Abstract
Cancer cells depend on glutamine to sustain their increased proliferation and manage oxidative stress, yet glutamine is often depleted at tumor sites due to excessive cellular consumption and poor vascularization. We have previously reported that p53 protein, while a well-known tumor suppressor, can contribute to cancer cell survival and adaptation to low glutamine conditions. However, the TP53 gene is frequently mutated in tumors, and the role of mutant p53 (mutp53) in response to metabolic stress remains unclear. Here, we demonstrate that tumor-associated mutp53 promotes cancer cell survival upon glutamine deprivation both in vitro and in vivo. Interestingly, cancer cells expressing mutp53 proteins are more resistant to glutamine deprivation than cells with wild type p53 (wtp53). Depletion of endogenous mutp53 protein in human lymphoma cells leads to cell sensitivity to glutamine withdrawal, while expression of mutp53 in p53 null cells results in resistance to glutamine deprivation. Furthermore, we found that mutp53 proteins hyper-transactivate p53 target gene CDKN1A upon glutamine deprivation, thus triggering cell cycle arrest and promoting cell survival. Together, our results reveal an unidentified mechanism by which mutp53 confers oncogenic functions by promoting cancer cell adaptation to metabolic stresses.
Keywords: Glutamine deprivation, p53 mutation, cancer metabolism, cell survival
Introduction
Glutamine is essential for survival and proliferation of most malignant cells1. The amino acid sustains highly proliferative cells by contributing to the biosynthesis of amino acids and nucleotides2,3. Additionally, glutamine protects cells from oxidative stress by maintaining healthy glutathione levels4. To ensure sufficient energy production, some cancer cells also rely on glutamine to synthesize alpha-ketoglutarate (α-KG) to replenish TCA cycle intermediates5. Therefore, inhibition of glutaminolysis alone is sufficient to inhibit tumor growth in some cancers6. Even though glutamine directly supports tumorigenesis, developing tumors are often subject to severe glutamine restriction due to increased uptake and poor vascularization at tumor sites7,8. For example, glutamine falls to almost undetectable levels relative to normal tissues in vivo in hepatomas and sarcomas7. Consistently, a recent study using metabolomics analysis comparing paired pancreatic tumor patient samples with benign adjacent tissue specimens revealed that glutamine is one of the most strongly depleted metabolites in tumors8. Thus, tumors need to develop multiple strategies to survive and grow in the low glutamine conditions.
Tumor suppressor p53 has been commonly described as a transcription factor that contributes to cell death and cell cycle arrest in response to various stresses9. Interestingly, recent reports have established that p53 also contributes to cell survival upon metabolic stress10. For example, p53 induces CDKN1A expression to trigger reversible cell-cycle arrest upon serine depletion, which allows cancer cells to pause and manage oxidative stress thus leading to enhanced survival, whereas p53 deficient cells lacking the adaptive response display drastic cell death11. In addition, it was demonstrated that activation of p53 in response to low glucose levels promotes cell adaptation through a cell cycle arrest check point12. Similarly, we have reported that p53 is activated upon glutamine deprivation and is required for cell survival under low glutamine conditions both in vitro and in vivo13. Therefore, unlike numerous DNA-damage induced p53 signaling cascades that result in apoptosis, it appears that p53 is important for cell survival under broad nutrient restrictions. Besides promoting cell survival upon metabolic stress through induction of cell cycle arrest, p53 also activates other metabolic regulators in response to nutritional stress. For example, under glucose starvation, p53 promotes expression of Sco2 or Lpin1 to decrease the rate of glycolysis by upregulating mitochondrial oxidative phosphorylation or promoting fatty acid oxidation14,15. Moreover, by driving the expression of the metabolic enzyme GLS2, and the metabolic reprogramming protein TIGAR, p53 contributes to cell survival by enhancing antioxidant capacity in nutrient depleted cells16–18. Thus, depending on cellular context, p53 may promote multiple survival mechanisms in response to metabolic stress.
The p53 protein is mutated in over 50% of all human cancers, and is often associated with a poor prognosis19–21. The majority of these cancers carry a missense mutation in the DNA binding domain of the p53 protein, with arginine residue 248 or 273 among the most frequently mutated. These are commonly referred to as ‘hot spot’ mutations, and often result in a more stable full-length protein with gain-of-function activity22. Intriguingly, several phenotypes of tumor-associated mutp53 suggest that tumors acquire the ability to promote cancer aggression and invasion23. Despite evidence supporting the role of wild type p53 in cell survival upon metabolic stress, it remains unclear whether mutp53 proteins could confer a survival advantage in response to metabolic stress.
In this study, we investigate the cellular response of mutp53 proteins upon glutamine withdrawal. We found that mutp53 promotes cell survival upon glutamine deprivation and glutaminase inhibitor treatment both in vitro and in vivo. Our study demonstrates that mutp53 amplifies the pro-survival effects of wtp53 protein to protect cancer cells from low glutamine conditions.
Results
Cancer cells with mutp53 are more resistant to glutamine deprivation than cells with wtp53
To determine whether p53 status can impact the survival of cancer cells upon glutamine starvation, we analyzed a panel of lymphoma cell lines with known p53 status by withdrawing glutamine over time. Strikingly, we found cells expressing mutp53 (CA46, DB and SupT1) were significantly more resistant to glutamine deprivation than cells expressing wtp53 (EB3, LY3 and DOHH2) (Figure 1a). Consistent with this, we found increased apoptosis, as measured by cleaved-PARP and cleaved-caspase 3 in EB3 cells expressing wtp53 compared to CA46 cells expressing mutp53 upon glutamine starvation (Figure 1b). Moreover, using annexin-V and PI staining, we confirmed substantial induction of early and late apoptosis in wtp53 cells, but not in mutp53 cells (Figure 1c–d). To further confirm resistance to cell death in glutamine restricted conditions, we measured cell viability of EB3 and CA46 cells after treatment with the glutaminase inhibitor 6-Diazo-5-oxo-L-norleucine (L-DON). While L-DON, a glutamine analogue, has been shown to directly bind and inhibit human glutaminase, the compound also affects the activity of other glutamine utilizing enzymes24. Therefore, we also treated cells with other identified glutaminase inhibitors such as Bis-2-(5-phenylacetamido-1,3,4-thiadiazol-2-yl)ethyl sulfide (BPTES)25 and Compound 9686. Consistently, we found that L-DON, BPTES and Compound 968 treatments induced cell death in a dose-dependent manner in wtp53 expressing EB3 cells, while having no effect on cell viability of CA46 cells harboring mutp53 (Figure 1e). Together, these results demonstrated that mutp53 status correlates with better survival in glutamine deprived conditions and upon glutaminase inhibitor treatment.
Figure 1. Cancer cells with mutp53 are more resistant to glutamine deprivation than cells with wtp53.
(a) Lymphoma cell lines with known p53 status were cultured in complete or glutamine (Gln) free medium for 2 days. Viability was determined by propidium iodide (PI) exclusion and normalized to cells cultured in complete medium. Mutp53 cells: CA46, SupT1, DB. Wtp53 cells: EB3, DOHH2, LY3. (b) EB3 and CA46 cells were cultured in Gln free medium for 24 hrs. Western blot was performed using antibodies as indicated. (c,d) EB3 and CA46 cells were cultured in complete or Gln free medium for 48 hrs. Apoptosis was assessed by staining cells with annexin-V and PI. Positive staining was measured by flow cytometry and analyzed with FlowJo. Representative graphs and percentage of late apoptotic cells (PI and annexin-V positive cells) are shown. Data represent mean ± S.D. of three independent experiments (***P≤.001, Student’s t test). (e) EB3 and CA46 cells were treated with L-DON for 3 days, Compound 968 and BPTES for 4 days. Cell viability was assessed by PI exclusion, and normalized to control treated cells. Data represents mean ± S.D. of three independent experiments (**P<.01, ***P≤.001, Student’s t test).
Loss of mutp53 sensitizes cancer cells to glutamine deprivation and glutaminase inhibitor treatment
To determine whether loss of mutp53 could sensitize cancer cells to glutamine starvation, we used shRNA to knock down mutp53 in CA46 cells. We generated stable cell lines with p53 knocked down (Figure 2a and 2b), and found that mutp53 deficiency significantly sensitized CA46 cells to glutamine deprivation compared to control (Figure 2c). Based on the annexin V/PI staining, glutamine deprivation resulted in more dramatic apoptotic cell death in mutp53 knockdown cells compared to vector control cells (Figure 2d and 2e). In addition, we found that knockdown of mutp53 promoted cell sensitivity to different glutaminase inhibitor treatment, such as L-DON, BPTES and Compound 968 (Figure 2f). These results demonstrated that mutp53 is required for cell survival under glutamine deprivation and glutaminase inhibitor treatment.
Figure 2. Loss of mutp53 sensitizes cancer cells to glutamine deprivation and glutaminase inhibitor treatment.

(a–b) p53 mRNA levels (a) relative to actin and protein levels (b) in control vector and p53 shRNA transduced CA46 cells was assessed using qRT-PCR and Western blot. (c) The indicated control vector or shRNA p53 transduced cells were cultured in complete or Gln free medium for the indicated time points. Viability was assessed by PI exclusion and normalized to cells cultured in complete medium. Data represent mean ± S.D. of three independent experiments (***P≤.001, Student’s t test). (d–e) Annexin-V and PI staining of stably transfected CA46 cells cultured in Gln free medium or complete medium for 24 hrs. Representative graphs of cells in late apoptosis (annexin-V and PI positive) are shown. Data represent mean ± S.D. of three independent experiments (***P≤.001, Student’s t test). (f) The control vector transduced cells and p53 shRNA transduced cells were treated with 12.5µM L-DON for two days, 40µM Compound 968 and 50µM BPTES for four days. Cell viability was assessed by PI exclusion and normalized to control treated cells. Data represent mean ± S.D. of three independent experiments (*P<.05, **P<.01, ***P≤.001, Student’s t test).
Expression of mutp53 promotes cell survival upon glutamine deprivation and glutaminase inhibitor treatment
To directly test whether mutp53 promotes cell survival upon glutamine deprivation, we stably transduced p53 deleted colorectal carcinoma HCT116 cells with retroviral vectors expressing tumor-associated p53 hotspot mutants R248Q or R273H (Figure 3a). Expression of either p53 mutant in HCT116 p53−/− cells largely protected cells upon glutamine deprivation compared to control (Figure 3b). In agreement, cells expressing mutp53 displayed less apoptosis as measured by cleaved-PARP and cleaved-caspase 3 upon glutamine deprivation compared to vector control cells (Figure 3c). In addition, we compared survival upon glutamine deprivation in HCT116 cells with p53 deletion (Vec), and cells expressing either endogenous wtp53 or exogenous mutant p53 (R248Q, R273H). Consistent with previous reports, cells with wtp53 are more resistant to glutamine deprivation than p53 null cells (Figure 3d)13. More interestingly, we found that cells expressing mutp53 displayed better survival than cells expressing wtp53, indicating mutp53 may acquire additional function to promote cell survival in response to glutamine deprivation. Consistent with the cellular response to glutamine deprivation, cells expressing mutp53 are significantly more resistant to both L-DON treatment and Compound 968 treatment than vector control cells (Figure 3e). Interestingly, expression of mutp53 had no significant protection upon treatment of different genotoxic agents including camptothecin (CPT), doxorubicin (DOX) and docetaxel (DTX) (Figure 3f), suggesting that mutp53 might specifically promote cell survival upon metabolic stress. Altogether, these results strongly support a robust survival role of mutp53 proteins in protecting cancer cells from glutamine starvation.
Figure 3. Expression of mutp53 promotes cell survival upon glutamine deprivation and glutaminase inhibitor treatment.
(a) Mutp53 R248Q or R273H were stably expressed in p53 −/− HCT116 cells. Protein levels were assessed by Western blot using antibodies against p53 (DO-1) and actin. (b) HCT116 p53 −/− cells expressing an empty vector or mutp53 protein were cultured in either Gln free or complete medium for the indicated time points. Cell viability was determined by Trypan blue exclusion and normalized to cells cultured in complete medium. Data represent mean ± S.E.M. of three independent experiments. (c) HCT116 p53−/− cells expressing an empty vector or mutp53 protein were cultured in Gln free medium or complete medium for 4 days. Lysate was extracted to perform Western blot analysis using antibodies as indicated. (d) HCT116 p53−/− cells expressing R248Q, R273H, or empty vector and HCT116 p53+/+ cells were cultured in Gln free medium or complete medium for three days. (e) HCT116 p53−/− cells expressing R248Q, R273H, or empty vector were treated with 50µM L-DON for four days, or 40µM Compound 968 for five days. (f) HCT116 p53−/− cells expressing R248Q, R273H, or empty vector were treated with 5µM camptothecin (CPT) for three days, 10µM doxorubicin (DOXO) for four days, or 10 nM docetaxel (DTX) for two days. (d–f) Cell viability was determined by Trypan blue exclusion and normalized to cells cultured in complete medium. Data represent mean ± S.E.M. of three independent experiments (**P<.01, ***P≤.001, Student’s t test).
Mutp53 induces expression of p53 target genes upon glutamine deprivation
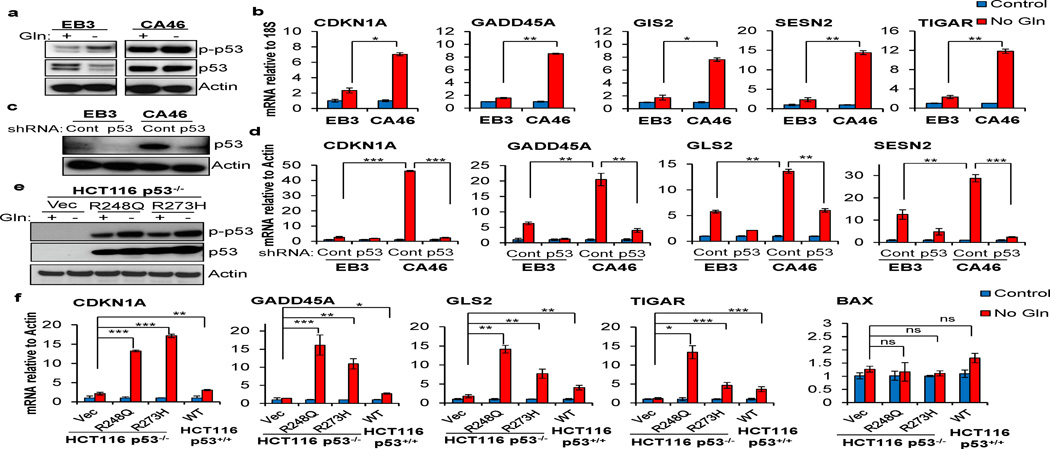
It has been reported that cell survival promoted by wtp53 upon metabolic stress is often mediated by the induction of cell cycle arrest and metabolic reprogramming genes including CDKN1A, GADD45A, GLS2 and TIGAR11,16,17. Therefore, we asked whether mutp53 contributes to the induction of these genes upon glutamine deprivation. Consistent with previous investigations, we found p53 was phosphorylated on Serine 15, a critical site for p53 activation, upon glutamine deprivation in EB3 cells with wild type p53 (Figure 4a)13. Moreover, mutp53 in CA46 cells was also phosphorylated upon glutamine deprivation (Figure 4a). Consistent with the phosphorylation, we found that p53-targeted pro-survival genes were significantly induced in CA46 cells upon glutamine deprivation (Figure 4b). Interestingly, the induction of these genes was more robust in CA46 cells harboring mutp53 than EB3 cells with wtp53 (Figure 4b). Importantly, we found that knockdown of p53 in both wtp53 and mutp53 cells dramatically inhibited the induction of the survival genes upon glutamine deprivation, demonstrating that, similar to wtp53, mutp53 contributes to the upregulation of the p53 target genes (Figure 4c and 4d). To further confirm that mutp53 promotes the induction of these p53-target genes, we evaluated the effect of ectopically expressed mutp53 on gene expression upon glutamine deprivation. Consistent with previous investigations, expression of CDKN1A, GLS2, GADD45A and TIGAR are significantly higher in wtp53 expressing cells compared with p53 deleted cells (Figure 4e and 4f). Strikingly, we found that expression of R248Q or R273H mutp53 in HCT116 cells robustly induced CDKN1A, GLS2, GADD45A and TIGAR expression upon glutamine starvation compared with p53 deleted cells or wtp53 expressing cells (Figure 4f). Conversely, no significant changes were found in the expression of pro-apoptotic gene, BAX (Figure 4f). Together, these results suggest that mutp53 not only retains, but also exaggerates the transactivation activity of the wtp53 protein toward pro-survival genes to promote cell survival in response to glutamine deprivation.
Figure 4. Mutp53 induces expression of p53 target genes upon glutamine deprivation.
(a) EB3 and CA46 cells were cultured in complete or Gln free medium for 24 hrs. Cells were lysed for Western blot using antibodies as indicated. (b) EB3 and CA46 cells were cultured in complete or Gln free medium overnight. mRNA expression of p53 target genes relative to 18S was determined using qRT- PCR and normalized to the complete medium. (c) Cells were transduced with lenti-viral particles followed by puromycin selection to generate stable knockdown of wtp53 in EB3 cells and mutp53 in CA46. p53 protein levels were determined by Western blot. (d) EB3 and CA46 cells infected with virus containing control vector or shRNA against p53 were cultured in Gln free medium overnight. mRNA expression of p53 target genes relative to actin was determined using qRT- PCR and normalized to the complete control medium. (e) HCT116 p53−/− cells expressing R248Q, R273H, or empty vector were cultured in Gln free medium for three days. p53 activation and total p53 expression was determined by Western blot analysis using anti-phospho-p53 (Ser15) and anti-p53 antibody. (f) HCT116 p53+/+ cells and HCT116 p53−/− cells expressing R248Q, R273H, or empty vector were cultured in complete or Gln free medium overnight. mRNA expression of p53 target genes relative to actin was determined using qRT-PCR and normalized to the complete medium. Data represent mean ± S.D. of duplicates from two independent experiments (*P<.05, **P<.01, ***P≤.001, Student’s t test).
Mutp53 directly binds to the promoter of p53 target genes upon glutamine deprivation
To investigate whether mutp53 can regulate the expression of the pro-survival genes via direct binding to DNA, we performed chromatin immunoprecipitation assays (ChIP) to assess p53 occupancy on promoter regions of target genes upon glutamine deprivation. We found that binding of endogenous mutp53 R248Q to the promoters of CDKN1A, GLS2 and BAX in CA46 cells was very weak in complete medium (Figure 5a). However, the binding of mutp53 to the promoter of CDKN1A and GLS2 dramatically increased upon glutamine deprivation (Figure 5a), consistent with the increased gene expression as shown in Figure 4f. Interestingly, no noticeable binding of mutp53 to the promoter of the pro-apoptotic gene BAX was found, even upon glutamine deprivation, supporting that mutp53 gains transactivation activity toward “survival” genes, but not “death” genes in response to metabolic stress, consistent with BAX expression (Figure 4f) and previously published reports26–28. To further confirm this, HCT116 p53−/− cells expressing either mutp53 or vector control were subjected to glutamine deprivation overnight before the chromatin complexes were harvested for ChIP analysis (Figure 5b). Consistently, we found that binding of mutp53 to promoters of CDKN1A, GADD45A, TIGAR and GLS2 increased strikingly upon glutamine withdrawal. No binding of mutp53 to the promoter region of pro-apoptotic gene BAX was found in cells cultured in either complete or glutamine free medium, while binding of wtp53 with BAX promoter was detected and increased upon glutamine deprivation (Figure 5b). Together, these results suggest that mutp53 can directly bind to p53 response elements in the promoter region of the “survival” genes in response to glutamine withdrawal with possible selectivity towards pro-survival but not pro-death gene activation.
Figure 5. Mutp53 directly binds to the promoter of p53 target genes upon glutamine deprivation.

(a) CA46 cells (R248Q) were cultured in complete or Gln free medium for 16 hrs. ChIP analysis was performed to determine p53 binding to the promoter of p53 target genes in response to Gln deprivation. (b) HCT116 p53+/+ cells or HCT116 p53−/− cells expressing R248Q, R273H, or vector control were cultured in complete or Gln free medium for 16 hrs. ChIP analysis was performed to determine p53 binding to the promoter of p53 target genes. DNA-protein complexes were pulled down using total p53 antibody (DO-1) or isotype matched IgG. All p53 binding sites were assessed by PCR. PCR products were separated using agarose electrophoresis.
Mutp53 promotes cell survival upon glutamine deprivation through p21 induction
It has been reported that wtp53-dependent p21 activation and cell cycle arrest promoted cell survival upon serine starvation11. To determine if mutp53 promotes cell survival upon glutamine deprivation via a similar mechanism, we first examined the cell cycle profile of HCT116 p53−/− cells with ectopic mutp53 or control vector in response to glutamine deprivation. We found that glutamine deprivation led to significant cell cycle arrest at G1/S phase in cells expressing mutp53 (Figure 6a and 6b). On the other hand, glutamine deprivation did not result in G1/S arrest of p53 deleted cells, indicating that cell cycle arrest may contribute to cell survival upon glutamine deprivation in mutp53 cells. The cell cycle arrest upon glutamine deprivation seen in mutp53 expressing cells is likely to be mediated by p21 induction, due to the role of p21 in inhibiting G1/S cell cycle progression29,30. To further test this, we examined p21 levels in HCT116 p53−/− cells expressing mutp53 or an empty vector cultured in either complete or glutamine free medium using Western blot analysis. Consistent with Figure 4f and Figure 5b, p21 protein was significantly induced upon glutamine deprivation in cells expressing mutp53 R248Q, but not in vector control cells (Figure 6c). To further determine if the induction of p21 in mutp53 cells contributes to cell survival in low glutamine conditions, we used siRNA to transiently knock down p21 in HCT116 p53−/− cells expressing either mutp53 or vector control. Importantly, we found that p21 depletion increased cell sensitivity in response to glutamine deprivation in cells expressing mutp53, suggesting that the protective effects of mutp53 are mediated through p21 expression (Figure 6d). In addition, this increased sensitivity correlated with an increase in apoptosis, as indicated by Western blot analysis of cleaved-PARP (Figure 6e). Conversely, p21 depletion had no effect on cell viability upon glutamine deprivation in p53 deleted cells expressing vector control (Figure 6d and 6e). Taken together, these results demonstrate that mutp53 induces p21-dependent cell cycle arrest, which promotes cell survival in low glutamine conditions.
Figure 6. Mutp53 promotes cell survival upon glutamine deprivation through p21 induction.

(a,b) HCT116 p53−/− cells expressing mutp53 (R248Q) or empty vector were cultured in complete or Gln free medium for 24 hrs. PI staining followed by Flow Cytometry was performed to assess cell cycle profile. Representative graphs of three independent experiments are shown. Data represent the mean ± S.D. (c) HCT116 p53−/− cells expressing mutp53 R248Q or empty vector were cultured in complete or Gln free medium for 24 hours. Western blots were performed using antibodies as indicated. (d) p21 was transiently knocked down in HCT116 p53−/− cells expressing mutp53 R248Q or empty vector using siRNA (20nM). 48 hrs after the siRNA transfection, cells were cultured in complete or Gln free medium for four days. Cell viability was determined by Trypan blue exclusion. Data represent the mean ± S.D. of four independent experiments, (***P≤.001, Student’s t-test). (e) Western blot was performed using antibodies as indicated.
Tumors expressing mutp53 are more resistant to glutaminase inhibitor treatment in vivo
To further investigate whether mutp53 can protect cancer cells from glutamine starvation in vivo, we established xenograft tumors using HCT116 p53−/− cells expressing either an empty vector or mutp53 R248Q. Once tumors were established, mice were treated with the glutaminase inhibitor L-DON, and tumor volume was measured over time (Figure 7a and 7b). As expected, inhibition of glutamine metabolism by L-DON dramatically suppressed tumor growth in mice with p53 null tumors. In contrast, L-DON treatment had no significant effect on tumor growth in mice harboring tumors with mutp53 R248Q (Figure 7a and 7b). Consistent with our previous results, L-DON treatment induced apoptosis as indicated by cleaved-PARP in p53 null tumors, but not in tumors expressing mutp53 (Figure 7c). Together, these results show that mutp53 may play a role in tumor resistance to glutamine deprivation in vivo.
Figure 7. Tumors expressing mutant p53 are more resistant to glutaminase inhibitor treatment in vivo.
(a,b) Athymic Nude mice at 7 weeks old were injected with HCT116 p53 −/− cells on the left flank. HCT116 p53 −/− cells expressing mutp53 R248Q were injected on the right flank. Once the tumor size reached an average of 60 mm3, the mice were treated with 15 mg/kg of L-DON every other day by i.p. injection. Tumor size was measured over time. Data represent the mean ± S.D. (n=5 or 6 tumors as indicated), ***P≤.001, Student’s t-test. (c) Tumors with L-DON or vehicle treatment were harvested at day 11. Western blot was performed using the indicated antibodies.
Discussion
More than half of human cancers carry mutations in the TP53 gene. These tumor-associated mutp53 proteins have been previously shown to drive aggressive cancer growth, metastasis and chemotherapeutic drug resistance31. However, whether mutp53 proteins can protect cancer cells from metabolic stress that commonly occurs in the tumor microenvironment is not well established. Our results demonstrate that tumor-associated mutp53 proteins promote cancer cell survival in response to glutamine starvation both in vitro and in vivo. Altogether, the data reveals a previously unidentified mechanism by which mutp53 promotes tumorigenesis through resistance to metabolic stresses.
In this paper, we found that mutp53 can directly bind to the promoters of p53-target genes that regulate either cell cycle or metabolism and induce their expression upon glutamine deprivation (Figure 4 and Figure 5). While mutp53 proteins acquire non-canonical transactivation activity to promote cancer growth and invasion, recent studies demonstrate that mutp53 can bind to the response elements of wtp5332. Studies in both yeast and mammalian systems reveal that mutp53 proteins have differential transactivation activity towards canonical p53 target genes including loss of function, reduced transactivation activity, and super-transactivation activity33. Moreover, mutp53 proteins can differentially regulate canonical p53 target genes to modulate cellular response to stress. For example, mutp53 may confer a survival advantage through its ability to transactivate the cell cycle arrest gene CDKN1A, but not the pro-apoptotic gene BAX 26–28. These evidences support our findings that mutp53 can transactivate the expression of p53 target genes to support cell survival and adaptation during metabolic stress. However, it still remains unclear how these mutp53 proteins are recruited to promoters of the target genes in response to metabolic stress. Based on our ChIP analysis, the binding of mutp53 to the target promoters remained minimal in normal conditions, but dramatically increased in response to glutamine withdrawal. These results suggest that other factors or pathways induced by glutamine deprivation might be required for the transactivation activity of mutp53. For example, upon glucose deprivation, PGC-1 protein can bind and preferentially promote p53 transactivation activity toward genes associated with cell cycle arrest and metabolic regulation34. Therefore, it will be of interest to determine the molecular mechanisms that modulate the transactivation of mutp53 towards pro-survival genes in response to metabolic stress.
Our data also demonstrate that induction of CDKN1A (p21) is required for mutp53-mediated cell survival under low glutamine conditions. It has been well established that activation of wtp53 upon metabolic stress can contribute to survival through induction of cell cycle arrest genes. For example, AMPK-dependent p53 activation upon glucose starvation promotes cell cycle arrest at the G1/S phase transition leading to improved cellular survival12. In addition, p53 in serine depleted cells can promote CDKN1A–dependent cell cycle arrest and antioxidant biosynthesis, which together support adaptation and cellular survival11. Here, we found that mutp53 proteins not only retain, but also amplify the survival effect of wtp53 to protect cancer cells from glutamine deprivation via induction of p21. Specifically, mutp53 hyper-induces p21 expression to promote G1/S cell cycle arrest to protect cells from glutamine deprivation. Moreover, we observed that p21 knockdown is sufficient to restore cell sensitivity of mutp53 cells to glutamine deprivation. Cell cycle arrest upon metabolic stress can serve as a critical initial response that stops the energy-demanding process of cell proliferation to allow other cellular repair and adaptive processes to occur. Therefore, elimination of cell cycle arrest in glutamine deprived cells leads to unchecked proliferation and failure to adapt to metabolic stress, causing decreased cell viability. Besides p21, other p53 target genes involved in cell cycle arrest or metabolism are also highly induced in cells expressing mutp53 upon glutamine deprivation. Although our data support that p21 is critical for mutp53-mediated survival, we cannot exclude that other mutp53-regulated pathways also contribute to cell survival under glutamine deprivation. For example, it was recently reported that mutp53 proteins can directly bind to the metabolic protein sensor, AMPK, and inhibit its function to promote cancer cell growth35. In addition, tumor-associated mutp53 has been shown to stimulate the Warburg effect36.
The resistance to glutamine restriction in mutp53-expressing cells can impact tumor physiology and therapeutic response. First, glutamine levels at tumor sites can drop to nearly undetectable levels, partially because tumors heavily use exogenous glutamine to sustain their growth and survival. As glutamine is depleted in poorly vascularized tumor microenvironment, cancer cells expressing mutp53 protein are able to adapt and survive the metabolic stress, whereas p53 deficient cells and wtp53 expressing cells will experience impaired proliferation and increased cell death. Therefore, the glutamine depleted environment within the tumors is more favorable for the development of cancer cells expressing mutp53 proteins. Second, targeting glutamine metabolism underscores a therapeutic benefit against cancers37–39. Recently, there is an increasing effort to inhibit anabolic glutamine metabolism using small molecule inhibitors targeting glutaminase. For examples, BPTES has been shown to inhibit growth of lymphoma xenografts with minimal toxicity, and a more potent glutaminase inhibitor CB-839 is currently in clinical trial for triple negative breast cancer40,41. However, we found that mutp53 expressing cells are strongly resistant to glutamine depletion and glutaminase inhibitor treatment compared to p53 deficient cells. These findings suggest a potential benefit in using glutaminase inhibitors to treat patients with p53-deficient tumors, but not those with tumors harboring mutp53.
Materials and methods
Cell culture and reagents
HCT116 p53+/+ and HCT116 p53−/− cells were cultured in Dulbecco's modified Eagle medium (DMEM, Corning) supplemented with 10% fetal bovine serum (FBS, Gemini Bio-Products), 100 units/mL of penicillin, and 100 µg/mL of streptomycin (Gemini Bio-Products). These cells have been generated previously42. EB3, SUPT1, DB (purchased from ATTC) and DOHH2 (purchased from DSMZ) lymphoma cell lines were cultured in RPMI 1640 medium (Corning) with 10% FBS, 100 units/mL of penicillin, and 100 µg/mL of streptomycin (Gemini Bio-Products). CA46 (purchased from ATTC) and LY3 (purchased from DSMZ) cells were cultured in RPMI medium with 20% FBS. All cells were cultured at 37°C with 5% CO2. All cells were routinely tested for mycoplasma contamination using MycoAlert™ Mycoplasma Detection Kit (Lonza). L-DON, BPTES and camptothecin were purchased from Sigma. Docetaxel was purchased from Selleckchem, and Compound 968 was purchased from Calbiochem.
Glutamine starvation
For glutamine deprivation experiments, cells were washed once with 1× PBS and cultured in glutamine free medium or complete medium. To make glutamine free medium, DMEM and RPMI without glutamine (Corning) were supplemented with 10% dialyzed FBS (Gemini). To make the complete medium, 2 mM L-Glutamine (Corning) was added back to the glutamine free medium.
Flow Cytometry
To assess cell viability, cells were washed once with PBS and stained with 1 µg/mL propidium iodide (PI, Molecular Probes) for 10 minutes at room temperature. To assess apoptotic cell death, cells were washed once with PBS and stained with PI and Annexin-V (eBioscience) and processed according to the manufacturer’s protocol. Flow analysis was carried out using the 9-color CyAn™ ADP from Beckman Coulter (Miami, FL). To assess cell viability by Trypan Blue exclusion, cells were washed once with PBS and stained with Trypan Blue solution (Sigma). Cell viability was determined using TC20 automated cell counter (BioRad).
Cell cycle staining
To assess cell cycle profiles, cells were washed once with PBS and fixed with ice-cold 70% ethanol overnight. Fixed cells were washed twice with PBS and then treated with RNase A (Sigma) for 10 minutes at 37°C. The cells were then stained with PI (Molecular Probes) and analyzed by flow cytometry on the CyAn™ ADP.
RT-PCR
RNA extractions were performed using Trizol (Invitrogen) according to the manufacturer’s guidelines. 1 µg of RNA was used for each cDNA synthesis reaction (Quanta Biosciences). Real time reverse transcription PCR (qRT-PCR) reactions were performed with SYBR Green PCR reagents (Quanta Biosciences) using an iQ5 thermal cycler (Bio-Rad). The Ct values of the target genes were normalized to Ct values of Actin or 18S. Forward and reverse primers were generated to check gene expression. 18S F: 5’-CGCTTCCTTACCTGGTTGAT-3’ R: 5’-GAGCGACCAA AGGAACCATA-3’ ACTIN- F: 5’-CACCAACTGGGAGGACAT-3’ R: 5’GCACAGCCT GGATAGCAAC-3’ CDKN1A- F: 5’-AGGTGGACCTGGAGACTCTCAG-3’ R: 5’TCCTCTTGGAGAAGATCA GCCG-3’ GADD45A- F: 5’-CTGGAGGAAGTGC TCAGCAAAG-3’R: 5’-AGAGCCACA TCTCTGTCGTCGT-3’ GLS2- F: 5’-CAGAAGG CACAGACATGGTTGG-3’ R: 5’-GGCA GAAACCACCATTAGCCAG-3’ TIGAR- F: 5’-TTCGGGAAAGGAAATACGGGG-3’ R: 5’-CCACGCATTTTCACCTGGTC-3’ SESN2- F: 5’-GCGCTTTCATTCCAGTGGAAGAG-3’ R: 5’-CAGAAGCTGCTAAGGTAGTCCG-3’ BAX- F: 5’-CCCGAGAGGTCTTTTTCCGAG-3’ R: 5’-CCAGCCCATGATGGTTCTGAT −3’
Western Blotting
Immunoblotting was carried out as described previously43. Briefly, cells were washed 3 times with PBS and lysed in ice-cold RIPA lysis buffer supplemented with protease inhibitor (Roche) for 5 minutes. Immunoblotting was performed with the following antibodies: p53 (Santa Cruz, DO-1, SC126), p-p53 Ser-15 (Cell Signaling,9284), total PARP and cleaved PARP (Cell Signaling, 9542 and 5625), caspase 3 and cleaved caspase 3 (Cell Signaling, 9664 and 9665), Beta Actin (Sigma), p21 (Cell Signaling,2947).
shRNA knockdown
p53 shRNA constructs (TRCN0000003753) were purchased from GE Dharmacon. shRNA lentiviral particles were generated in 293T cells as described previously43. Briefly, 293T cells were co-transfected with pLKO.1 empty vector or p53 shRNA vector, pMDL, pCMV-VSV-G and pRSV-Rev at a ratio of 4:2:1:1. After the transfection, lentiviral particles were collected at day 2 and day 3. Lymphoma cells were spin-infected twice with the lentiviral particles for 50 minutes at 2000 rpm at 30°C. Cells were selected with 0.25 µg/ml puromycin.
siRNA knockdown
On-target plus human CDKN1A siRNA from Dharmacon (SMARTpool L-003471) was used to transiently knock down p21 protein in HCT116 cells. Cells were transfected with CDKN1A siRNA or control siRNA (Dharmacon) using RNAi max lipofectamine reagent (Invitrogen). Glutamine deprivation experiments were performed 48 hours post siRNA transfection.
Generation of stable cell lines expressing mutp53 proteins
pLPCX retroviral vectors expressing mutp53 R248Q or R273H were generous gifts from Dr. Zhaohui Feng’s Laboratory (Rutgers University, New Jersey). 293T cells were transfected with the mutp53 expressing vectors and the pLPCX empty vectors as previously described to generate retroviral particles43. HCT116 p53−/− cells were cultured with the virus containing medium with 10 µg/ml polybrene overnight followed by puromycin selection (1 µg/ml).
Chromatin immunoprecipitation assay
The chromatin immunoprecipitation assay was performed using a ChIP assay kit (Millipore) according to the manufacturer’s guideline. In brief, cells were cultured in complete medium or glutamine free medium overnight. Cells were cross-linked with 1% formaldehyde for 10 minutes at room temperature. Cells were washed 3 times with PBS and lysed in ice cold lysis buffer with protease inhibitors on ice for 5 minutes. Samples were sonicated to yield 200–1000bp DNA fragments. After centrifugation, cell supernatant was diluted in immunoprecipitation buffer with protease inhibitors and pre-cleared with Salmon Sperm DNA/ Protein A Agarose for 1 hour at 4°C with rotation. 1 µg of p53 (DO-1, Santa Cruz) or IgG antibody was used for each overnight immunoprecipitation with rotation at 4°C. Binding sites of p53 were amplified by 30–35 cycles of PCR using Hotstart Taq DNA polymerase (Bioneer). The PCR products were detected using agarose gel electrophoresis. PCR primers for the ChIP assays: CDKN1A- F: 5’-GCTGTGGCTCTGATTGGCTTT-3’ R: 5’-ACAGGCAGCCCAAGGACAA A-3’ GADD45A- F: 5’-AGCGGAAGAGATCCCTGTGA-3’ R: 5’-CGGGAGGCAGGCAGA TG-3’ GLS2- F: 5’-GGCCTCCCAAGTCACCAGTTCA-3’ R:5’TGTTTTTGCTTGTTTTCG CCTTCT-3’ TIGAR- F: 5’-GCTTCAGACGTATATATAGA-3’ R: 5’GGGGCTATTCT TGGTAGTAA-3’ BAX F 5’-GGGTTATCTCTTGGGCTCACAA-3’ R 5’-GAGCTCTCCCCAGCGCA-3’ Negative primer: - F: 5’-TAAATGGGACAGGTAGGACC-3’ - R: 5’- TCCACCGCTTCTTGTCCTGC-3’
Mouse Xenografts
All animal procedures were approved by the Institutional Animal Care and Use Committee at City of Hope Cancer Center in compliance with ethical regulations. Animal were randomized before treatments. Sample size was generally chosen based on preliminary data indicating the variance within each group and the differences between groups. 7 week old Athymic Nude male mice (Taconic Laboratories) were injected subcutaneously with 1×106 cells. HCT116 p53−/− cells expressing vector control were injected in the left flank and HCT116 p53−/− cells expressing mutp53 R248Q were injected in the right flank. When the tumor size reached an average of 60 mm3, mice were treated with 15 mg/kg of L-DON or PBS 3 times per week. The tumor size was measured every other day as described previously44. Eleven days after drug treatment, the mice were sacrificed and the tumors were harvested for Western Blot analysis.
Statistical analysis
Results are shown as averages; error bars represent either the standard error of the mean (S.E.M.) or standard deviation (S.D.) as indicated. The unpaired Student's t test was used to determine the statistical significance of differences between means (*P<.05, **P<.01, ***P≤.001). All western blot experiments were repeated at least three times with a representative gel being shown. For all other experiments, each was repeated independently at least two times with similar results.
Acknowledgments
We thank members of the Kong laboratory for helpful comments on the manuscript. We also thank Dr. Zhaohui Feng (Rutgers University, New Jersey) for the mutp53 constructs. This work was supported by National Institutes of Health (NIH)/R01CA183989 to M.K. and the V Foundation for Cancer Research to MK. M.K. is the Pew Scholar in the Biomedical Sciences and American Cancer Society Research Scholar (RSG-16-085-01-TBE). Research reported here includes work carried out in Core Facilities supported by the NIH/NCI under grant number P30CA33572.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
References
- 1.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell metabolism. 2008 Jan;7(1):11–20. doi: 10.1016/j.cmet.2007.10.002. PubMed PMID: 18177721. [DOI] [PubMed] [Google Scholar]
- 2.Young VR, Ajami AM. Glutamine: the emperor or his clothes? The Journal of nutrition. 2001 Sep;131(9 Suppl):2449S–2459S. doi: 10.1093/jn/131.9.2449S. discussion 86S-7S. PubMed PMID: 11533293. [DOI] [PubMed] [Google Scholar]
- 3.Wise DR, Thompson CB. Glutamine addiction: a new therapeutic target in cancer. Trends in biochemical sciences. 2010 Aug;35(8):427–433. doi: 10.1016/j.tibs.2010.05.003. PubMed PMID: 20570523. Pubmed Central PMCID: 2917518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mates JM, Perez-Gomez C, Nunez de Castro I, Asenjo M, Marquez J. Glutamine and its relationship with intracellular redox status, oxidative stress and cell proliferation/death. The international journal of biochemistry & cell biology. 2002 May;34(5):439–458. doi: 10.1016/s1357-2725(01)00143-1. PubMed PMID: 11906817. [DOI] [PubMed] [Google Scholar]
- 5.Le A, Lane AN, Hamaker M, Bose S, Gouw A, Barbi J, et al. Glucose-independent glutamine metabolism via TCA cycling for proliferation and survival in B cells. Cell metabolism. 2012 Jan 4;15(1):110–121. doi: 10.1016/j.cmet.2011.12.009. PubMed PMID: 22225880. Pubmed Central PMCID: 3345194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang JB, Erickson JW, Fuji R, Ramachandran S, Gao P, Dinavahi R, et al. Targeting mitochondrial glutaminase activity inhibits oncogenic transformation. Cancer cell. 2010 Sep 14;18(3):207–219. doi: 10.1016/j.ccr.2010.08.009. PubMed PMID: 20832749. Pubmed Central PMCID: 3078749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roberts E, Caldwell AL, et al. Amino acids in epidermal carcinogenesis in mice. Cancer research. 1949 Jun;9(6):350–353. PubMed PMID: 18144236. [PubMed] [Google Scholar]
- 8.Kamphorst JJ, Nofal M, Commisso C, Hackett SR, Lu W, Grabocka E, et al. Human pancreatic cancer tumors are nutrient poor and tumor cells actively scavenge extracellular protein. Cancer research. 2015 Feb 1;75(3):544–553. doi: 10.1158/0008-5472.CAN-14-2211. PubMed PMID: 25644265. Pubmed Central PMCID: 4316379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vousden KH, Lu X. Live or let die: the cell's response to p53. Nature reviews Cancer. 2002 Aug;2(8):594–604. doi: 10.1038/nrc864. PubMed PMID: 12154352. [DOI] [PubMed] [Google Scholar]
- 10.Maddocks OD, Vousden KH. Metabolic regulation by p53. Journal of molecular medicine. 2011 Mar;89(3):237–245. doi: 10.1007/s00109-011-0735-5. PubMed PMID: 21340684. Pubmed Central PMCID: 3043245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maddocks OD, Berkers CR, Mason SM, Zheng L, Blyth K, Gottlieb E, et al. Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells. Nature. 2013 Jan 24;493(7433):542–546. doi: 10.1038/nature11743. PubMed PMID: 23242140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones RG, Plas DR, Kubek S, Buzzai M, Mu J, Xu Y, et al. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Molecular cell. 2005 Apr 29;18(3):283–293. doi: 10.1016/j.molcel.2005.03.027. PubMed PMID: 15866171. [DOI] [PubMed] [Google Scholar]
- 13.Reid MA, Wang WI, Rosales KR, Welliver MX, Pan M, Kong M. The B55alpha subunit of PP2A drives a p53-dependent metabolic adaptation to glutamine deprivation. Molecular cell. 2013 Apr 25;50(2):200–211. doi: 10.1016/j.molcel.2013.02.008. PubMed PMID: 23499005. [DOI] [PubMed] [Google Scholar]
- 14.Mauro C, Leow SC, Anso E, Rocha S, Thotakura AK, Tornatore L, et al. NF-kappaB controls energy homeostasis and metabolic adaptation by upregulating mitochondrial respiration. Nature cell biology. 2011 Oct;13(10):1272–1279. doi: 10.1038/ncb2324. PubMed PMID: 21968997. Pubmed Central PMCID: 3462316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Assaily W, Rubinger DA, Wheaton K, Lin Y, Ma W, Xuan W, et al. ROS-mediated p53 induction of Lpin1 regulates fatty acid oxidation in response to nutritional stress. Molecular cell. 2011 Nov 4;44(3):491–501. doi: 10.1016/j.molcel.2011.08.038. PubMed PMID: 22055193. [DOI] [PubMed] [Google Scholar]
- 16.Hu W, Zhang C, Wu R, Sun Y, Levine A, Feng Z. Glutaminase 2, a novel p53 target gene regulating energy metabolism and antioxidant function. Proceedings of the National Academy of Sciences of the United States of America. 2010 Apr 20;107(16):7455–7460. doi: 10.1073/pnas.1001006107. PubMed PMID: 20378837. Pubmed Central PMCID: 2867677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R, et al. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. cell. 2006 Jul 14;126(1):107–120. doi: 10.1016/j.cell.2006.05.036. PubMed PMID: 16839880. [DOI] [PubMed] [Google Scholar]
- 18.Budanov AV. The role of tumor suppressor p53 in the antioxidant defense and metabolism. Sub-cellular biochemistry. 2014;85:337–358. doi: 10.1007/978-94-017-9211-0_18. PubMed PMID: 25201203. Pubmed Central PMCID: 4206257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muller PA, Vousden KH. p53 mutations in cancer. Nature cell biology. 2013 Jan;15(1):2–8. doi: 10.1038/ncb2641. PubMed PMID: 23263379. [DOI] [PubMed] [Google Scholar]
- 20.Gaidano G, Ballerini P, Gong JZ, Inghirami G, Neri A, Newcomb EW, et al. p53 mutations in human lymphoid malignancies: association with Burkitt lymphoma and chronic lymphocytic leukemia. Proceedings of the National Academy of Sciences of the United States of America. 1991 Jun 15;88(12):5413–5417. doi: 10.1073/pnas.88.12.5413. PubMed PMID: 2052620. Pubmed Central PMCID: 51883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013 Oct 17;502(7471):333–339. doi: 10.1038/nature12634. PubMed PMID: 24132290. Pubmed Central PMCID: 3927368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng T, Wang J, Zhao Y, Zhang C, Lin M, Wang X, et al. Spliced MDM2 isoforms promote mutant p53 accumulation and gain-of-function in tumorigenesis. Nature communications. 2013;4:2996. doi: 10.1038/ncomms3996. PubMed PMID: 24356649. Pubmed Central PMCID: 3960723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muller PA, Caswell PT, Doyle B, Iwanicki MP, Tan EH, Karim S, et al. Mutant p53 drives invasion by promoting integrin recycling. cell. 2009 Dec 24;139(7):1327–1341. doi: 10.1016/j.cell.2009.11.026. PubMed PMID: 20064378. [DOI] [PubMed] [Google Scholar]
- 24.Thangavelu K, Chong QY, Low BC, Sivaraman J. Structural basis for the active site inhibition mechanism of human kidney-type glutaminase (KGA) Scientific reports. 2014;4:3827. doi: 10.1038/srep03827. PubMed PMID: 24451979. Pubmed Central PMCID: 4929687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robinson MM, McBryant SJ, Tsukamoto T, Rojas C, Ferraris DV, Hamilton SK, et al. Novel mechanism of inhibition of rat kidney-type glutaminase by bis-2-(5-phenylacetamido-1,2,4-thiadiazol-2-yl)ethyl sulfide (BPTES) The Biochemical journal. 2007 Sep 15;406(3):407–414. doi: 10.1042/BJ20070039. PubMed PMID: 17581113. Pubmed Central PMCID: 2049044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Campomenosi P, Monti P, Aprile A, Abbondandolo A, Frebourg T, Gold B, et al. p53 mutants can often transactivate promoters containing a p21 but not Bax or PIG3 responsive elements. Oncogene. 2001 Jun 14;20(27):3573–3579. doi: 10.1038/sj.onc.1204468. PubMed PMID: 11429705. [DOI] [PubMed] [Google Scholar]
- 27.Friedlander P, Haupt Y, Prives C, Oren M. A mutant p53 that discriminates between p53-responsive genes cannot induce apoptosis. Molecular and cellular biology. 1996 Sep;16(9):4961–4971. doi: 10.1128/mcb.16.9.4961. PubMed PMID: 8756655. Pubmed Central PMCID: 231498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ryan KM, Vousden KH. Characterization of structural p53 mutants which show selective defects in apoptosis but not cell cycle arrest. Molecular and cellular biology. 1998 Jul;18(7):3692–3698. doi: 10.1128/mcb.18.7.3692. PubMed PMID: 9632751. Pubmed Central PMCID: 108951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. cell. 1993 Nov 19;75(4):805–816. doi: 10.1016/0092-8674(93)90499-g. PubMed PMID: 8242751. [DOI] [PubMed] [Google Scholar]
- 30.Waldman T, Kinzler KW, Vogelstein B. p21 is necessary for the p53-mediated G1 arrest in human cancer cells. Cancer research. 1995 Nov 15;55(22):5187–5190. PubMed PMID: 7585571. [PubMed] [Google Scholar]
- 31.Muller PA, Vousden KH. Mutant p53 in cancer: new functions and therapeutic opportunities. Cancer cell. 2014 Mar 17;25(3):304–317. doi: 10.1016/j.ccr.2014.01.021. PubMed PMID: 24651012. Pubmed Central PMCID: 3970583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Strano S, Dell'Orso S, Di Agostino S, Fontemaggi G, Sacchi A, Blandino G. Mutant p53: an oncogenic transcription factor. Oncogene. 2007 Apr 2;26(15):2212–2219. doi: 10.1038/sj.onc.1210296. PubMed PMID: 17401430. [DOI] [PubMed] [Google Scholar]
- 33.Resnick MA, Inga A. Functional mutants of the sequence-specific transcription factor p53 and implications for master genes of diversity. Proceedings of the National Academy of Sciences of the United States of America. 2003 Aug 19;100(17):9934–9939. doi: 10.1073/pnas.1633803100. PubMed PMID: 12909720. Pubmed Central PMCID: 187891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sen N, Satija YK, Das S. PGC-1alpha, a key modulator of p53, promotes cell survival upon metabolic stress. Molecular cell. 2011 Nov 18;44(4):621–634. doi: 10.1016/j.molcel.2011.08.044. PubMed PMID: 22099309. [DOI] [PubMed] [Google Scholar]
- 35.Zhou G, Wang J, Zhao M, Xie TX, Tanaka N, Sano D, et al. Gain-of-function mutant p53 promotes cell growth and cancer cell metabolism via inhibition of AMPK activation. Molecular cell. 2014 Jun 19;54(6):960–974. doi: 10.1016/j.molcel.2014.04.024. PubMed PMID: 24857548. Pubmed Central PMCID: 4067806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang C, Liu J, Liang Y, Wu R, Zhao Y, Hong X, et al. Tumour-associated mutant p53 drives the Warburg effect. Nature communications. 2013;4:2935. doi: 10.1038/ncomms3935. PubMed PMID: 24343302. Pubmed Central PMCID: 3969270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Korangath P, Teo WW, Sadik H, Han L, Mori N, Huijts CM, et al. Targeting Glutamine Metabolism in Breast Cancer with Aminooxyacetate. Clinical cancer research : an official journal of the American Association for Cancer research. 2015 Jul 15;21(14):3263–3273. doi: 10.1158/1078-0432.CCR-14-1200. PubMed PMID: 25813021. Pubmed Central PMCID: 4696069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chakrabarti G, Moore ZR, Luo X, Ilcheva M, Ali A, Padanad M, et al. Targeting glutamine metabolism sensitizes pancreatic cancer to PARP-driven metabolic catastrophe induced by ss-lapachone. Cancer & metabolism. 2015;3:12. doi: 10.1186/s40170-015-0137-1. PubMed PMID: 26462257. Pubmed Central PMCID: 4601138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lukey MJ, Wilson KF, Cerione RA. Therapeutic strategies impacting cancer cell glutamine metabolism. Future medicinal chemistry. 2013 Sep;5(14):1685–1700. doi: 10.4155/fmc.13.130. PubMed PMID: 24047273. Pubmed Central PMCID: 4154374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiang Y, Stine ZE, Xia J, Lu Y, O'Connor RS, Altman BJ, et al. Targeted inhibition of tumor-specific glutaminase diminishes cell-autonomous tumorigenesis. The Journal of clinical investigation. 2015 Jun;125(6):2293–2306. doi: 10.1172/JCI75836. PubMed PMID: 25915584. Pubmed Central PMCID: 4497742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gross MI, Demo SD, Dennison JB, Chen L, Chernov-Rogan T, Goyal B, et al. Antitumor activity of the glutaminase inhibitor CB-839 in triple-negative breast cancer. Molecular cancer therapeutics. 2014 Apr;13(4):890–901. doi: 10.1158/1535-7163.MCT-13-0870. PubMed PMID: 24523301. [DOI] [PubMed] [Google Scholar]
- 42.Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP, et al. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998 Nov 20;282(5393):1497–1501. doi: 10.1126/science.282.5393.1497. PubMed PMID: 9822382. [DOI] [PubMed] [Google Scholar]
- 43.Rosales KR, Reid MA, Yang Y, Tran TQ, Wang WI, Lowman X, et al. TIPRL Inhibits Protein Phosphatase 4 Activity and Promotes H2AX Phosphorylation in the DNA Damage Response. PloS one. 2015;10(12):e0145938. doi: 10.1371/journal.pone.0145938. PubMed PMID: 26717153. Pubmed Central PMCID: 4696667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hernandez-Davies JE, Tran TQ, Reid MA, Rosales KR, Lowman XH, Pan M, et al. Vemurafenib resistance reprograms melanoma cells towards glutamine dependence. Journal of translational medicine. 2015;13:210. doi: 10.1186/s12967-015-0581-2. PubMed PMID: 26139106. Pubmed Central PMCID: 4490757. [DOI] [PMC free article] [PubMed] [Google Scholar]