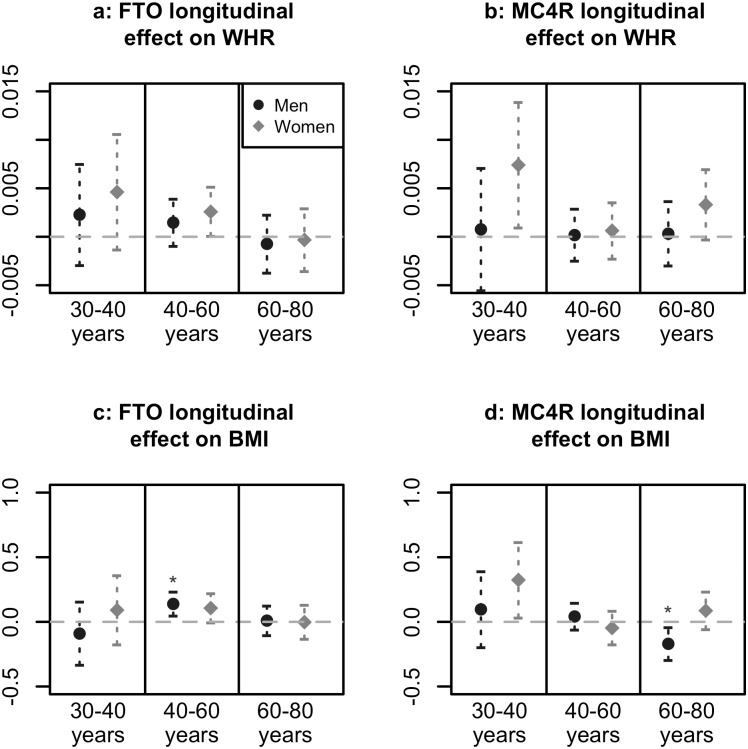
Fig 2. Longitudinal genetic effects.
Longitudinal effects of FTO and MC4R with 95% confidence intervals, for men and women in three age groups (30–40 years as youngest age group because individuals younger than 30 years in HUNT3 did not participate in HUNT2). Asterisk indicates FDR-adjusted p-value below 0.05 for testing H0: βSNP = 0 against H: βSNP ≠ 0.
Table 2. Interactions with physical activity.
| Trait | SNP | Age group | Gender | βPA | βSNP | βSNP*PA | 95% CI for βSNP*PA | Adjusted p-value |
|---|---|---|---|---|---|---|---|---|
| WHR | FTO | 20–40 years | Men | -0.0026 | 0.017 | -0.0029 | -0.0048, -0.0011 | 0.029 |
| Women | 6.9e-05 | 0.017 | -0.0028 | -0.0048, -0.00073 | 0.048 | |||
| 40–60 years | Men | -0.0066 | 0.0024 | 0.00028 | -0.001, 0.0016 | 0.86 | ||
| Women | -0.0038 | 0.0018 | 0.00034 | -0.0012, 0.0019 | 0.86 | |||
| 60–80 years | Men | -0.007 | 0.0044 | -0.00051 | -0.0023, 0.0013 | 0.81 | ||
| Women | -0.0088 | -0.00081 | 0.00064 | -0.0017, 0.003 | 0.86 | |||
| MC4R | 20–40 years | Men | -0.0026 | 0.0085 | -0.0013 | -0.0034, 0.00071 | 0.51 | |
| Women | 6.9e-05 | 0.01 | -0.001 | -0.0032, 0.0012 | 0.7 | |||
| 40–60 years | Men | -0.0066 | 0.0042 | -0.00043 | -0.0019, 0.0011 | 0.84 | ||
| Women | -0.0038 | 0.0051 | -0.00043 | -0.0022, 0.0013 | 0.86 | |||
| 60–80 years | Men | -0.007 | -0.0066 | 0.0016 | -0.00033, 0.0036 | 0.3 | ||
| Women | -0.0088 | 0.0068 | -0.00014 | -0.0027, 0.0025 | 1 | |||
| BMI | FTO | 20–40 years | Men | -0.041 | 1.0 | -0.15 | -0.28, -0.018 | 0.11 |
| Women | 0.053 | 1.4 | -0.2 | -0.34, -0.049 | 0.048 | |||
| 40–60 years | Men | -0.22 | 0.36 | -0.0088 | -0.1, 0.082 | 1 | ||
| Women | -0.30 | 0.84 | -0.078 | -0.2, 0.04 | 0.45 | |||
| 60–80 years | Men | -0.40 | -0.078 | 0.075 | -0.043, 0.19 | 0.46 | ||
| Women | -0.66 | 0.14 | 0.027 | -0.14, 0.19 | 0.95 | |||
| MC4R | 20–40 years | Men | -0.041 | 0.26 | -0.0075 | -0.15, 0.14 | 1 | |
| Women | 0.053 | 1.7 | -0.24 | -0.41, -0.078 | 0.025 | |||
| 40–60 years | Men | -0.22 | 0.46 | -0.031 | -0.13, 0.073 | 0.81 | ||
| Women | -0.3 | 0.024 | 0.048 | -0.085, 0.18 | 0.78 | |||
| 60–80 years | Men | -0.4 | -0.6 | 0.14 | 0.015, 0.27 | 0.11 | ||
| Women | -0.66 | 0.64 | -0.046 | -0.23, 0.14 | 0.86 |
Gene-environment interaction effects for physical activity (PA); estimated main effect of physical activity (βPA), estimated main effect of SNPs (βSNP), estimated GEI effects (βSNP*PA), 95% confidence intervals for the GEI effects and FDR-adjusted p-value for testing H0: βSNP*PA = 0 against H0: βSNP*PA ≠ 0.
Table 3. Interactions with artificially sweetened beverages.
| Trait | SNP | Age group | Gender | βASB | βSNP | βSNP*ASB | 95% CI for βSNP*ASB | Adjusted p-value |
|---|---|---|---|---|---|---|---|---|
| WHR | FTO | 20–40 years | Men | 0.0013 | 0.0029 | 0.00037 | -0.00018, 0.00091 | 0.43 |
| Women | 0.0013 | 0.005 | -4.3e-05 | -0.00056, 0.00047 | 1 | |||
| 40–60 years | Men | 0.0011 | 0.0024 | 0.00043 | -6.1e-05, 0.00093 | 0.43 | ||
| Women | 0.0012 | 0.0025 | 3e-04 | -2e-04, 0.00081 | 0.66 | |||
| 60–80 years | Men | 8e-04 | 0.00059 | 0.00094 | 6.3e-05, 0.0018 | 0.15 | ||
| Women | 0.0015 | 0.0013 | 0.00032 | -0.00067, 0.0013 | 0.87 | |||
| MC4R | 20–40 years | Men | 0.0013 | 0.0046 | -0.00041 | -0.001, 0.00019 | 0.43 | |
| Women | 0.0013 | 0.0051 | 0.00013 | -0.00043, 7e-04 | 0.89 | |||
| 40–60 years | Men | 0.0011 | 0.0028 | -0.00016 | -0.00072, 0.00039 | 0.72 | ||
| Women | 0.0012 | 0.0026 | 0.00027 | -3e-04, 0.00084 | 0.72 | |||
| 60–80 years | Men | 8e-04 | 0.0017 | -7e-04 | -0.0016, 0.00018 | 0.19 | ||
| Women | 0.0015 | 0.0068 | -0.00042 | -0.0015, 7e-04 | 0.72 | |||
| BMI | FTO | 20–40 years | Men | 0.085 | 0.13 | 0.059 | 0.021, 0.097 | 0.024 |
| Women | 0.14 | 0.52 | 0.002 | -0.037, 0.041 | 1 | |||
| 40–60 years | Men | 0.056 | 0.19 | 0.053 | 0.019, 0.086 | 0.024 | ||
| Women | 0.17 | 0.46 | 0.013 | -0.025, 0.052 | 0.81 | |||
| 60–80 years | Men | 0.074 | 0.15 | 0.059 | -0.00049, 0.12 | 0.15 | ||
| Women | 0.19 | 0.35 | -0.069 | -0.14, 0.0046 | 0.2 | |||
| MC4R | 20–40 years | Men | 0.085 | 0.39 | -0.038 | -0.082, 0.0049 | 0.22 | |
| Women | 0.14 | 0.62 | 0.0028 | -0.041, 0.046 | 1 | |||
| 40–60 years | Men | 0.056 | 0.33 | -0.00019 | -0.039, 0.039 | 1 | ||
| Women | 0.17 | 0.3 | -0.024 | -0.067, 0.02 | 0.59 | |||
| 60–80 years | Men | 0.074 | 0.044 | -0.021 | -0.08, 0.038 | 0.62 | ||
| Women | 0.19 | 0.41 | 0.025 | -0.064, 0.11 | 0.78 |
Gene-environment interaction effects for artificially sweetened beverages (ASB; estimated main effect of artificially sweetened beverages (βASB), estimated main effect of SNPs (βSNP), estimated GEI effects (βSNP*ASB), 95% confidence intervals for the GEI effects and FDR-adjusted p-value for testing H0: βSNP*ASB = 0 against H1: βSNP*ASB ≠ 0.
Table 4. Interactions with pack years.
| Trait | SNP | Age group | Gender | βPCYR | βSNP | βSNP*PCYR | 95% CI for βSNP*PCYR | Adjusted p-value |
|---|---|---|---|---|---|---|---|---|
| WHR | FTO | 20–40 years | Men | 0.001 | 0.0044 | 2e-05 | -0.00071, 0.00075 | 1 |
| Women | 0.0016 | 0.0039 | 0.00042 | -0.00036, 0.0012 | 0.59 | |||
| 40–60 years | Men | 0.00045 | 0.0013 | 3e-04 | 0.00011, 0.00048 | 0.81 | ||
| Women | 0.00079 | 0.0018 | 0.00023 | -8.8e-06, 0.00046 | 1 | |||
| 60–80 years | Men | 0.00036 | -3.2e-5 | 0.00019 | 1.7e-05, 0.00036 | 1 | ||
| Women | -0.00011 | -0.0015 | 0.00036 | 7.3e-05, 0.00065 | 0.73 | |||
| MC4R | 20–40 years | Men | 0.001 | 0.0017 | 5e-04 | -0.00035, 0.0013 | 0.5 | |
| Women | 0.0016 | 0.0057 | -3.3e-06 | -0.00078, 0.00077 | 1 | |||
| 40–60 years | Men | 0.00045 | 0.0022 | 1.8e-05 | -0.00019, 0.00023 | 1 | ||
| Women | 0.00079 | 0.0042 | -0.00013 | -0.00039, 0.00013 | 0.42 | |||
| 60–80 years | Men | 0.00036 | -0.0015 | 0.00014 | -2.9e-05, 0.00031 | 1 | ||
| Women | -0.00011 | 0.0063 | -7e-05 | -0.00039, 0.00025 | 1 | |||
| BMI | FTO | 20–40 years | Men | 0.076 | 0.37 | 0.00098 | -0.051, 0.053 | 1 |
| Women | 0.018 | 0.4 | 0.063 | 0.0056, 0.12 | 0.1 | |||
| 40–60 years | Men | 0.0012 | 0.2 | 0.016 | 0.0041, 0.028 | 0.048 | ||
| Women | 0.033 | 0.63 | -0.019 | -0.036, -0.0023 | 0.1 | |||
| 60–80 years | Men | 0.023 | 0.31 | -0.0053 | -0.016, 0.0055 | 0.73 | ||
| Women | -0.036 | 0.11 | 0.02 | 0.0015, 0.039 | 0.11 | |||
| MC4R | 20–40 years | Men | 0.076 | 0.24 | -0.0045 | -0.066, 0.057 | 1 | |
| Women | 0.018 | 0.56 | 0.026 | -0.03, 0.083 | 0.5 | |||
| 40–60 years | Men | 0.0012 | 0.34 | -0.0014 | -0.015, 0.012 | 1 | ||
| Women | 0.033 | 0.37 | -0.018 | -0.037, 0.00021 | 0.15 | |||
| 60–80 years | Men | 0.023 | 0.099 | -0.0078 | -0.018, 0.0026 | 0.42 | ||
| Women | -0.036 | 0.54 | -0.011 | -0.033, 0.011 | 0.5 |
Gene-environment interaction effects for pack-years (PCYR); estimated main effect of smoking (βPCYR), estimated main effect of SNPs (βSNP) and estimated GEI effects (βSNP*PCYR), 95% confidence intervals for the GEI effects and FDR-adjusted p-value for testing H0: βSNP*PCYR = 0 against H1: βSNP*PCYR ≠ 0.
Over-all genetic effects
Over-all effects of the FTO and MC4R SNPs on WHR are presented in Fig 1. The FTO minor-allele was associated with higher WHR in 20–40 and 40–60 year olds (Fig 1a). For both genders, the effect sizes of FTO tended to be highest in the youngest age group. Similar results were seen for BMI (Fig 1c). The MC4R minor-allele was associated with higher WHR in all female age groups (Fig 1b), and with BMI also in the male 40–60 year olds (Fig 1d). Among women the greatest effect sizes of MC4R were found among 20–40 and 60–80 year olds while in men the association decreased with age.
Longitudinal effects
In the HUNT population, average WHR and BMI generally increased in the decade between HUNT2 and HUNT3 in all age and gender groups (S3 Table). Our hypothesis was then that this increase was more (or less) rapid depending on the FTO or MC4R genotype. Estimated longitudinal effects of FTO and MC4R on WHR and BMI are presented in Fig 2. Two significant associations (FDR-adjusted p-value < 0.05) were found; the FTO minor-allele was associated with a more rapid increase in BMI in 40–60 year old men, and the MC4R minor-allele was associated with a less rapid increase in BMI among 60–80 year old men. Although significantly different from zero, these effect sizes are negligible compared to the over-all effects presented in Fig 1. Sample sizes were low for the youngest HUNT3 individuals (Table 1), but longitudinal effect size estimates and confidence intervals indicated that the MC4R minor-allele was associated with a more rapid increase in WHR and BMI in young women between HUNT2 and HUNT3 (Fig 2b and 2d). These effect sizes are comparable to the over-all effects.
Interactions with physical activity
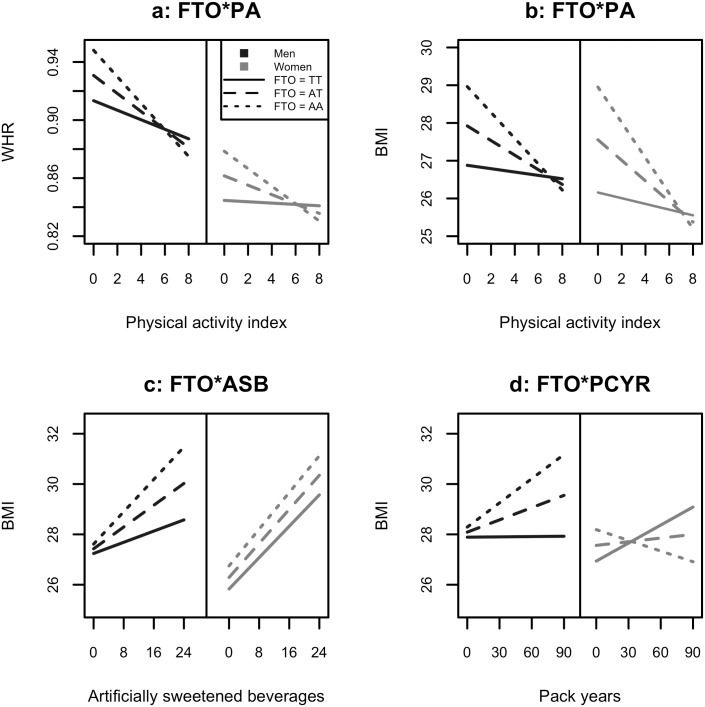
Estimated parameters from modelling interaction effects between FTO and MC4R and physical activity are presented in Table 2. For both men and women 20–40 years, we found that the interaction between physical activity and FTO was associated with WHR (FDR-adjusted p-value < 0.05). For women 20–40 years, we also found that both the interactions between FTO and physical activity, and MC4R and physical activity were associated with BMI. Consider a man 20–40 years with one copy of the FTO minor-allele. In our regression model for WHR with interactions (Model (3)) we estimated βFTO = 0.017 and βFTO*PA = −0.0029 (Table 2). Thus, with physical activity index 5 or greater, we see that the effect of one FTO minor-allele was cancelled out by the interaction between FTO and physical activity. In Fig 3a we have plotted estimated WHR against increasing levels of physical activity for an average man and woman 20–40 years for the three genotypes of FTO. Similarly, estimated BMI is presented in Fig 3b. Estimated WHR or BMI was quite similar for individuals with high levels of physical activity, regardless of FTO genotype. For individuals with low levels of physical activity, those with one or two copies of the FTO minor-allele had higher estimated WHR or BMI than those with no minor-allele. In other words, the over-all effect of FTO on obesity was negligible in highly active individuals aged between 20 and 40 years.
Fig 3. Gene-environment interaction effects.
Estimated WHR (a) and BMI (b) in the 20–40 years age group for all FTO genotypes and for increasing levels of physical activity (PA) for an average man (age 31, ASB 4.2, PCYR 1.8) and woman (age 31, ASB 4.7, PCYR 2.1). Estimated BMI in the 40–60 years age group for all FTO genotypes for increasing levels of intake of artificially sweetened beverages (c) and increasing number of pack years (d) for an average man (age 51, PA 4.2, ASB 2.7, PCYR 7.4) and woman (age 49, PA 4.3, ASB 2.7, PCYR 7.1).
Interactions with artificially sweetened beverages
Estimated parameters from modelling interaction effects between FTO and MC4R and weekly intake of artificially sweetened beverages are presented in Table 3. Among men, aged 20–40 and 40–60 years, the interaction between artificially sweetened beverages and FTO was significantly associated with BMI. Estimated BMI in the 40–60 year age group (for an average man and woman) for all genotypes of FTO and with respect to increasing intake of such beverages is presented in Fig 3c. Among both men and women the estimated BMI increased as the weekly intake of artificially sweetened beverages increased. In men, the increase was more rapid in individuals with one or more copies of the FTO minor-allele. In other words, a high intake of artificially sweetened beverages was associated with a higher BMI in men with the FTO minor-allele, compared to men with no FTO minor-allele. Among individuals with a low intake of such beverages, the estimated BMI was similar for all men, regardless of FTO genotype. In women, no significant GEI effect was found and estimated BMI increased with the same rate for increasing intake, regardless of FTO genotype.
Interactions with smoking (pack years)
Estimated parameters from modelling interaction effects between FTO and MC4R and pack years of smoking are presented in Table 4. The direction of the over-all smoking effects and GEI effects tended to vary between age groups, genders and between WHR and BMI.
For men 40–60 years, the interaction between smoking and FTO was associated with increased BMI. We estimated βFTO = 0.201 and βFTO*PCYR = 0.016. For women 40–60 years the direction of the GEI effect seemed the opposite of that found in men (Fig 3d) but was not significant. Here, for men, estimated BMI was not affected by smoking in individuals with no FTO minor-alleles, while for men with one or more FTO minor-allele, an increase in pack years was associated with higher BMI. For women, estimated BMI increased with pack years for individuals with no FTO minor-allele, but decreased with pack years for individuals with two copies of the FTO minor-allele. In women 20–40 and 60–80 years, the estimated GEI effect was in the opposite direction of that found in women 40–60 years (Table 4).
Discussion
Here we have assessed gene-environment interaction effects of FTO and MC4R on obesity, taking into account any modulation due to age and gender. Our contribution to this field seems important since our data material comes from a large homogeneous population- and area-based study (HUNT). Anthropometric measurements in HUNT were taken by trained nurses and not self-reported. We have carefully selected environmental covariates to represent various lifestyle aspects: physical activity, smoking and diet. Furthermore we used the extreme phenotype sampling design in order to increase the statistical power of our study, as compared to randomly sampling study participants.
We expectedly confirm an overall obesity-promoting effect of the FTO and MC4R minor-alleles in our study population. Of note, the effect appeared age-dependent with the strongest effect seen in the 20–40 year olds. Also our longitudinal findings indicate a diminished or absent influence of the FTO and MC4R minor-alleles at older age. An age-dependent attenuation of the FTO effect in adults has also been reported in other populations, albeit in smaller [17] and non-population-based [18] studies which did not include the highest age group of the present study [29–32]. (Some studies indicate a biphasic development with increasing effect during childhood and adolescence, followed by decreasing influence. Our study population did not include non-adults). In our analysis, MC4R effects seemed more pronounced in women than in men. Gender related effects on the impact of genetic factors on obesity have also previously been recognized [17, 18] but not characterized in detail. Gender-related differences can tentatively be ascribed to hormonal influences and gender-dependent differences in regional depots of adipose tissue in which the regulation of metabolism is known to differ [33, 34], and on which genetic influences may vary [35–37].
Genetic interactions with physical activity and in particular the age- and gender-dependent modulations of these effects were the main findings of our study. A previous study based on HUNT data did not find an interaction [38], possibly due to a much smaller study population than the present one. Also, we constructed here a variable to represent physical activity that accounted for frequency, intensity and duration of exercise, in addition to overall daily physical inactivity. Importantly, significant interactions with physical activity were observed only in the 20–40 year age group. In this group, the interaction between FTO and physical activity was associated with WHR and BMI in both genders, and the interaction between MC4R and physical activity was associated with BMI in women. Our results indicate that high physical activity diminishes the obesity-promoting effect of FTO and MC4R. From these results it could seem possible that interactions with physical activity can be generalized to the totality of the genetic load predisposing for obesity. However, the modulating influences of factors such as age and gender may differ between genetic risk factors as further highlighted by our results on diet and smoking, which are discussed below.
For interactions with diet we chose a somewhat unconventional parameter, namely weekly intake of artificially sweetened beverages. The choice of this parameter was based on its strong association with central obesity, as reported by us [22]. Other studies have also reported an obesity-promoting effect of artificially sweetened beverages [39–41]. Reverse causality, i.e. increased intake of artificially sweetened beverages in obese people as an attempt to decrease weight, could certainly be at play. That we find an interaction effect with FTO in men 20–40 and 40–60 years old indicates that the over-all effect of drinking artificially sweetened beverages is at least not wholly explained by reverse causality. In support of this notion several recent studies report increasing appetite [42, 43] and obesity-favouring microbiotic effects [44] by artificially sweetened beverages. We speculate that a likely effect of FTO on food intake may synergize with an effect of artificially sweetened beverages on appetite. In our data material, artificially sweetened beverages was the dietary variable that best described central obesity. It was therefore selected as the variable that would carry most power to detect gene-diet interactions. A more powerful study for gene-diet interactions could perhaps be based on a dietary variable combining several aspects of participants’ diet. In this context it should be recognized as a limitation that all dietary data from HUNT are based on self-reporting.
Smoking has well recognized effects on body size, including associations to obesity. It was therefore of interest to test for interactions between smoking and genetic variants with respect to obesity. We found little evidence of interactions (only in men 40–60 years old), and effect sizes were in varying directions between age groups and genders. Smoking behaviour may differ between the genders and age groups with regard to cigarette brand choices, daily vs. occasional smoking, and association or not with intake of alcoholic beverages; such putative differences may impact the present results. Also, the diverging effects of smoking on body size should be taken into account when interpreting our results; a decreasing effect due to effects of nicotine on metabolism and appetite, increasing effects due to other factors [45]. The pack year variable was chosen to capture the totality of the smoking load and is widely used in epidemiological research on tobacco. Self-reporting is however an obvious limitation for this variable.
Interpretations of our results are subject to limitation by the sizable number of non-participants. The total rate of non-participation in the HUNT3 survey was 46%. Data on non-participants has been assembled and analyzed [46] and the analysis indicates that obesity was more prevalent in non-participants than in participants. Non-participation combined with increased obesity could indicate a different impact of lifestyle factors vs. genetic factors in non-participants than in the present study population.
In conclusion, the present data from an all-adult population survey demonstrate that age and gender importantly modify interactions between lifestyle factors and the obesity-promoting effects of FTO and MCR4. These findings appear relevant when designing individualized treatment for and prophylaxis against obesity.
Supporting information
(PDF)
(PDF)
(PDF)
Estimated over-all effects (βSNP) with 95% confidence intervals for men and women in age groups 20–40 years, 40–60 years and 60–80 years. FDR-adjusted p-values for testing H0: βSNP = 0 against H1: βSNP ≠ 0.
(PDF)
Estimated longitudinal effects (βSNP) with 95% confidence intervals for men and women in age groups 30–40 years (individuals younger than 30 years in HUNT3 did not participate in HUNT2), 40–60 years and 60–80 years. FDR-adjusted p-values for testing H0: βSNP = 0 against H1: βSNP ≠ 0.
(PDF)
Average differences in WHR and BMI between HUNT3 and HUNT2 for men and women in three age groups. Positive differences indicate increase from HUNT2 to HUNT3.
(PDF)
Acknowledgments
The Nord-Trøndelag Health Study (the HUNT study) is collaboration between the HUNT Research Centre (Faculty of Medicine, Norwegian University of Science and Technology), the Nord-Trøndelag County Council, the Central Norway Health Authority, and the Norwegian Institute of Public Health.
Data Availability
Data are third party data owned by the The Nord-Trøndelag Health Study (HUNT) (http://www.ntnu.edu/hunt/). Any research group with a Principal Investigator affiliated with a Norwegian research institute can apply for access to analyse HUNT data. Projects must have recommendations from The Regional Committee for Medical Research in Norway (REK) and be registered with The Norwegian Social Science Data Services (NSD). Interested researchers may apply for access to these data here: http://www.ntnu.edu/hunt/data.
Funding Statement
ILM received funding from the Liaison Committee between the Central Norway Regional Health Authority and the Norwegian University of Science and Technology (https://helse-midt.no/samarbeidsorganet). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316(5826):889–94. Epub 2007/04/17. 10.1126/science.1141634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fall T, Ingelsson E. Genome-wide association studies of obesity and metabolic syndrome. Molecular and cellular endocrinology. 2014;382(1):740–57. 10.1016/j.mce.2012.08.018 [DOI] [PubMed] [Google Scholar]
- 3.Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518(7538):197–206. Epub 2015/02/13. 10.1038/nature14177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kilpelainen TO, Qi L, Brage S, Sharp SJ, Sonestedt E, Demerath E, et al. Physical activity attenuates the influence of FTO variants on obesity risk: a meta-analysis of 218,166 adults and 19,268 children. PLoS medicine. 2011;8(11):e1001116 10.1371/journal.pmed.1001116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahmad S, Rukh G, Varga TV, Ali A, Kurbasic A, Shungin D, et al. Gene x physical activity interactions in obesity: combined analysis of 111,421 individuals of European ancestry. PLoS genetics. 2013;9(7):e1003607 10.1371/journal.pgen.1003607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andreasen CH, Stender-Petersen KL, Mogensen MS, Torekov SS, Wegner L, Andersen G, et al. Low physical activity accentuates the effect of the FTO rs9939609 polymorphism on body fat accumulation. Diabetes. 2008;57(1):95–101. Epub 2007/10/19. 10.2337/db07-0910 [DOI] [PubMed] [Google Scholar]
- 7.Rampersaud E, Mitchell BD, Pollin TI, Fu M, Shen H, O'Connell JR, et al. Physical activity and the association of common FTO gene variants with body mass index and obesity. Archives of Internal Medicine. 2008;168(16):1791–7. Epub 2008/09/10. 10.1001/archinte.168.16.1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vimaleswaran KS, Li S, Zhao JH, Luan J, Bingham SA, Khaw KT, et al. Physical activity attenuates the body mass index-increasing influence of genetic variation in the FTO gene. American Journal of Clinical Nutrition. 2009;90(2):425–8. Epub 2009/06/26. 10.3945/ajcn.2009.27652 [DOI] [PubMed] [Google Scholar]
- 9.Berentzen T, Kring SI, Holst C, Zimmermann E, Jess T, Hansen T, et al. Lack of association of fatness-related FTO gene variants with energy expenditure or physical activity. Journal of Clinical Endocrinology and Metabolism. 2008;93(7):2904–8. Epub 2008/05/01. 10.1210/jc.2008-0007 [DOI] [PubMed] [Google Scholar]
- 10.Hetherington MM, Cecil JE. Gene-environment interactions in obesity. Forum Nutr. 2010;63:195–203. Epub 2009/12/04. 10.1159/000264407 [DOI] [PubMed] [Google Scholar]
- 11.Speakman JR, Rance KA, Johnstone AM. Polymorphisms of the FTO gene are associated with variation in energy intake, but not energy expenditure. Obesity (Silver Spring). 2008;16(8):1961–5. Epub 2008/06/14. [DOI] [PubMed] [Google Scholar]
- 12.den Hoed M, Westerterp-Plantenga MS, Bouwman FG, Mariman EC, Westerterp KR. Postprandial responses in hunger and satiety are associated with the rs9939609 single nucleotide polymorphism in FTO. American Journal of Clinical Nutrition. 2009;90(5):1426–32. Epub 2009/10/02. 10.3945/ajcn.2009.28053 [DOI] [PubMed] [Google Scholar]
- 13.Hasselbalch AL, Angquist L, Christiansen L, Heitmann BL, Kyvik KO, Sorensen TI. A variant in the fat mass and obesity-associated gene (FTO) and variants near the melanocortin-4 receptor gene (MC4R) do not influence dietary intake. Journal of Nutrition. 2010;140(4):831–4. Epub 2010/02/26. 10.3945/jn.109.114439 [DOI] [PubMed] [Google Scholar]
- 14.Hakanen M, Raitakari OT, Lehtimaki T, Peltonen N, Pahkala K, Sillanmaki L, et al. FTO genotype is associated with body mass index after the age of seven years but not with energy intake or leisure-time physical activity. Journal of Clinical Endocrinology and Metabolism. 2009;94(4):1281–7. Epub 2009/01/23. 10.1210/jc.2008-1199 [DOI] [PubMed] [Google Scholar]
- 15.Qi Q, Kilpelainen TO, Downer MK, Tanaka T, Smith CE, Sluijs I, et al. FTO genetic variants, dietary intake and body mass index: insights from 177,330 individuals. Human molecular genetics. 2014;23(25):6961–72. 10.1093/hmg/ddu411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fesinmeyer MD, North KE, Lim U, Buzkova P, Crawford DC, Haessler J, et al. Effects of smoking on the genetic risk of obesity: the population architecture using genomics and epidemiology study. BMC medical genetics. 2013;14:6 10.1186/1471-2350-14-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kvaloy K, Kulle B, Romundstad P, Holmen TL. Sex-specific effects of weight-affecting gene variants in a life course perspective—The HUNT Study, Norway. International journal of obesity. 2013;37(9):1221–9. 10.1038/ijo.2012.220 [DOI] [PubMed] [Google Scholar]
- 18.Hubacek JA, Pitha J, Adamkova V, Lanska V, Poledne R. A common variant in the FTO gene is associated with body mass index in males and postmenopausal females but not in premenopausal females. Czech post-MONICA and 3PMFs studies. Clinical chemistry and laboratory medicine. 2009;47(4):387–90. 10.1515/CCLM.2009.109 [DOI] [PubMed] [Google Scholar]
- 19.Krokstad S, Langhammer A, Hveem K, Holmen T, Midthjell K, Stene T, et al. Cohort Profile: The HUNT Study, Norway. International journal of epidemiology. 2012. Epub 2012/08/11. [DOI] [PubMed] [Google Scholar]
- 20.Loos RJ, Lindgren CM, Li S, Wheeler E, Zhao JH, Prokopenko I, et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nature genetics. 2008;40(6):768–75. 10.1038/ng.140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Razquin C, Marti A, Martinez JA. Evidences on three relevant obesogenes: MC4R, FTO and PPARgamma. Approaches for personalized nutrition. Mol Nutr Food Res. 2011;55(1):136–49. Epub 2011/01/06. 10.1002/mnfr.201000445 [DOI] [PubMed] [Google Scholar]
- 22.Mostad IL, Langaas M, Grill V. Central obesity is associated with lower intake of whole-grain bread and less frequent breakfast and lunch: results from the HUNT study, an adult all-population survey. Applied physiology, nutrition, and metabolism = Physiologie appliquee, nutrition et metabolisme. 2014:1–10. [DOI] [PubMed] [Google Scholar]
- 23.Huang BE, Lin DY. Efficient association mapping of quantitative trait loci with selective genotyping. American journal of human genetics. 2007;80(3):567–76. 10.1086/512727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wong MY, Day NE, Luan JA, Chan KP, Wareham NJ. The detection of gene-environment interaction for continuous traits: should we deal with measurement error by bigger studies or better measurement? Int J Epidemiol. 2003;32(1):51–7. [DOI] [PubMed] [Google Scholar]
- 25.Luan JA, Wong MY, Day NE, Wareham NJ. Sample size determination for studies of gene-environment interaction. Int J Epidemiol. 2001;30(5):1035–40. [DOI] [PubMed] [Google Scholar]
- 26.Boks MP, Schipper M, Schubart CD, Sommer IE, Kahn RS, Ophoff RA. Investigating gene environment interaction in complex diseases: increasing power by selective sampling for environmental exposure. Int J Epidemiol. 2007;36(6):1363–9. 10.1093/ije/dym215 [DOI] [PubMed] [Google Scholar]
- 27.R_Development_Core_Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing, 2010. ISBN 3-900051-07-0. [Google Scholar]
- 28.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the royal statistical society. 1995;Series B(Methodological):289–300. [Google Scholar]
- 29.Hardy R, Wills AK, Wong A, Elks CE, Wareham NJ, Loos RJ, et al. Life course variations in the associations between FTO and MC4R gene variants and body size. Human molecular genetics. 2010;19(3):545–52. 10.1093/hmg/ddp504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rukh G, Ahmad S, Ericson U, Hindy G, Stocks T, Renstrom F, et al. Inverse relationship between a genetic risk score of 31 BMI loci and weight change before and after reaching middle age. International journal of obesity. 2016;40(2):252–9. 10.1038/ijo.2015.180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jacobsson JA, Almen MS, Benedict C, Hedberg LA, Michaelsson K, Brooks S, et al. Detailed analysis of variants in FTO in association with body composition in a cohort of 70-year-olds suggests a weakened effect among elderly. PloS one. 2011;6(5):e20158 10.1371/journal.pone.0020158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qi L, Kang K, Zhang C, van Dam RM, Kraft P, Hunter D, et al. Fat mass-and obesity-associated (FTO) gene variant is associated with obesity: longitudinal analyses in two cohort studies and functional test. Diabetes. 2008;57(11):3145–51. 10.2337/db08-0006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kloting N, Schleinitz D, Ruschke K, Berndt J, Fasshauer M, Tonjes A, et al. Inverse relationship between obesity and FTO gene expression in visceral adipose tissue in humans. Diabetologia. 2008;51(4):641–7. Epub 2008/02/06. 10.1007/s00125-008-0928-9 [DOI] [PubMed] [Google Scholar]
- 34.Pinnick KE, Nicholson G, Manolopoulos KN, McQuaid SE, Valet P, Frayn KN, et al. Distinct developmental profile of lower-body adipose tissue defines resistance against obesity-associated metabolic complications. Diabetes. 2014;63(11):3785–97. 10.2337/db14-0385 [DOI] [PubMed] [Google Scholar]
- 35.Zillikens MC, Yazdanpanah M, Pardo LM, Rivadeneira F, Aulchenko YS, Oostra BA, et al. Sex-specific genetic effects influence variation in body composition. Diabetologia. 2008;51(12):2233–41. 10.1007/s00125-008-1163-0 [DOI] [PubMed] [Google Scholar]
- 36.Karpe F, Pinnick KE. Biology of upper-body and lower-body adipose tissue—link to whole-body phenotypes. Nature reviews Endocrinology. 2015;11(2):90–100. 10.1038/nrendo.2014.185 [DOI] [PubMed] [Google Scholar]
- 37.Winkler TW, Justice AE, Graff M, Barata L, Feitosa MF, Chu S, et al. The Influence of Age and Sex on Genetic Associations with Adult Body Size and Shape: A Large-Scale Genome-Wide Interaction Study. PLoS genetics. 2015;11(10):e1005378 10.1371/journal.pgen.1005378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cuypers KF, Loos RJ, Kvaloy K, Kulle B, Romundstad P, Holmen TL. Obesity-susceptibility loci and their influence on adiposity-related traits in transition from adolescence to adulthood—the HUNT study. PloS one. 2012;7(10):e46912 10.1371/journal.pone.0046912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brown RJ, de Banate MA, Rother KI. Artificial sweeteners: a systematic review of metabolic effects in youth. International journal of pediatric obesity: IJPO: an official journal of the International Association for the Study of Obesity. 2010;5(4):305–12. Epub 2010/01/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang Q. Gain weight by "going diet?" Artificial sweeteners and the neurobiology of sugar cravings: Neuroscience 2010. The Yale journal of biology and medicine. 2010;83(2):101–8. Epub 2010/07/01. [PMC free article] [PubMed] [Google Scholar]
- 41.Fowler SP, Williams K, Resendez RG, Hunt KJ, Hazuda HP, Stern MP. Fueling the obesity epidemic? Artificially sweetened beverage use and long-term weight gain. Obesity (Silver Spring). 2008;16(8):1894–900. [DOI] [PubMed] [Google Scholar]
- 42.Wardle J, Carnell S, Haworth CM, Farooqi IS, O'Rahilly S, Plomin R. Obesity associated genetic variation in FTO is associated with diminished satiety. Journal of Clinical Endocrinology and Metabolism. 2008;93(9):3640–3. Epub 2008/06/28. 10.1210/jc.2008-0472 [DOI] [PubMed] [Google Scholar]
- 43.Tanofsky-Kraff M, Han JC, Anandalingam K, Shomaker LB, Columbo KM, Wolkoff LE, et al. The FTO gene rs9939609 obesity-risk allele and loss of control over eating. American Journal of Clinical Nutrition. 2009;90(6):1483–8. Epub 2009/10/16. 10.3945/ajcn.2009.28439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suez J, Korem T, Zeevi D, Zilberman-Schapira G, Thaiss CA, Maza O, et al. Artificial sweeteners induce glucose intolerance by altering the gut microbiota. Nature. 2014;514(7521):181–6. 10.1038/nature13793 [DOI] [PubMed] [Google Scholar]
- 45.Thorgeirsson TE, Gudbjartsson DF, Sulem P, Besenbacher S, Styrkarsdottir U, Thorleifsson G, et al. A common biological basis of obesity and nicotine addiction. Translational psychiatry. 2013;3:e308 10.1038/tp.2013.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Langhammer A, Krokstad S, Romundstad P, Heggland J, Holmen J. The HUNT study: participation is associated with survival and depends on socioeconomic status, diseases and symptoms. BMC medical research methodology. 2012;12(1):143. Epub 2012/09/18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
(PDF)
Estimated over-all effects (βSNP) with 95% confidence intervals for men and women in age groups 20–40 years, 40–60 years and 60–80 years. FDR-adjusted p-values for testing H0: βSNP = 0 against H1: βSNP ≠ 0.
(PDF)
Estimated longitudinal effects (βSNP) with 95% confidence intervals for men and women in age groups 30–40 years (individuals younger than 30 years in HUNT3 did not participate in HUNT2), 40–60 years and 60–80 years. FDR-adjusted p-values for testing H0: βSNP = 0 against H1: βSNP ≠ 0.
(PDF)
Average differences in WHR and BMI between HUNT3 and HUNT2 for men and women in three age groups. Positive differences indicate increase from HUNT2 to HUNT3.
(PDF)
Data Availability Statement
Data are third party data owned by the The Nord-Trøndelag Health Study (HUNT) (http://www.ntnu.edu/hunt/). Any research group with a Principal Investigator affiliated with a Norwegian research institute can apply for access to analyse HUNT data. Projects must have recommendations from The Regional Committee for Medical Research in Norway (REK) and be registered with The Norwegian Social Science Data Services (NSD). Interested researchers may apply for access to these data here: http://www.ntnu.edu/hunt/data.