Abstract
Resveratrol contributes to a plant’s tolerance of various abiotic and biotic stresses and is highly beneficial to human health. A search for elite alleles affecting resveratrol production was undertaken to find useful grapevine germplasm resources. Resveratrol levels in both berry skins and leaves were determined in 95 grapevine accessions (including 50 wild Chinese grapevine accessions and 45 cultivars) during two consecutive years. Resveratrol contents were higher in berry skins than in leaves and in wild Chinese grapevines than in grapevine cultivars. Using genotyping data, 79 simple sequence repeat (SSR) markers linked to 44 stilbene synthase (STS) genes were detected in the 95 accessions, identifying 40 SSR markers with higher polymorphisms. Eight SSR marker loci, encompassing 19 alleles, were significantly associated with resveratrol content on (P < 0.001), and 5 SSR loci showed repeated associations. Locus Sh5 had four associations: three positive for allele 232 (including leaves in the 2 years) and one negative for allele 236 in four environments. Loci Sh9 and Sh56 for a total of 7 alleles exhibited positive effects in berry skins in the 2 years. In berry skins, locus Sh56 with positive effects was closely linked to VvSTS27, and locus Sh77 with negative effects to VvSTS17, importantly, the two candidate genes both were located on Chromosome 16. The SSR marker loci and candidate genes identified in this study will provide a useful basis for future molecular breeding for increased production of natural resveratrol and its derivatives.
Keywords: resveratrol, stilbene synthase, elite allele, grape, association analysis, SSR
Introduction
Resveratrol (trans-3, 5, 4′-trihydroxystilbene) is a natural phytoalexin occuring in a limited number of plant species, including Vitis spp. (Langcake and Pryce, 1976). Stilbenes in grapevine are very complex, and 18 stilbene derivatives were also identified in two grape samples, including resveratrol and piceid (Flamini et al., 2013). Resveratrol and piceid, in both cis and trans have been characterized in wine and grape berry (Pezet et al., 1994; Lamuela-Raventos et al., 1995; Romero-Pérez et al., 2001; Vitrac et al., 2005). These compounds are formed by oligomerization of trans-resveratrol in grape tissues under stress conditions such as exogenous attack or pathogen infections (Cichewicz et al., 2000; Romero-Pérez et al., 2001). It is interesting to note that trans-resveratrol showed either lower or higher concentration in wine and berry using different determination methods, compared with trans-piceid (Lamuela-Raventos et al., 1995; Ribeiro de Lima et al., 1999; Romero-Pérez et al., 2001; Vian et al., 2005; Vitrac et al., 2005; Flamini et al., 2013).
Table grapes and wines are the main food sources of resveratrol. The studies have focused on trans-resveratrol due to its various physiological functions in consumers, including antioxidative, anti-tumor, anti-inflammatory activities and reduction of cardiovascular disease and obesity (Jang et al., 1997; Alonso et al., 2002; Frombaum et al., 2012; Konings et al., 2014). The accumulation of resveratrol in plant tissue is induced by exogenous hormone, pathogen attack and UV-C irradiation (Zheng et al., 2009; Shi et al., 2014; Wang et al., 2015, 2016; Yin et al., 2016).
Stilbene synthase (STS), a key enzyme in the biosynthesis pathway of resveratrol, belongs to the polyketide synthase family (Rupprich and Kindl, 1978). Experiments aimed at the generation of transgenic plants with increased resveratrol content or improved resistance to fungal pathogens have focused on inserting foreign STS genes, which were mostly from Vitis vinifera (Leckband and Lorz, 1998; Zhu et al., 2004; Serazetdinova et al., 2005; Cheng et al., 2016). Additionally, inserting a foreign STS gene also influenced piceid accumulation in transgenic lines (Ruhmann et al., 2006; Liu et al., 2011; Carlos-Hilario et al., 2015). Recent studies showed that the STS gene family from grapevine included 40 or so members (Parage et al., 2012; Vannozzi et al., 2012; Shi et al., 2014). A very recent report characterized the function of an STS allele (Jiao et al., 2016).
Although the identity and/or function of some members of the STS gene family have been demonstrated, little information is available on how allelic diversities among STS genes contribute to variation in resveratrol accumulation in Vitis germplasm. In our previous study, members of the STS gene family showed one of two expression patterns and different expression levels in response to powdery mildew (Shi et al., 2014). Examination of allelic variation and linkage disequilibrium by a candidate gene-based approach would help to decipher the genetic basis of resveratrol biosynthesis. To do this, a representative sample of 95 grapevine accessions were selected, comprising both wild Chinese and cultivated grapevines, both green- and red-skin berries, and both seedless and seeded berries. SSR markers (79 pairs) distributed over the known STS genes from the grapevine PN40024 genotype were designed. Association analysis between STS genes and resveratrol content was performed on this wide collection of wild Chinese grapevines and cultivated European grapevines in order to find the elite alleles responsible for resveratrol accumulation. The results identify grapevine resources that can be used to obtain new grapevine cultivars with high levels of resveratrol in their berries, and can provide useful information for further research on resveratrol biosynthesis.
Materials and Methods
Plant Materials and Treatments
Grape accessions, including 50 wild Chinese grapevine species and 45 cultivars from the European species V. vinifera or the American species V. labrusca (Table 1), were grown under natural field conditions at the National Grape Germplasm Resources Repository of Zhengzhou Fruit Research Institute, Chinese Academy of Agricultural Sciences. Warm temperate continental climate of Zhengzhou has clear four seasons. The average annual precipitation is about 630 mm and mean temperature is 14.4°C. The details of climatic data were shown in Supplementary Table S1. The experiment vines were planted 9 or 10 years ago in sandy fluvo-aquic soil. And no special cultural practices were taken. All of the vines were in good condition. Grape berries were collected from June to September and leaves were picked at the end of June in 2013 and 2014. Samples were harvested from three grape vines for each accession. For the berries, three grape clusters on each plant were picked, one from the top, middle, and bottom of the canopy, respectively. To ensure that all berries were harvested at their full ripeness, we checked the seeds in the berries every 2 days from June till September. When the seeds completely ripened, the size of berries was no longer increasing, and the red grapes were fully colored, the berries were sampled from that accession. For the leaves, the second or third leaves (depending on healthiness) from the bottom of three different branches with more than 10 leaves were picked in the end of June. Unhealthy berries (cracking, smaller and other underdeveloped fruits) were removed before the samples were quickly frozen in liquid nitrogen and held at -80°C until use.
Table 1.
Fifty wild Chinese grapevine accessions and 45 cultivars were used in this study.
| No. | Species | Accession or cultivar | No | Species | Accession or cultivar |
|---|---|---|---|---|---|
| 1 | V. labrusca | Champion | 49 | V. adenoclada | Shuangxi 01 |
| 2 | V. vinifera | Zhengguo 6 | 50 | Shuangxi 03 | |
| 3 | ′′ | Jan-87 | 51 | Zhijiangshui | |
| 4 | ′′ | Amilia | 52 | V. davidii | Huitong No.1 |
| 5 | ′′ | Guifeimeigui | 53 | ′′ | Huitong No.2 |
| 6 | ′′ | Irsay Oliver | 54 | ′′ | Wuhan |
| 7 | ′′ | Olimpia | 55 | ′′ | Dongxiangjiao |
| 8 | ′′ | Baijixin | 56 | ′′ | Hongjiangyanlong 05 |
| 9 | ′′ | Bolgar | 57 | ′′ | Hongjiangtongmu 07 |
| 10 | ′′ | Pink varieties Taipei | 58 | ′′ | Zhijiang 01 |
| 11 | ′′ | Mathias Aromatic | 59 | ′′ | Hongjiang 04 |
| 12 | ′′ | Fenghuang 51 | 60 | ′′ | Hongjiang 08 |
| 13 | ′′ | Guibao | 61 | ′′ | Hongjiang 09 |
| 14 | ′′ | Red Globe | 62 | ′′ | Hongjiang 10 |
| 15 | ′′ | Malaga Rose | 63 | ′′ | Fuan |
| 16 | ′′ | Huangmisi | 64 | ′′ | Tangwei seedling |
| 17 | ′′ | Jingxiu | 65 | ′′ | Zhejiangtianmushan No.2 |
| 18 | ′′ | Muscat Hamburg | 66 | ′′ | Zhejiangtianmushan No.3 |
| 19 | ′′ | Manai | 67 | ′′ | Xiangzhenzhuhongye |
| 20 | ′′ | Munage | 68 | ′′ | Xiangzhenzhulvye |
| 21 | ′′ | Senio de Malingre | 69 | ′′ | Hunan |
| 22 | ′′ | Miskat Plevenski | 70 | ′′ | Gaoshan No.1 |
| 23 | ′′ | Queen of Vineyard | 71 | ′′ | Gaoshan No.2 |
| 24 | ′′ | Zhengguo 5 | 72 | V. amurensis | S48-3 |
| 25 | ′′ | Xiangfei | 73 | ′′ | N43-3 |
| 26 | ′′ | Shenyangmeigui | 74 | ′′ | Changbai No.9 |
| 27 | ′′ | Ribier | 75 | ′′ | Shuangyou |
| 28 | ′′ | Yangputao | 76 | V. ficifolia | 946 |
| 29 | ′′ | Yalishanda | 77 | ′′ | 943 |
| 30 | ′′ | Muscat MathiaszJanosne | 78 | ′′ | Qinling No.2 |
| 31 | ′′ | Xiabai | 79 | ′′ | Wugang |
| 32 | ′′ | Italia | 80 | ′′ | Xinyang 01 |
| 33 | ′′ | Zaomanao | 81 | ′′ | Fengjugou 02 |
| 34 | ′′ | Zaotianmeiguixiang | 82 | ′′ | Fengjugou 03 |
| 35 | ′′ | Zhengzhouzaoyu | 83 | ′′ | Shibanyan 02 |
| 36 | V. vinifera x V. labrusca | Zifeng | 84 | ′′ | Shibanyan 05 |
| 37 | V. vinifera | Zexiang | 85 | ′′ | Shibanyan 06 |
| 38 | ′′ | Zijixin | 86 | ′′ | Shibanyan 08 |
| 39 | ′′ | Jingzaojing | 87 | ′′ | Luoning 06 |
| 40 | ′′ | Thompson Seedless | 88 | ′′ | Qinling 03 |
| 41 | ′′ | Pinot Noir | 89 | ′′ | Jiuligou |
| 42 | ′′ | Cabernet Sauvignon | 90 | V. betulifolia | Songxian |
| 43 | V. vinifera x V. amurensis | Beimei | 91 | V. romanetii | Lingbao |
| 44 | ′′ | Beichun | 92 | V. pseudoreticulata | Huadong |
| 45 | V. vinifera | Zhengguodawuhe | 93 | ′′ | 1057 |
| 46 | V. quinquangularis | Guizhou | 94 | V. yeshanensis | Yanshan |
| 47 | V. amurensis | Baitianman 03 | 95 | V. adstricta | Yingyu |
| 48 | V. wilsonae | Baotianman |
Numbers 1–45 were Vitis cultivars, and Numbers 46–95 were wild grapevine species.
Determination of Trans-resveratrol Content by HPLC Method
Trans-resveratrol levels in berry skins and leaves were measured using HPLC as described by Li et al. (2006) with some modifications, in 95 grapevine accessions in 2013 and 2014. The standard for trans-resveratrol was purchased from Sigma–Aldrich (USA). Fruits were peeled and juice was soaked up using filter paper.
Three gram samples were ground to powder using a porcelain mortar and pestle in liquid nitrogen, extracted by 15 mL ethyl acetate in the dark at 25°C for 48 h, and centrifuged at 10,000 r⋅min-1 for 10 min. The supernatants were transferred into a tube containing 5 mL ethyl acetate, followed by centrifugation at 10,000 r⋅min-1 for 10 min. All supernatants were evaporated to dryness by Nitrogen blowing instrument (DCY-12S, Qingdao Haike, China) at 40°C. Dried samples were then dissolved in 2 mL of methanol and stored at -80°C. The samples were filtered through a 0.22 μm PTFE membrane filter before resveratrol analysis. Extractable amounts of resveratrol were analyzed using a Waters e2695 HPLC system (USA). Elution was carried out with a mobile phase delivered using a Waters C18 HPLC pump at a flow rate 0.8 mL⋅min-1. A Waters 2996 UV detector was used at 306 nm. Mean values and standard deviations were obtained from three biological replicates. An HPLC chromatogram of resveratrol was made with a standard solution. The resveratrol content was analyzed by Excel 2003 (Microsoft, USA) and SPSS 17.0 software (IBM, USA).
DNA Isolation and PCR Amplification
Genomic DNA was extracted using Ezup Column Plant Genomic DNA Purification Kit following the manufacturer’s protocol (Sangon Biotech, Shanghai, China). The concentration of the extracted DNA was assessed using a Thermo ND 2000 spectrophotometer (ThermoFisher, USA). Genomic DNA was adjusted to a final concentration 50 ng/μL and was used for PCR amplification.PCR reactions were carried out in a final volume of 20 μL. Amplification reactions were carried out on a ABI Veriti thermal cycler (USA) using the following cycling profile: 95°C for 5 min, followed by 35 cycles at 95°C for 45 s, 48–56°C for 45 s, and 72°C for 1 min, and a final extension step at 72°C for 10 min. The amplification products were separated through polyacrylamide gel electrophoresis.
Analysis of SSR Markers
Based on predicted STS gene sequences in the 12x grapevine PN40024 genome1 and the gene positions of these 44 STS genes (Shi et al., 2014), a total of 79 pairs of SSR primers on chromosomes 10 and 16 were designed using GRAMENE ssrtool2. Parameter settings were as follows: tetramer for the maximum motif-length group, and 4 for the minimum number of repeats.
Allelic variation was analyzed by calculating the number of alleles (Na), effective number of alleles (Ne), observed heterozygosity (Ho), and expected heterozygosity (He) using Popgene software. Polymorphism information content (PIC) was calculated using PIC-CALC.
Genetic distance matrices were obtained using SSR data in DPS software3. A phylogenetic tree was constructed by the unweighted pair-group method with arithmetic averages (UPGMA) with MEGA 6.0 software4.
Population Structure and Association Analysis
Using 40 STS-gene-associated SSR markers, the genetic population structure of the 95 accessions was determined by Structure 2.15. A burn-in phase of 10,000 iterations was followed by 100,000 Monte Carlo Markov Chain iterations. The optimal population number k (from 1 to 10 assumed in this study) was estimated (Evanno et al., 2005). Ten replicates were performed for each cluster, k. When an inflection emerged in the LnP (D) curve, the corresponding k value was adopted as the optimal group number. The values of the estimated membership probability (Q) were calculated to serve as covariates in the association analysis with general linear model (GLM) in Tassel 2.16. Phenotypic effect values of some marker alleles were evaluated according to null allele as suggested by Breseghello and Mark (2006).
Results
Trans-resveratrol Content
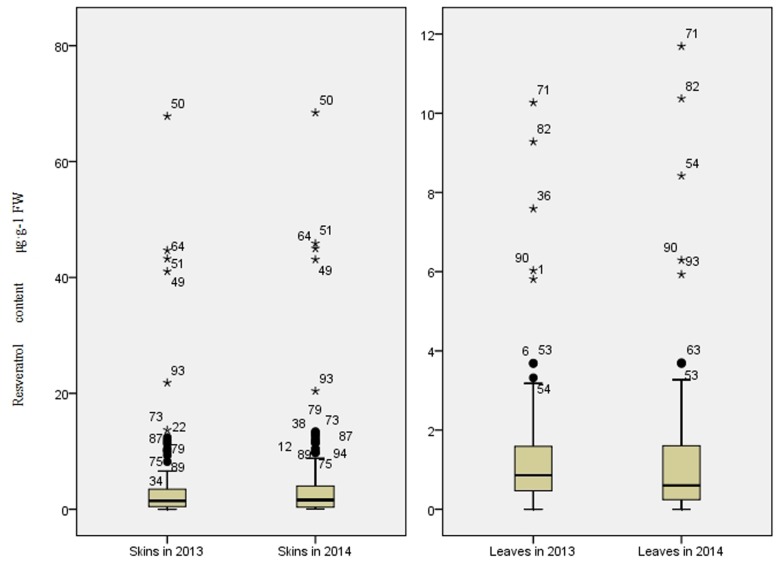
The trans-resveratrol levels in skin and in leaf collected from all accessions were determined by HPLC (Figure 1). The trans-resveratrol content in berry skins ranged from 0.05 to 67.82 μg⋅g-1 FW in 2013 and from 0.03 to 68.44 μg⋅g-1 FW in 2014. For both seasons, the highest levels were from the wild Chinese grapevine V. adenoclada accession Shuangxi 03. In leaves, the trans-resveratrol content ranged from 0.04 to 10.27 μg⋅g-1 FW in 2013 and from 0.09 to 11.69 μg⋅g-1 FW in 2014. The highest levels for both years were in leaves from wild Chinese grapevine V. amurensis accession Gaoshan No.2. Resveratrol contents were higher in berry skins than in leaves for each genotype.
FIGURE 1.
Range and distribution of trans-resveratrol content in skins and leaves of 95 grapevine accessions (50 wild Chinese accessions and 45 grapevine cultivars) in 2013 and 2014.
Between the 2 years, the variation of resveratrol content was more stable in wild grapevine accessions than that of cultivated ones. More of the cultivated accessions (51%) showed year-to-year variations of resveratrol content in skin greater than 50%, compared to only 8% of wild grapevine ones, showing such large variations. Similarly, in leaves, 22% of wild accessions and 67% of cultivated ones showed resveratrol content variations greater than 50% (Supplementary Table S2). The results suggested that wild ones retained stable resveratrol biosynthetic capacity.
Polymorphisms of Molecular Markers
Based on the predicted STS gene sequences of the 12x grapevine PN40024 genome, 79 SSR primers were designed. These 79 markers were analyzed in the 95 grapevine accessions. Forty SSR markers showed higher polymorphism, and 123 alleles were identified. The PICs of the SSR loci ranged from 0.0206 to 0.6712, with an average of 0.2877 (Supplementary Table S3).
SSR Analysis
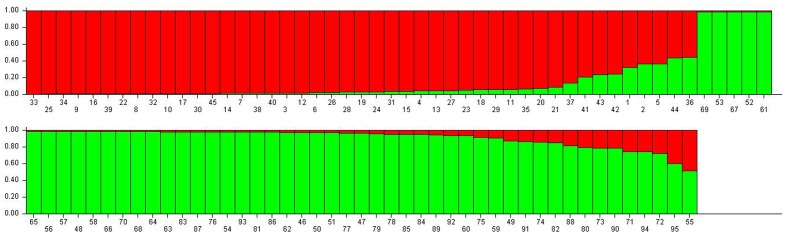
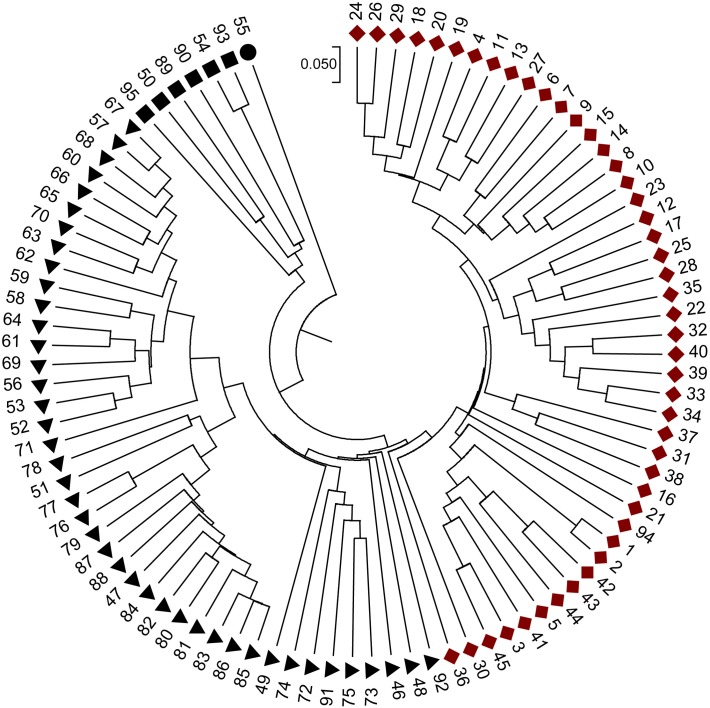
When the STRUCTURE software was run using all 95 grapevine accessions, the delta k showed a significant peak when k = 2; thus the grapevine accessions were divided into two populations, termed P1 and P2 (Figure 2). This division of the population was supported by statistical probability and could ensure the accuracy of association analysis with minimum false association. P1 included 45 grapevine cultivars, both table and wine grapes, whereas P2 included 50 accessions, all of which were wild Chinese grapevine accessions (Figure 2). A phylogenetic tree was constructed by UPGMA analysis based on genetic distances calculated from the SSR data of the 95 accessions (Figure 3). Due to sufficient variability, all selected accessions were discriminated. The accessions clustered into two main groups, with six accessions (Nos. 50, 54, 89, 90, 93, and 95) forming a third, distinct cluster (black square). All accessions formed a branch with other accessions and cultivars, except two, namely V. davidii accession Dongxiangjiao (No. 55, black circle), which did fall in close to another branch, and V. yeshanensis accession Yanshan (No. 94), which did not sort into near wild grapevines. This corresponded to the evaluated populations with STRUCTURE software, with a few exceptions. The above SSR analysis generally agreed with the geographic origins and pedigree of the grapevine accessions.
FIGURE 2.
Population structure of the 95 grapevine accessions. The numbers represent plant material according to Table 1. Population one (P1, red) included 45 table and wine grapes, whereas Population 2 (P2, green) included 50 wild Chinese grapevine accessions.
FIGURE 3.
Phylogenetic relationships of the accessions based on genetic distances calculated using SSR data and UPGMA clustering constructed using MEGA 6.0 software. Four icons,  , represent four sub-divisions. Red and black represent two main groups, respectively.
, represent four sub-divisions. Red and black represent two main groups, respectively.
Association Analysis between Resveratrol and SSR Marker Loci
Linkage disequilibrium (LD) among genes was the basis of the association analysis. Distribution of LD among the 40 SSR loci in the two groups (according to Figure 2) was shown as Figure 4A. Loci with high LD values (D′ > 0.7; upper right corner) were Sh13, Sh16, Sh22, Sh31, Sh37, Sh68, and Sh78.The LD among the wild Chinese grapevines (Figure 4B) was significantly higher than those of the grapevine cultivars (Figure 4C, including table grapes and wine grapes). The mean frequency distribution of the D′ value (P < 0.001) was 0.5329 for all experimental samples (Table 2A), 0.6046 for the V. vinifera cultivars, and 0.7037 for the wild Chinese accessions (Table 2B). The higher D′ in the wild population indicates more variation. In addition, the number of LD loci among the grapevine cultivars was fewer than in the wild Chinese accessions (Table 2B).
FIGURE 4.
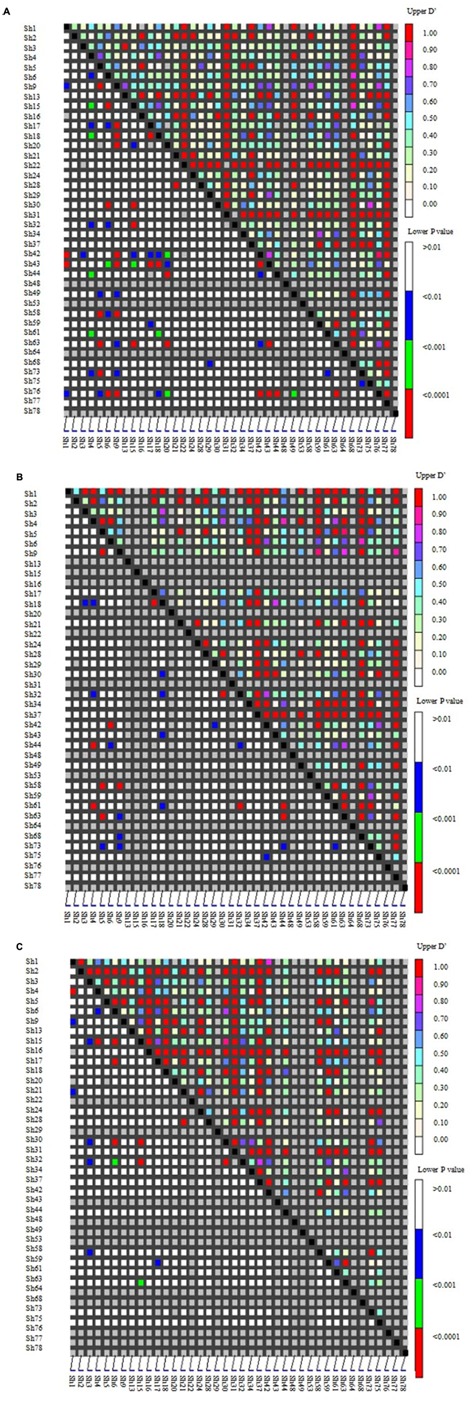
Distribution of Linkage disequilibrium (LD) among 40 SSR loci in two groups of 95 grapevine accessions. SSR markers are organized in linkage groups marked along the X- and Y-axis; each pixel above the diagonal indicates the D′value of the corresponding marker pair as shown in the color code at the upper right, while each pixel below the diagonal indicates the p-value size of the testing LD of the corresponding marker pairs as shown in the color code at the lower right. (A) Distribution of LD in 95 grapevine accessions. (B) 50 wild Chinese accessions. (C) 45 grapevine cultivars.
Table 2.
The frequency distribution of D′ value.
| (A) Linkage disequilibrium (LD) for pairwise SSR loci among all 95 grapevine accessions. | |||||||
|---|---|---|---|---|---|---|---|
| Number of LD locus pairs |
Frequency distribution of D′ (P < 0.001) |
||||||
| 0–0.2 | 0.2–0.4 | 0.4–0.6 | 0.6–0.8 | 0.8–1.0 | Mean of D′ | ||
| 68 (8.72%) | 0 | 14 | 32 | 20 | 2 | 0.5329 | |
| (B) Comparison of LD values for pairwise SSR loci between V. vinifera cultivars and wild Chinese grapes. | |||||||
| Population | Number of LD locus pairs |
Frequency distribution of D′ (P < 0.001) |
|||||
| 0–0.2 | 0.2–0.4 | 0.4–0.6 | 0.6–0.8 | 0.8–1.0 | Mean of D′ | ||
| Vitis cultivars | 18 (2.44%) | 0 | 2 | 6 | 8 | 2 | 0.6046 |
| Wild Chinese grapes | 28 (3.59%) | 0 | 0 | 9 | 11 | 8 | 0.7037 |
Based on LD analysis and the current suitable population, association analysis was performed with candidate markers using Tassel 2.1 software. Eight SSR loci, namely Sh5, Sh9, Sh21, Sh28, Sh56, Sh63, Sh76, and Sh77, were significantly (P < 0.001) associated with resveratrol content and their explained phenotypic variation (EPV) were all higher than 10% (Table 3). Loci Sh5, Sh21, Sh28, Sh63, and Sh76 were associated with high resveratrol content in the leaves, whereas loci Sh5, Sh9, Sh56, and Sh77 were associated with high resveratrol in berry skins (Table 4). Moreover, these associations were independent of the year. Locus Sh5 was associated with high resveratrol content in both tissues in both seasons.
Table 3.
Marker loci associated with resveratrol content and their explained phenotypic variation (significance at P < 0.001).
| Trait | Locus | p_Marker | EPV (%) |
|---|---|---|---|
| Leaf in 2013 | Sh5 | 0.00044317 | 0.1891 |
| Sh21 | 0.00034856 | 0.1916 | |
| Sh28 | 0.00008824 | 0.1922 | |
| Leaf in 2014 | Sh5 | 0.00080000 | 0.1429 |
| Sh63 | 0.00001241 | 0.2018 | |
| Sh76 | 0.00000073 | 0.2760 | |
| Skin in 2013 | Sh5 | 0.00000027 | 0.3121 |
| Sh9 | 0.00098624 | 0.1939 | |
| Sh56 | 0.00069694 | 0.1187 | |
| Sh77 | 0.00000003 | 0.2850 | |
| Skin in 2014 | Sh5 | 0.00000028 | 0.3033 |
| Sh9 | 0.00050000 | 0.1800 | |
| Sh56 | 0.00062219 | 0.1172 | |
| Sh77 | 0.00000001 | 0.2930 |
Table 4.
Phenotypic effects of some marker alleles at loci significantly associated with resveratrol content.
| Trait | Locus | Allele size (bp) | Phenotypic effect |
|---|---|---|---|
| Leaf in 2013 | Sh5 | 232 | 17.49 |
| 236 | -1.81 | ||
| Sh21 | 264 | -4.48 | |
| 266 | -4.92 | ||
| Sh28 | 220 | -4.09 | |
| 222 | -3.93 | ||
| Leaf in 2014 | Sh5 | 232 | 10.43 |
| 236 | -10.11 | ||
| Sh63 | 120 | -6.17 | |
| 122 | -5.82 | ||
| 124 | -5.89 | ||
| Sh76 | 113 | -3.34 | |
| 115 | -4.02 | ||
| Skin in 2013 | Sh5 | 232 | 19.28 |
| 236 | -0.19 | ||
| Sh9 | 239 | 18.05 | |
| 243 | 0.24 | ||
| 247 | 2.77 | ||
| 253 | 0.10 | ||
| 256 | 1.30 | ||
| Sh56 | 125 | 1.45 | |
| 129 | 8.60 | ||
| Sh77 | 117 | -38.45 | |
| Skin in 2014 | Sh5 | 232 | -1.50 |
| 236 | -3.12 | ||
| Sh9 | 239 | 20.06 | |
| 243 | 2.49 | ||
| 247 | 4.23 | ||
| 253 | 2.08 | ||
| 256 | 2.99 | ||
| Sh56 | 125 | 1.28 | |
| 129 | 8.99 | ||
| Sh77 | 117 | -39.90 |
The phenotypic effects of the different alleles of the eight loci significantly associated with resveratrol content were evaluated (Table 4). Allele 236 at locus Sh5 produced negative effects four times. On the other hand, allele 232 produced positive effects three times, including in leaves in the 2 years. Loci Sh9 and Sh56, through seven alleles, exhibited only positive effects in berry skins, whereas one allele of locus Sh77 created negative effects in berry skins in the 2 years. The rest of the loci showed negative effects at least once.
The eight loci significantly associated with resveratrol content were mapped to the 12x grapevine PN40024 genome. This revealed that locus Sh56 (location 16506665–16506789 on Chromosome 16) was closely linked to VvSTS27 (16507444-16503155) and that locus Sh77 (16:366055-16:366171) was closely linked to VvSTS17 (16372414-16366426) (Table 5). The other six loci were not very closed to known STS genes. However, future investigation of predicted genes at these loci may reveal their functions in secondary metabolism.
Table 5.
Repeat motif and physical location of eight SSR loci significantly associated with resveratrol (P < 0.001) on the 12x grapevine PN40024 genome.
| Primer name | Motif | No. of Repeats | PN40024 12 X location |
|---|---|---|---|
| Sh5 | at | 13 | 16323230 16323465 |
| Sh9 | tat | 7 | 16320838 16321080 |
| Sh21 | at | 26 | 16247793 16248056 |
| Sh28 | ga | 5 | 16257727 16257946 |
| Sh56 | at | 4 | 16506665 16506789 |
| Sh63 | ag | 6 | 16630877 16631000 |
| Sh76 | tc | 10 | Chr16:363088 Chr16: 363201 |
| Sh77 | tc | 4 | Chr16: 366055 Chr16: 366171 |
Discussion
Grapevine is one of the most important fruits in the world. Table grapes are a healthy snack, grape leaves are a staple in some diets, and wine grapes produce a favorite beverage. Resveratrol in both berries and leaves benefit human health, an attribute which has attracted widespread interest. Breeders aim to select and improve the content of resveratrol and other secondary metabolites, such as stilbenes, in grape. Moreover, stilbene concentrations vary depending on multiple factors, including grape cultivar, fungal infection, and climate condition (Jeandet et al., 1995; Mattivi et al., 1995; Ribeiro de Lima et al., 1999). In the present study, the resveratrol contents in 95 accessions were determinated by HPLC method in two growing seasons. Trans-resveratrol content ranged from 0.03 to 68.44 μg⋅g-1FW in berry skins and from 0.04 to 11.69 μg⋅g-1 FW in leaves. A previous study found that resveratrol was significantly higher (1) in berry skin of seeded cultivars than of seedless ones; (2) in berry skin and seeds in wine grapes than in table grapes; (3) and in red grapes than in green (Li et al., 2006). A recent study reported that an STS allele from the wild Chinese grapevine V. pseudoreticulata could confer accumulation of stilbenes and resistance against powdery mildew in an Arabidopsis heterologous system, whereas the allele from V. vinifera ‘Carigane’ could not be expressed (Jiao et al., 2016). Together these results demonstrate a wide range of resveratrol content in wild, table and wine grapes, which also suggests the existence of potential genetic variation for resveratrol biosynthesis. Therefore, the use of a wide collection of 95 grapevine accessions in our study lays a foundation for finding elite alleles for resveratrol production.
STS genes encode key enzymes in the last stage of resveratrol biosynthesis. In grapevine, the STS gene family contains at least 40 members, although most relevant studies thus far have focused on only one or two STS genes from grapevines and peanuts. Overexpression of STS genes can improve resistance against a fungal pathogen and other abiotic stresses and increase either resveratrol accumulation (Zhu et al., 2004; Kiselev and Aleynova, 2016), or piceid accumulation (Ruhmann et al., 2006; Liu et al., 2011; Carlos-Hilario et al., 2015). The expression of 32 STS genes was analyzed after exposure to UV light, and function of nine STS genes of them was characterized (Parage et al., 2012). Our previous findings also showed that about 40 STS genes had different expression patterns in different tissues and environments (Shi et al., 2014). Members of the STS gene family were analyzed for differences in their molecular structure and transcript accumulation (Vannozzi et al., 2012). In the present study, 40 SSR loci with high polymorphism (an average of 0.2877) were located on Chromosome 16 of the grapevine PN40024 genome, suggesting that Chromosome 16 may be more responsible for resveratrol biosynthesis than STS genes on other chromosomes.
Through correlation analysis, all representative samples of the population and the polymorphisms of the SSR markers link an associated locus to several allelic variants. If the corresponding allelic variation tends to phenotypic diversity, it might be selected as optimal allelic variation. In the present study, 8 SSR loci were significantly (P < 0.001) associated with resveratrol content, with EPV higher than 10%. Of them, four loci showed repeated associations in four environments. Locus Sh5 associated with high resveratrol content four times, with allele 232 linked three times for positive effects, including in leaves in the 2 years. But allele 236 showed negative effects four times. For resveratrol content in berry skins, loci Sh9 and Sh56, with a combined seven alleles, exhibited positive effects. Recently, many studies using molecular markers have amplified multiple bands, identified relationships, mapped markers to chromosomes, and analyzed the association between molecular markers and agronomic traits (Abdurakhmonov et al., 2008; Jahnke et al., 2011; Lorenzis et al., 2013; Liu et al., 2014; Cai et al., 2016). However, there have not been many studies on the markers of selected genes (Jin et al., 2016).
As resveratrol is directly catalyzed by STS, correlation between known STS alleles, our SSR markers, and resveratrol content were sought. We found eight loci with significant association to resveratrol content in a wide grapevine germplasm collection, while controlling false positives potentially deriving from population structure and multiple testing. Three SSR loci in berry skins with positive effects were mapped onto Chromosome 16. These loci were close to VvSTS17 or VvSTS27. These findings can inform future use of grapevine germplasm resources in breeding for production of resveratrol and its derivatives.
Author Contributions
XZ and JS contributed equally to this work. JF, XZ, and JS: conceived and designed the experiments. JS, YY, and YS: performed the experiments and analyzed the data. XZ, JS, and YY: contributed reagents/materials/analysis tools. JF, BT, XY, and JL: provided guidance for the entire study. JS: wrote the manuscript. All authors approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Prof. Chonghuai Liu and Dr. Ying Zhang at Zhengzhou Fruit Research Institute, Chinese Academy of Agricultural Sciences, for great assistance in collecting samples.
Funding. This work was supported by the National Natural Science Foundation of China (Grant No. 31201591), Open Project of Key Laboratory of National Genetic Engineering (20150321), Major Project Science and Technology in Henan Province (151100110900), MATS of Henan Province (S2014-11-G02), and Key Scientific Project of College and University of Henan Province (15A210034).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00487/full#supplementary-material
References
- Abdurakhmonov I. Y., Kohel R. J., Yu J. Z., Pepper A. E., Abdullae A. A., Kushanov F. N., et al. (2008). Molecular diversity and association mapping of fiber quality traits in exotic G. hirsutum L. germplasm. Genomics 92 478–487. 10.1016/j.ygeno.2008.07.013 [DOI] [PubMed] [Google Scholar]
- Alonso A. M., Domingue C., Guille D. A., Barroso C. G. (2002). Determination of antioxidant power of red and white wines by a new electrochemical method and its correlation with polyphenolic content. J. Agric. Food Chem. 50 3112–3115. 10.1021/jf0116101 [DOI] [PubMed] [Google Scholar]
- Breseghello F., Mark E. S. (2006). Association mapping of kernel size and milling quality in wheat (Triticum aestivum L.) cultivars. Genetics 172 1165–1177. 10.1534/genetics.105.044586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S., Han Z., Huang Y., Hu H., Dai F., Zhang G. (2016). Identification of quantitative trait loci for the phenolic acid contents and their association with agronomic traits in Tibetan wild barley. J. Agric. Food Chem. 64 980–987. 10.1021/acs.jafc.5b05441 [DOI] [PubMed] [Google Scholar]
- Carlos-Hilario L., Shimshock R., Ng C., Bingham J., Christopher D. A. (2015). Screening Carica papaya native promoters driving stilbene synthase expression in Arabidopsis thaliana for resveratrol glucoside (piceid) synthesis. Plant Biotechnol. Rep. 9 307–317. 10.1007/s11816-015-0367-2 [DOI] [Google Scholar]
- Cheng S., Xie X., Xu Y., Zhang C., Wang X., Zhang J., et al. (2016). Genetic transformation of a fruit-specific, highly expressed stilbene synthase gene from Chinese wild Vitis quinquangularis. Planta 243 1041–1053. 10.1007/s00425-015-2459-1 [DOI] [PubMed] [Google Scholar]
- Cichewicz R. H., Kouzi S. A., Hamann M. T. (2000). Dimerization of resveratrol by the grapevine pathogen Botrytis cinerea. J. Nat Prod. 63 29–33. 10.1021/np990266n [DOI] [PubMed] [Google Scholar]
- Evanno G., Regnaut S., Goudet J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14 2611–2620. 10.1111/j.1365-294X.2005.02553.x [DOI] [PubMed] [Google Scholar]
- Flamini R., De Rosso M., De Marchi F., Vedova A. D., Panighel A., Gardiman M. (2013). An innovative approach to grape metabolomics: stilbene profiling by suspect screening analysis. Metabolomics 9 1243–1253. 10.1007/s11306-013-0530-0 [DOI] [Google Scholar]
- Frombaum M., Clanche S. L., Bonnefont-Rousselot D., Borderi D. (2012). Antioxidant effects of resveratrol and other stilbene derivatives on oxidative stress and NO bioavailability, potential benefits to cardiovascular diseases. Biochimie 94 269–276. 10.1016/j.biochi.2011.11.001 [DOI] [PubMed] [Google Scholar]
- Jahnke G., Májer J., Varga P., Szoke B. (2011). Analysis of clones of Pinots grown in Hungary by SSR markers. Sci. Hortic. 129 32–37. 10.1016/j.scienta.2011.03.004 [DOI] [Google Scholar]
- Jang M., Cai L., Udeani G. O., Slowing K. V., Thomas C. F., Beecher C. W. W., et al. (1997). Cancer chemopreventive activity of resveratrol, a natural product derived from grapes. Science 275 218–220. 10.1126/science.275.5297.218 [DOI] [PubMed] [Google Scholar]
- Jeandet P., Bessis R., Sbaghi M., Meunier P. (1995). Production of the phytoalexin resveratrol by grapes as a response to Botrytis attack under natural conditions. J. Phytopathol. 143 135–139. 10.1111/j.1439-0434.1995.tb00246.x [DOI] [Google Scholar]
- Jiao Y., Xu W., Duan D., Wang Y., Nick P. (2016). A stilbene synthase allele from a Chinese wild grapevine confers resistance to powdery mildew by recruiting salicylic acid signalling for efficient defence. J. Exp. Bot. 67 5841–5856. 10.1093/jxb/erw351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J. Q., Yao M. Z., Ma C. L., Ma J. Q., Chen L. (2016). Association mapping of caffeine content with TCS1 in tea plant and its related species. Plant Physiol. Biochem. 105 251–259. 10.1016/j.plaphy.2016.04.032 [DOI] [PubMed] [Google Scholar]
- Kiselev K. V., Aleynova O. A. (2016). Influence of overexpression of stilbene synthase VaSTS7 gene on resveratrol production in transgenic cell cultures of grape Vitis amurensis Rupr. Appl. Biochem. Microbiol. 52 56–60. 10.1134/S0003683815060071 [DOI] [Google Scholar]
- Konings E., Timmers S., Boekschoten M. V., Goossens G. H., Jocken J. W., Afman L. A., et al. (2014). The effects of 30 days resveratrol supplementation on adipose tissue morphology and gene expression patterns in obese men. Int. J. Obes. 38 470–473. 10.1038/ijo.2013.155 [DOI] [PubMed] [Google Scholar]
- Lamuela-Raventos R. M., Romero-Pérez A. I., Waterhouse A. L., de la Torre-Boronat M. C. (1995). Direct HPLC analysis of cis–and trans-resveratrol and piceid isomers in Spanish red Vitis vinifera wines. J. Agric. Food Chem. 43 281–283. 10.1021/jf00050a003 [DOI] [Google Scholar]
- Langcake P., Pryce R. J. (1976). The production of resveratrol by Vitis vinifera and other members of the Vitaceae as a response to infection or injury. Physiol. Plant Pathol. 9 77–86. 10.1016/0048-4059(76)90077-1 [DOI] [Google Scholar]
- Leckband G., Lorz H. (1998). Transformation and expression of a stilbene synthase gene of Vitis vinifera L. in barley and wheat for increased fungal resistance. Theor. Appl. Genet. 96 1004–1012. 10.1007/s001220050832 [DOI] [Google Scholar]
- Li X., Wu B., Wang L., Li S. (2006). Extractable amounts of trans-resveratrol in seed and berry skin in Vitis evaluated at the germplasm level. J. Agric. Food Chem. 54 8804–8811. 10.1021/jf061722y [DOI] [PubMed] [Google Scholar]
- Liu Y., Liu S., Liu D., Wei Y., Liu C., Yang Y., et al. (2014). Exploiting EST databases for the development and characterization of EST-SSR markers in blueberry (Vaccinium) and their cross-species transferability in Vaccinium spp. Sci. Hortic. 176 319–329. 10.1016/j.scienta.2014.07.026 [DOI] [Google Scholar]
- Liu Z., Zhuang C., Sheng S., Shao L., Zhao W., Zhao S. (2011). Overexpression of a resveratrol synthase gene (PcRS) from Polygonum cuspidatum in transgenic Arabidopsis causes the accumulation of trans-piceid with antifungal activity. Plant Cell Rep. 30 2027–2036. 10.1007/s00299-011-1110-2 [DOI] [PubMed] [Google Scholar]
- Lorenzis G. D., Imazio S., Rusjan D., Vouillamoz J. F., Nikolaou N., Failla O., et al. (2013). Genetic investigation of grapevine varieties ‘RibollaGialla’ (Italy), ‘Rebula’ (Slovenia) and ‘Robola’ (Ionian Islands). Sci. Hortic. 150 425–431. 10.1016/j.scienta.2012.12.003 [DOI] [Google Scholar]
- Mattivi F., Reniero F., Korhammer S. (1995). Isolation, characterization, and evolution in red wine vinification of resveratrol monomers. J. Agric. Food Chem. 43 1820–1823. 10.1021/jf00055a013 [DOI] [Google Scholar]
- Parage C., Tavares R., Réty S., Baltenweck-Guyot R., Poutaraud A., Renault L., et al. (2012). Structural, functional, and evolutionary analysis of the unusually large stilbene synthase gene family in grapevine. Plant Physiol. 160 1407–1419. 10.1104/pp.112.202705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezet R., Pont V., Cuenat P. (1994). Method to determine resveratrol and pterostilbene in grape berries and wines using high performance liquid chromatography and highly sensitive fluorimetric detection. J. Chromatogr. A 663 191–197. 10.1016/0021-9673(94)85245-6 [DOI] [Google Scholar]
- Ribeiro de Lima M. T., Waffo-Téguo P., Teissedre P. L., Pujolas A., Vercauteren J., Cabanis J. C. (1999). Determination of stilbenes (trans-astringin, cis- and trans-piceid, and cis- and trans-resveratrol) in Portuguese wines. J. Agric. Food Chem. 47 2666–2670. 10.1021/jf9900884 [DOI] [PubMed] [Google Scholar]
- Romero-Pérez A. I., Lamuela-Raventós R. M., Andrés-Lacueva C., de la Torre-Boronat M. C. (2001). Method for the quantitative extraction of resveratrol and piceid isomers in grape berry skins. Effect of powdery mildew on the stilbene content. J. Agric. Food Chem. 49 210–215. 10.1021/jf000745o [DOI] [PubMed] [Google Scholar]
- Ruhmann S., Treutter D., Fritsche S., Briviba K., Szankowski I. (2006). Piceid (resveratrol glucoside) synthesis in stilbene synthase transgenic apple fruit. J. Agric. Food Chem. 54 4633–4640. 10.1021/jf060249l [DOI] [PubMed] [Google Scholar]
- Rupprich N., Kindl H. (1978). Stilbene synthases and stilbenecarboxylate synthases, I enzymatic synthesis of 3,5,-4-trihydroxy stilbene from p-coumaroyl coenzyme A and malonyl coenzyme A, Hoppe Seylers Z. Physiol. Chem. 359 165–172. [PubMed] [Google Scholar]
- Serazetdinova L., Oldach K. H., Lorz H. (2005). Expression of transgenic stilbene synthases in wheat causes the accumulation of unknown stilbene derivatives with antifungal activity. J. Plant Physiol. 162 985–1002. 10.1016/j.jplph.2004.11.005 [DOI] [PubMed] [Google Scholar]
- Shi J., He M., Cao J., Wang H., Ding J., Li R., et al. (2014). The comparative analysis of the potential relationship between resveratrol and stilbene synthase gene family in the development stages of grapes (Vitis quinquangularis and Vitis vinifera). Plant Physiol. Biochem. 74 24–32. 10.1016/j.plaphy.2013.10.021 [DOI] [PubMed] [Google Scholar]
- Vannozzi A., Dry L. B., Fasoli M., Zenoni S., Lucchin M. (2012). Genome-wide analysis of the grapevine stilbene synthase multigenic family, genomic organization and expression profiles upon biotic and abiotic stresses. BMC Plant Biol. 12:130 10.1186/1471-2229-12-130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vian M. A., Tomao V., Gallet S., Coulomb P. O., Lacombe J. M. (2005). Simple and rapid method for cis- and trans-resveratrol and piceid isomers determination in wine by high-performance liquid chromatography using Chromolith columns. J. Chromatogr A 1085 224–229. 10.1016/j.chroma.2005.05.083 [DOI] [PubMed] [Google Scholar]
- Vitrac X., Bornet A., Vanderlinde R., Valls J., Richard T., Delaunay J., et al. (2005). Determination of stilbenes (δ-viniferin, trans-astringin, trans-piceid, cis- and trans-resveratrol, e-viniferin) in Brazilian wines. J. Agric. Food Chem. 53 5664–5669. 10.1021/jf050122g [DOI] [PubMed] [Google Scholar]
- Wang J., Ma L., Xi H., Wang L., Li S. (2015). Resveratrol synthesis under natural conditions and after UV-C irradiation in berry skin is associated with berry development stages in ‘Beihong’ (V. vinifera × V. amurensis). Food Chem. 168 430–438. 10.1016/j.foodchem.2014.07.025 [DOI] [PubMed] [Google Scholar]
- Wang J., Wang S., Liu G., Edwards E. J., Duan W., Li S., et al. (2016). The synthesis and accumulation of resveratrol are associated with veraison and abscisic acid concentration in Beihong (Vitis vinifera ×Vitis amurensis) Berry Skin. Front. Plant Sci. 7:1605 10.3389/fpls.2016.01605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X., Singer S. D., Qiao H., Liu Y., Jiao C., Wang H., et al. (2016). Insights into the mechanisms underlying ultraviolet-c induced resveratrol metabolism in grapevine (V. amurensis Rupr.) cv. “Tonghua-3”. Front. Plant Sci. 19:503 10.3389/fpls.2016.00503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Li X., Wu B., Wang L., Li S. (2009). Effects of UV-C irradiation on resveratrol and its glycosides content in leaves and the berries of neighboring clusters of potted Beifeng grapevines (Vitist hunbergii ×Vitis vinifera). J. Fruit Sci. 26 461–465. [Google Scholar]
- Zhu Y. J., Agbayani R., Jackson M. C., Tang C. S., Moore P. H. (2004). Expression of the grapevine stilbene synthase gene VST1 in papaya provides increased resistance against diseases caused by Phytophthora palmivora. Planta 220 241–250. 10.1007/s00425-004-1343-1 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.