Abstract
Fibroblasts are cells with a structural function, synthesizing components of the extracellular matrix. They are accordingly associated with various forms of connective tissue. During cardiac development fibroblasts originate from different sources. Most derive from the epicardium, some derive from the endocardium, and a small population derives from the neural crest. Cardiac fibroblasts have important functions during development, homeostasis, and disease. However, since fibroblasts are a very heterogeneous cell population no truly specific markers exist. Therefore, studying them in detail is difficult. Nevertheless, several lineage tracing models have been widely used. In this review, we describe the developmental origins of cardiac fibroblasts, comment on fibroblast markers and related lineage tracing approaches, and discuss the cardiac cell composition, which has recently been revised, especially in terms of non-myocyte cells.
Keywords: Cardiac fibroblasts, embryonic development, lineage tracing, fibroblast markers, endothelial cells, smooth muscle cells, cardiomyocytes, cardiac cell composition
Introduction
Fibroblasts are cells of mesenchymal origin with a structural function, synthesizing interstitial collagen (mainly collagen types I, III, and VI) and fibronectin, two of the main components of the extracellular matrix [e.g., (1,2)]. By this they maintain structural integrity within connective tissues throughout the body and are widely distributed in most vertebrate organisms [e.g., (1,2)]. Fibroblasts also play an important role in the repair process during wound healing. They migrate to the site of damage, undergo differentiation (i.e., conversion to myofibroblasts/activated fibroblasts), and by this promote structural and functional repair. Inactive fibroblasts that are involved in maintenance and tissue metabolism are sometimes referred to as fibrocytes.
Fibroblast cells display extensive phenotypic heterogeneity among different tissues and even within the same tissue under different physiological conditions (2,3). Chang et al. (4) analyzed the gene expression of cultured fetal and adult skin fibroblasts from different locations of the human body. They found that fibroblasts from different anatomical sites presented distinct gene expression patterns and even proposed to consider them as different cell types.
In the heart, cardiac fibroblasts are essential for maintaining structural, mechanical, and electrical functions (2,5). In addition to cardiac fibroblasts, the heart is composed of cardiomyocytes, smooth muscle cells, and endothelial cells. Recent studies quantified and mapped various cell lineages that are involved in cardiac maintenance (2,6,7). However, the fibroblast cell population is still the least characterized cell type in the heart compared with other cell populations because no specific markers of fibroblasts exist. Nevertheless, lineage tracing approaches try to genetically trace the fate of cardiac fibroblasts.
The objective of this review is to understand the origins of cardiac fibroblasts and the ways in which they contribute to embryonic cardiac development in mammals. Typical markers of cardiac fibroblasts, also in terms of lineage tracing approaches, are furthermore critically discussed. Finally, the cellular composition of the healthy/homeostatic adult mammalian heart is reviewed.
Fibroblasts in cardiac development: origins and functions
Together with the overall cardiovascular system, the heart is the first organ to develop. It is essential for the distribution of nutrients and oxygen in the embryo (8). In mice, embryonic cardiac development begins with formation of the precardiac mesoderm, which emerges from the anterior primitive streak (8,9). On embryonic day 6.5 (E6.5), during gastrulation, these cells migrate to form bilaterally paired heart fields (9). The two heart fields build the so-called cardiac crescent (E7.75), the first anatomically distinct heart structure and location of the first heart field progenitor cells (9). These progenitor cells express such markers as Nkx2.5, Gata4/5/6, Mef2b/c, Hand1/2, Tbx5/20, and Myocardin and mainly contribute to the left ventricle and atria (10,11). The second heart field derives from the pharyngeal mesoderm and contributes primarily to the right ventricle, outflow tract, and atria (12,13). Progenitor cells of the second heart field are delineated by the expression of Islet1, Nkx2.5, and Flk1 (14). By E8.0, both halves of the cardiac crescent migrate medially to form the linear heart tube, which consists of an inner endocardial cell layer (comprising non-contractile endothelial-like cells) and an outer myocardial cell layer that is capable of contractility (9). The heart tube subsequently undergoes looping (~E9.5) and differential growth that transforms the linear arrangement of the future chambers into a configuration in which the atria are cranial to the ventricles. The emergence of a four-chambered structure appears by E10.5 (9). The process of forming the multichambered heart is first outlined by the migration of neural crest cells to the heart. These progenitor cells mainly contribute to the conotruncus and great vessels (9). The development of the conduction system, septa, atria, and ventricles then progresses as a result of cells that derive from cardiac muscle cells (3). Together with smooth muscle cells, the venous and arterial vasculature is modeled. Finally, the endocardium and valves derive from endothelial cells (11).
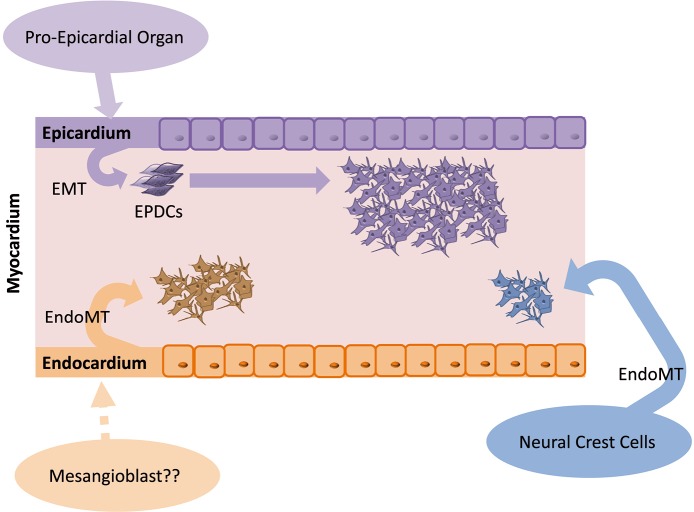
Cardiac fibroblasts derive from different progenitor cell populations, including the epicardium, endothelium/endocardium, and neural crest (15-17) (Figure 1). However, epicardial lineages are the main origin of cardiac fibroblasts (15,18). During embryonic development, the majority of cardiac fibroblasts derive from the pro-epicardial organ, a transient structure that is located near the base of the developing heart and a source of cardiovascular progenitor cells (3,10,19). These progenitor cells are able to differentiate into various cardiac cell lineages, such as cardiac fibroblasts and smooth muscle cells (17,19). By E9.5 in the mouse, immediately after heart looping, pro-epicardial cells migrate and cover the entire embryonic heart as a single cell layer beginning from the apex to form the embryonic epicardium (19). A subset of cells from the epicardium undergoes epithelial-to-mesenchymal transition (EMT) and migrates to the myocardium, referred to as epicardial-derived cells (EPDCs) (3,19). EPDCs further migrate to the compact myocardium and differentiate into interstitial and adventitial cardiac fibroblasts and coronary vascular smooth muscle cells (3,19). Interstitial cardiac fibroblasts are first detected on E12.5 in the mouse embryo (3,20). At this stage of development, heart chambers enlarge, however, the four-chambered heart is not fully developed because septation has not yet occurred (3). The transcription factor 21 (Tcf21) is essential for determination of the fate of cells that are destined to become cardiac fibroblasts in the epicardium before EMT (21). Tcf21-null murine hearts fail to form cardiac fibroblasts, but they are able to generate coronary vascular smooth muscle cells. Lineage tracing of Tcf21-null cells in these mice demonstrated their inability to undergo EMT (21). Subsequent processes, such as EMT and migration to the myocardium, require interactions with further growth and transcription factors, including Ets factors, fibroblast growth factors (FGFs), platelet-derived growth factor-β, Sox9, Tbx5, Thymosin β4, and transforming growth factors (TGFs) (3). Fgf10 and its receptor FGFR2b, for example, are responsible for regulating the migration of EPDCs to the compact myocardium. By inactivating this signaling pathway, fewer EPDCs are found in the myocardium, and the size of the heart decreases (19). According to Moore-Morris et al. (15), ~85% of all Col1a1-positive fibroblasts in the myocardium of the adult heart are also Wt1-positive and thus should have an epicardial origin.
Figure 1.
Embryonic development of cardiac fibroblasts. During embryonic development, cardiac fibroblasts derive from three different pools of progenitor cells. The majority of fibroblasts derive from the epicardium (~80%), whereas ~18% derive from the endocardium, and only a few derive from the neural crest. EMT, epithelial-to-mesenchymal transition; EPDCs, epicardial derived cells; EndoMT, endothelial-to-mesenchymal transition.
In addition to the epicardial layer, the embryonic heart is formed by the myocardium and endocardium. The myocardium promotes cardiac muscle tissue, and the endocardium lines the inner lumen of the heart, consisting of the endothelium (22). Subsets of cells from the endocardium undergo endothelial-to-mesenchymal transition (EndMT) and give rise to mesenchymal cells that act as primitive valves during early development and will later contribute to cardiac valves (23). Fibroblasts that are generated by this process further contribute to the interventricular septum (15). Moore-Morris et al. (15) detected Tie2-positive cardiac fibroblasts that were adjacent to the atrioventricular canal cushions on E12.5. Approximately 18% of myocardial Col1a1-positive fibroblasts originated from the endothelial lineage (the marker Tie2 is widely used for the determination of endothelial origin). The authors also detected Col1a1-positive cardiac fibroblasts throughout the valve mesenchyme and septum labeled with VE-cadherin, another endothelial marker (15).
A small subset of cardiac fibroblasts arises from the neural crest, a heterogeneous progenitor population that originates from the dorsal aspect of the neural tube. By undergoing EndMT, neural crest cells can develop into neurons, glial cells, melanocytes, or mesenchymal cells (16). The neural crest plays a key role in the development of the outflow region of the heart. However, cardiac neural crest cells also contribute to the valve mesenchyme, and a very minor subset of neural crest-derived fibroblasts was found within the myocardium of the right atrium (16). Ali et al. (24) confirmed the existence of neural crest-derived fibroblasts in the great vessels of the outflow tract region of the heart by tracing neural crest cells with Pax3. Neither Ali et al. (24) nor Moore-Morris et al. (15) identified a contribution of bone marrow-derived or hematopoietic cell-derived fibroblasts to the fibroblast pool of the heart.
Links between development and pathology have been frequently reported. Therefore, embryonic and early postnatal interactions between cardiac fibroblasts and cardiomyocytes and the ways in which they contribute to the overall cardiac milieu are particularly interesting. Embryonic cardiac fibroblasts build a three-dimensional network throughout the embryonic heart (1). They are thought to stimulate surrounding cardiomyocytes to grow and proliferate through ß1-integrin signaling and secretory factors, such as fibronectin, heparin-binding epidermal growth factor-like factor, and periostin (20). After birth, there is a substantial increase in systolic pressure that must be compensated by an increase in ventricular thickness and tensile strength. Cardiac fibroblasts play an important role in this process. The number of cardiac fibroblasts doubles postnatally to enable sufficient remodeling of the extracellular matrix, thus distributing the mechanical stress more efficiently (3). However, this dynamic period of development is restricted to the first weeks after birth in the mouse (3). A similar pattern of morphogenesis is assumed to exist in the human neonatal heart, but this possibility remains to be investigated.
Fate mapping and specificity of cardiac fibroblast markers
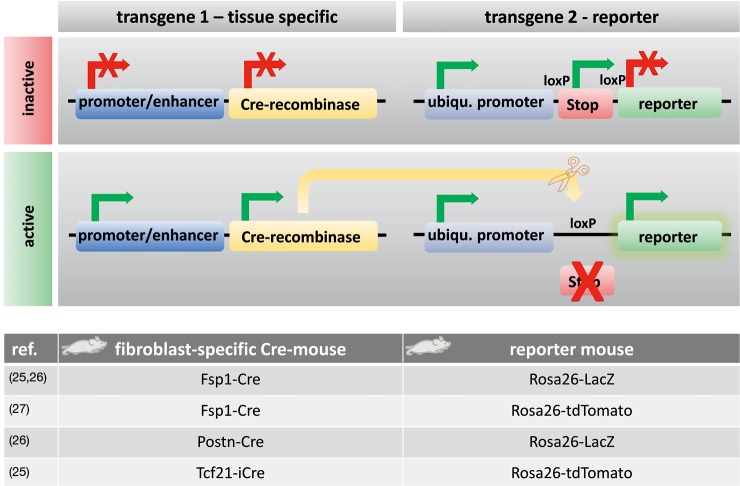
Tagging a specific cell and tracing its fate is called “lineage tracing” or “fate mapping”. Such techniques were first developed to study cell lineages during embryonic development and follow their developmental history in the adult body (5). Fate mapping techniques include the injection of dyes into cells, the creation of chimeric tissues of different species, or the use of a genetic reporter that is under the control of a cell-specific promoter (5) (Figure 2). Improvements in gene targeting and the identification of tissue-specific promoters or enhancers have enabled the irreversible genetic tagging of a given cell type at various stages (28). A double transgenic system is usually used for lineage tracing. One transgene expresses a site-specific recombinase in a cell-specific manner. The other transgene contains a reporter gene that is designed to permanently label the cell of interest after site-specific recombination (5). Cre-recombinase (i.e., an enzyme that cuts DNA at specific sites; e.g., loxP) is used for lineage tracing techniques predominantly (5). Figure 2 shows how this mechanism works. Cre-recombinase is driven by a cell-specific promoter or enhancer (e.g., a fibroblast-specific promoter) and thus is expressed in every cell of a specific type (e.g., fibroblast). Reporter genes, such as fluorescent proteins or ß-galactosidase behind a loxP site-flanked stop cassette (e.g., an antibiotic resistance gene), are driven by ubiquitous promoters. Ubiquitous promoters are active in every cell, regardless of their origin, although the expression of the reporter gene is initially blocked by the stop cassette before recombination. If Cre-recombinase is expressed, then it excises the stop cassette in front of the reporter only in the cell-type of interest, which can then be expressed, thus irreversibly marking the cell of interest (e.g., fibroblast) (5).
Figure 2.
Lineage tracing with transgenic systems. In this example, lineage tracing is explained by a double transgenic system. Transgene 1 expresses a site-specific Cre-recombinase in a cell-specific manner. Transgene 2 contains a reporter gene that is designed to permanently label the cell of interest after site-specific recombination. The mechanism works as the following. Cre-recombinase is driven by a cell-specific promoter or enhancer (e.g., a putative fibroblast-specific promoter) and is expressed in every cell of a specific type (e.g., fibroblast). Reporter genes, such as fluorescent proteins or ß-galactosidase behind a loxP site-flanked stop cassette (e.g., an antibiotic resistance gene), are driven by ubiquitous promoters. Ubiquitous promoters are active in every cell, regardless of their origin, although the expression of the reporter gene is initially blocked by the stop cassette before recombination. If Cre-recombinase is expressed, then it excises the stop cassette only in the cell type of interest because it recognizes the loxP sites in front of the reporter, which can then be expressed and irreversibly mark the cell of interest (e.g., fibroblast) (25-27).
However, fate-mapping experiments must be interpreted critically because Cre-recombinase can be nonspecifically expressed in cells of uncertain origin, which can result in false positives, especially in the context of disease (16). Furthermore, Cre-dependent reporter susceptibility has been reported to significantly influence fate-mapping results (29). Another issue is that a “ubiquitous” promoter that is used for reporter gene expression is not always equally active in every cell type; it may also be affected by epigenetic silencing (5). Such cases can result in false negatives. Additionally, the potential toxicity of Cre-recombinase has been reported in some cell types (30). Finally, the detection of reporter activity may be challenging. β-galactosidase, for example, is naturally expressed endogenously in some tissues, which may also yield false positives (5). In conclusion, both false positives and false negatives should be considered when working with transgenic lineage-tracing systems. Some limitations, such as timing issues, can be overcome using tamoxifen- or doxycycline-inducible Cre-drivers, but the efficiency of excision may be much lower (16). However, none of the aforementioned techniques can be used for human tissues.
Fibroblast markers
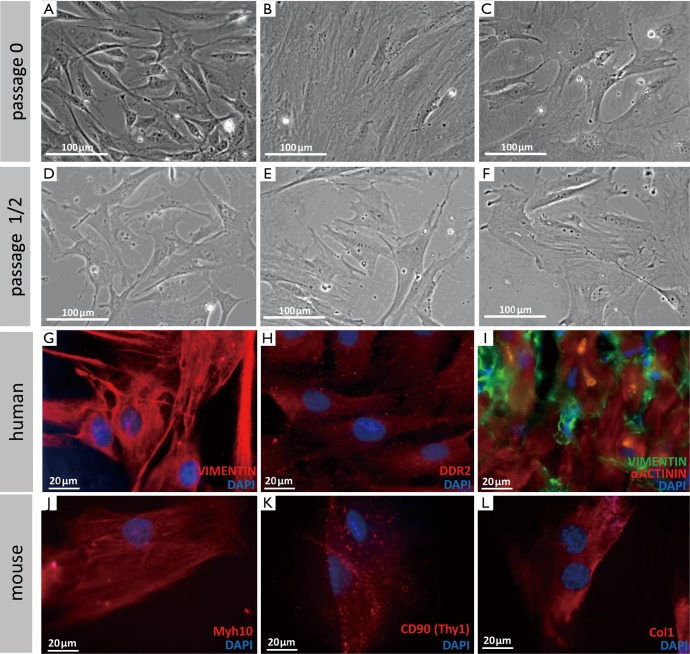
Cardiac fibroblasts were usually defined by their morphological appearance and ability to synthesize and secrete extracellular matrix proteins (21). They lack a basement membrane, exhibit a branched cytoplasm with multiple processes, and have an oval, speckled nucleus that typically has one or two nucleoli (1,2,5). The absence of a basement membrane distinguishes fibroblasts from other main cardiac cell types, which all contain one (2). In in vitro culture, fibroblasts are flat and often spindle-shaped, and they have the ability to adhere to plastic cell culture plates (see Figure 3A-F for examples). However, they remain poorly characterized in molecular terms (5). Various molecular tools have been used, but most markers are either nonspecific or insufficiently sensitive. Additional cell types or only subpopulations of fibroblasts are detected (5). A main issue in identifying fibroblasts is that they are a highly heterogeneous cell population, depending on their origin, location, physiological state, and specific function (5). Notably, cell culture can alter cell physiology (31,32). However, numerous studies of cultured fibroblasts have been performed, instead of in situ cell populations. Future investigations need to determine which markers are truly specific to cardiac fibroblasts in vivo.
Figure 3.
Morphology and markers of different types of fibroblasts. (A-F) Morphology of different types of fibroblasts grown in cell culture for different times (phase contrast). (A,D) Mouse tailtip fibroblasts. (B,E) Human adipose tissue-derived fibroblasts. (C,F) Human cardiac fibroblasts. (G-L) Immunofluorescent staining with different widely used markers of human and mouse fibroblasts. Vimentin (G) and DDR2 (H) were used to stain cultured human cardiac fibroblasts. (I) Vimentin staining combined with α-Actinin (cardiomyocyte) staining on a cryosection of a human right ventricular biopsy. Myh10 (J), CD90 (K), and Col1 (L) were used to stain cultured murine tail tip fibroblasts.
Below we discuss markers that are widely used to detect cardiac fibroblasts and provide some examples of their use in lineage tracing studies (see Table 1 and examples in Figure 3G-L).
Table 1. Cardiac fibroblast markers.
| Fibroblast marker | Sensitivity (expressed in all CFs or only in subpopulations?) | Specificity (expressed only in CFs or also in other cells?) | Accessibility for antibody detection (cellular location) | Mouse models (Cre-mouse for lineage tracing/reporter mouse) | Notes |
|---|---|---|---|---|---|
| Ddr2 | Low (only small percentage of CFs) | Medium (also by lymphocytic lineages, SMCs, ECs) | Good (surface) | No/No | – |
| CD90 | Medium (subset of fibroblasts: 65–85%) | Medium (also on immune cells and endothelial lineages) | Good (surface) | Yes (Thy1.2-Cre)/No | – |
| Pdgfrα | High | High-Medium (disease setting: also in SMCs, CMs; development: also in cardiovascular progenitor cells) | Good (surface) (however, antibodies seem to work only for IHC not for FACS) | Yes (tamoxifen-inducible)/Yes | Emerging as a definitive marker of CFs in adult tissues |
| Vimentin | High (in all CFs) | Low (also in the entire endothelial lineage, SMCs, pericytes) | Medium (intracellular) | No/No | – |
| Fsp1 | Low (only small subset of CFs → perivascular, not interstitial); Fsp1+ CFs rare in the normal heart | Low (also in inflammatory leukocytes and vascular cells after MI and TAC; marks monocytes, SMCs, ECs, CMs) | Medium (intracellular) | Yes (Fsp1-Cre mouse)//Yes (Fsp1-GFP reporter mouse) | In resting and activated CFs |
| Tcf21 | Medium (subset of fibroblasts) | High-medium (also in other proepicardial lineages) | Medium-low (nucleus → transcription factor) | Yes (Tcf21-iCre)/ Yes (Tcf21-LacZ) | Unknown how robust the expression of Tcf21 is in normal or injured adult hearts |
| Postn | Low (only small percentage of CFs; ~10% in the homeostatic heart) | High (specifically by epicardial derived myofibroblasts) | Low (secreted → ECM protein) | Yes (Postn-Cre mouse)/Yes (Postn-LacZ) | Robustly up-regulated after injury → useful marker of myofibroblasts |
| Col1a1 | High (Col1a1-CFs are positive for Vim and Pdgfrα) | Medium (also in osteoblasts, odontoblasts, SMCs, mesenchymal cells) | Low (secreted → ECM protein) | Yes/Yes | – |
| MEFSK4 | Not analyzed | High-medium (also detects pericytes, granulocytes) | Good (surface) | No/No | ONLY in mouse! |
CF, cardiac fibroblasts; SMCs, smooth muscle cells; ECs, endothelial cells; CMs, cardiomyocytes; IHC, immunohistochemical staining; FACS, fluorescent activated cell sorting; MI, myocardial infarction; TAC, transverse aortic constriction; ECM, extracellular matrix; Cre, Cre-recombinase.
Discoidin domain receptor 2
Discoidin domain receptor 2 (Ddr2) is a tyrosine kinase receptor that is expressed on the surface of cells of mesenchymal origin (33,34). It plays a marked regulatory role in EMT and is associated with fibrotic diseases because it is a collagen-specific receptor (34). Ddr2 was long believed to be a specific marker of cardiac fibroblasts. It was argued to not be expressed in cardiomyocytes, endothelial cells, or smooth muscle cells (1), and it seemed to be expressed in specific bone marrow-derived cells, termed fibrocytes and activated epithelial cells (2,35). However, Ddr2 was recently reported to mark smooth muscle cells, hepatic stellate cells, and endothelial cells (36). Furthermore, it seems to be only expressed in a small percentage of cardiac fibroblasts (5,16). Nonetheless, the authors of recent studies believe that Ddr2 in the heart is selectively expressed in cardiac fibroblasts (34,37).
CD90 (Thy1)
Thy1 cell surface antigen (also called CD90) is a membrane glycoprotein that is expressed on the cell surface. It is commonly used to identify cardiac fibroblasts (3,16). CD90 plays roles in inflammation and wound healing by mediating cell-matrix and cell-cell adhesion (38). CD90 was originally described as a thymocyte differentiation marker, but it is also expressed in the brain (39). Its expression differs among species. In humans, for example, CD90 is expressed in fibroblasts, mesenchymal stem cells, hematopoietic stem cells, neurons, and activated endothelial cells (36,38), thus limiting its use as an unambiguous marker of cardiac fibroblasts. Mice have two alleles of CD90: Thy1.1 and Thy1.2 (39). Campsall et al. generated several Thy 1.2-Cre mice, which were described to be neuron-specific (40). However, a study about established Cre-driver lines clearly showed that Cre-recombinase was also expressed in several cells in other organs, including the heart (41). Furthermore, CD90 does not mark all fibroblasts. Furtado et al. found that CD90 stained ~65% of mouse cardiac fibroblasts in short-term culture and ~86% of mouse tail tip fibroblasts (31).
Platelet-derived growth factor receptor α
Platelet-derived growth factor receptor α (Pdgfrα) and platelet-derived growth factor receptor β (Pdgfrβ) belong to the family of receptor tyrosine kinases (42). Pdgfrα and Pdgfrβ play important roles in forming the vasculature during embryonic development and are expressed in cardiovascular and mesenchymal progenitor cells (43,44). Pdgfrβ regulates the development of vascular smooth muscle cells from epicardium-derived cells (45). Pdgfrα appears to be involved in the formation of cardiac fibroblasts from the epicardium (42). Moore-Morris et al. reported that Pdgfrα is a definitive and comprehensive marker of fibroblasts in adult tissues (16). In addition to cardiac fibroblasts, it is also strongly expressed in collagen-producing fibroblasts in skeletal muscles and the lungs (46,47). Limited expression of Pdgfrα has also been detected in endothelial cells and cardiomyocytes in the adult heart, albeit only in a disease setting (43). However, a recent study reported that Pdgfrα marked a stem cell antigen-1 (Sca1)-positive cardiogenic clonogenic resident stem cell population in the adult heart, which also expressed Tcf21, Gata4/6, Tbx5/20, and Hand2 (48).
Pdgfrα is a surface receptor that is easily detectable by immunohistochemical techniques. However, for whatever reason, Pdgfrα antibodies that have been designed for flow cytometry applications are not sufficiently robust for the detection of cardiac fibroblasts (7). Surface epitopes can be influenced by digestion techniques for tissue samples to achieve single cell suspensions that are necessary for flow cytometry analyses.
Vimentin
Vimentin is an intermediate filament protein. It is a typical marker of mesenchymal lineages (49). Vimentin is the most sensitive marker of cardiac fibroblasts (3,16). However, the entire endothelial lineage also expresses vimentin (16,49). Moreover, smooth muscle cells, myoepithelial cells, pericytes, and neurons contain intermediate filaments and therefore also express vimentin (1,36). Thus, it is not very useful as a specific marker of cardiac fibroblasts (3).
Fibroblast-specific protein 1
Fibroblast-specific protein 1 (Fsp1; also known as S100A4) is a cytoplasmic filament-associated Ca2+ binding protein (50,51) that is highly expressed in activated fibroblasts (2). Fsp1 was first identified in different fibroblast cell lines (50). Although it is currently considered a reliable marker of fibroblasts (16), it is only expressed in a subset of cardiac fibroblasts in the normal/homeostatic heart (52). Furthermore, Fsp1 also marks immune cells, smooth muscle cells, endothelial cells, and myocytes, at least after injury (5,16,36,51). It is therefore neither sensitive nor specific. Moore-Morris et al. found only rare Fsp1-positive cells in the adult homeostatic myocardium (15). Fsp1-positive cells also were not positive for another fibroblast marker, Col1α1, but they co-localized with the pan-hematopoietic marker CD45, supporting other studies that reported that Fsp1 is an immune cell marker (53). Kong et al. found that ~50% of Fsp1-positive cells in the murine heart after myocardial infarction were leukocytes (53). In the pressure-overloaded myocardium, a large number of Fsp1-positive endothelial cells (~17%) were detected.
Transcription factor 21 (Tcf21)
Tcf21 is a member of the class A basic helix-loop-helix (bHLH) family of transcription factors that manage cell-fate specification. It has been successfully used to trace cardiac resident fibroblasts from development until adulthood (21). Tcf21 is required for cell fate determination during development, but it is still active in adult resident fibroblasts (21). However, it is a marker of the proepicardium and may also mark cell populations other than cardiac fibroblasts (54). CD45-positive immune cells were not positive for Tcf21 (54). Acharya et al. used an inducible Tcf21-Cre mouse to irreversibly label Tcf21-positive cells and their progeny (21). Co-staining with markers of endothelial cells (CD31), smooth muscle cells (Pdgfrβ, SM22α), cardiomyocytes (Acta1), and fibroblasts (Pdgfrα, Col1) revealed no double staining with endothelial cells, smooth muscle cells, or cardiomyocytes. However, Tcf21 overlapped to a certain extent with Pdgfrα and Col1 (although this was not quantified), indicating that it labels a subpopulation of cardiac fibroblasts. Tcf21 is a transcription factor that acts in the nucleus. Therefore, it is difficult to detect using antibodies in immunohistochemistry or flow cytometry studies (32).
Periostin
Periostin (Postn) is a TGF-ß inducible secreted matricellular protein that is involved in cell adhesion (55). The majority of Postn is secreted, which limits antibody staining (5,21). The level of expression of Postn is high during cardiac development, but it is only moderately expressed in resting fibroblasts in the adult heart (3). Furtado et al. analyzed the expression of Postn in homeostatic adult hearts in Postn-Cre-Rosa26R mice and found that only ~10% of quiescent cardiac fibroblasts expressed Postn (31). However, after myocardial infarction, this percentage increased significantly. Postn-positive fibroblasts were mainly detected in the injury/scar area. Therefore, it may be a good marker of activated fibroblasts (32). A recent study reported that Postn-positive myofibroblasts derive from Tcf21-positive cardiac-resident fibroblasts upon injury stimulation (56). Therefore, these authors generated a tamoxifen-inducible Postn-Cre knock-in mouse. There is also a widely used reporter mouse with LacZ knock-in in the first exon of Postn (57). Furthermore, another Postn-Cre mouse contains a regulatory genomic region 3.9 kb upstream of Postn (Postn promoter) that drives an eGFP-Cre fusion gene (58).
Collagen1α1 (Col1α1)
Col1α1 is an extracellular matrix structural protein with a major fibrillar component that selectively and comprehensively marks fibroblasts in the normal heart (15,51). Moore-Morris et al. used a Col1α1-GFP reporter mouse, confocal microscopy, and flow cytometry and found that all collagen1α1-GFP double-positive cells in the adult left ventricular myocardium were also positive for vimentin and Pdgfrα (15). As described above, Pdgfrα is emerging as a definitive marker of cardiac fibroblasts in adult tissues (16). According to Moore-Morris et al., interstitial myocardial and perivascular Col1α1-positive cells were negative for αSma (smooth muscle cell marker), Pdgfrβ (pericyte marker), Pecam1 (pan-endothelial cell marker), and CD45 (pan-leukocyte marker) (15). However, other authors claimed that Col1α1 is also synthesized by valve interstitial cells, vascular smooth muscle cells, pericytes, and other mesenchymal cells (32,36,51). Therefore, the true specificity of Col1α1 remains open to debate. Because this protein is secreted by cells, detection by antibodies using immunohistochemical techniques is technically difficult (32).
MEFSK4
MEFSK4 is a monoclonal antibody that identifies an antigen that is expressed by Pdgfrα- Col1α1 double-positive murine cardiac fibroblasts (32). Originally, MEFSK4 was developed to remove murine embryonic fibroblast (MEF) feeder cells from embryonic and induced pluripotent stem cell cultures (7). However, MEFSK4 antibodies also detect surface antigens on pericytes and granulocytes as shown by staining of a subpopulation with CD11b (leukocyte marker) and NG2 (mesenchymal cell marker) (7). Pinto et al. nevertheless recommended that MEFSK4 can be used as a surrogate marker of cardiac fibroblasts, at least in combination with CD31 (Pecam1) and CD45 (Ptprc) to exclude endothelial and hematopoietic cells (7). The authors demonstrated that cells that are identified as fibroblasts by Pdgfrα, Col1a1, MEFSK4, and Tcf21 are relatively uniform originating from different transgenic mouse models. Fibroblast markers, such as CD90 and Sca1, however, were suggested to underrepresent the cardiac resident fibroblast population. Unfortunately, MEFSK4 antibody is only suitable for mice and not for rats or humans.
Paradigm change: cardiac fibroblasts express stem cell markers and cardiac transcription factors
Mouse cardiac fibroblasts were recently reported to express mesenchymal stem cell markers, such as Sca1, CD49e, CD51, and CD29 (31). Seventy-nine percent of mouse cardiac fibroblasts were positive for Sca1 (determined by fluorescence-activated cell sorting), and 87% of murine tail tip fibroblasts. Sca1 is a cell surface protein that is used as a common biological marker to identify hematopoietic stem cells, among other markers (7). CD49e, CD51, and CD29 were expressed in more than 90% of heart and tail fibroblasts (31). Furthermore, murine cardiac fibroblasts presented the unexpected expression of cardiogenic genes compared with tail tip fibroblasts, especially Tbx20, Gata4, Gata6, and Hand2 (31). Quantitative polymerase chain reaction revealed the strongest expression for Gata4 and Tbx20. This result was verified by immunofluorescent staining (31). The authors demonstrated that this signature was also conserved in human cardiac fibroblasts in the atria and ventricles; therefore, they claimed that cardiac fibroblasts have an evolutionarily conserved cardiogenic profile. Careful bioinformatic analysis even confirmed that murine cardiac fibroblasts were transcriptionally more similar to cardiomyocytes than to tail tip fibroblasts (31). Additionally, Tcf21 and another epicardial transcription factor, Wt1, were strongly upregulated in cardiac fibroblasts (31).
Fibroblast origin tracing in development and disease
In this review, we focus on fibroblast populations in the homeostatic heart. The use of lineage tracers has enabled researchers to trace the developmental origin of different fibroblast populations. In the homeostatic adult heart, ~80% of cardiac fibroblasts are of epicardial origin, indicated by the epicardial markers Wt1 and Tbx18 (15,24). Only 16% of cardiac fibroblasts have an endocardial/endothelial origin, indicated by Tie2 and Nfatc1 labeling (15,24).
Where do fibroblasts derive from in the adult heart after injury? De novo EMT was initially believed to be the primary mechanism for the generation of new fibroblasts after injury. Moreover, bone marrow cells or fibrocytes might contribute to the cardiac fibroblast pool after injury. However, this paradigm has recently been challenged. After the induction of pressure overload by transverse aortic constriction, newly formed cardiac fibroblasts appeared to be generated mainly through the proliferation of preexisting fibroblasts and not by de novo EMT (15,24). Furthermore, both of these studies (15,24) excluded the contribution of fibroblasts that were derived from hematopoietic cells. Ali et al. (24) performed transplantation and parabiosis studies with bone marrow and hematopoietic stem cells that were labeled with fluorochromes. Moore-Morris et al. (15) used bone marrow cells that were marked by hematopoietic-specific Vav-Cre.
Activated fibroblasts are widely referred to as myofibroblasts after injury but represent a distinct population of cardiac fibroblasts that are not discussed in further detail herein. Recent reviews on this topic have been published by Davis and Molkentin (59) and Hermans et al. (60).
Direct cardiac reprogramming approaches in vivo using fibroblast lineage tracing
Fibroblasts have received attention recently in regenerative medicine because they have been successfully reprogrammed/transdifferentiated directly into different lineages without passing through a pluripotent state. This includes direct reprogramming from fibroblasts into “functional” cardiomyocytes [e.g., (25-27,61)]. This further demonstrates the enormous plasticity of fibroblasts. The direct transdifferentiation of fibroblasts into other cell types reveals the possibility of directly reprogramming emerging scar tissue after myocardial infarction back into functional myocardium. Direct reprogramming techniques require good lineage tracing approaches to clearly identify fibroblasts that are transdifferentiated into cardiac cells, such as cardiomyocytes or cardiac progenitor cells. Lineage tracing is especially necessary to trace the fate of fibroblasts after injecting “reprogramming” factors/cocktails in vivo after cardiac injury to determine whether fibroblasts can truly transdifferentiate. Most researches have used the Fsp1-Cre mouse [e.g., (25-27)], but Postn-Cre (26) and Tcf21-Cre (25) have also been used for in vivo lineage tracing (see Figure 2).
Qian et al. applied their direct reprogramming strategy in infarcted Postn-Cre/β-galactosidase transgenic mice, in which Postn-positive cells were irreversibly marked by β-galactosidase (26). β-galactosidase activity was found in many, but not all, cardiac fibroblasts and also some endocardial and endothelial cells but not in cardiomyocytes or bone marrow cells. However, because this research group used retroviruses to deliver their reprogramming factors [i.e., Gata4, Mef2c, and Tbx5 (GMT)], they only infected proliferating cells that appeared in the border zone of the ischemic area after infarction. As mentioned above, Postn is a marker of actively dividing myofibroblasts that appear after myocardial infarction, and these cells should be well targeted by retroviruses. Four weeks after inducing myocardial injury by coronary ligation and delivering reprogramming factors, they detected β-galactosidase-positive cells that were also positive for the cardiomyocyte marker α-Actinin, indicating that myofibroblasts were directly reprogrammed into cardiomyocyte-like cells. However, Qian et al. did not provide exact numbers for these events (26). Furthermore, they found a similar amount of β-galactosidase-α-Actinin double-positive cells using a mouse in which Cre-recombinase was under the control of the Fsp1 promoter. Notably, Fsp1 only marks a very small subset of fibroblasts and according to Kong et al., more than 50% of Fsp1-positive cells were leukocytes after myocardial infarction (53). Qian et al. (26) also applied the three reprogramming factors GMT in Tie2-Cre/β-galactosidase mice, in which endothelial circulating hematopoietic cells should be marked by β-galactosidase. Four weeks after injury, however, these authors did not detect β-galactosidase-α-actinin double-positive cells, indicating that Tie2-positive endothelial or hematopoietic cells were not reprogrammed into cardiomyocytes by GMT. One reason may be that endothelial or hematopoietic cells did not proliferate at the time of retrovirus delivery. Another possibility is that the three factors were not sufficient to reprogram endothelial or hematopoietic cells into cardiomyocytes. Yet another possibility is that not many endothelial cells appeared in the border zone of the myocardial infarction where the authors injected the reprogramming factors; thus, endothelial cells were not targeted.
Another group used four factors [Gata4, Mef2c, Tbx5, Hand2 (GMTH)] in retroviruses for the direct reprogramming of fibroblasts into cardiomyocytes (25). They applied GMTH directly after coronary ligation to the myocardium in Fsp1-Cre/β-galactosidase mice and detected 6.5%±1.2% β-galactosidase-positive cardiomyocytes in the injured area. Using this approach, Fsp1 could again mark a significant amount of leukocytes. Song et al. (25) used a tamoxifen-inducible Tcf21-iCre/tdTomato mouse as another approach. They reported that Tomato was predominantly expressed in cardiac fibroblasts and to a lesser extent in endothelial cells in this mouse line. Four weeks after the induction of myocardial infarction and injection of GMTH factors, 2.4%±1.5% Tomato-positive cardiomyocytes from the whole heart were detected. The authors concluded that Tcf21 marks a more restricted fibroblast population than Fsp1. Based on the information above, it is more presumable that a significant portion of leukocytes were reprogrammed in the Fsp1 mouse, leading to a higher amount of double-positive cells, whereas Tcf21 only marked epicardial-derived cardiac cells.
Jayawardena et al. employed a lentiviral approach and injected four candidate microRNAs into infarcted hearts in triple transgenic αMHC-CFP/Fsp1-Cre/tdTomato mice (27). In this transgenic mouse, Fsp1-positive cells (“fibroblasts”) were irreversibly marked with red fluorescent Tomato protein. Additionally, αMHC-expressing cardiomyocytes express cyan fluorescent protein (CFP). CFP-positive cardiomyocytes that were also positive for Tomato should have developed from transdifferentiating fibroblasts/leukocytes in this transgenic mouse. Jayawardena et al. (27) found that 1% of cardiomyocytes were double-positive for CFP and Tomato 6 weeks after induction of myocardial infarction and injection of their micro-RNA reprogramming cocktail. A similar percentage of double-positive cells was reported by Song et al. (25) using the Tcf21-iCre mouse. However, Jayawardena et al. did not use a second transgenic mouse model to determine the efficiency of Fsp1-fibroblast labeling.
Overall, the efficiency of direct reprogramming is very low, even if the percentage of newly generated induced cardiomyocytes is higher with in vivo approaches than with in vitro approaches (62). Pioneering research by Lalit et al. sought to address this issue by reprogramming fibroblasts into induced cardiac progenitor cells that retain a certain proliferative capacity and are able to differentiate into all three cardiac lineages: cardiomyocytes, smooth muscle cells, and endothelial cells (63).
Cardiac cellular composition
Heart cells are unitary elements that define cardiac function and disease. The most abundant permanent cellular constituents are cardiomyocytes, fibroblasts, endothelial cells, and smooth muscle cells. Transient cell populations, such as lymphocytes, mast cells, and macrophages, mainly appear during various disease states and interact with permanent cell types to affect cardiac function (2).
Although cardiomyocytes occupy 70–85% of the volume of the mammalian heart (64,65), non-myocytes are equally important for normal heart homeostasis and function. They provide the extracellular matrix, mediate intercellular communication, and provide the vascular supply that is needed for cardiomyocyte contraction and long-term survival (64).
Early studies from the 1970s and 1980s suggested that the adult rat and human hearts comprise 30% cardiomyocytes and 70% non-myocytes (66,67). Since 2016, the current dogma was that fibroblasts represent the largest cell population of non-myocytes in the heart, whereas smooth muscle cells and endothelial cells account for a comparatively small proportion of the non-myocytes (1,2). However, species-specific differences have been found in the composition of cardiac cell populations between rats, mice, and humans (2). Banerjee et al. confirmed that the adult rat heart comprises ~30% cardiomyocytes and ~70% non-myocytes (68). In the mouse heart, up to 55% cardiomyocytes and 45% non-myocytes were detected. The authors explained that this difference was attributable to greater wall tension in the rat heart because its radius is larger. Therefore, the rat heart requires higher structural integrity, which is favored by more fibroblasts. The cellular composition during cardiac development was analyzed in mice from E14.5 to the early postnatal period (postnatal day 15, P15) (68) (see also Table 2). The proportion of fibroblasts slightly increased from the neonatal to adult periods, whereas the proportion of cardiomyocytes decreased. Banerjee et al. reported that there were no significant differences in cell composition in different locations of the adult heart (e.g., left and right ventricular free wall and intraventricular septum) (68).
Table 2. Cardiac cellular composition.
| Ref. | CMs | Non-myocyte cells | Species | Dev. stage | Method | Heart location | |||
|---|---|---|---|---|---|---|---|---|---|
| CFs | ECs | SMCs | Other cells | ||||||
| (68) | 62% (αMHC) | 10% (Ddr2) | n.d. | n.d. | n.d. | Mouse | P1 | FACS | Whole heart |
| (68) | 63% (αMHC) | 18% (Ddr2) | n.d. | n.d. | n.d. | Mouse | P15 | FACS | Whole heart |
| (68) | 56% (αMHC) | 27% (Ddr2) | 7% (CD31) | 10% (α-SMA) | n.d. | Mouse | Adult | FACS | Whole heart |
| (68) | 52% (αMHC) | 37% (Ddr2) | 11% non-CM/non-CF population | Mouse | Adult | Mouse | Adult | ||
| (68) | 51% (αMHC) | 37% (Ddr2) | 12% non-CM/non-CF population | Mouse | Adult | Mouse | Adult | ||
| (7) | 31%±4% (WGA) | n.d. | 44%±4% (IB4) | n.d. | 5%±1.5% leucocytes (Lin1 cocktail) | Mouse | Adult | IHC | Ventricles + interventricular septum |
| (7) | 32%±5% (ACTN2) | 13%±2.5% (Pdgfrα-GFP); 13%±3.6% (Col1a1-GFP) | 55%±6% (IB4); 52%±3.9% (DACH1) | 6%±1.1% (Pdgfrβ) (SMCs and pericytes) | 8.5%±1.5% leukocytes (Lin1 cocktail) | Mouse (transgenic: Pdgfrα-GFP or Col1a1-GFP) | Adult | IHC | Left ventricle |
| (68) | 62% (αMHC) | 30% (DDR2) | n.d. | n.d. | n.d. | Rat | P1 | FACS | Whole heart |
| (68) | 30% (αMHC) | 64% (DDR2) | n.d. | n.d. | n.d. | Rat | P15 | FACS | Whole heart |
| (66) | 30–35% | 65–70% (ECs, CFs, pericytes, SMCs, Macrophages) | Rat | ||||||
| (6) | 18%±3% (PCM-1) | 58% mesenchymal cells (non-PCM-1, non UEA-I), (CFs, SMCs, pericytes) | 24%±5% (UEA-I) | see CFs | Human | Adult | IHC | ||
| (6) | 33%±15% (PCM-1) | 43%±10% mesenchymal cells (non-PCM-1, non UEA-I), (CFs, SMCs, pericytes) | 24%±9.5% (UEA-I) | see CFs | Human | Adult | FACS | ||
| (7) | 31%±5.6% (ACTN2) | n.d. | 54%±6.4% (CD31); 51%±2.9% (DACH1) | n.d. | 3%±1.2% leukocytes (CD45) | Human | Adult | IHC | Ventricles + interventricular septum |
Ref., reference; CMs, cardiomyocytes; CFs, cardiac fibroblasts; ECs, endothelial cells; SMCs, smooth muscle cells; dev. stage, developmental stage; GFP, green fluorescent protein; P1, postnatal day 1; P15, postnatal day 15; n.d., not determined; IHC, immunohistochemical analyses; FACS, flow cytometric analysis; TEM, transmission electron microscopy.
Bergmann et al. recently analyzed human postmortem hearts from 29 subjects (1–73 years of age) who did not have a history or signs of cardiac pathology (6). To quantify the number of cells, these authors used PCM-1 (Pericentriolar Material 1) to stain cardiomyocyte nuclei and UEA-I (Ulex Europaeus lectin I) to mark endothelial cells. All of the cells that were negative for one of these markers were considered mesenchymal cells, including fibroblasts, pericytes, and smooth muscle cells. In young adults, an average of 18% cardiomyocytes, 24% endothelial cells, and 58% mesenchymal cells was detected by stereological analysis. Flow cytometry detected ~33% cardiomyocytes, 24% endothelial cells, and ~43% mesenchymal cells, which was stated to be consistent with the stereological data. During ageing, the number of cardiomyocytes remained constant, whereas the fractions of endothelial and mesenchymal cells increased.
Pinto et al. recently postulated that fibroblasts comprise a much lower cell population than previously thought, and endothelial cells appear to be the most abundant cell type in the non-myocyte cell population in the heart (7). These authors used newly available genetic tracers and enhanced flow cytometry techniques to analyze human and mouse hearts. Immunohistochemistry revealed that the non-myocyte cell population in the heart comprised >60% endothelial cells, 5–10% hematopoietic-derived cells, and ≤20% fibroblasts (see also Table 2). They confirmed their results by flow cytometry. The authors used different digestion and isolation protocols to generate single cell solutions from cardiac tissue because different enzymatic digestion techniques can influence markers on the cell surface and thus change the outcome. The authors applied collagenase type II, a mixture of collagenase type IV and dispase, or a mixture of collagenase type XI, hyaluronidase type I-s and DNase I. The collagenase type IV and dispase combination worked the best. The lack of a clear marker of fibroblasts may explain the difficulties that have been seen in quantifying and tracking them in vivo, which may be another reason for the disparate outcomes in different studies. Depending on the marker that is used, different amounts of fibroblasts were found. Ddr2, for example, detected ~30% fibroblasts in adult mice (68), whereas Pdgfrα and Col1a1 both detected ~15% fibroblasts (7). Furthermore, some studies have sought to quantify different types of cells by flow cytometry, and other studies employed stereological techniques (e.g., immunohistochemical staining and subsequent counting).
Despite these differences, the results of several studies have been similar in terms of the amount of cardiomyocytes. Three studies found ~30% cardiomyocytes in adults of three different species (i.e., mouse, rat, human), although different markers were used (αMHC, WGA, ACTN2, PCM-1) (6,7,68). The only difference was the distribution of non-myocyte cells, which may be partially attributable to nonspecific markers or different techniques. Future studies will clarify these issues.
Conclusions
The role of fibroblasts in the heart has long been underappreciated. However, in recent years, cardiac fibroblasts have been shown to play essential roles in cardiac development, cardiac homeostasis, and several disease states. For a detailed understanding of the underlying mechanisms, the molecular basis of cardiac fibroblasts must be elucidated. However, the main problem that has been encountered in such studies is the lack of a specific marker of fibroblasts, largely because of their heterogeneous nature. During embryonic development, cardiac fibroblasts derive from three different pools of progenitor cells. The majority of fibroblasts derive from the epicardium (~80%), whereas ~18% derive from the endocardium, and only a few derive from the neural crest.
Recently, a paradigm shift occurred, in which several stem cell markers and typical cardiac development markers were identified in cardiac fibroblasts (31). Such scientific progress revealed that cardiac fibroblasts are more similar to cardiomyocytes than to tail tip fibroblasts (31). However, still, no specific marker of cardiac fibroblasts has been identified, and still unknown is whether such a marker exists.
For a complete understanding of cardiac cellular composition, the unequivocal identification of non-myocyte cell populations (e.g., endothelial cells, smooth muscle cells, and fibroblasts) would be a tremendous advance. The number of cardiomyocytes can be reliably estimated. Even when using different markers or techniques, the proportion of cardiomyocytes (~30%) has been found to be generally similar in different species and different studies. For non-myocyte cell populations, however, highly variable results have been reported, suggesting that different markers only label subpopulations of a specific cell type or nonspecifically label additional cell types. Future studies will need to determine the cardiac cell composition in different species of different ages.
Acknowledgements
We thank the native English-speaking experts from BioMed Proofreading LLC for editing the manuscript.
Funding: MA Deutsch is supported by the Dr. Rusche Forschungsprojekt [2014] of the DSHF and DGTHG. R Lange is supported by the Bayerische Forschungsstiftung (AZ-1012-12). M Krane is supported by the Deutsche Stiftung für Herzforschung (F/37/11), Deutsches Zentrum für Herz Kreislauf Forschung (DZHK B 15-005, DZHK B 15-039SE) and Deutsche Forschungsgemeinschaft—Sachmittelantrag (KR3770/7-1, KR3770/9-1).
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Camelliti P, Borg TK, Kohl P. Structural and functional characterisation of cardiac fibroblasts. Cardiovasc Res 2005;65:40-51. 10.1016/j.cardiores.2004.08.020 [DOI] [PubMed] [Google Scholar]
- 2.Souders CA, Bowers SL, Baudino TA. Cardiac fibroblast: the renaissance cell. Circ Res 2009;105:1164-76. 10.1161/CIRCRESAHA.109.209809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lajiness JD, Conway SJ. Origin, development, and differentiation of cardiac fibroblasts. J Mol Cell Cardiol 2014;70:2-8. 10.1016/j.yjmcc.2013.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang HY, Chi JT, Dudoit S, et al. Diversity, topographic differentiation, and positional memory in human fibroblasts. Proc Natl Acad Sci U S A 2002;99:12877-82. 10.1073/pnas.162488599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zeisberg EM, Kalluri R. Origins of cardiac fibroblasts. Circ Res 2010;107:1304-12. 10.1161/CIRCRESAHA.110.231910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bergmann O, Zdunek S, Felker A, et al. Dynamics of Cell Generation and Turnover in the Human Heart. Cell 2015;161:1566-75. 10.1016/j.cell.2015.05.026 [DOI] [PubMed] [Google Scholar]
- 7.Pinto AR, Ilinykh A, Ivey MJ, et al. Revisiting Cardiac Cellular Composition. Circ Res 2016;118:400-9. 10.1161/CIRCRESAHA.115.307778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buckingham M, Meilhac S, Zaffran S. Building the mammalian heart from two sources of myocardial cells. Nat Rev Genet 2005;6:826-35. 10.1038/nrg1710 [DOI] [PubMed] [Google Scholar]
- 9.Latif S, Masino A, Garry DJ. Transcriptional pathways direct cardiac development and regeneration. Trends Cardiovasc Med 2006;16:234-40. 10.1016/j.tcm.2006.04.004 [DOI] [PubMed] [Google Scholar]
- 10.Harvey RP. Patterning the vertebrate heart. Nat Rev Genet 2002;3:544-56. 10.1038/nrg843 [DOI] [PubMed] [Google Scholar]
- 11.Garry DJ, Olson EN. A common progenitor at the heart of development. Cell 2006;127:1101-4. 10.1016/j.cell.2006.11.031 [DOI] [PubMed] [Google Scholar]
- 12.Masino AM, Gallardo TD, Wilcox CA, et al. Transcriptional regulation of cardiac progenitor cell populations. Circ Res 2004;95:389-97. 10.1161/01.RES.0000138302.02691.be [DOI] [PubMed] [Google Scholar]
- 13.Tzahor E, Evans SM. Pharyngeal mesoderm development during embryogenesis: implications for both heart and head myogenesis. Cardiovasc Res 2011;91:196-202. 10.1093/cvr/cvr116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moretti A, Caron L, Nakano A, et al. Multipotent embryonic isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell 2006;127:1151-65. 10.1016/j.cell.2006.10.029 [DOI] [PubMed] [Google Scholar]
- 15.Moore-Morris T, Guimaraes-Camboa N, Banerjee I, et al. Resident fibroblast lineages mediate pressure overload-induced cardiac fibrosis. J Clin Invest 2014;124:2921-34. 10.1172/JCI74783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moore-Morris T, Guimaraes-Camboa N, Yutzey KE, et al. Cardiac fibroblasts: from development to heart failure. J Mol Med (Berl) 2015;93:823-30. 10.1007/s00109-015-1314-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lighthouse JK, Small EM. Transcriptional control of cardiac fibroblast plasticity. J Mol Cell Cardiol 2016;91:52-60. 10.1016/j.yjmcc.2015.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deb A, Ubil E. Cardiac fibroblast in development and wound healing. J Mol Cell Cardiol 2014;70:47-55. 10.1016/j.yjmcc.2014.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vega-Hernández M, Kovacs A, De Langhe S, et al. FGF10/FGFR2b signaling is essential for cardiac fibroblast development and growth of the myocardium. Development 2011;138:3331-40. 10.1242/dev.064410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ieda M, Tsuchihashi T, Ivey KN, et al. Cardiac fibroblasts regulate myocardial proliferation through beta1 integrin signaling. Dev Cell 2009;16:233-44. 10.1016/j.devcel.2008.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Acharya A, Baek ST, Huang G, et al. The bHLH transcription factor Tcf21 is required for lineage-specific EMT of cardiac fibroblast progenitors. Development 2012;139:2139-49. 10.1242/dev.079970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Milgrom-Hoffman M, Harrelson Z, Ferrara N, et al. The heart endocardium is derived from vascular endothelial progenitors. Development 2011;138:4777-87. 10.1242/dev.061192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Combs MD, Yutzey KE. Heart valve development: regulatory networks in development and disease. Circ Res 2009;105:408-21. 10.1161/CIRCRESAHA.109.201566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ali SR, Ranjbarvaziri S, Talkhabi M, et al. Developmental heterogeneity of cardiac fibroblasts does not predict pathological proliferation and activation. Circ Res 2014;115:625-35. 10.1161/CIRCRESAHA.115.303794 [DOI] [PubMed] [Google Scholar]
- 25.Song K, Nam YJ, Luo X, et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature 2012;485:599-604. 10.1038/nature11139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qian L, Huang Y, Spencer CI, et al. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature 2012;485:593-8. 10.1038/nature11044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jayawardena TM, Egemnazarov B, Finch EA, et al. MicroRNA-mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to cardiomyocytes. Circ Res 2012;110:1465-73. 10.1161/CIRCRESAHA.112.269035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin-Puig S, Wang Z, Chien KR. Lives of a heart cell: tracing the origins of cardiac progenitors. Cell Stem Cell 2008;2:320-31. 10.1016/j.stem.2008.03.010 [DOI] [PubMed] [Google Scholar]
- 29.Ma Q, Zhou B, Pu WT. Reassessment of Isl1 and Nkx2-5 cardiac fate maps using a Gata4-based reporter of Cre activity. Dev Biol 2008;323:98-104. 10.1016/j.ydbio.2008.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmidt-Supprian M, Rajewsky K. Vagaries of conditional gene targeting. Nat Immunol 2007;8:665-8. 10.1038/ni0707-665 [DOI] [PubMed] [Google Scholar]
- 31.Furtado MB, Costa MW, Pranoto EA, et al. Cardiogenic genes expressed in cardiac fibroblasts contribute to heart development and repair. Circ Res 2014;114:1422-34. 10.1161/CIRCRESAHA.114.302530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ivey MJ, Tallquist MD. Defining the Cardiac Fibroblast. Circ J 2016;80:2269-76. 10.1253/circj.CJ-16-1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olaso E, Labrador JP, Wang L, et al. Discoidin domain receptor 2 regulates fibroblast proliferation and migration through the extracellular matrix in association with transcriptional activation of matrix metalloproteinase-2. J Biol Chem 2002;277:3606-13. 10.1074/jbc.M107571200 [DOI] [PubMed] [Google Scholar]
- 34.George M, Vijayakumar A, Dhanesh SB, et al. Molecular basis and functional significance of Angiotensin II-induced increase in Discoidin Domain Receptor 2 gene expression in cardiac fibroblasts. J Mol Cell Cardiol 2016;90:59-69. 10.1016/j.yjmcc.2015.12.004 [DOI] [PubMed] [Google Scholar]
- 35.Ebihara Y, Masuya M, Larue AC, et al. Hematopoietic origins of fibroblasts: II. In vitro studies of fibroblasts, CFU-F, and fibrocytes. Exp Hematol 2006;34:219-29. 10.1016/j.exphem.2005.10.008 [DOI] [PubMed] [Google Scholar]
- 36.Furtado MB, Nim HT, Boyd SE, et al. View from the heart: cardiac fibroblasts in development, scarring and regeneration. Development 2016;143:387-97. 10.1242/dev.120576 [DOI] [PubMed] [Google Scholar]
- 37.Cowling RT, Yeo SJ, Kim IJ, et al. Discoidin domain receptor 2 germline gene deletion leads to altered heart structure and function in the mouse. Am J Physiol Heart Circ Physiol 2014;307:H773-81. 10.1152/ajpheart.00142.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kisselbach L, Merges M, Bossie A, et al. CD90 Expression on human primary cells and elimination of contaminating fibroblasts from cell cultures. Cytotechnology 2009;59:31-44. 10.1007/s10616-009-9190-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gordon JW, Chesa PG, Nishimura H, et al. Regulation of Thy-1 gene expression in transgenic mice. Cell 1987;50:445-52. 10.1016/0092-8674(87)90498-3 [DOI] [PubMed] [Google Scholar]
- 40.Campsall KD, Mazerolle CJ, De Repentingy Y, et al. Characterization of transgene expression and Cre recombinase activity in a panel of Thy-1 promoter-Cre transgenic mice. Dev Dyn 2002;224:135-43. 10.1002/dvdy.10092 [DOI] [PubMed] [Google Scholar]
- 41.Heffner CS, Herbert Pratt C, Babiuk RP, et al. Supporting conditional mouse mutagenesis with a comprehensive cre characterization resource. Nat Commun 2012;3:1218. 10.1038/ncomms2186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith CL, Baek ST, Sung CY, et al. Epicardial-derived cell epithelial-to-mesenchymal transition and fate specification require PDGF receptor signaling. Circ Res 2011;108:e15-26. 10.1161/CIRCRESAHA.110.235531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chong JJ, Reinecke H, Iwata M, et al. Progenitor cells identified by PDGFR-alpha expression in the developing and diseased human heart. Stem Cells Dev 2013;22:1932-43. 10.1089/scd.2012.0542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miwa H, Era T. Generation and characterization of PDGFRalpha-GFPCreERT2 knock-In mouse line. Genesis 2015;53:329-36. 10.1002/dvg.22853 [DOI] [PubMed] [Google Scholar]
- 45.Mellgren AM, Smith CL, Olsen GS, et al. Platelet-derived growth factor receptor beta signaling is required for efficient epicardial cell migration and development of two distinct coronary vascular smooth muscle cell populations. Circ Res 2008;103:1393-401. 10.1161/CIRCRESAHA.108.176768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen L, Acciani T, Le Cras T, et al. Dynamic regulation of platelet-derived growth factor receptor alpha expression in alveolar fibroblasts during realveolarization. Am J Respir Cell Mol Biol 2012;47:517-27. 10.1165/rcmb.2012-0030OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Uezumi A, Ito T, Morikawa D, et al. Fibrosis and adipogenesis originate from a common mesenchymal progenitor in skeletal muscle. J Cell Sci 2011;124:3654-64. 10.1242/jcs.086629 [DOI] [PubMed] [Google Scholar]
- 48.Noseda M, Harada M, McSweeney S, et al. PDGFRalpha demarcates the cardiogenic clonogenic Sca1+ stem/progenitor cell in adult murine myocardium. Nat Commun 2015;6:6930. 10.1038/ncomms7930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lane EB, Hogan BL, Kurkinen M, et al. Co-expression of vimentin and cytokeratins in parietal endoderm cells of early mouse embryo. Nature 1983;303:701-4. 10.1038/303701a0 [DOI] [PubMed] [Google Scholar]
- 50.Strutz F, Okada H, Lo CW, et al. Identification and characterization of a fibroblast marker: FSP1. J Cell Biol 1995;130:393-405. 10.1083/jcb.130.2.393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goldsmith EC, Bradshaw AD, Zile MR, et al. Myocardial fibroblast-matrix interactions and potential therapeutic targets. J Mol Cell Cardiol 2014;70:92-9. 10.1016/j.yjmcc.2014.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zeisberg EM, Tarnavski O, Zeisberg M, et al. Endothelial-to-mesenchymal transition contributes to cardiac fibrosis. Nat Med 2007;13:952-61. 10.1038/nm1613 [DOI] [PubMed] [Google Scholar]
- 53.Kong P, Christia P, Saxena A, et al. Lack of specificity of fibroblast-specific protein 1 in cardiac remodeling and fibrosis. Am J Physiol Heart Circ Physiol 2013;305:H1363-72. 10.1152/ajpheart.00395.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Braitsch CM, Kanisicak O, van Berlo JH, et al. Differential expression of embryonic epicardial progenitor markers and localization of cardiac fibrosis in adult ischemic injury and hypertensive heart disease. J Mol Cell Cardiol 2013;65:108-19. 10.1016/j.yjmcc.2013.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Snider P, Standley KN, Wang J, et al. Origin of cardiac fibroblasts and the role of periostin. Circ Res 2009;105:934-47. 10.1161/CIRCRESAHA.109.201400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kanisicak O, Khalil H, Ivey MJ, et al. Genetic lineage tracing defines myofibroblast origin and function in the injured heart. Nat Commun 2016;7:12260. 10.1038/ncomms12260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rios H, Koushik SV, Wang H, et al. periostin null mice exhibit dwarfism, incisor enamel defects, and an early-onset periodontal disease-like phenotype. Mol Cell Biol 2005;25:11131-44. 10.1128/MCB.25.24.11131-11144.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lindsley A, Snider P, Zhou H, et al. Identification and characterization of a novel Schwann and outflow tract endocardial cushion lineage-restricted periostin enhancer. Dev Biol 2007;307:340-55. 10.1016/j.ydbio.2007.04.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Davis J, Molkentin JD. Myofibroblasts: trust your heart and let fate decide. J Mol Cell Cardiol 2014;70:9-18. 10.1016/j.yjmcc.2013.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hermans KC, Daskalopoulos EP, Blankesteijn WM. The Janus face of myofibroblasts in the remodeling heart. J Mol Cell Cardiol 2016;91:35-41. 10.1016/j.yjmcc.2015.11.017 [DOI] [PubMed] [Google Scholar]
- 61.Ieda M, Fu JD, Delgado-Olguin P, et al. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 2010;142:375-86. 10.1016/j.cell.2010.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Doppler SA, Deutsch MA, Lange R, et al. Direct Reprogramming-The Future of Cardiac Regeneration? Int J Mol Sci 2015;16:17368-93. 10.3390/ijms160817368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lalit PA, Salick MR, Nelson DO, et al. Lineage Reprogramming of Fibroblasts into Proliferative Induced Cardiac Progenitor Cells by Defined Factors. Cell Stem Cell 2016;18:354-67. 10.1016/j.stem.2015.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhou P, Pu WT. Recounting Cardiac Cellular Composition. Circ Res 2016;118:368-70. 10.1161/CIRCRESAHA.116.308139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vliegen HW, van der Laarse A, Cornelisse CJ, et al. Myocardial changes in pressure overload-induced left ventricular hypertrophy. A study on tissue composition, polyploidization and multinucleation. Eur Heart J 1991;12:488-94. 10.1093/oxfordjournals.eurheartj.a059928 [DOI] [PubMed] [Google Scholar]
- 66.Nag AC. Study of non-muscle cells of the adult mammalian heart: a fine structural analysis and distribution. Cytobios 1980;28:41-61. [PubMed] [Google Scholar]
- 67.Zak R. Development and proliferative capacity of cardiac muscle cells. Circ Res 1974;35:suppl II:17-26. [PubMed] [Google Scholar]
- 68.Banerjee I, Fuseler JW, Price RL, et al. Determination of cell types and numbers during cardiac development in the neonatal and adult rat and mouse. Am J Physiol Heart Circ Physiol 2007;293:H1883-91. 10.1152/ajpheart.00514.2007 [DOI] [PubMed] [Google Scholar]