Abstract
Adoptive cellular therapy using chimeric antigen receptor (CAR) T cell therapies have produced significant objective responses in patients with CD19+ hematological malignancies, including durable complete responses. Although the majority of clinical trials to date have used autologous patient cells as the starting material to generate CAR T cells, this strategy poses significant manufacturing challenges and, for some patients, may not be feasible because of their advanced disease state or difficulty with manufacturing suitable numbers of CAR T cells. Alternatively, T cells from a healthy donor can be used to produce an allogeneic CAR T therapy, provided the cells are rendered incapable of eliciting graft versus host disease (GvHD). One approach to the production of these cells is gene editing to eliminate expression of the endogenous T cell receptor (TCR). Here we report a streamlined strategy for generating allogeneic CAR T cells by targeting the insertion of a CAR transgene directly into the native TCR locus using an engineered homing endonuclease and an AAV donor template. We demonstrate that anti-CD19 CAR T cells produced in this manner do not express the endogenous TCR, exhibit potent effector functions in vitro, and mediate clearance of CD19+ tumors in an in vivo mouse model.
Keywords: chimeric antigen receptor, gene editing, homology-directed repair
MacLeod and colleagues use a homing endonuclease and an AAV vector to insert an anti-CD19 CAR into the TRAC gene to create universal allogeneic CAR T cells. Targeted insertion of a CAR transgene may allow the generation of more homogeneous CAR T cell products with more predictable safety and efficacy profiles.
Introduction
Adoptive transfer of engineered chimeric antigen receptor (CAR) T cells has yielded promising results in clinical studies for multiple cancer indications, with the greatest success in hematological malignancies.1 CARs combine an extracellular antigen recognition domain and an intracellular T cell activation domain to redirect the specificity of T cells to recognize and lyse target cells bearing an antigen of interest.2 Numerous CAR T cell studies have utilized CARs recognizing CD19, enabling these engineered cells to target and kill CD19-expressing B cell tumors.3, 4 Remarkable responses have been observed, most notably in acute lymphoblastic leukemia, where it has been reported that up to 90% of patients treated with anti-CD19 CAR T cells achieve complete remission.5, 6
Despite the remarkable efficacy of CAR T cells, there are significant challenges associated with making the treatment widely available. To date, the vast majority of studies have used autologous cells, which impose significant manufacturing, logistical, and cost issues.7, 8 Each autologous therapy is, by definition, unique, and the resulting product heterogeneity and lack of reference standards complicate the evaluation of safety and efficacy. Importantly, many patients may not be eligible for treatment because of low T cell numbers and poor T cell quality or because the risk of undergoing apheresis is too great.9 Patients who are eligible for treatment must also wait weeks for the procedure to be scheduled and for the CAR T product to be generated and made available, and any difficulties encountered in the manufacturing of cells could prove to be devastating.
The use of allogeneic CAR T cells derived from healthy donors could potentially address many of the challenges associated with autologous CAR T therapy.10 However, allogeneic CAR T cells expressing a diverse repertoire of endogenous T cell receptors (TCRs) cannot be safely administered to human leukocyte antigen (HLA)-mismatched patients because they have the capacity to induce graft versus host disease (GvHD).11 Eliminating expression of the endogenous TCR through gene editing is one approach that has been described to generate “universal” donor cells incapable of mediating GvHD.12 Gene editing of T cells to eliminate TCR expression has been reported using site-specific endonucleases, including TALENs, Mega-TALs, zinc-finger nucleases (ZFNs), and CRISPR/Cas9.12, 13, 14, 15, 16
To achieve CAR expression, the majority of studies have utilized lentiviral or γ-retroviral vectors to stably insert a CAR expression cassette into the T cell genome, resulting in semi-random integration, variable copy number, heterogeneous expression, and the potential for insertional mutagenesis.17, 18, 19, 20 Exploiting cellular homology-directed repair (HDR) mechanisms to “knock in” a CAR transgene to a defined location in the genome could result in a more consistent and safe product. HDR with an exogenous DNA sequence has been described previously in T cells using short oligonucleotides paired with CRISPR/Cas9.21 Other groups have shown that adeno-associated virus (AAV) vectors can be used as a template in conjunction with a site-specific nuclease to achieve high levels of gene insertion via HDR.22, 23, 24 Two groups have also reported the targeted insertion of gene expression cassettes in T cells using MegaTAL nucleases25 and ZFNs26 in combination with an AAV6 HDR template. Notably, Sather et al. report the targeted insertion of CAR transgenes into the native CCR5 locus using a MegaTAL.25
Here we describe, for the first time, a gene editing approach to target the insertion of a CAR expression cassette while simultaneously knocking out the native TCR in activated T cells. We demonstrate that an anti-CD19 CAR transgene encoded on an AAV6 vector can be targeted directly to the TCR alpha constant (TRAC) locus using an engineered homing endonuclease, thereby preventing cell surface expression of the TCR complex. The resulting gene-edited CAR T cells produced using this streamlined process exhibit potent anti-tumor activity in vitro and in vivo in preclinical models, suggesting that these cells have potential for safe and efficacious use as adoptive cellular therapy in unrelated patients with CD19+ hematological malignancies.
Results
Efficient and Specific Editing of the TRAC Locus by TRC1-2 Nuclease Disrupts Endogenous TCR Expression
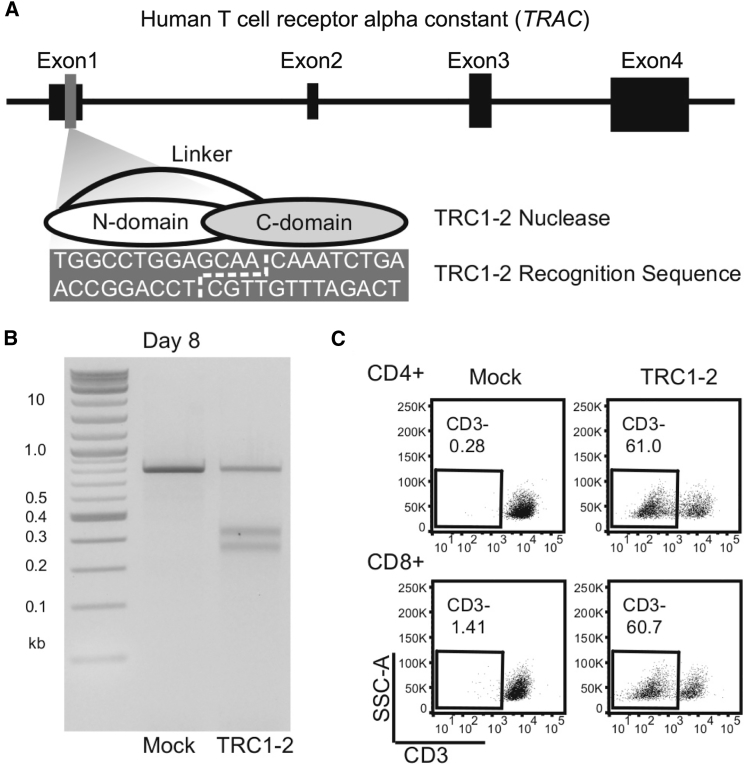
To enable editing of the TRAC gene, we produced an engineered, site-specific endonuclease based on the I-CreI homing endonuclease from Chlamydomonas reinhardtii. Our group and others have reported previously that I-CreI can be engineered to recognize DNA sequences that deviate significantly from its native target site in the algae genome.27, 28, 29, 30 We developed a single-chain variant of I-CreI, called TRC1-2, that recognizes a 22-base pair (bp) sequence in exon 1 of the TRAC gene (Figure 1A). To evaluate nuclease function, activated T cells were electroporated with mRNA encoding TRC1-2. Site-specific cleavage of genomic DNA in the absence of a suitable HDR template frequently results in variable insertion/deletion mutations (indels) at the intended target site, caused by mutagenic repair via non-homologous end joining. Indels at the TRC1-2 target site were identified by a T7 endonuclease 1 assay (Figure 1B) and DNA sequencing (Figure S1). Many of these indels frameshift the gene and should eliminate expression of the TCR. Indeed, by day 8 post-electroporation, >60% of TRC1-2 treated T cells did not express a TCR, as demonstrated by staining for CD3, a component of the TCR complex (Figure 1C). Knockout efficiency was equivalent in both CD4+ and CD8+ cells. As anticipated, unedited CD3+ T cells proliferated strongly in response to alloantigens; however, cells treated with TRC1-2 and depleted of the majority of remaining CD3+ cells exhibited minimal allo-reactivity (Figure S2). Finally, to evaluate the specificity of the TRC1-2 nuclease, we identified the 15 sites in the genome that deviate from the intended recognition site by less than four base pairs using COSMID31 and performed deep sequencing to analyze off-targeting (Figure S3). Indel frequencies did not exceed background levels for all but one of the potential off-target sites. The one off-target site where activity was observed (site 8) was cut and mutated in ∼1% of cells and is >250 kb from any known gene coding region. Thus, the TRC1-2 nuclease induces DNA breaks with high frequency at the TRAC locus to efficiently knock out expression of the TCR and prevent allo-reactivity, and the nuclease exhibits a favorable specificity profile.
Figure 1.
Characterization of TRC1-2 Nuclease Activity in T Cells
(A) Diagram of the TRC1-2 nuclease and recognition site within the TRAC locus. The TRC1-2 nuclease is a single-chain protein consisting of an N-terminal domain (N-domain) and C-terminal domain (C-domain) connected by a flexible linker. The recognition site consists of 9-bp half-sites recognized by each of the two nuclease domains, separated by a 4-bp central sequence. A broken white line in the recognition sequence denotes the overhangs generated following cleavage by the TRC1-2 nuclease. (B) A T7 endonuclease (T7E) assay was performed on mock-electroporated T cells and T cells treated with TRC1-2 nuclease on day 8 post-electroporation to confirm editing at the TRAC locus. (C) Flow cytometry staining of CD3 expression in CD4+ and CD8+ T cells on day 8 post-electroporation with TRC1-2 nuclease. Reduction of cell surface expression of CD3, a component of the TCR complex, is a functional marker of disruption of TCRα expression.
Targeted Insertion of a GFP Transgene into the TRAC Locus
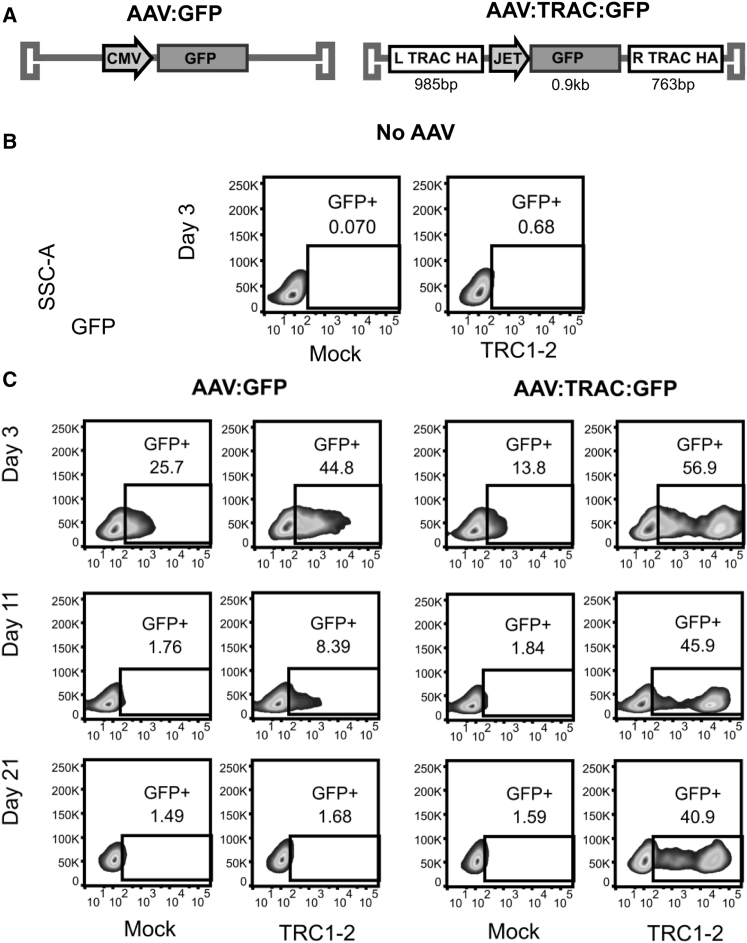
We next sought to determine whether DNA breaks in the TRAC locus could be used to target gene insertion via HDR. To test HDR-mediated gene insertion using the TRC1-2 nuclease, we produced a pair of AAV6 vectors carrying a GFP expression cassette either alone or flanked by “homology arm” sequences homologous to the TRAC locus (AAV:GFP or AAV:TRAC:GFP, respectively) (Figure 2A). Activated T cells were electroporated with mRNA encoding TRC1-2 (or mock-electroporated as a control) and then transduced with one of the two AAV vectors or mock-transduced. In the absence of either of the AAV vectors, no GFP expression was observed, as expected (Figure 2B). GFP expression was observed in cells transduced with AAV:GFP on day 3 in mock-electroporated (25.7% GFP+) and TRC1-2-electroporated cells (44.8% GFP+) but, by day 21, had declined to essentially baseline levels (1.49% and 1.68% GFP+, respectively) (Figure 2C, third and fourth columns from the left). These findings suggest that non-homologous capture of the vector at the TRC1-2 target site and/or random vector integration was infrequent and that early GFP expression was due to episomal AAV that was diluted through cell division. In mock-electroporated cells, AAV:TRAC:GFP exhibited a similar pattern of expression, indicating infrequent integration into the genome (Figure 2C, third column from the left). In sharp contrast, when TRC1-2 electroporated cells were transduced with AAV:TRAC:GFP, a distinct cell population (>40% of cells) was observed in which GFP expression persisted at a high level for the duration of the experiment (Figure 2C, fourth column from the left). Furthermore, the GFP intensity for this sub-population was >10-fold higher at all time points compared with the control cells. Taken together, these data demonstrate that both a DNA break at the TRC1-2 site and homology arms on the AAV donor are required for long-term expression of the GFP transgene. This strongly suggests that the GFP transgene in the stable, “high-expressing” cell population was targeted to the TRAC locus via HDR with the vector.
Figure 2.
Stable Expression of GFP following Integration into the T Cell Genome by HDR
(A) Diagrams of AAV vectors used to transduce cells. AAV:GFP is a vector enabling transient expression of GFP in transduced cells driven by a cytomegalovirus (CMV) promoter. AAV:TRAC:GFP contains a GFP transgene with expression driven by the JeT promoter and flanked by homology arms on the 5′ (L TRAC HA) and 3′ (R TRAC HA) sides to enable targeted integration. (B and C) T cells were mock-electroporated or electroporated with TRC1-2 mRNA and then mock transduced (No AAV) (B) or immediately transduced with AAV:GFP or AAV:TRAC:GFP at an MOI of 1e5 vector genomes (vg)/cell (C). Cells were cultured with 10 ng/mL IL-2 until 7 days post-electroporation, at which point they were cultured in medium containing 10 ng/mL IL-7 and 10 ng/mL IL-15 for the duration of the experiment. Flow cytometry was used to evaluate GFP expression 3, 11, and 21 days post-transduction.
Targeted Insertion of an Anti-CD19 CAR Transgene into the TRAC Locus
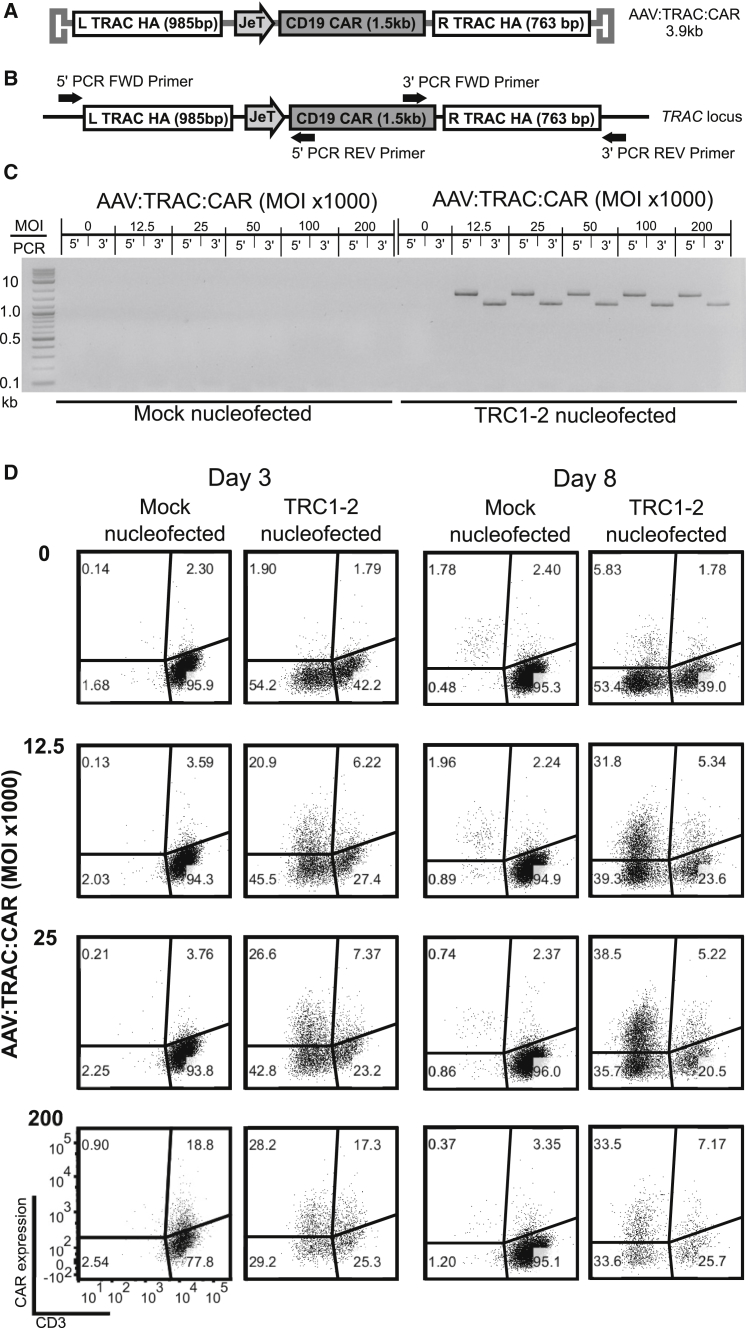
Next, we sought to combine the disruption of the endogenous TRAC gene with simultaneous homology-directed insertion of an anti-CD19 CAR transgene. We generated an AAV6 vector called AAV:TRAC:CAR, comprising an anti-CD19-BB-zeta CAR32 expression cassette flanked by TRAC homology arms (Figure 3A). We transduced mock-electroporated and TRC1-2-electroporated T cells at multiple vector doses and evaluated the cells by PCR and flow cytometry. To confirm site-specific integration, we designed PCR primers to amplify products spanning the homology arms on the 5′ and 3′ sides of the CAR transgene so that PCR products could only be generated by properly targeted events (Figure 3B). No PCR products were amplified from mock-electroporated cells, consistent with earlier observations that targeted gene integration in the absence of a DNA break is exceedingly rare, or in TRC1-2-electoporated mock-transduced cells (Figure 3C). In contrast, PCR products were amplified from all cells electroporated with TRC1-2 mRNA and transduced with AAV:TRAC:CAR, confirming targeted integration into the TRAC locus in these cells. Analysis of CD3 and CAR expression by flow cytometry showed a high frequency of CAR+ cells in the CD3− population (Figure 3D). At all MOIs, the frequency of CD3− cells increased between days 3 and 8 post-transduction, reflecting turnover of the TCR complex. In this experiment, integration efficiency approached 40% of total cells expressing the CAR but not CD3. Furthermore, under these conditions, >50% of the CD3− cells expressed the CAR. These data indicate that the TRC1-2 nuclease can efficiently target the insertion of a CAR transgene into the TRAC locus.
Figure 3.
Combining TRC1-2 and an AAV Donor Template Results in Highly Efficient HDR-Mediated Insertion of an Anti-CD19 CAR into the TRAC Locus and Simultaneous Disruption of TCR Expression
(A) Diagram of the AAV vector used to transduce cells. AAV:TRAC:CAR contains a CD19 CAR transgene with expression driven by the JeT promoter and flanked by homology arms on the 5′ (L TRAC HA) and 3′ (R TRAC HA) sides to enable targeted integration. (B) Diagram of the PCR used to confirm CAR integration by amplification with one primer located within the CAR and one primer in TRAC outside of the homology arms at both the 5′ and 3′ ends to generate 1,872-bp and 1107-bp products, respectively. (C and D) T cells were mock-electroporated or electroporated with TRC1-2 mRNA and then immediately transduced with the indicated amounts of AAV:TRAC:CAR. (C) PCR was used to confirm the presence of the CAR transgene integrated in the TRAC locus on day 3 post-electroporation and transduction as outlined in (B). (D) CAR and CD3 expression were evaluated by flow cytometry on days 3 and 8 post-electroporation and transduction.
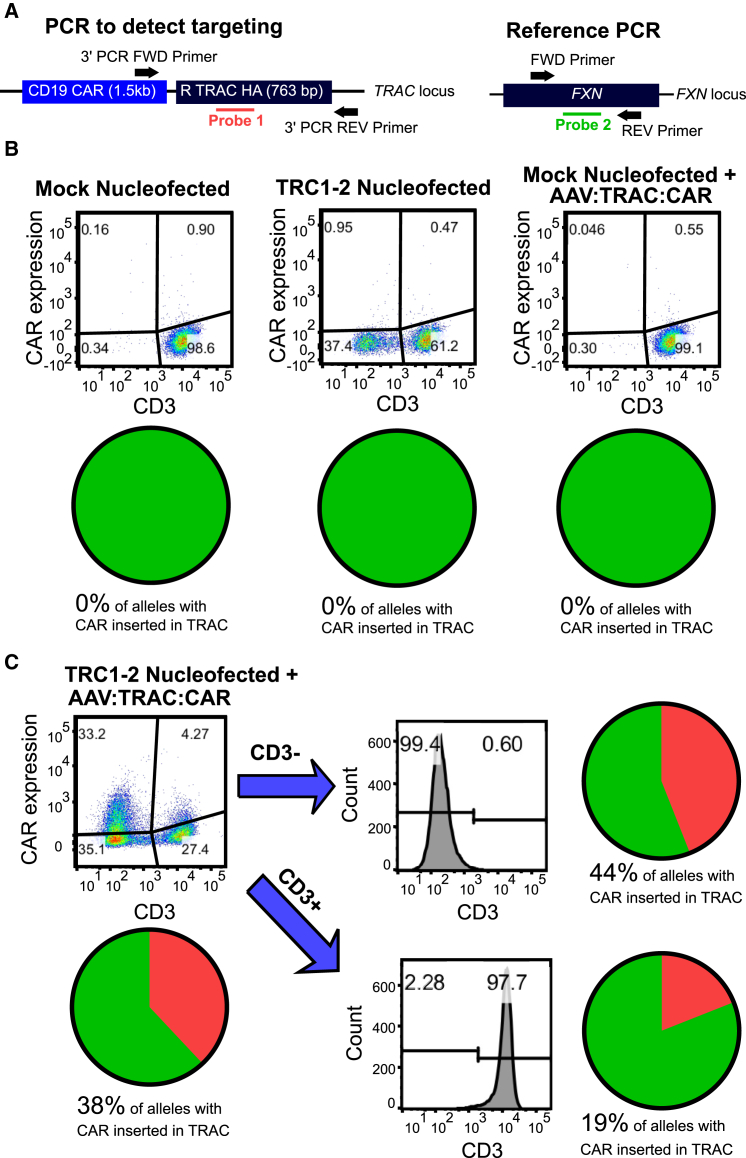
To further quantify the efficiency of CAR transgene integration into the TRAC locus, we developed a digital droplet PCR (ddPCR) assay (Figure 4A). In this assay, two primer sets are used, each in conjunction with a labeled TaqMan probe: the first set spans a portion of the 3′ end of the CAR transgene and the 3′ homology arm flanking the CAR, and the second set amplifies an unrelated gene sequence (in the FXN gene) and serves as a reference sequence to control for template number. Consistent with the conventional PCR results shown above, cells that were mock-electroporated or that were not transduced with AAV:TRAC:CAR showed no sign of gene integration in this assay (Figure 4B). However, ddPCR measured 38% targeted gene integration in DNA from cells that were both electroporated with mRNA encoding TRC1-2 and transduced with the AAV:TRAC:CAR vector (Figure 4C). When this same population of cells was analyzed by flow cytometry, total CAR expression was just over 37%, suggesting that detectable CAR expression was due to insertion of the CAR expression cassette into the TRC1-2 recognition sequence (Figure 4C). Of the 37% of CAR+ cells detected by flow cytometry, a small percentage was also CD3+. We hypothesized that this was due to targeted integration of the CAR expression cassette into the unpaired TRAC allele (which does not contribute to forming a functional TCRαβ pair), which allowed expression of both the CAR and a functional TCR. To test this hypothesis, the cells were separated into CD3− and CD3+ fractions and assayed individually by ddPCR. Indeed, the total CD3+ cell population was 13.5% CAR+ by flow cytometry, and 19% of the TRAC alleles in this CD3+ population were found to harbor an integrated CAR transgene by ddPCR. Importantly, the total CD3− population was 48.6% CAR+ by flow cytometry, and 44% of alleles in this therapeutically relevant CD3− population harbored an integrated CAR transgene by ddPCR. Taken together, the flow cytometry and ddPCR results confirm highly efficient targeted insertion of an anti-CD19 CAR into the TRAC locus, most frequently with concomitant knockout of the endogenous TCR.
Figure 4.
Confirmation of Targeted Insertion of the CAR Transgene by Digital Droplet PCR
(A) Diagram showing digital PCR strategy. Two primer pairs and probes are used: one to detect the CAR transgene inserted in TRAC and another to detect FXN and serve as a reference standard for genomic DNA. (B) Activated CD3+ T cells were mock-electroporated, electroporated with TRC1-2 nuclease mRNA, or mock-electroporated and transduced with 25,000 vg/cell AAV:TRAC:CAR. Digital PCR was used to quantify targeted integration of the CAR transgene in TRAC 11 days post-electroporation/transduction. (C) Activated CD3+ T cells were electroporated with TRC1-2 mRNA and transduced with 50,000 vg/cell AAV:TRAC:CAR. CD3+ and CD3− groups were magnetically separated on day 8 post-transduction. Cells were stained for CD3 expression and CAR expression in the pre-separation samples and CD3 expression post-separation to confirm purity. Digital PCR was used to quantify targeted integration of the CAR transgene in TRAC in pre-separation, CD3+, and CD3− populations.
Gene-Edited TCR Knockout Anti-CD19 CAR T Cells Exhibit Potent In Vitro and In Vivo Responses to CD19-Bearing Tumor Cells
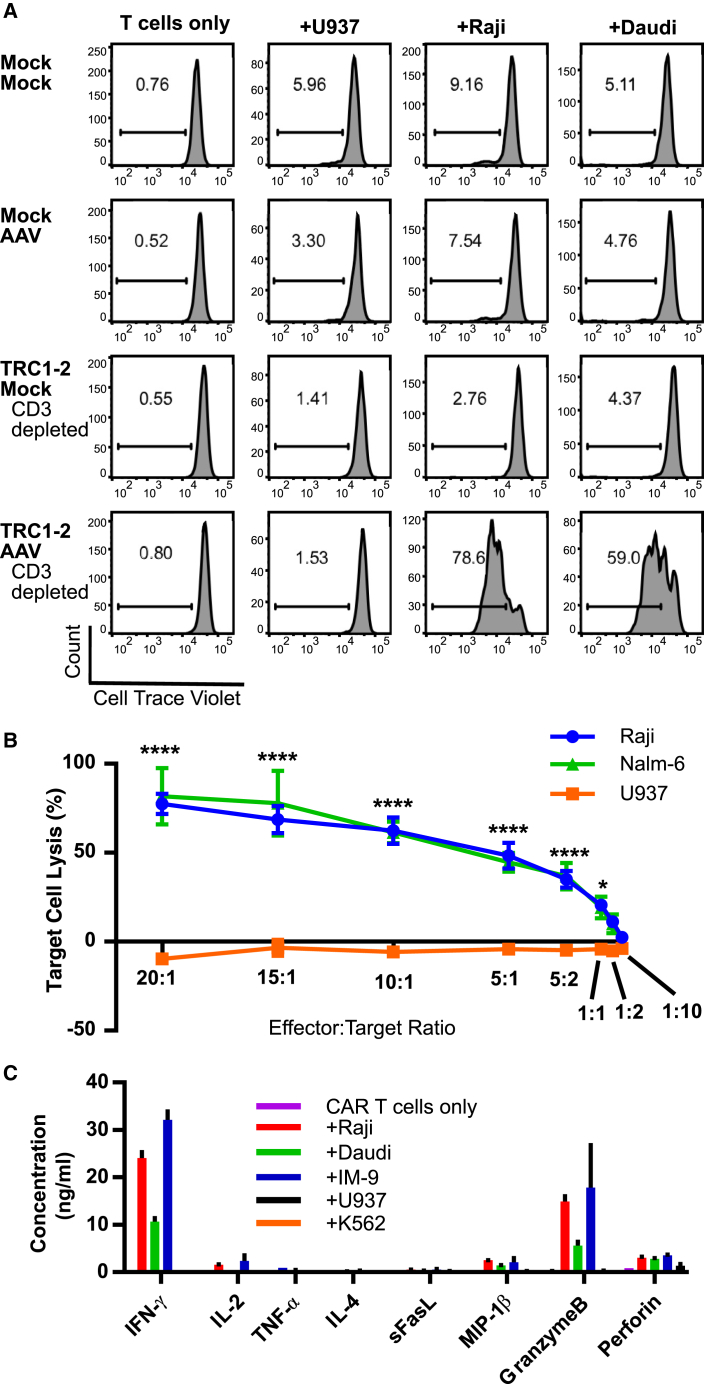
Next, anti-CD19 CAR T cells were characterized for functional activity in vitro. Proliferation in response to antigenic stimulation is an important measure of CAR T cell function. Control cells that were not treated with TRC1-2 or AAV:TRAC:CAR failed to proliferate specifically in response to CD19+ Raji or Daudi cells (Figure 5A). In contrast, anti-CD19 CAR+ T cells created by treatment with both the TRC1-2 nuclease and the AAV vector responded specifically to co-culture with CD19+ target cells but did not proliferate when exposed to control CD19− U937 cells (Figure 5A, bottom). Importantly, CD3− CAR T cells did not demonstrate reduced proliferative capacity compared with CD3+ CAR T cells (Figure S4), indicating that the lack of endogenous TCR expression does not impair function. Furthermore, anti-CD19 CAR+ T cells exhibited potent cytolytic activity and released IFN-γ and other pro-inflammatory cytokines when co-cultured with CD19+ cells but not CD19− cells, confirming the specific functional activity of these cells against relevant targets (Figures 5B and 5C).
Figure 5.
In Vitro Activity of Gene-Edited TCR Knockout Anti-CD19 CAR+ Cells
(A) Cells were either mock-electroporated or electroporated with TRC1-2 mRNA (TRC1-2) and then immediately split into two groups; one was mock-transduced, and one was transduced with AAV:TRAC:CAR (AAV) at an MOI of 50,000 vg/cell. CD3+ cells were depleted from both TRC1-2 mRNA-treated groups post-electroporation or post-electroporation and transduction with AAV. T cells from all four groups were labeled with Cell Trace Violet and then cultured alone or co-cultured at a ratio of 1:1 with control CD19− U937 cells or CD19+ Raji or Daudi cells. All cell lines were pre-treated with Mitomycin C to arrest cell growth and washed extensively prior to co-culture. After 3 days of co-culture in medium in the absence of exogenous cytokines, proliferation (dilution of Cell Trace Violet) was assessed by flow cytometry. (B and C) TCR knockout CAR+ T cells were produced by electroporation of T cells with TRC1-2 mRNA, followed immediately by transduction with AAV:TRAC:CAR at an MOI of 400,000. Cells were depleted of CD3+ cells 5 days post-electroporation and transduction. (B) TCR knockout CAR+ T incubated with Raji (CD19+), NALM-6 (CD19+), or U937 (CD19−) cells at various effector:target ratios. Cytolytic activity of the CAR T cells against the Raji, NALM-6, or U937 targets was measured by assessment of LDH release. Data are from n = 7 individual wells per sample per time point, mean ± SEM, ****p < 0.0001, *p < 0.05, two-way ANOVA with Tukey multiple comparisons test comparing Raji or NALM-6 to U937 samples at the same E:T ratio. (C) CAR T cells were incubated alone or co-cultured at a ratio of 10:1 with control CD19− U937 or K562 cells or CD19+ Raji, Daudi, or IM-9 cells for 24 hr in medium in the absence of exogenous cytokines. Cytokine production and release were quantified from culture supernatants (n = 3, mean ± SEM).
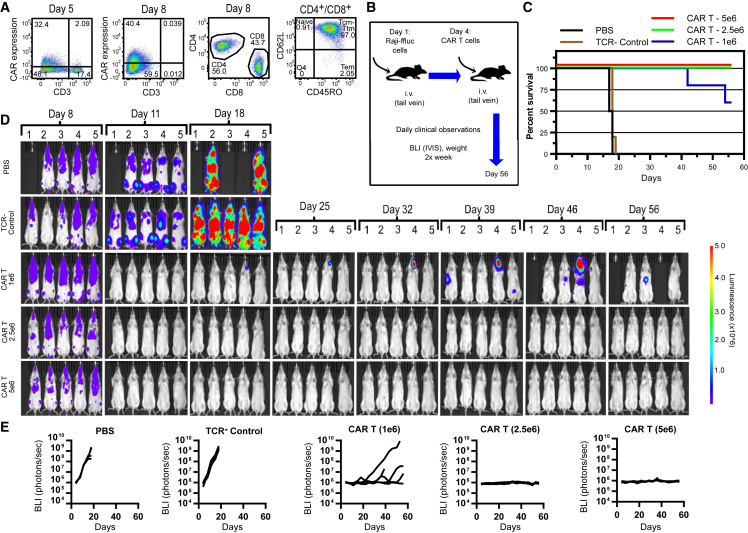
Finally, we tested the efficacy of gene-edited, TCR knockout anti-CD19 CAR T cells in a murine model of disseminated B cell lymphoma. Activated T cells were electroporated with TRC1-2 mRNA and transduced with AAV:TRAC:CAR. CD3− cells were then isolated on day 5 post-transduction using anti-CD3 magnetic beads, resulting in 99.9% purity as measured by flow cytometry 3 days later (Figure 6A). The purified CD3− population comprised 56% CD4+ and 44% CD8+ cells and primarily consisted of central memory (Tcm)/transitional memory (Ttm) phenotypes, determined by staining for CD62L and CD45RO (Figure 6A) and CCR7, CD27, and CD95 (Table S1). Mice were injected intravenously (i.v.) with firefly luciferase-expressing Raji cells (Raji-ffLuc) and, after 4 days, were injected i.v. with three different doses of CD3− anti-CD19 CAR T cells (Figures 6B–6E). Control mice were injected with TCR− T cells or PBS. Engraftment and growth of Raji-ffLuc cells was evident in all mice by day 4 and increased significantly in untreated and TCR− control groups by day 11 (Figures 6D and 6E). Peak CAR T cell frequencies in the blood of treated mice were observed on day 8, reaching ∼10% of cells in peripheral blood in the high-dose group (Figure S5A). Frequencies of human T cells in peripheral blood of animals in all three CAR T dose groups were significantly higher than TCR− control cell-treated mice, indicating antigen-driven expansion of the CAR T cells (Figure S5B). By days 17–19, all mice in control groups showed evidence of significant tumor burden, especially in the spine and bone marrow, resulting in complete hindlimb paralysis, and were euthanized (Figure 6C). In contrast, all groups of mice treated with anti-CD19 CAR T cells showed no evidence of tumor growth by day 11 and, with the exception of a single mouse in the low-dose group, remained tumor-free through day 32 of the study (Figures 6C–6E). Tumor re-growth was observed in three mice (mice 1, 4, and 5) in the low-dose cohort on day 39. One of the three mice (mouse 1) was found dead on day 42. Imaging revealed only a low level of tumor at a single site in this animal, and there were no clinical observations of duress or weight loss, so it is unlikely that the death was tumor-related. Interestingly, the apparent recurrence observed for mouse 5 on day 39 was no longer evident by bioluminescent imaging at all time points through day 56, possibly because of re-expansion of CAR T memory cells upon re-exposure to tumor antigen. On day 56, three of five mice in the low-dose CAR T cohort remained alive, with only one mouse showing evidence of tumor recurrence. All mice in the mid- and high-dose cohorts showed no evidence of tumor re-growth, and all were alive and gaining weight as of day 56. These results show potent in vivo clearance of CD19+ tumor cells by gene-edited CD3− CAR T cells and support further preclinical development of this platform for allogeneic CAR T cell therapy.
Figure 6.
Gene-Edited TCR Knockout Anti-CD19 CAR+ Cells Are Highly Efficacious in a Murine Model of Disseminated Lymphoma
(A) Activated T cells were electroporated with TRC1-2 mRNA and transduced with AAV:TRAC:CAR at an MOI of 400,000 vg/cell and cultured for 5 days in the presence of IL-2. Five days post-transduction, cells were stained for expression of the CAR using a biotinylated CD19-Fc reagent and CD3, with TRC1-2-treated, mock-transduced cells used as a control for gating of CAR expression. CD3+ cells were then depleted. Enriched CD3− cells were cultured for 3 additional days in the presence of IL-15 and IL-21 and then analyzed again by flow cytometry for CD3 and CAR expression and CD4 and CD8 expression. Total T cells (gated on CD4+ and CD8+) were further analyzed for CD62L and CD45RO expression. (B) 2 × 105 Raji-ffluc cells were injected i.v. into 5- to 6-week-old female NSG mice on day 1. On day 4, mice were injected i.v. with 0.2 mL PBS (PBS), 0.2 mL of PBS containing 5 × 106 gene-edited, mock-transduced TCR− cells (TCR− control), or 0.2 mL PBS containing 1 × 106, 2.5 × 106, or 5 × 106 gene-edited, AAV-transduced TCR− CAR+ (CAR+) T cells produced as described in (A). (C) Mice were monitored throughout the course of the study and euthanized according to pre-defined criteria. Kaplan-Meier survival curves are displayed. (D and E) On the indicated days, luciferin substrate (150 mg/kg in saline) was administered to live mice by intraperitoneal injection, mice were anesthetized, and luciferase activity measured using IVIS SpectrumCT (PerkinElmer). Bioluminescence images (D) are displayed for all mice in the study at the indicated time points, with bioluminescence values for each mouse displayed as individual curves plotted over time (E).
Discussion
We have developed a streamlined and technically straightforward method for developing allogeneic CAR T cells. The method is based on a single gene-editing step in which a CAR expression cassette is targeted into the TRAC locus, thereby knocking out the native TCR while simultaneously knocking in the CAR. The process consists of isolating and activating T cells from a leukapheresis product obtained from a healthy donor, electroporating the cells with mRNA encoding an engineered nuclease specific for the TRAC gene, immediately transducing cells with an AAV6 vector carrying the CAR expression cassette, and isolating and expanding the CD3− cell population. The process is scalable and compatible with good manufacturing practices (GMP)-compliant manufacturing. With the exception of the AAV vector, the process relies on growth media, reagents, and equipment that are commonly used in the manufacture of CAR T products that are currently being evaluated in phase I and phase II clinical trials.8, 33, 34, 35 Fortunately, because of their prominent role in gene therapy, AAV vectors have a well-established regulatory history and safety profile and are readily produced under GMP conditions.36
Allogeneic CAR T therapies have a number of potential advantages over autologous CAR T approaches. First, the cells are derived from a healthy donor, and the donor can be pre-screened for the desirable number, CD4:CD8 ratio, and phenotype of T cells. In the current study, the donor was pre-screened for T cells with primarily naive and central memory phenotypes (CD62L+/CCR7+/CD45RA+ or CD62L+/CCR7+/CD45RO+). In an autologous setting, one is limited to the cell number and phenotypes that are present in the patient at the time of apheresis. Furthermore, for many patients, it is not possible to generate an autologous CAR T product at all because of the health of the patient, his or her chemotherapeutic regimen, and/or difficulty manufacturing a sufficient number of CAR T cells. From a manufacturing perspective, the ability to define the starting material for an allogeneic CAR T product should allow greater control of the process, generation of a more consistent and homogeneous product, and the ability to better qualify and release individual batches. Moreover, it should be possible to generate hundreds of therapeutic doses from a single GMP manufacturing run (the process used in this report would yield >1010 CAR T cells from a standard leukapheresis product). Allogeneic CAR T also has a timing advantage in that the product would be available “off the shelf,” eliminating the need for patient apheresis and minimizing time to treatment.
It is presently unknown whether allogeneic CAR T cells will persist long enough in a patient to maintain a durable response. It is expected that the engineered cells will be cleared in an immunocompetent patient via recognition of alloantigens, leading to rejection of the allogeneic cells, although it is not known how quickly this will happen in an immunosuppressed patient or how long the cells need to persist to be effective. It may be possible to prolong in vivo persistence by using gene editing to eliminate expression of certain alloantigens, such as HLA class I, in the CAR T cells.37 In all likelihood, it is only possible to address the issue of persistence time and durable clinical benefit with clinical studies, and the first allogeneic CAR T clinical investigations are currently underway.38
The ability to target the insertion of a CAR transgene into a defined location in the genome has a number of potential advantages, including reduced risk of insertional mutagenesis and greater control over CAR transgene expression because of the minimization of positional epigenetic effects. Accomplishing this requires a very specific gene-editing reagent. We use an engineered homing endonuclease to achieve a high level of targeted gene insertion. Homing endonucleases are less prevalent than other gene-editing tools such as ZFNs, TALENs, and CRISPR/Cas9 because they are difficult to engineer. Despite these engineering challenges, our group has focused on homing endonucleases because they have a number of structural and mechanistic features that are attractive for in vivo and ex vivo gene editing. Homing endonucleases are extremely specific.39 They can be engineered as small, single-chain enzymes (TRC1-2 has 310 amino acids), which makes them easy to deliver to cells. Additionally, and of particular relevance to the current study, engineered homing endonucleases cleave DNA to leave 4-bp 3′ overhangs on both strands at the site of the double-strand break. It has been noted that 3′ overhangs contribute to HDR by enabling strand invasion, and this may explain, in part, the high rates of gene insertion we observed in this study.40 Indeed, Sather, et al. observed significantly higher rates of gene insertion mediated by a MegaTAL, which generates 3′ overhangs, compared with a TALEN, which generates 5′ overhangs.25
Consistent with the observations of numerous other groups, we found AAV to be a particularly effective template for HDR.22, 23, 24, 41 When we attempted to target CAR expression cassette insertion using plasmid DNA or PCR products as the template, the efficiency of gene integration was >10-fold lower than with AAV (data not shown). It is not known whether this is due to the ability of AAV to achieve higher intracellular concentrations or whether the virus interacts with host factors involved in HDR.24, 41 Also consistent with other reports is our observation of a striking increase in gene expression following integration into the genome.25, 26 Figures 2B and 3D show marked increases in gene expression 3 days post-transduction in cells that were electroporated with TRC1-2 mRNA compared with cells that were mock-electroporated. This suggests that the AAV vector itself contributes very little to total gene expression despite likely being present in many copies per cell at this early time point. This might be due to suppression of the virus-encoded gene by host factors or could be an intrinsic property of the vector itself. Alternatively, the TRAC locus may be a particularly transcriptionally active region of the genome, and any transgene integrated into this location would be highly expressed. Nonetheless, we observed stable gene expression following integration into the genome.
We did not observe a high frequency of non-homologous capture of the AAV vector, either at the TRC1-2 cut site or at other sites in the genome. Figure 2 shows that both a nuclease-induced DNA break and homology arms on the AAV donor are required for stable gene expression, suggesting that HDR is the predominant mechanism of gene integration. Not surprisingly, we found that the CAR expression cassette was able to integrate into both alleles of the TRAC gene, resulting in CAR+ cells in both the CD3− and CD3+ fractions (Figure 4C). However, there is a strong tendency for CAR+ cells to be CD3−, suggesting a preference for the selected TRAC allele. The reason for this preference is unclear; however, it is advantageous for our process because the CD3− population contains the therapeutically relevant cells.
In summary, we demonstrate that potent allogeneic CAR T cells can be produced in an efficient and streamlined process using an engineered homing endonuclease and an AAV HDR template. The process was used to generate anti-CD19 CAR T cells that were effective in a disseminated lymphoma model, eradicating tumors in the mid- and high-dose cohorts. This general approach can be combined with additional gene modifications, such as knockout of the PD-1 gene to facilitate resistance to suppression by PD-L1-expressing cells, in the next generation of CAR T therapeutics.
Materials and Methods
Cells and Media
CD3+ T cells were isolated from apheresis products (Hemacare and Key Biologics) using the EasySep human T cell enrichment kit or EasySep human CD3 positive selection kit (STEMCELL Technologies). T cells were cultured in X-VIVO-15 medium (Lonza) supplemented with 5% pooled human AB serum (Innovative Research), 10 mM HEPES (Thermo Fisher Scientific), and 2 mM L-glutamine (Thermo Fisher Scientific). The Raji and Daudi (CD19+ Burkitt lymphoma), IM-9 (CD19+ Epstein-Barr virus (EBV)-transformed B lymphoblastoid), K562 (CD19− chronic myelogenous leukemia), and U937 (CD19− histiocytic lymphoma) cell lines were purchased from the ATCC. NALM-6 cells were purchased from the Leibniz Institute DSMZ (Deutsche Sammlung von Mikroorganismen und Zellkulturen [German Collection of Microorganisms and Cell Cultures]). Raji cells stably transduced to express firefly luciferase (Raji-ffluc) were a kind gift from Drs. Gianpietro Dotti and Barbara Savoldo of the Lineberger Cancer Center at the University of North Carolina. For proliferation assays, Raji, Daudi, and U937 cells were treated with freshly reconstituted Mitomycin C (Sigma-Aldrich) for 60 min at a concentration of 50 μg/mL to arrest growth, followed by five washes with X-VIVO-15 medium to remove residual Mitomycin C.
Reagents
Interleukin-2 (IL-2), IL-7, IL-15, and IL-21 were purchased from Thermo Fisher Scientific and used at concentrations of 10 ng/mL. Red blood cell (RBC) lysis buffer was purchased from BioLegend.
mRNA Production and T Cell Electroporation
The TRC1-2 nuclease sequence was cloned into the pRNA2 vector, which includes both 5′ and 3′ UTRs and a polyT repeat to serve as the template for a polyA tail of defined length. This vector was linearized, gel-purified using the NucleoSpin gel and PCR clean-up column kit (Macherey-Nagel), and used as a DNA template for in vitro mRNA transcription. Transcription of mRNA from the DNA template was performed using the HiScribe T7 quick high-yield RNA synthesis kit (New England Biolabs) with 1 μg of gel-purified DNA template and 10 mM 3′-0-Me-m7G(5′)ppp(5′)G RNA cap structure analog (New England Biolabs). DNase treatment was performed using the manufacturer’s protocol for capped RNA synthesis. Purification of mRNA was performed using the Promega SV total RNA isolation system following the manufacturer’s protocol and eluted into nuclease-free water. RNA concentration was determined was by UV absorption using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific). The quality of RNA was assessed by visualization of samples run on a FlashGel RNA cassette (Lonza) using the FlashGel system (Lonza) to check for the absence of degradation. All purified mRNA samples were stored at −80°C prior to use.
T cells were stimulated for 2 or 3 days with Dynabeads human T cell activator CD3/CD28 (Thermo Fisher Scientific) prior to bead removal or Immunocult human CD3/CD28/CD2 T cell activator (STEMCELL Technologies) for 3 days in supplemented X-VIVO-15 containing IL-2, and, on day 3 post-activation, cells were mixed with mRNA and electroporated using the P3 primary cell kit and 4D-Electroporator according to the manufacturer’s instructions (Lonza).
AAV Production and Characterization
A CD19 chimeric antigen receptor sequence containing the FMC63 scFv, hinge and transmembrane domain of CD8α, and intracellular domains of 4-1BB and CD3ζ32 was synthesized (Integrated DNA Technologies). This sequence was cloned into a plasmid that contained a pair of inverted terminal repeat (ITR) sequences for production of AAV. The AAV plasmid was initially prepared by cloning two homology arms from the TRAC genomic sequence sequentially in between the ITRs. The first homology arm included half of the nuclease target site and 985 bp of upstream genomic sequence. The second homology arm included the other half of the nuclease target site and 763 bp of downstream genomic sequence. A JeT promoter was synthesized from overlapping oligonucleotides in a PCR reaction and cloned between the homology arms. An SV40 polyadenylation sequence was amplified by PCR and cloned downstream of the JeT promoter. Finally, the synthesized CAR was inserted between the JeT promoter and the SV40 polyadenylation sequence. All AAV6 vectors used in Figures 1, 2, 3, 4, and 5A were produced using a triple transfection protocol and were purified using a CsCl gradient before dialysis in 1× PBS.42 The AAVs used in Figures 5B, 5C, and 6 were produced by Virovek. Vectors were aliquoted and stored at −80°C until use.
Flow Cytometry
For evaluation of CAR expression, cells were stained with a goat anti-mouse Fab antibody conjugated with Alexa Fluor 647 (Jackson ImmunoResearch) or biotinylated CD19-Fc (ACRO Biosystems) for 15 min at room temperature. Cells were thoroughly washed before staining with antibodies for additional surface markers. Anti-CD3-BB515 and anti-CD62L-BB515 were purchased from BD Biosciences. Anti-CD4-BV785, anti-CD8-BV711, anti-CD45RA-BV421, anti-CD45RO-PECy7, streptavidin-PE, anti-CD27-PE, anti-CD95-allophycocyanin (APC)Cy7, anti-CCR7-APC, anti-CD45-BV711, and anti-CD19-PE, anti-CD3-BV711 were purchased from BioLegend and used to stain surface antigens. Cell Trace Violet was purchased from Thermo Fisher Scientific and used at a concentration of 1 μM to label cells for 10 min. All data were acquired using a BD Fortessa flow cytometer (BD Biosciences) and analyzed using FlowJo software.
Cytotoxic T Lymphocyte Assay
In a 96-well, V-bottom plate, gene-edited CAR+ T cells (effectors) and NALM-6, Raji, or U937 cells (targets) were cultured together at 37°C for 4 hr at various effector to target (E:T) ratios. Cells were washed, and lactate dehydrogenase (LDH) release was measured using the CytoTox 96 non-radioactive cytotoxicity assay according to the manufacturer’s instructions (Promega) and a SpectraMax i3x multi-mode microplate reader (Molecular Devices). Percent lysis was calculated using the following formula: ((experimental value – effector spontaneous release – target spontaneous release) / (target maximum release – target spontaneous release) × 100.
Multiplexed Cytokine Assay
Cytokine release was evaluated using a custom MILLIPLEX multiplex assay and MAGPIX instrument (EMD Millipore) according to the manufacturer’s instructions.
T7 Endonuclease Assay
T cells were pelleted and lysed in a buffer containing water, Phusion GC reaction buffer, and proteinase K (New England Biolabs). A sequence spanning the TRC1-2 target site was PCR-amplified from cell lysates using the following primers: TRC 1-2 forward (GAGCAGCTGGTTTCTAAGATGC) and TRC1-2 reverse (GGAGAGGCAACTTGGAGAAGG). The PCR was set up using 200 μM deoxynucleoside triphosphate (dNTP) mix, Q5 reaction buffer, Q5 high GC enhancer, 0.5 μM of each primer, and Q5 hot start high-fidelity DNA polymerase (New England Biolabs). PCR conditions were as follows: 98°C 4 min for initial denaturation, 35 cycles at 98°C for 10 s, 67°C for 20 s, 72°C for 31 s, and then a final elongation at 72°C for 5 min. A 25-μL aliquot of the PCR reaction was transferred to new tubes, and the DNA was denatured and slowly rehybridized to allow the formation of heteroduplex DNA sequences and then digested with 20 units of T7 endonuclease (New England Biolabs), which recognizes and cleaves heteroduplex DNA sequences. Digested products were run on 2% agarose gel containing EtBr (VWR) and visualized using a Fotodyne gel dock system.
CAR Insertion PCR
T cells were pelleted and lysed in a buffer containing water, Phusion GC reaction buffer, and proteinase K (New England Biolabs). Sequences spanning each end of the TRC1-2 homology arms were amplified from the cell lysate by PCR using the following primers: for 5′ insertions, forward (GATAGACGCTGTGGCTCTGCATGAC) and reverse (GCCTGAGTGTAATCTCGACGTGTGG) and for 3′ insertions, forward (GCTGCACATGCAAGCCTTACCACCTC) and reverse (GCGTACTTAGAATACTGTCTACCCTCTCATGGC). PCR reactions were performed containing 200 μM dNTP mix, Q5 reaction buffer, Q5 high GC enhancer, 0.5 μM of each primer, and Q5 hot start high-fidelity DNA polymerase (New England Biolabs). Both primer sets used the same PCR conditions as follows: 98°C for 4 min for initial denaturation, 30 cycles at 98°C for 10 s, 65°C for 20 s, 72°C for 1:35 min, and then a final elongation at 72°C for 5 min. PCR products were run on a 1.5% gel containing EtBr (VWR International) and visualized using a Fotodyne gel dock system.
Digital PCR
Genomic DNA from T cells was isolated using the FlexiGene DNA kit (QIAGEN). Briefly, 2 × 106 cultured T cells were pelleted and incubated in lysis buffer. This lysate was then neutralized and resuspended in denaturation buffer containing QIAGEN protease. DNA was then precipitated, recovered by centrifugation, washed with 70% ethanol, and resuspended in nuclease-free water (Thermo Fisher Scientific). Digital PCR was performed on a QX200 droplet digital PCR system (Bio-Rad) according to the manufacturer’s instructions. Briefly, 50 ng of purified genomic DNA was combined with ddPCR Supermix for Probes (no dUTP) (Bio-Rad), 900 nM of each primer, 250 nM of a fluorophore-conjugated probe, 4 U of the restriction endonuclease HindIII (New England Biolabs) and nuclease-free water to a final reaction volume of 20 μL. Samples were partitioned into approximately 20,000 nL-sized droplets using a QX200 droplet generator. PCR was performed on a C1000 Touch thermal cycler (Bio-Rad) using the following three-step cycling profile: 10 min at 95°C; 45 times 30 s at 94°C, 30 s at 59°C, and 3 min 72°C; and 10 min at 98°C. Droplets were analyzed individually on a QX200 droplet reader and, based on their fluorescence amplitude, counted as positive or negative. Assuming random template partitioning, the QuantaSoft software converts positive and negative droplet counts into the absolute copy number of the targeted nucleic acid. Genomic integration of CAR molecules was quantified using the primers CARFWD (GGACACGACGGCTTATAC), which binds near the 3′ end of the CAR sequence, and CARREV (TGCTGCTCTTCTCCTTTC), which binds in the T cell receptor alpha chain locus downstream of the region homologous to the donor template. Amplicons specific for CAR molecules integrated at the desired genomic location were detected using the probes CARFAM1 (/56-FAM/AACTCCTCT/ZEN/GATTGGTGGTCTCGG/3IABkFQ/) and CARFAM2 (/56-FAM/ATGCAAGCC/ZEN/TTACCACCTCGATGA/3IABkFQ/). The total number of genome equivalents interrogated was determined, in a separate well, by a reference copy number assay targeting the first intron of the human frataxin gene (Thermo Fisher Scientific, AIBJZBE). The CAR integration efficiency is calculated as the ratio of the number of CAR molecules per total number of genomes interrogated.
Raji Disseminated Lymphoma Model
All animal studies were conducted by Charles River Laboratories following review and approval by the CRL, Inc. Institutional Animal Care and Use Committee (IACUC). Raji cells stably expressing ffluc43 were injected i.v. into 5- to 6-week-old female NSG mice on day 1 at a dose of 2.0 × 105 cells/mouse. On day 4, mice were injected i.v. with PBS, PBS containing gene-edited control TCR knockout (KO) T cells prepared from healthy donor peripheral blood mononuclear cells (PBMCs), or PBS containing the indicated doses of CAR T cells prepared from the same donor using TRC1-2 and AAV:TRAC:CAR (Virovek) as described in Figure 6A. On the indicated days, live mice were injected intraperitoneally (i.p.) with luciferin substrate (150 mg/kg in saline) and anesthetized, and luciferase activity was measured after 7 min using IVIS SpectrumCT (PerkinElmer). Data were analyzed and exported using Living Image software 4.5.1 (PerkinElmer). Luminescence signal intensity is represented by radiance in photons per second per centimeter squared per steradian (p/sec/cm2/sr).
Statistics
All statistical analyses were performed using GraphPad Prism version 7 (GraphPad).
Author Contributions
D.T.M., J.A., and A.J.M. conceived and performed the experiments and wrote the manuscript. R.J.M, A.H., K.J.W., A.E.B., M.A.T., J.A.H., C.D.P., V.V.B., C.A.T., J.L., S.K., C.W.B., J.S., and M.L.H., conceived and/or performed the experiments and reviewed the manuscript. M.G.N., D.J., and B.M. conceived the experiments and wrote the manuscript.
Acknowledgments
All authors, with the exception of R.J.M. and M.L.H., receive compensation as employees of Precision BioSciences, Inc. R.J.M. and M.L.H. are supported by RO1AI072176-06A1, RO1AR064369-01A1, and Research to Prevent Blindness. The authors would like to thank all members of the Precision BioSciences Core Technology team for their efforts on the production of the engineered nuclease used in this manuscript.
Footnotes
Supplemental Information includes Supplemental Materials and Methods, five figures, and one table and can be found with this article online at http://dx.doi.org/10.1016/j.ymthe.2017.02.005.
Contributor Information
Daniel T. MacLeod, Email: dan.macleod@precisionbiosciences.com.
Bruce McCreedy, Email: bruce.mccreedy@precisionbiosciences.com.
Supplemental Information
References
- 1.Qasim W., Thrasher A.J. Progress and prospects for engineered T cell therapies. Br. J. Haematol. 2014;166:818–829. doi: 10.1111/bjh.12981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sadelain M., Brentjens R., Rivière I. The basic principles of chimeric antigen receptor design. Cancer Discov. 2013;3:388–398. doi: 10.1158/2159-8290.CD-12-0548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lorentzen C.L., Straten P.T. CD19-chimeric antigen receptor t cells for treatment of chronic lymphocytic leukaemia and acute lymphoblastic leukaemia. Scand. J. Immunol. 2015;82:307–319. doi: 10.1111/sji.12331. [DOI] [PubMed] [Google Scholar]
- 4.Porter D.L., Hwang W.-T., Frey N.V., Lacey S.F., Shaw P.A., Loren A.W., Bagg A., Marcucci K.T., Shen A., Gonzalez V. Chimeric antigen receptor T cells persist and induce sustained remissions in relapsed refractory chronic lymphocytic leukemia. Sci. Transl. Med. 2015;7:303ra139. doi: 10.1126/scitranslmed.aac5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maude S.L., Frey N., Shaw P.A., Aplenc R., Barrett D.M., Bunin N.J., Chew A., Gonzalez V.E., Zheng Z., Lacey S.F. Chimeric antigen receptor T cells for sustained remissions in leukemia. N. Engl. J. Med. 2014;371:1507–1517. doi: 10.1056/NEJMoa1407222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turtle C.J., Hanafi L.-A.A., Berger C., Gooley T.A., Cherian S., Hudecek M., Sommermeyer D., Melville K., Pender B., Budiarto T.M. CD19 CAR-T cells of defined CD4+:CD8+ composition in adult B cell ALL patients. J. Clin. Invest. 2016;126:2123–2138. doi: 10.1172/JCI85309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abou-El-Enein M., Bauer G., Medcalf N., Volk H.-D., Reinke P. Putting a price tag on novel autologous cellular therapies. Cytotherapy. 2016;18:1056–1061. doi: 10.1016/j.jcyt.2016.05.005. [DOI] [PubMed] [Google Scholar]
- 8.Wang X., Rivière I. Clinical manufacturing of CAR T cells: foundation of a promising therapy. Mol Ther Oncolytics. 2016;3:16015. doi: 10.1038/mto.2016.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kebriaei P., Singh H., Huls M.H., Figliola M.J., Bassett R., Olivares S., Jena B., Dawson M.J., Kumaresan P.R., Su S. Phase I trials using Sleeping Beauty to generate CD19-specific CAR T cells. J. Clin. Invest. 2016;126:3363–3376. doi: 10.1172/JCI86721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Torikai H., Cooper L.J. Translational Implications for Off-the-shelf Immune Cells Expressing Chimeric Antigen Receptors. Mol. Ther. 2016;24:1178–1186. doi: 10.1038/mt.2016.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Afzali B., Lechler R.I., Hernandez-Fuentes M.P. Allorecognition and the alloresponse: clinical implications. Tissue Antigens. 2007;69:545–556. doi: 10.1111/j.1399-0039.2007.00834.x. [DOI] [PubMed] [Google Scholar]
- 12.Poirot L., Philip B., Schiffer-Mannioui C., Le Clerre D., Chion-Sotinel I., Derniame S., Potrel P., Bas C., Lemaire L., Galetto R. Multiplex genome-edited T-cell manufacturing platform for “off-the-shelf” adoptive T-cell immunotherapies. Cancer Res. 2015;75:3853–3864. doi: 10.1158/0008-5472.CAN-14-3321. [DOI] [PubMed] [Google Scholar]
- 13.Torikai H., Reik A., Liu P.-Q.Q., Zhou Y., Zhang L., Maiti S., Huls H., Miller J.C., Kebriaei P., Rabinovich B. A foundation for universal T-cell based immunotherapy: T cells engineered to express a CD19-specific chimeric-antigen-receptor and eliminate expression of endogenous TCR. Blood. 2012;119:5697–5705. doi: 10.1182/blood-2012-01-405365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Valton J., Guyot V., Marechal A., Filhol J.M., Juillerat A., Duclert A., Duchateau P., Poirot L. A Multidrug-resistant Engineered CAR T Cell for Allogeneic Combination Immunotherapy. Mol. Ther. 2015;23:1507–1518. doi: 10.1038/mt.2015.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boissel S., Jarjour J., Astrakhan A., Adey A., Gouble A., Duchateau P., Shendure J., Stoddard B.L., Certo M.T., Baker D., Scharenberg A.M. megaTALs: a rare-cleaving nuclease architecture for therapeutic genome engineering. Nucleic Acids Res. 2014;42:2591–2601. doi: 10.1093/nar/gkt1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Osborn M.J., Webber B.R., Knipping F., Lonetree C.-L.L., Tennis N., DeFeo A.P., McElroy A.N., Starker C.G., Lee C., Merkel S. Evaluation of TCR Gene Editing Achieved by TALENs, CRISPR/Cas9, and megaTAL nucleases. Mol. Ther. 2016;24:570–581. doi: 10.1038/mt.2015.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hacein-Bey-Abina S., Von Kalle C., Schmidt M., McCormack M.P., Wulffraat N., Leboulch P., Lim A., Osborne C.S., Pawliuk R., Morillon E. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 18.Hacein-Bey-Abina S., Garrigue A., Wang G.P., Soulier J., Lim A., Morillon E., Clappier E., Caccavelli L., Delabesse E., Beldjord K. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J. Clin. Invest. 2008;118:3132–3142. doi: 10.1172/JCI35700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Braun C.J., Boztug K., Paruzynski A., Witzel M., Schwarzer A., Rothe M., Modlich U., Beier R., Göhring G., Steinemann D. Gene therapy for Wiskott-Aldrich syndrome--long-term efficacy and genotoxicity. Sci. Transl. Med. 2014;6:227ra33. doi: 10.1126/scitranslmed.3007280. [DOI] [PubMed] [Google Scholar]
- 20.Ellis J. Silencing and variegation of gammaretrovirus and lentivirus vectors. Hum. Gene Ther. 2005;16:1241–1246. doi: 10.1089/hum.2005.16.1241. [DOI] [PubMed] [Google Scholar]
- 21.Schumann K., Lin S., Boyer E., Simeonov D.R., Subramaniam M., Gate R.E., Haliburton G.E., Ye C.J., Bluestone J.A., Doudna J.A., Marson A. Generation of knock-in primary human T cells using Cas9 ribonucleoproteins. Proc. Natl. Acad. Sci. USA. 2015;112:10437–10442. doi: 10.1073/pnas.1512503112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirsch M.L., Samulski R.J. AAV-mediated gene editing via double-strand break repair. Methods Mol. Biol. 2014;1114:291–307. doi: 10.1007/978-1-62703-761-7_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller D.G., Petek L.M., Russell D.W. Human gene targeting by adeno-associated virus vectors is enhanced by DNA double-strand breaks. Mol. Cell. Biol. 2003;23:3550–3557. doi: 10.1128/MCB.23.10.3550-3557.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moser R.J., Hirsch M.L. AAV Vectorization of DSB-mediated Gene Editing Technologies. Curr. Gene Ther. 2016;16:207–219. doi: 10.2174/1566523216666160602213738. [DOI] [PubMed] [Google Scholar]
- 25.Sather B.D., Romano Ibarra G.S., Sommer K., Curinga G., Hale M., Khan I.F., Singh S., Song Y., Gwiazda K., Sahni J. Efficient modification of CCR5 in primary human hematopoietic cells using a megaTAL nuclease and AAV donor template. Sci. Transl. Med. 2015;7:307ra156. doi: 10.1126/scitranslmed.aac5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J., DeClercq J.J., Hayward S.B., Li P.W., Shivak D.A., Gregory P.D., Lee G., Holmes M.C. Highly efficient homology-driven genome editing in human T cells by combining zinc-finger nuclease mRNA and AAV6 donor delivery. Nucleic Acids Res. 2016;44:e30. doi: 10.1093/nar/gkv1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seligman L.M., Chisholm K.M., Chevalier B.S., Chadsey M.S., Edwards S.T., Savage J.H., Veillet A.L. Mutations altering the cleavage specificity of a homing endonuclease. Nucleic Acids Res. 2002;30:3870–3879. doi: 10.1093/nar/gkf495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Djukanovic V., Smith J., Lowe K., Yang M., Gao H., Jones S., Nicholson M.G., West A., Lape J., Bidney D. Male-sterile maize plants produced by targeted mutagenesis of the cytochrome P450-like gene (MS26) using a re-designed I-CreI homing endonuclease. Plant J. 2013;76:888–899. doi: 10.1111/tpj.12335. [DOI] [PubMed] [Google Scholar]
- 29.Ménoret S., Fontanière S., Jantz D., Tesson L., Thinard R., Rémy S., Usal C., Ouisse L.H., Fraichard A., Anegon I. Generation of Rag1-knockout immunodeficient rats and mice using engineered meganucleases. FASEB J. 2013;27:703–711. doi: 10.1096/fj.12-219907. [DOI] [PubMed] [Google Scholar]
- 30.Grizot S., Smith J., Daboussi F., Prieto J., Redondo P., Merino N., Villate M., Thomas S., Lemaire L., Montoya G. Efficient targeting of a SCID gene by an engineered single-chain homing endonuclease. Nucleic Acids Res. 2009;37:5405–5419. doi: 10.1093/nar/gkp548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cradick T.J., Qiu P., Lee C.M., Fine E.J., Bao G. COSMID: A Web-based Tool for Identifying and Validating CRISPR/Cas Off-target Sites. Mol. Ther. Nucleic Acids. 2014;3:e214. doi: 10.1038/mtna.2014.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Imai C., Mihara K., Andreansky M., Nicholson I.C., Pui C.-H., Geiger T.L., Campana D. Chimeric receptors with 4-1BB signaling capacity provoke potent cytotoxicity against acute lymphoblastic leukemia. Leukemia. 2004;18:676–684. doi: 10.1038/sj.leu.2403302. [DOI] [PubMed] [Google Scholar]
- 33.Dykes J.H., Toporski J., Juliusson G., Békássy A.N., Lenhoff S., Lindmark A., Scheding S. Rapid and effective CD3 T-cell depletion with a magnetic cell sorting program to produce peripheral blood progenitor cell products for haploidentical transplantation in children and adults. Transfusion. 2007;47:2134–2142. doi: 10.1111/j.1537-2995.2007.01438.x. [DOI] [PubMed] [Google Scholar]
- 34.Schumm M., Lang P., Bethge W., Faul C., Feuchtinger T., Pfeiffer M., Vogel W., Huppert V., Handgretinger R. Depletion of T-cell receptor alpha/beta and CD19 positive cells from apheresis products with the CliniMACS device. Cytotherapy. 2013;15:1253–1258. doi: 10.1016/j.jcyt.2013.05.014. [DOI] [PubMed] [Google Scholar]
- 35.Sahin U., Karikó K., Türeci Ö. mRNA-based therapeutics--developing a new class of drugs. Nat. Rev. Drug Discov. 2014;13:759–780. doi: 10.1038/nrd4278. [DOI] [PubMed] [Google Scholar]
- 36.Hastie E., Samulski R.J. Adeno-associated virus at 50: a golden anniversary of discovery, research, and gene therapy success--a personal perspective. Hum. Gene Ther. 2015;26:257–265. doi: 10.1089/hum.2015.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Torikai H., Reik A., Soldner F., Warren E.H., Yuen C., Zhou Y., Crossland D.L., Huls H., Littman N., Zhang Z. Toward eliminating HLA class I expression to generate universal cells from allogeneic donors. Blood. 2013;122:1341–1349. doi: 10.1182/blood-2013-03-478255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.ClinicalTrials.gov (2016). Study of UCART19 in Pediatric Patients with Relapsed/Refractory B Acute Lymphoblastic Leukemia (PALL). https://clinicaltrials.gov/ct2/show/NCT02808442.
- 39.Stoddard B.L. Homing endonucleases from mobile group I introns: discovery to genome engineering. Mob. DNA. 2014;5:7. doi: 10.1186/1759-8753-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Si W., Mundia M.M., Magwood A.C., Mark A.L., McCulloch R.D., Baker M.D. A strand invasion 3′ polymerization intermediate of mammalian homologous recombination. Genetics. 2010;185:443–457. doi: 10.1534/genetics.110.115196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vasileva A., Jessberger R. Precise hit: adeno-associated virus in gene targeting. Nat. Rev. Microbiol. 2005;3:837–847. doi: 10.1038/nrmicro1266. [DOI] [PubMed] [Google Scholar]
- 42.Grieger J.C., Choi V.W., Samulski R.J. Production and characterization of adeno-associated viral vectors. Nat. Protoc. 2006;1:1412–1428. doi: 10.1038/nprot.2006.207. [DOI] [PubMed] [Google Scholar]
- 43.Hoyos V., Savoldo B., Quintarelli C., Mahendravada A., Zhang M., Vera J., Heslop H.E., Rooney C.M., Brenner M.K., Dotti G. Engineering CD19-specific T lymphocytes with interleukin-15 and a suicide gene to enhance their anti-lymphoma/leukemia effects and safety. Leukemia. 2010;24:1160–1170. doi: 10.1038/leu.2010.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.