ABSTRACT
We have recently identified the first human lysine (K) acetyltransferase 2A and 2B (called KAT2A/2B; known also as GCN5/PCAF, respectively)-dependent acetylome and revealed a mechanism by which KAT2A/2B-mediated acetylation of serine/threonine polo-like kinase 4 (PLK4) maintains correct centrosome number in human cells, therefore contributing to the maintenance of genome stability.1
Keywords: Acetylation, cell cycle, centrosome, GCN5, genome stability, inhibition, kinase activity, PCAF, polo like kinase 4 (PLK4), proteomic analysis
Genomic instability refers to the increased predisposition to alterations in the genome. Those alterations include mutations in the DNA of specific genes, amplifications, deletions, or rearrangements of chromosomes fragments as well as gain, loss, or translocation of an entire chromosome. Several processes have been proposed to maintain genome stability, including timely cell cycle progression and high-fidelity DNA replication, error-free repair of DNA damage, accurate distribution of chromosomes to daughter cells during mitosis, as well as checkpoint controls. Defects in these processes lead to the accumulation of genomic alterations after cell division and can contribute to tumorigenesis. Genomic instability is recognized as a hallmark of cancer, such that the identification of molecular mechanisms preventing this to occur could reveal novel avenues for cancer therapy.
Post-translational modifications of proteins, such as phosphorylation, ubiquitylation, or acetylation, are key for proper modulation of protein function, enabling precise regulation of processes targeted by these modifications. Lysine acetylation is a conserved and widespread post-translational modification, targeting more than 6000 proteins in human cells (www.phosphosite.org). Lysine acetylation is regulated by lysine (K) acetyltransferases (KATs) and lysine deacetylases (KDACs). The human KAT2A and its paralog KAT2B are well-characterized histone acetyltransferases and essential regulators of gene transcription. Additionally, KAT2A/2B has roles in cell cycle regulation,2 DNA replication,3 and DNA repair,4 highlighting these KATs as key players in the maintenance of genome stability. Accordingly, human cells depleted of the adaptor protein ADA3, which regulates KAT2A/2B enzymatic activity and specificity, exhibit defective DNA repair and enhanced chromosomal aberrations.5 Mechanisms by which KAT2 regulates these cellular processes have mostly been studied to date in the context of histone acetylation and gene transcription. However, recent reports suggest that KAT2A and KAT2B can also acetylate non-histone proteins.2,6 To get more insights into how KAT2A/2B regulates cellular processes, we have uncovered a comprehensive list of their physiologic targets in human cells using shotgun proteomics, which contains ∼400 KAT2A/2B potential acetylated proteins targets.1 These KAT2A/2B target proteins belong to diverse cellular pathways including chromatin remodeling, transcriptional regulation, DNA replication, DNA repair, cell death, and cell cycle progression (see Fig. 1A), pathways in which KAT2A/2B were already suggested to play a role. Additionally, we identified novel pathways in which KAT2A/2B are involved, notably protein phosphorylation and centrosome duplication (see Fig. 1A).1 Our findings reveal that the action of these KAT2s is more widespread than previously suspected and that KAT2A/2B might regulate cellular pathways by the coordinated acetylation of histone and non-histone proteins.
Figure 1.
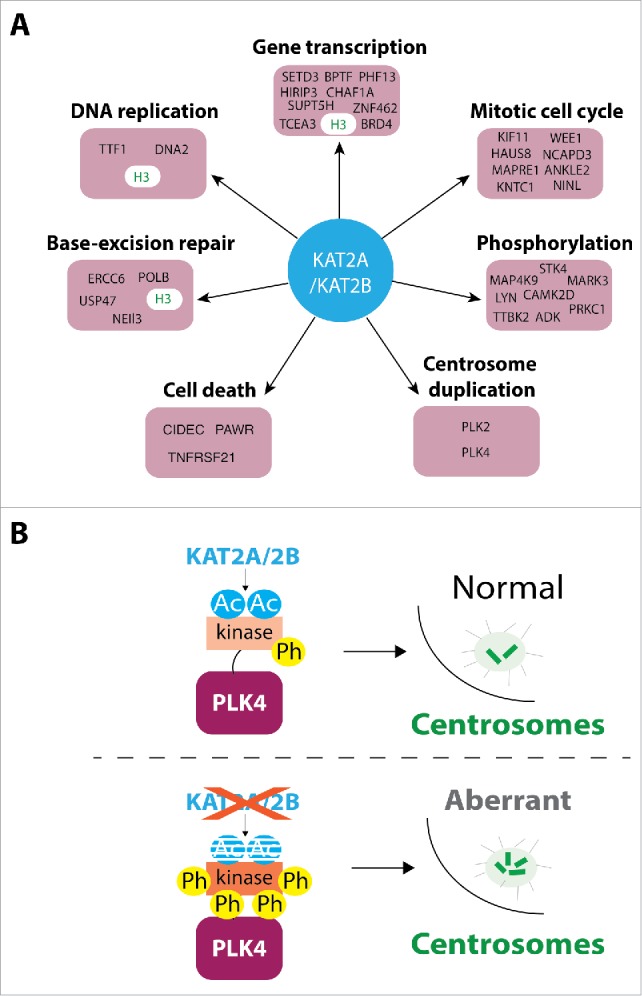
KAT2A/KAT2B-dependent acetylome reveals how PLK4 preserves centrosome number in human cells. (A) Examples of proteins acetylated by KAT2A (GCN5) and/or KAT2B (PCAF) are highlighted from the about 400 proteins identified in the KAT2A/2B-dependent acetylome1 and categorized by their putative cellular functions. H3: histone H3. (B) Model of prevention of centrosome amplification by KAT2A/2B: KAT2A- or KAT2B-containing acetyl transferase complexes acetylate the kinase domain of PLK4 to control its kinase activity, thus preventing PLK4 hyperactivity and centrosome overamplification.1 Ac, acetylation; Ph, phosphorylation. PLK4 is depicted with 2 domains: its N-terminal kinase domain (kinase) and the remaining of the protein. The red X indicates knock down, or knock out, of KAT2A/2B activity.
The identification of potential novel KAT2A/2B-acetylated targets expands the list of possible mechanisms by which these enzymes maintain genome stability. Thus, we reveal a mechanism by which KAT2A/2B acetylate the serine/threonine polo-like kinase 4 (PLK4), a central regulator of centrosome duplication, and show that PLK4 acetylation prevents centrosome amplification in human cells. Centrosomes serve as microtubule-organizing centers in the cell and consist of a pair of centrioles surrounded by pericentriolar material, from which microtubules are nucleated. Centrosomes duplicate once per cell cycle. Aberrant centrosome number has catastrophic consequences for cell division, as too few, or too many, centrosomes can cause chromosome segregation defects.7,8 Centrosome amplification is frequently observed in cancer cells and has been proposed to promote tumor progression.7 Therefore, the number of centrosomes has to be tightly regulated to maintain genome integrity and contribute to proliferation control. Depletion of PLK4 leads to failure in centrosome duplication, whereas PLK4 overexpression results in centrosome overamplification. Consequently, PLK4 level and kinase activity have to be properly controlled during cell cycle progression. Although the ubiquitin-mediated proteasomal degradation serves to control PLK4 protein level during cell cycle progression,8 the mechanisms by which PLK4 kinase activity is regulated remained unclear until now. In this study, we reveal a novel mechanism by which the acetylation of PLK4 by KAT2A/2B controls its kinase activity to prevent centrosome amplification: KAT2A/B acetylates PLK4 in its kinase domain, which negatively regulates its kinase activity by stabilizing the inactive form of PLK4 over its active form. Accordingly, the depletion of KAT2A/B acetyltransferase activity results in increased PLK4 phosphorylation of PLK4 and centrosome overamplification in human cells (see Fig. 1B). Furthermore, in contrast to the overexpression of wild-type PLK4, overexpression of the non-acetylable kinase domain mutant of PLK4 prevents centrosome amplification. While we have described a role for the acetyltransferases KAT2A/2B in the regulation of centrosome numbers, other studies have reported roles for lysine deacetylases (KDACs) in regulating centrosome function,9 supporting a significant role of lysine acetylation in centrosome biology. In the future, it would be interesting to investigate whether the centrosome amplification phenotype observed in cancer cells correlates with altered PLK4 acetylation pattern, perhaps due to decreased expression of KATs or overexpression of KDACs. Moreover, as the ubiquitin-mediated proteasomal degradation of PLK4 and the acetylation-dependent inhibition of kinase activity may act in concert to achieve a tight control of centrosome duplication, it will be interesting to investigate whether these PTMs are deregulated during cell cycle progression in cancer cells to contribute to genome instability.
Because acetylation patterns are often altered in cancer, small molecule inhibition of enzymes that regulate acetylation and deacetylation may offer novel potential for cancer therapy. Thus, inhibitors of KDACs including panabinostat and belinostat, are currently in clinical trial for cancer therapy.6 Moreover, the disruption of interactions between acetylated residues and acetyl “reader” proteins such as bromodomain inhibitors targeting bromodomain and extra-terminal domain family are also in clinical trials.10 In conclusion, our study has revealed a role for lysine acetylation in preventing centrosome amplification, supporting its role in the maintenance of genome stability and suggests the use of small-molecule inhibitors targeting regulators of lysine acetylation as a strategy for cancer therapy.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors thank P. Gönczy for his helpful comments on our present manuscript and all the authors of the original paper for their contribution to the present Author's View.
Funding
M.F. was supported by a European Commission (EC) Marie Curie fellowship (PIIF-GA-2009–255266, SAGATAC). This work was supported by funds from Agence Nationale de Recherche (ANR-13-BSV6–0001–02 COREAC) and European Research Council Advanced grant (ERC-2013–340551, Birtoaction).
References
- 1.Fournier M, et al.. KAT2A/KAT2B-targeted acetylome reveals a role for PLK4 acetylation in preventing centrosome amplification. Nat Commun 2016; 7:13227; PMID:27796307; http://dx.doi.org/ 10.1038/ncomms13227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Orpinell M, et al.. The ATAC acetyl transferase complex controls mitotic progression by targeting non-histone substrates. EMBO J 2010; 29:2381-94; PMID:20562830; http://dx.doi.org/ 10.1038/emboj.2010.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Espinosa MC, Rehman MA, Chisamore-Robert P, Jeffery D, Yankulov K. GCN5 is a positive regulator of origins of DNA replication in Saccharomyces cerevisiae. PLoS ONE 2010; 5:e8964; PMID:20126453; http://dx.doi.org/ 10.1371/journal.pone.0008964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nagy Z, Tora L. Distinct GCN5/PCAF-containing complexes function as co-activators and are involved in transcription factor and global histone acetylation. Oncogene 2007; 26:5341-57; PMID:17694077; http://dx.doi.org/ 10.1038/sj.onc.1210604 [DOI] [PubMed] [Google Scholar]
- 5.Mirza S, et al.. Alteration/deficiency in activation-3 (Ada3) plays a critical role in maintaining genomic stability. Cell Cycle 2012; 11:4266-74; PMID:23095635; http://dx.doi.org/ 10.4161/cc.22613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Di Martile M, Del Bufalo D, Trisciuoglio D. The multifaceted role of lysine acetylation in cancer: prognostic biomarker and therapeutic target. Oncotarget 2016; 7(34):55789-810; http://dx.doi.org/ 10.18632/oncotarget.10048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gönczy P. Centrosomes and cancer: revisiting a long-standing relationship. Nat Rev Cancer 2015; 15:639-52; PMID:26493645; http://dx.doi.org/ 10.1038/nrc3995 [DOI] [PubMed] [Google Scholar]
- 8.Sillibourne JE, Bornens M. Polo-like kinase 4: the odd one out of the family. Cell Div 2010; 5:25; PMID:20920249; http://dx.doi.org/ 10.1186/1747-1028-5-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ling H, Peng L, Seto E, Fukasawa K. Suppression of centrosome duplication and amplification by deacetylases. Cell Cycle 2012; 11:3779-91; PMID:23022877; http://dx.doi.org/ 10.4161/cc.21985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Filippakopoulos P, Knapp S. Targeting bromodomains: epigenetic readers of lysine acetylation. Nat Rev Drug Discov 2014; 13:337-56; PMID:24751816; http://dx.doi.org/ 10.1038/nrd4286 [DOI] [PubMed] [Google Scholar]


