Background
Probiotics are living microorganisms providing health beneficial effect to the host (1). Probiotics have been used for the treatment or prevention of various diseases related to diarrhea (2), cholesterol (3) immune function (4), and inflammatory bowel disease (5). In addition, recent study also presents that probiotic bacteria in the Bifidobacterium and Lactobacillus genera are able to have therapeutic effects in the patients of psychological disorders, such as depression, anxiety, and memory (6).
Lactobacillus casei is a Gram-positive bacterium that naturally inhabits the human and animal gastrointestinal and mouth organs (7). As its name implies, this heterofermentative microorganism is the dominant species present in ripening cheddar cheese (8). In probiotic aspects, L. casei showed beneficial roles in the activation of the gut mucosal immune system (9), treatment of diabetics (10), and chronic constipation (11). In the previous study, we isolated L. casei LC5 strain from fermented dairy products, which showed immune regulatory functions, especially, therapeutic effect on atopic dermatitis as a member of complex probiotics (12–14).
In order to gain better insight of the probiotic effect on atopic dermatitis, we analyzed the genome sequence of L. casei LC5. According to the report of NCBI Genome,1 more than two hundreds of Lactobacillus organisms are sequenced and their beneficial properties derived from genomic information are used in the food industry. However, the available genomes of L. casei strains as members of health promoting probiotics are still insufficient. Furthermore, L. casei strains are frequently confused with the closely related strains such as Lactobacillus paracasei and Lactobacillus rhamnosus. Therefore, comparative study in a whole genome scale is required to clarify taxonomic association of L. casei LC5 as well as its functional characteristics. The availability of the genomic information of L. casei LC5 will aid as a basis for further in-depth analysis of the probiotic function of L. casei strains.
Materials and Methods
Bacterial Strains and DNA Preparation
Lactobacillus casei LC5 was isolated from fermented dairy products and commercially used as probiotics in Korea (15). L. casei LC5 was cultured aerobically in MRS medium (Difco, USA) at 37°C for 18 h. Genomic DNA from L. casei LC5 was extracted and purified using a QIAamp DNA Mini Kit (Qiagen, Germany). The concentration of genomic DNA was qualified with NanoDrop 2000 UV–vis spectrophotometer (Thermo Scientific, USA) and Qubit 2.0 fluorometer (Life Technology, USA).
Genome Sequencing, Assembly, and Annotation
Whole genome sequencing of L. casei LC5 was carried out by using PacBio RS II platform. A 20 kb DNA library was constructed according to the manufacturer’s instruction and sequenced using single molecule real-time (SMRT) sequencing technology with the P6 DNA polymerase and C4 chemistry. A total of 138,180 subreads (1.04 Gb) were obtained with 400-fold coverage. The average length of subreads was 7,550 bp and N50 was 10,940 bp. Genome assembly was performed using HGAP 3.0 (16) with default options. The annotation was carried out with NCBI Prokaryotic Genome Annotation Pipeline (17) through NCBI Genome submission portal (GenomeSubmit at http://ncbi.nlm.nih.gov). The chromosome topology was drawn using DNAPlotter (18). Clusters of orthologous groups (COG) categories were assigned to the coding genes using BLASTP (e-value: 1e−3) against COG database (19).
Phylogenetic Analysis and Comparative Genomic Analysis
For phylogenetic and comparative study, we downloaded 19 genome sequences of L. casei group (10 of L. casei, 8 of L. paracasei, 1 of Lactobacillus zeae, and 1 of L. rhamnosus) from NCBI genome database.2 A list of the reference genomes are as follows: L. casei Zhang (NC_014334), L. casei BL23 (NC_010999), L. casei BD-II (NC_017474), L. casei LC2W (NC_017473), L. casei 12A (NZ_CP006690), L. casei W56 (NC_018641), L. casei LcY (NZ_CM001848), L. casei LcA (NZ_CM001861), L. casei LOCK919 (NC_021721), L. casei ATCC 393 (NZ_AP012544), L. paracasei ATCC 334 (NC_008526), L. paracasei 362.5013889 (NC_022112), L. paracasei N1115 (NZ_CP007122), L. paracasei JCM (NZ_AP012541), L. paracasei CAUH35 (NZ_CP012187), L. paracasei L9 (NZ_CP012148), L. paracasei KL1 (NZ_CP013921), L. zeae DSM 20178 (NZ_AZCT01000001), and L. rhamnosus GG (NC_013198). The assembly levels of all genomes are “complete genome” or chromosome except L. zeae DSM 20178 (includes 55 scaffolds). Because we failed to fetch full-length 16S rRNA gene from the genome of L. zeae DSM 20178, we alternatively used a 16S rRNA gene of L. zeae RIA 482 (NR_037122), the closest sequence of DSM 20178 (sequence identity = 99.9%), in the phylogenetic analysis.
The evolutionary history was inferred by using the maximum likelihood method based on the Tamura–Nei model (20). All positions containing gaps and missing data were eliminated. There were a total of 1521 positions in the final dataset. Those phylogenetic analyses were conducted in MEGA6 (21). To compute genomic distance, we first computed orthologous average nucleotide identity (OrthoANI) values using orthologous average nucleotide identity tool (22). The OrthoANI values were converted to distance values by following formula: distance = 1 − (OrthoANI/100). The evolutionary distance was computed using the neighbor-joining method of MEGA6 (21). The tree is drawn to scale with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The resulting phylogenetic tree was produced using MEGA6. Pan-genomic study using Panseq (23) was performed to investigate the genomic conservation and finding novel region in the sequenced genome.
Results
Genome Characteristics of L. casei LC5
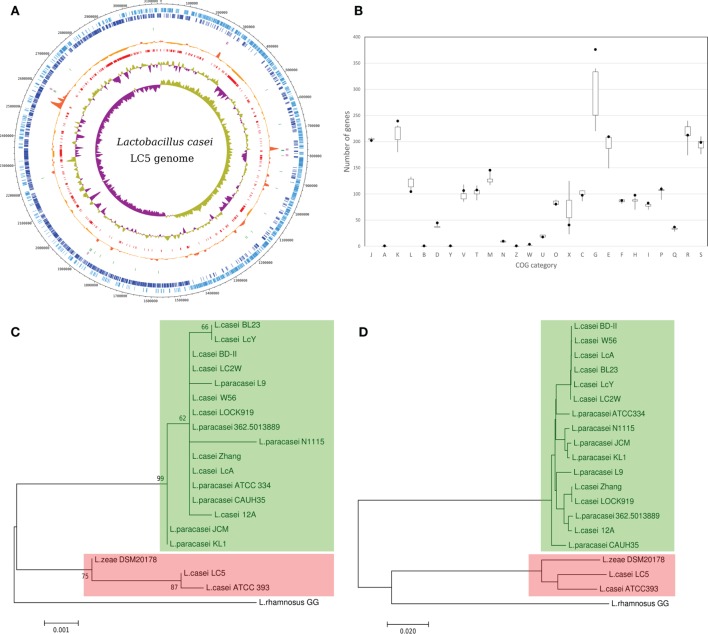
We obtained a complete genome sequence of L. casei LC5 using SMRT sequencing. This genome has a chromosome and no organelle sequences. The total size of the genome is 3,132,867 bp and its GC content is 47.9%. A total of 2,925 genes were detected from the genome sequence. The number of coding CDS is 2,817 and pseudogenes is 31. Seventy seven RNAs (15 rRNAs, 59 tRNAs, and 3 non-coding RNAs) were also identified. Repeating region or CRISPR array was not identified. Genomic features of L. casei LC5 are shown in Figure 1A.
Figure 1.
Genome characteristics of Lactobacillus casei LC5 genome. (A) Circular map of genomic features; eight tracks were plotted in the map. Track 1 (light blue; outermost), forward-stranded coding CDS; Track 2 (blue), reverse-stranded coding CDS; Track 3 (light purple), rRNA including 5S, 16S, and 23S; Track 4 (green), Trna; Track 5 (orange), peak of pan-genomic conservation; Track 6 (red), novel regions (below 85% similarities with the other genomes); Track 7 (light green and purple), GC content; and Track 8 (light green and purple), GC skew. (B) Abundance of clusters of orthologous groups (COG) categories; black point indicates the abundance of LC5 for each category. A box and whisker plot indicates a statistical distribution of the COG categories of 19 L. casei genomes. (C) Phylogenetic tree of L. casei group based on 16S rRNA genes and (D) phylogenetic tree of L. casei group based on average nucleotide identity. The value 0.02 of the ruler in (D) indicates 2% of genomic dissimilarity. Red boxes on the (C,D) indicate the genomes associated to the high-GC group and green boxes indicate the genomes associated to the low-GC group.
Although L. casei LC5 was identified as a strain of L. casei, it showed different genomic features compared to the other published L. casei strains; According to the summary of 37 L. casei genomes deposited in NCBI Assembly, the median length is 3.01993 Mb, the median of coding genes is 2,712, and the median of GC contents is 46.4%. An interesting point is that those genomes can be split into two groups by the difference of GC contents, high-GC group (47.7–47.9%) and low-GC group (46.2–46.6%). Five genomes (ATCC 393, N87, 867_LCAS, Lbs2, JCM 1134) and L. casei LC5 belong to the high-GC group and the other genomes belong to the low-GC group (Table 1).
Table 1.
Genome summary of Lactobacillus casei group.
| Organism/name | Strain | Clade | Assembly level | Size (Mb) | GC% | GC group |
|---|---|---|---|---|---|---|
| L. casei LC5 | LC5 | L. casei | Complete genome | 3.13 | 47.9 | High |
| L. casei str. Zhang | Zhang | L. casei | Complete genome | 2.90 | 46.4 | Low |
| L. casei BL23 | BL23 | L. casei | Complete genome | 3.08 | 46.3 | Low |
| L. casei BD-II | BD-II | L. casei | complete genome | 3.13 | 46.3 | Low |
| L. casei LC2W | LC2W | L. casei | Complete genome | 3.08 | 46.4 | Low |
| L. casei 12A | 12A | L. casei | Complete genome | 2.91 | 46.4 | Low |
| L. casei W56 | W56 | L. casei | Complete genome | 3.13 | 46.3 | Low |
| L. casei LOCK919 | LOCK919 | L. casei | Complete genome | 3.14 | 46.2 | Low |
| L. casei subsp. casei ATCC 393 | ATCC 393 | L. casei | Complete genome | 2.95 | 47.9 | High |
| L. casei LcY | LcY | L. casei | Chromosome | 3.10 | 46.3 | Low |
| L. casei LcA | LcA | L. casei | Chromosome | 3.13 | 46.3 | Low |
| L. casei A2-362 | A2-362 | L. casei | Scaffold | 3.19 | 46.2 | Low |
| L. casei | KL1-Liu | L. casei | Scaffold | 2.85 | 46.6 | Low |
| L. casei DSM 20011 = JCM 1134 | DSM 20011 | L. casei | Scaffold | 2.82 | 46.5 | Low |
| L. casei 21/1 | 21/1 | L. casei | Contig | 3.22 | 46.2 | Low |
| L. casei 32G | 32G | L. casei | Contig | 3.01 | 46.4 | Low |
| L. casei A2-362 | A2-362 | L. casei | Contig | 3.36 | 46.1 | Low |
| L. casei CRF28 | CRF28 | L. casei | Contig | 3.04 | 46.3 | Low |
| L. casei M36 | M36 | L. casei | Contig | 3.15 | 46.3 | Low |
| L. casei T71499 | T71499 | L. casei | Contig | 3.00 | 46.2 | Low |
| L. casei UCD174 | UCD174 | L. casei | Contig | 3.07 | 46.4 | Low |
| L. casei UW1 | UW1 | L. casei | Contig | 2.87 | 46.4 | Low |
| L. casei UW4 | UW4 | L. casei | Contig | 2.76 | 46.4 | Low |
| L. casei Lc-10 | Lc-10 | L. casei | Contig | 2.95 | 46.4 | Low |
| L. casei Lpc-37 | Lpc-37 | L. casei | Contig | 3.08 | 46.3 | Low |
| L. casei UW4 | UW4 | L. casei | Contig | 2.63 | 46.4 | Low |
| L. casei 12A | 12A | L. casei | Contig | 2.93 | 46.3 | Low |
| L. casei 5b | 5b | L. casei | Contig | 3.02 | 46.3 | Low |
| L. casei | N87 | L. casei | Contig | 3.00 | 47.9 | High |
| L. casei | 867_LCAS | L. casei | Contig | 3.09 | 47.9 | High |
| L. casei | DPC6800 | L. casei | Contig | 3.05 | 46.4 | Low |
| L. casei | Lc1542 | L. casei | Contig | 2.92 | 46.5 | Low |
| L. casei | 1316.rep1_LPAR | L. casei | Scaffold | 2.86 | 46.5 | Low |
| L. casei | 1316.rep2_LPAR | L. casei | Scaffold | 2.79 | 46.4 | Low |
| L. casei | 844_LCAS | L. casei | Scaffold | 2.79 | 46.4 | Low |
| L. casei | BM-LC14617 | L. casei | Scaffold | 3.04 | 46.3 | Low |
| L. casei | Lbs2 | L. casei | Scaffold | 3.27 | 47.9 | High |
| L. casei DSM 20011 = JCM 1134 | JCM 1134 | L. casei | Contig | 2.78 | 47.7 | High |
| Lactobacillus paracasei ATCC 334 | ATCC 334 | L. paracasei | Complete genome | 2.92 | 46.6 | Low |
| L. paracasei subsp. paracasei 8700:2 | 8700:2 | L. paracasei | Complete genome | 3.03 | 46.3 | Low |
| L. paracasei N1115 | N1115 | L. paracasei | Complete genome | 3.06 | 46.5 | Low |
| L. paracasei subsp. paracasei JCM 8130 | JCM 8130 | L. paracasei | Complete genome | 3.02 | 46.6 | Low |
| L. paracasei | CAUH35 | L. paracasei | Complete genome | 2.97 | 46.3 | Low |
| L. paracasei | L9 | L. paracasei | Complete genome | 3.08 | 46.3 | Low |
| L. paracasei | KL1 | L. paracasei | Complete genome | 2.92 | 46.6 | Low |
| Lactobacillus zeae DSM 20178 = KCTC 3804 | DSM 20178 | L. zeae | Scaffold | 3.12 | 47.7 | High |
| Lactobacillus rhamnosus GG | GG (ATCC 53103) | L. rhamnosus | Complete genome | 3.01 | 46.7 | Low |
Comparative Study of L. casei Group
Comparative study of both 16S rRNA genes and whole genome sequences revealed that the closest genome of L. casei LC5 was L. casei ATCC 393 and second closest one was L. zeae DSM 20178. The three genomes which showed distinguishable differences on the comparative study, LC5, ATCC 393, and L. zeae DSM 20178, belong to the high-GC group as described in the above section. In contrast to the phylogenetic distances based on 16S rRNA gene among the high-GC group (below 0.001), the distances between the high-GC group and the low-GC group were above 0.003 (Figure 1C). It was also supported by the estimation result of the whole genomic comparison. Average nucleotide identity (ANI) values among the high-GC group were above 94% whereas ANI values between two groups were below 80% (Figure 1D). All the L. casei strains and L. paracasei strains belonging to the low-GC group showed the high genomic similarity of 98% or higher.
Functional Classification
Functional classification based on COG assigned the 2,334 CDSs into the 1,309 COG numbers. From the comparison of functional categories against the 19 L. casei group genomes, we found that L. casei LC5 contains the high number of proteins which associate with “carbohydrate transport and metabolism (G)” (376 proteins) and “transcription (K)” (239 proteins) excluding two unknown categories, “general function prediction only (R)” and “function unknown (S)” as shown in Figure 1B. L. casei LC5 has at least 36 more proteins than the other genomes on the category G and has at least 8 more proteins than the other genomes on the category K. The gene expansion of those two functional categories in the LC5 genome is not found on the other members of high-GC group. Although the genomes belonging to high-GC group showed high similarities to each other and the genomes belonging to the high-GC group do not have excessive proteins on the categories, G and K, when compared to those belonging to the low-GC group. Moreover, L. casei ATCC 393 which is the most similar genome of LC5 has fewer proteins than the average number of those categories, 223 proteins for the category G and 192 proteins for the category K.
In the previous study, probiotic LC5 strain isolated from Korean fermented dairy product showed great therapeutic effect on atopic dermatitis. Here, we report a genomic overview and distinguishing gene features of LC5 by comparative genomic analysis of 20 related strains. The genomic data presented in this report will broaden our knowledge about roles and mechanisms of microorganisms ameliorating symptoms of immune diseases and help developing functional probiotics for the treatment of immune disorders.
Data Access
The L. casei LC5 genome sequencing project has been deposited at GenBank under the accession number CP017065. The BioProject and BioSample designation for this project is PRJNA340077 and SAMN05631198, respectively. This strain has been deposited in the Korean Collection for Type Cultures (deposit ID: KCTC 12398BP).
Author Contributions
Y-DN and SL designed and coordinated all the experiments. T-JL and JK performed cultivation and DNA preparation. JK and W-HC performed genome assembly, gene prediction, gene annotation, and comparative genomic analysis. Y-DN, W-HC, TW, and JK wrote the manuscript. All authors have read the manuscript and approved.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding
This work was supported by a grant from Korea Food Research Institute (project no. E0170602-01).
References
- 1.Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B, et al. Expert consensus document: the International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol (2014) 11:506–14. 10.1038/nrgastro.2014.66 [DOI] [PubMed] [Google Scholar]
- 2.McFarland LV. Meta-analysis of probiotics for the prevention of antibiotic associated diarrhea and the treatment of Clostridium difficile disease. Am J Gastroenterol (2006) 101:812–22. 10.1111/j.1572-0241.2006.00465.x [DOI] [PubMed] [Google Scholar]
- 3.Sanders ME. Considerations for use of probiotic bacteria to modulate human health. J Nutr (2000) 130:384S–90S. [DOI] [PubMed] [Google Scholar]
- 4.Reid G, Jass J, Sebulsky MT, Mccormick JK. Potential uses of probiotics in clinical practice. Clin Microbiol Rev (2003) 16:658–72. 10.1128/CMR.16.4.658-672.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saez-Lara MJ, Gomez-Llorente C, Plaza-Diaz J, Gil A. The role of probiotic lactic acid bacteria and bifidobacteria in the prevention and treatment of inflammatory bowel disease and other related diseases: a systematic review of randomized human clinical trials. Biomed Res Int (2015) 2015:15. 10.1155/2015/505878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang H, Lee I-S, Braun C, Enck P. Effect of probiotics on central nervous system functions in animals and humans: a systematic review. J Neurogastroenterol Motil (2016) 22:589–605. 10.5056/jnm16018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai H, Rodríguez BT, Zhang W, Broadbent JR, Steele JL. Genotypic and phenotypic characterization of Lactobacillus casei strains isolated from different ecological niches suggests frequent recombination and niche specificity. Microbiology (2007) 153:2655–65. 10.1099/mic.0.2007/006452-0 [DOI] [PubMed] [Google Scholar]
- 8.Banks JM, Williams A. The role of the nonstarter lactic acid bacteria in Cheddar cheese ripening. Int J Dairy Technol (2004) 57:145–52. 10.1111/j.1471-0307.2004.00150.x [DOI] [Google Scholar]
- 9.Galdeano CM, Perdigon G. The probiotic bacterium Lactobacillus casei induces activation of the gut mucosal immune system through innate immunity. Clin Vaccine Immunol (2006) 13:219–26. 10.1128/CVI.13.2.219-226.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yadav H, Jain S, Sinha PR. Antidiabetic effect of probiotic dahi containing Lactobacillus acidophilus and Lactobacillus casei in high fructose fed rats. Nutrition (2007) 23:62–8. 10.1016/j.nut.2006.09.002 [DOI] [PubMed] [Google Scholar]
- 11.Koebnick C, Wagner I, Leitzmann P, Stern U, Zunft HF. Probiotic beverage containing Lactobacillus casei shirota improves gastrointestinal symptoms in patients with chronic constipation. Can J Gastroenterol (2003) 17:655–9. 10.1155/2003/654907 [DOI] [PubMed] [Google Scholar]
- 12.Hee YJ, Kim DH, Ku JK, Kang Y, Kim M-Y, Kim HO, et al. Therapeutic effects of probiotics in patients with atopic dermatitis. J Microbiol Biotechnol (2006) 16:1699–705. [Google Scholar]
- 13.Seo J-G, Chung M-J, Lee H-G. Alleviation of atopic dermatitis through probiotic and mixed-probiotic treatments in an atopic dermatitis model. Korean J Food Sci Animal Resour (2011) 31:420–7. 10.5851/kosfa.2011.31.3.420 [DOI] [Google Scholar]
- 14.Yang H-J, Min TK, Lee HW, Pyun BY. Efficacy of probiotic therapy on atopic dermatitis in children: a randomized, double-blind, placebo-controlled trial. Allergy Asthma Immunol Res (2014) 6:208–15. 10.4168/aair.2014.6.3.208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cha YS, Seo J-G, Chung M-J, Cho CW, Youn HJ. A mixed formulation of lactic acid bacteria inhibits trinitrobenzene-sulfonic-acid-induced inflammatory changes of the colon tissue in mice. J Microbiol Biotechnol (2014) 24:1438–44. 10.4014/jmb.1403.03064 [DOI] [PubMed] [Google Scholar]
- 16.Chin C-S, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods (2013) 10:563–9. 10.1038/nmeth.2474 [DOI] [PubMed] [Google Scholar]
- 17.Tatusova T, Dicuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, et al. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res (2016) 44(14):6614–24. 10.1093/nar/gkw569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carver T, Thomson N, Bleasby A, Berriman M, Parkhill J. DNAPlotter: circular and linear interactive genome visualization. Bioinformatics (2009) 25:119–20. 10.1093/bioinformatics/btn578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galperin MY, Makarova KS, Wolf YI, Koonin EV. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res (2015) 43:D261–9. 10.1093/nar/gku1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol (1993) 10:512–26. [DOI] [PubMed] [Google Scholar]
- 21.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol (2013) 30:2725–9. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee I, Ouk Kim Y, Park S-C, Chun J. OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol (2016) 66:1100–3. 10.1099/ijsem.0.000760 [DOI] [PubMed] [Google Scholar]
- 23.Laing C, Buchanan C, Taboada EN, Zhang Y, Kropinski A, Villegas A, et al. Pan-genome sequence analysis using Panseq: an online tool for the rapid analysis of core and accessory genomic regions. BMC Bioinformatics (2010) 11:1. 10.1186/1471-2105-11-461 [DOI] [PMC free article] [PubMed] [Google Scholar]