ABSTRACT
Numerous techniques for isolating circulating tumor cells (CTCs) have been developed. Concurrently, single-cell techniques that can reveal molecular components of CTCs have become widely available. We discuss how the combination of isolation and multigene profiling of single CTCs in our platform can facilitate eventual translation to the clinic.
KEYWORDS: Circulating tumor cells, rare-cell sorting, single-cell analysis
Circulating tumor cells (CTCs) have garnered much attention as an established biomarker for tumor assessment ever since their use in the clinic was approved by the US Food and Drug Administration. As pertinent technologies develop, many exciting CTC isolation technologies have been introduced to promote higher-efficiency capture of CTCs. Unfortunately, they are overwhelmingly dependent on the “enumeration” of CTCs by immunocytochemistry to measure tumor burden, which clearly does not recapitulate their underlying tumor biology. To overcome this limitation, various downstream methods have also been introduced for molecular analysis of CTCs. As highlighted by Dive et al. in Nat. Rev. Clin. Oncol.1 and Pantel et al. in Clin. Chem.2 for CTC molecular analysis methods to be more useful clinically, they must address the following critical needs. First, molecular profiling at the single-cell level is necessary to overcome the significant hurdle of the presence of wild-type mRNA from white blood cells (WBCs) and other normal blood cells. Other cells of different origin are typically known to be confounding elements that obscure the identification of each CTC's true origin. In addition, quantification of the number of CTCs with multigene profiling capability, which is not possible with bulk reverse transcription-polymerase chain reaction (RT-PCR) methods, is also imperative. CTC identification beyond immunocytochemistry (based on DAPI [4′,6-diamidino-2-phenylindole] and cytokeratin staining) also addresses the challenge of minimizing the false detection of non-tumor cells. For example, certain inflammatory diseases such as Crohn's disease and diverticulosis also generate “circulating epithelial cells (CECs)” in the bloodstream at concentrations of up to 37 cells per mL. These CECs are identified with typical immunocytochemistry platforms. Since the identification of even a single CTC can be a clinically important indicator, these CECs may contribute to non-negligible false positives and confound tumor assessment. Lastly, lowering assay costs compared to whole genome amplification and next-generation sequencing methods is, indeed, a challenging hurdle to facilitate widespread research and clinical utility. These clinical needs are especially relevant for lung cancer, a disease that presents unique challenges of difficult accessibility by tumor biopsy, a lack of established biomarkers, significant molecular heterogeneity, and urgent need for an effective therapy prediction and monitoring. Particularly, gene mutation profiling is crucial to identify molecularly targeted agents for lung cancer treatment.
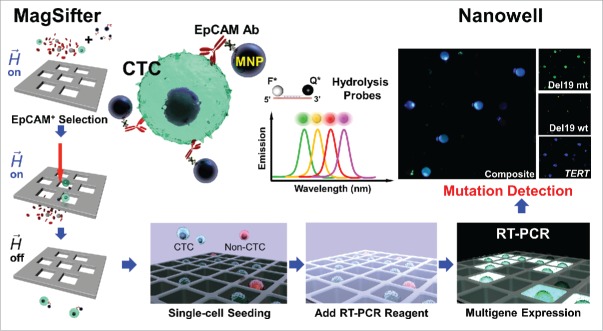
Accordingly, a completely new strategy for “liquid biopsy” of whole blood that enables disease assessment by mutation detection from the cellular fraction of whole blood was developed, which goes beyond just being another CTC isolation methodology.3 This entirely new work demonstrates high-throughput compartmentalization of individual CTCs and their subsequent transcriptomic characterization. We use an integrated nanoplatform that uses modular mRNA gene expression panels on single CTCs from lung cancer patients. By leveraging high-throughput single-cell multigene expression analysis by Nanowell after enrichment of CTCs from blood, we demonstrate comprehensive profiling of individual CTCs. As demonstrated in Fig. 1, we show how our nanoplatform technology enables single-CTC multigene expression profiling in humans after extensive preclinical validation. We address each of the above needs via (1) CTC detection and molecular characterization using tumor-specific multigene panels, rather than immunocytochemistry, (2) the Nanowell's compartmentalization of individual purified CTCs to achieve greatly enhanced sensitivity compared with “bulk” analysis via quantitative PCR, while also avoiding background signals from WBCs and other wild-type cells, (3) a simple yet robust platform with low fabrication and reagent costs for ease of research and clinical application, and (4) modular gene panels (currently up to a limit of 4 genes) for identification and mutational detection of putative CTCs.
Figure 1.
Integrated nanoplatform for single-CTC multigene profiling. Whole blood samples are processed for the isolation of CTCs by the MagSifter. Magnetic nanoparticles conjugated with antibodies are used to tag CTCs. Magnetically labeled CTCs are captured on the sifter when flowed through the sifter. All eluents are directly loaded without staining onto a Nanowell device and subjected to RT-PCR. Up to 4 genes can be identified from a single circulating cell. Adapted and modified from the publication.3
Our major findings can be summarized as follows. First, our integrated nanoplatform achieves highly sensitive detection of putative CTCs in non-small cell lung cancer (NSCLC) patients. Second, we demonstrate heterogeneity of individual CTCs with different modular multigene panels, which potentially yields insights into CTC subpopulations that possess more aggressive and metastatic phenotypes. Third, we have profiled various epidermal growth factor receptor (EGFR) mutations including exon 19 deletion, L858R, and de novo T790M mutation at the individual CTC level by using modular gene panels. We believe that this work is the first demonstration of single-CTC multigene expression analysis in lung cancer, a cancer type that is generally considered more challenging than breast cancer, prostate cancer, and colorectal cancer in the context of CTC-based disease assessment.
We envision that our strategy will facilitate the development of high-throughput single-cell multigene expression technologies for CTC identification and quantification toward cancer management. Our strategy is readily applicable to both research and clinical settings, given the low costs and ease of assaying blood samples over the course of management, treatment, and monitoring. In particular, it is well suited for longitudinal lung cancer management, since new molecularly targeted therapies currently available in the clinic make such monitoring clinically actionable and relevant. Our integrated nanoplatform is also broadly applicable to other cancer types, due to the use of modular multigene panels in Nanowell4,5 and flexibility in antibody selection for CTC capture by a magnetic sifter (MagSifter).6 However, it should be noted that extensive validation of the proposed CTC gene markers is also required for any new definition of putative CTCs, since there is currently no consensus on a set of point mutations or other genomic alterations that identifies a cell as malignant. Nevertheless, we believe that this is a powerful approach to CTC interrogation that will draw broad interest within the cancer research community and may serve as an integrated molecular diagnostic system7 for cancer assessment.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
This research was supported by the US National Institutes of Health (NIH) Awards U54CA151459 (Center for Cancer Nanotechnology Excellence and Translation) and R21CA185804 (to S.S.G. and S.X.W.), the Canary Foundation (to S.S.G. and V.S.N.), and the LUNGevity Foundation (to V.S.N.).
References
- 1.Krebs MG, Metcalf RL, Carter L, Brady G, Blackhall FH, Dive C. Molecular analysis of circulating tumour cells[mdash]biology and biomarkers. Nat Rev Clin Oncol 2014; 11(3):129-44; PMID:24445517; http://dx.doi.org/ 10.1038/nrclinonc.2013.253 [DOI] [PubMed] [Google Scholar]
- 2.Alix-Panabières C, Pantel K. Circulating tumor cells: Liquid biopsy of cancer. Clin Chem 2013; 59(1):110-8; PMID:23014601; http://dx.doi.org/ 10.1373/clinchem.2012.194258 [DOI] [PubMed] [Google Scholar]
- 3.Park SM, Wong DJ, Ooi CC, Kurtz DM, Vermesh O, Aalipour A, Suh S, Pian KL, Chabon JJ, Lee SH, et al.. Molecular profiling of single circulating tumor cells from lung cancer patients. Proc Natl Acad Sci 2016; 113(52):E8379-86; http://dx.doi.org/ 10.1073/pnas.1608461113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dimov IK, Lu R, Lee EP, Seita J, Sahoo D, Park SM, Weissman IL, Lee LP. Discriminating cellular heterogeneity using microwell-based RNA cytometry. Nat Commun 2014; 5:3451; PMID:24667995; http://dx.doi.org/ 10.1038/ncomms4451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park SM, Lee JY, Hong S, Lee SH, Dimov IK, Lee H, Suh S, Pan Q, Li K, Wu AM, et al.. Dual transcript and protein quantification in a massive single cell array. Lab Chip 2016; 16(19):3682-8; PMID:27546183; http://dx.doi.org/ 10.1039/C6LC00762G [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Earhart CM, Hughes CE, Gaster RS, Ooi CC, Wilson RJ, Zhou LY, Humke EW, Xu L, Wong DJ, Willingham SB, et al.. Isolation and mutational analysis of circulating tumor cells from lung cancer patients with magnetic sifters and biochips. Lab Chip 2014; 14(1):78-88; PMID:23969419; http://dx.doi.org/ 10.1039/C3LC50580D [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park SM, Sabour AF, Son JH, Lee SH, Lee LP. Toward integrated molecular diagnostic system (iMDx): Principles and applications. IEEE Trans Biomed Eng 2014; 61(5):1506-21; PMID:24759281; http://dx.doi.org/ 10.1109/TBME.2014.2309119 [DOI] [PMC free article] [PubMed] [Google Scholar]